Abstract
The extent to which ligand occupancy and dimerization contribute to erbB1 signaling is controversial. To examine this, we utilized two-color Quantum Dot tracking for visualization of erbB1 homodimerization and quantification of the dimer off rate (koff) on living cells. Kinetic parameters were extracted using a 3-state Hidden Markov Model to identify transition rates between free, co-confined, and dimerized states. We report that dimers composed of 2 ligand-bound receptors are long-lived and their koff is independent of kinase activity. By comparison, unliganded dimers have >4-fold faster koff. Transient co-confinement of receptors promotes repeated encounters and enhances dimer formation. Mobility decreases >6-fold when ligand-bound receptors dimerize. Blockade of erbB1 kinase activity or disruption of actin networks results in faster diffusion of receptor dimers. These results implicate both signal propagation and the cortical cytoskeleton in reduced mobility of signaling-competent erbB1 dimers.
Ligand-induced signaling by the epidermal growth factor receptor (EGFR or HER1 or erbB1) drives cell growth and survival, with roles in normal development and disease pathogenesis1. A wealth of structural knowledge supports a model of signal initiation through the formation of back-to-back erbB1 dimers2, 3. However, the size and ligand-occupancy of the erbB1 signaling complex remains controversial4-9. This is largely attributable to the range of methodologies applied to the problem, each with a unique set of advantages and limitations. For example, estimates of erbB1 dimerization parameters were originally derived from solution-based measurements and the use of recombinant domains10-13. Classical approaches have also included chemical crosslinking, small-angle x-ray scattering and tyrosine phosphorylation kinetics12-14. The field has relied on mathematical modeling for estimation of homodimer equilibrium dissociation constants in the absence and presence of ligand15, as well as the effects of receptor number and density on dimerization behavior16, 17.
Prior studies considered the ensemble, steady state characteristics of erbB1 but could not address the stochastic nature of receptors encountering each other in the fluid and dynamic landscape of the plasma membrane. The role of heterogeneous microdomains18 within membranes also remains unresolved, particularly the potential for transient confinement zones19, 20 to influence erbB1 interactions. Advances in fluorescence imaging techniques have provided some insights into intact erbB1’s behavior in live membranes, including receptor diffusion and clustering7, 9, 21-23. One important advance was the application of EGF-bound Quantum Dots (QDs), which captured filopodial transport and internalization of ligand-bound, erbB17, 24. QD nanoprobes have particularly useful photophysical properties, such as high photostability and narrow emission spectra, permitting extended periods of observation and facilitating simultaneous two-color single particle tracking (SPT)7, 25, 26.
In this work, we take advantage of QD probes to track erbB1 monomers on the surface of intact epithelial cell membranes and, further, to capture the dimerization process in real-time. Sophisticated analytical methods were developed that extract receptor reaction kinetics and discriminate between monomer and dimer state behavior. A novel feature of our analysis is the ability to detect receptor co-confinement and demonstrate its role in promoting repeated encounters between receptors. We show that “preformed” dimers formed between unoccupied receptors are highly transient. In the presence of ligand, dimers composed of 2 ligands:2 receptors are more stable than complexes having only one ligand-bound receptor. Thus, this study unequivocally links ligand occupancy to dimer stability.
RESULTS
Changes in erbB1 diffusion are linked to activation status
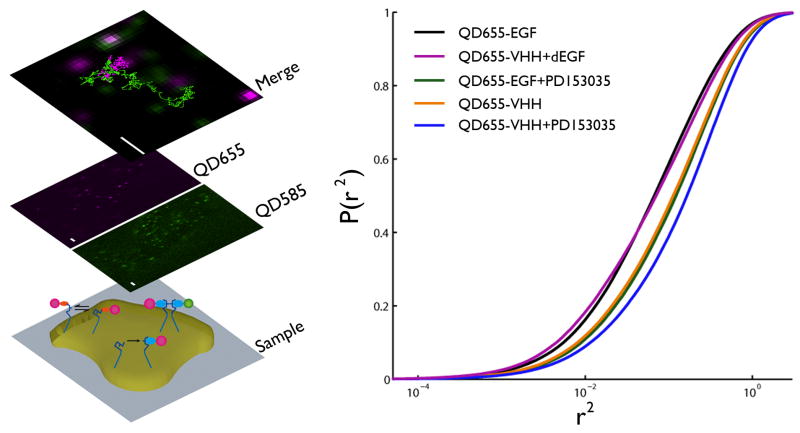
Labels for erbB1 SPT that minimally perturb individual receptor dynamics were essential to these studies. To monitor the ligand-bound receptors, we used QD-labeled EGF (QD-EGF), as described and characterized previously24. To follow resting receptors, we took advantage of the small, monovalent camelid anti-erbB1 antibody fragment (VHH) that is non-activating and does not compete for EGF binding27. Figure 1a illustrates our experimental approach using two-color QD tracking to follow the dynamics of endogenous erbB1 receptors on the apical surface of live cells (bottom plane). Simultaneous, two-color imaging was achieved using a beam splitter that projects QD655 (magenta) and QD585 (green) emissions onto an emCCD (middle plane). Finally, a custom-designed image registration method28 (see Supplementary Methods and Supplementary Fig. 1) is used to map the relative positions of the 585 and 655 QDs over the time course of data acquisition (top plane). Fast computation of single molecule trajectories was accomplished using a Graphics Processing Unit (GPU)-based approach, which identifies single molecule locations and builds trajectories using postprocessing algorithms29 (see Supplementary Methods and Supplementary Fig. 1). These data sets provided two types of information about receptor behavior. First, diffusion coefficients were extracted from the trajectories of individual receptors using well-established methods30 that provide global changes in diffusion but cannot distinguish between monomers and dimers. Second, novel computation analyses of two-color imaging data permitted measurement of dimerization and state-dependent behavior.
Figure 1.
Slowed diffusion as a function of receptor activation is revealed by single QD tracking on the apical surface of A431 cells. (a) Following serum starvation in the absence or presence of PD15035, the activated and resting erbB1 receptor were labeled and tracked on live A431 cells with either two-colors of QD-EGF, QD-VHH, or a combination of each, respectively. Bottom plane: ErbB1 receptor cartoons (blue, not to scale) on the surface of an adherent cell on a coverslip, with probes recognizing the tethered and extended conformations (EGF ligand, light blue; VHH, orange; QD655, magenta; QD585, green). Middle plane: Single molecules were visualized in a QD655 or QD585 channel. Top plane: Following SPT and image registration, trajectories are plotted on the same coordinate system. Scale bar, 2 μm. (b) Cumulative probability plot of squared displacement for QD655 tracking of each condition: QD-EGF (black), QD-VHH + non-fluorescent EGF (purple), QD-VHH (orange), QD-VHH + PD153035 (green), and QD-EGF + PD153035 (blue). A rightward shift in distribution indicates increased diffusion. Fits for two-component square displacement analyses are provided as Supplementary information (Supplementary Table 3).
Figure 1b reports changes in diffusional behavior of erbB1 expressed endogenously on A431 cells under different experimental conditions, represented as the probability distribution of squared displacements30. Each curve is calculated from thousands of receptor trajectories and shows the broad range of diffusional behaviors, with left-shifted plots being the slowest (see Supplementary Methods and Supplementary Table 1). These data show a strong relationship between receptor mobility and activation state, with the resting receptors (QD-VHH, orange) having a higher mobility than ligand-bound (QD-EGF, black) receptors. In the presence of PD153035, an erbB kinase inhibitor31,32, QD-EGF-bound receptors displayed increased mobility (green), similar to resting diffusion. Treatment with the kinase inhibitor also slightly enhances mobility of resting (QD-VHH bound) receptors in A431 cell. This suggests that basal signaling from erbB1, as the predominant erbB form in these cells, influences membrane properties or its associated cytoskeleton. Note that the addition of unlabeled EGF to QD-VHH-labeled cells results in slower diffusion of the receptors, confirming that the QD-VHH does not interfere with ligand binding. The slowed lateral motility of ligand-bound receptors is thus linked to tyrosine phosphorylation, possibly through protein scaffolding or signaling-induced changes in the local environment. Similar mobility trends are seen for endogenous erbB1 on HeLa cells, which express fewer receptors than A431 cells by more than an order of magnitude (Supplementary Fig. 1)7.
Capturing Dimerization with 2-color SPT
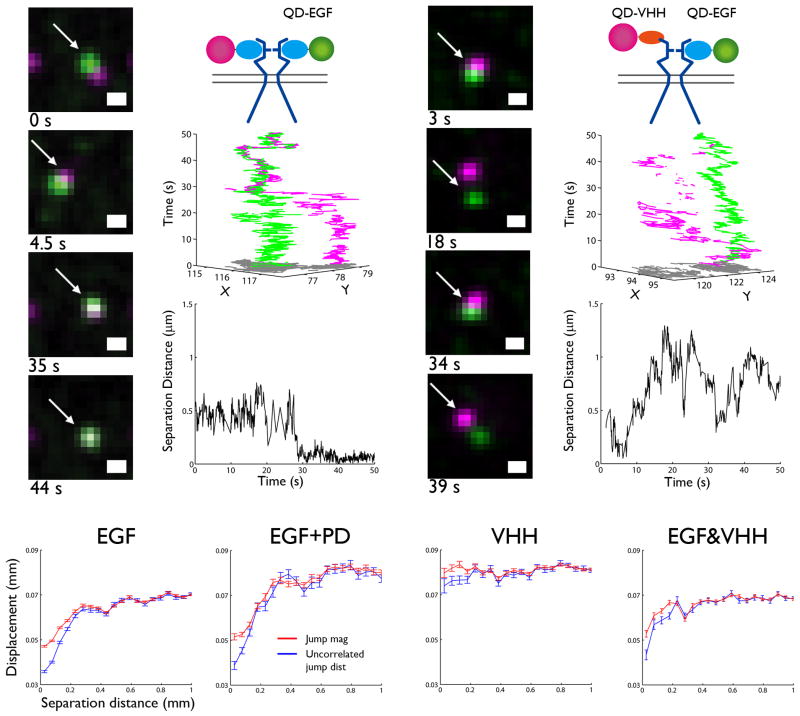
Figure 2 reports the direct observation of receptor interactions, based upon the simultaneous imaging of spectrally-distinct QD probes. As receptors approach each other, their relative motion reveals insights into their interactions and the constraints imposed by membrane architecture. In Fig. 2a, QD585-EGF-erbB1 (green) and QD655-EGF-erbB1 (magenta) complexes are observed to diffuse in close proximity for ~30 s before forming a stable dimer (white) that persists until the end of the acquisition (see Supplementary Methods and Supplementary Video 1). Plots in Fig. 2b show the trajectories of each receptor (middle), as well as their separation distance (bottom) over time. Note the sharp drop to a separation distance of ~50 nm that marks the dimerization event is accompanied by correlated motion of the trajectories starting around t=30 s. This 40-50 nm offset is consistent with an estimate of the spacing between the centers of two QDs attached to the back-to-back, ligand-bound erbB1 homodimer crystal structure (see Supplementary Methods and Supplementary Fig. 2). Figures 2c-d show a representative encounter between one ligand-bound and one resting erbB1. These receptors experience long durations of close proximity (<500 nm), demonstrating that membrane microdomains serve to co-confine receptors19, 25, 26. After 5-10 s of co-confinement, the receptors move far apart as they escape from the domain. These data show that dimerization events can be captured in real-time and also illustrate the need to develop analytical tools that distinguish between close approach and bona fide dimer events.
Figure 2.
Direct visualization of erbB1 dimerization is captured by two-color SPT. (a) Sample time series showing dimer formation (white) between QD585-EGF-erbB1 (green) and QD655-EGF-erbB1 (magenta). (b) Cartoon of tracking condition (top), 3D trajectories (middle), and distance between receptors (bottom) as a function of time are shown for the indicated receptors in (a). (c) Sample time series for QD655-EGF-erbB1 and QD585-VHH-erbB1 shows interactions for a 1:2 EGF:erbB1 dimer. (d) Cartoon of tracking condition (top), 3D trajectories (middle), and distance between receptors (bottom) as a function of time are shown for the receptors in (c). Scale bar, 0.5 μm for (a) and (c). (e-h) Ensemble correlated motion plots summarize all two-color data for EGF, EGF+PD, and VHH conditions. A decrease in uncorrelated jump distance (blue) at short separations indicates that receptors are moving together. A concurrent drop in jump magnitude (red) demonstrates decreased diffusion.
Dimerized receptor complexes are expected to move together, exhibiting correlated motion. Therefore, correlated motion analysis offers a means to validate interactions across large data sets of two-color trajectories25 (Fig. 2e-f; see Supplementary Methods and Supplementary Fig. 3). This method reports average receptor displacement (jump magnitude, red) and the degree of uncorrelated motion (blue) between receptors in the population as a function of separation distance. For ligand bound receptors (Fig. 2e,f), correlated motion is observed as a reduction in uncorrelated jump distance at small separation. This behavior is not altered by PD153035 treatment, confirming that EGF binding promotes dimer formation that is independent of kinase activity. We further address this interesting observation below. Of note, despite observations of close approach between QD-VHH-erbB1 complexes, there was no correlated motion observed in the absence of ligand (Fig. 2g). These data indicate that if dimerization occurs between unliganded receptors, it must be short-lived compared to the time-scale of these measurements (20 frames per second). In contrast, interactions between one EGF-bound and one unliganded receptor demonstrate correlated motion (Fig. 2h). This shows that 1EGF:2erbB1 dimers do form, as previously reported9.
Dimer stability is governed by ligand occupancy
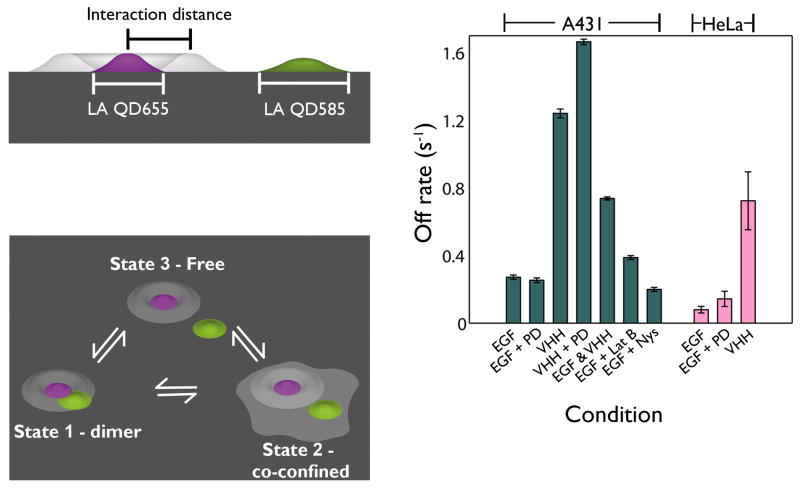
While correlated motion analysis can indicate the presence or absence of dimerization, it cannot quantify protein interaction kinetics. To extract dimerization kinetics, we developed a mathematical model based upon a Hidden Markov Model (HMM) approach33. This method generates a maximum likelihood estimate of the kinetic rate constants for transitions between states. For a set of observables, in this case separation between two receptor trajectories, the HMM is used to identify hidden states that reflect the underlying behaviors of the proteins. In order to accurately represent the data, we implemented a three-state model: free, co-confined and dimer (see Fig. 3a,b). The addition of a state representing receptors confined with the same domain was required in order to accurately fit the data (see Supplementary Methods “A two-state HMM is insufficient to describe observed erbB1 behaviors” and Supplementary Fig. 3). In the model, domains are considered to be cell surface regions that may be mobile and provide a barrier to free diffusion, causing receptors to deflect off the boundaries and facilitate repeated interactions between resident proteins. Such microdomains are consistent with prior work describing “actin corrals”19, 25, “lipid rafts”34 or “protein islands”35.
Figure 3.
Kinetics of homodimerization characterized by a three-state HMM reveal activation state dependent off rates. (a) Definition of a dimer. Two-color receptors are fit with localization accuracies (LA) for each channel; the white probability surface represents the area within which a dimer is identified. The interaction distance (ID) is defined by the crystal structure of the back-to-back erbB1, EGF bound dimer and the diameters of the QDs (40-50 nm). (b) The three states are defined as Free, Co-Confined, and Dimer, based on observed separation. Six kinetic rates are fit for the transitions between these states. (c) Off rates (s-1) for dimers fit using the three-state HMM. Off rate is defined as the sum of the rates of transitions between the dimer-to-domain confined and dimer-to-free states. Values obtained on both A431 and HeLa cells are shown.
Results in Figure 3c provide improved quantification of erbB1 dimer off rates afforded by the 2-color tracking and 3-state HMM approach. A summary of kinetic parameters derived from these data is reported in Supplementary Methods and Supplementary Table 2. Notably, koff for ligand-bound homodimers are similar, regardless of treatment with the kinase inhibitor. Dimers that form between receptors in the absence of ligand (QD-VHHs) exhibit a >4 fold higher off rate (see Supplementary Methods and Supplementary Video 2). We conclude that preformed dimers are highly transient, consistent with the lack of detectable correlated motion (Fig 2g). Dimers composed of one ligand-bound erbB1 and one unoccupied receptor were also relatively unstable, with a 2-fold larger off rate than dimers composed of two EGF-bound erbB1. Pretreatment of cells with Latrunculin B (LatB; disrupts actin) or Nystatin (Nys; sequesters cholesterol) did not dramatically alter dimer stability. Experiments in HeLa cells showed consistent trends, with similar off rates for ligand-bound dimers in the absence and presence of PD153035. Thus, EGF binding to both receptors is required for the most stable dimer formation.
State-dependent behaviors reflect changes in local environment
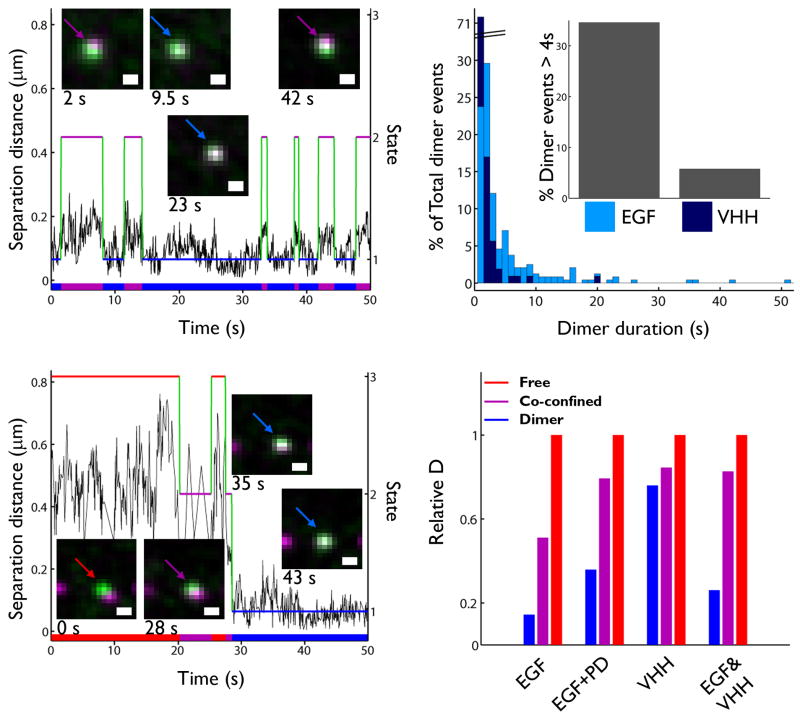
As a final analysis step, the Viterbi algorithm was used to identify states within candidate pair trajectories (see Supplementary Methods and Supplementary Fig. 4) based on the HMM estimation of the kinetic parameters36. An example of such a state path is shown in Fig. 4a. Here, two ligand-bound erbB1 are observed to explore the same domain (magenta line) for up to 50 seconds, interspersed by multiple 10-15 second periods of dimer formation (blue line). This type of behavior was observed in multiple experiments (see Supplementary Methods and Supplementary Fig. 4, Videos 3 and 4), underscoring the importance of co-confinement to foster repeated interactions between the same pair of receptors.
Figure 4.
State-dependent analysis distinguishes between free, co-confined and dimerized erbB1 characteristics. (a) An example distance trace (black) for two QD-EGF-bound receptors shows close approach punctuated by periods of excursion. Domain state (2, purple) and dimer state (1, blue) are connected by green segments to show state path. This path is projected onto the x-axis to show the timeline of states explored. Stills of the receptors involved in the interaction are inset. (b) Distance and state trace for two QD-EGF bound receptors shows formation of a dimer (Free state in red and others as aforementioned) that persists until the end of the acquisition. Scale bars, 0.5 μm for (a) and (b). (c) Normalized histogram of erbB1 dimer lifetimes for QD-EGF (red) or QD-VHH (yellow) homodimers, determined from Viterbi analysis. Inset shows the raw number of long-lived dimers for each condition. Note that the long-lived dimer duration may be underestimated due to the finite length of the time series. (d) Summary of normalized diffusion coefficients for QD-EGF, QD-EGF+PD, QD-VHH, and QD-EGF&QD-VHH conditions following characterization of states by HMM and Viterbi analyses. Each group of coefficients is normalized to the diffusivity of the free state within that condition.
A second type of behavior is shown in Fig. 4b, in which two receptors form a stable dimer at 28 s that lasts for the remaining 22 s of the acquisition period. We next determined the duration of individual dimer events observed in our entire data set. Fig. 4c reports the distribution of dimer lifetimes for ligand-bound (QD-EGF) versus unoccupied (QD-VHH) receptors. Note that less than 6% of dimers between unoccupied receptors last longer than 4 s (Fig. 4c inset). In contrast, long-lived dimers are a striking feature of two liganded receptors (>34% exceed 4 s; Supplementary Video 5). These examples demonstrate that dimers experience a range of interaction lifetimes, which can only be appreciated though single molecule observations.
Previous measurements of erbB1 diffusion using FRAP23, 37, 38 or single-color SPT4, 7, 9, 23, 39 approaches could not directly resolve differences in receptor motion upon dimerization. From our state information, we were able to calculate the diffusion coefficient for receptors identified as free monomers, co-confined monomers, or dimers under each experimental condition (Fig. 4d, see Supplementary Methods and Supplementary Table 3). Consistent with the correlated motion analysis (Fig. 2g), unoccupied receptors retained fast mobility in all states. The most dramatic change in mobility is seen with signaling competent QD-EGF-bound dimers. These complexes often become immobile, with an average >6-fold reduction in diffusion compared to the free monomer state. In the presence of PD153035, QD-EGF-erbB1 dimers only slow down by two-fold, reinforcing the concept that dimer slow down is dependent on tyrosine kinase activity (see Supplementary Methods and Supplementary Video 6). Note that analysis of receptor pairs composed of 1 ligand-bound receptor and one unoccupied receptor (EGF&VHH) fail to slow when co-confined, suggesting that the short durations of their interactions are not as efficient at propagating signals that influence their local environment.
Actin limits erbB1 diffusion and promotes repeated interactions
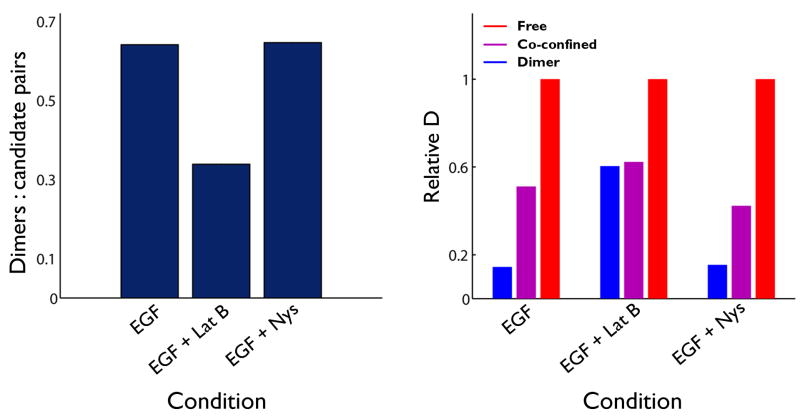
We next explored the influence of membrane architecture on erbB1 dynamics by disruption of actin polymerization25 or removal of cholesterol39. As expected, disruption of actin increased overall receptor diffusion while sequestration of cholesterol reduced mobility 23, 39(Fig S8). Since the HMM provides an objective fit for the size of confinement zones by optimizing error terms, we could compare their size between treatments. QD-EGF-bound receptors are the most constrained, with a Gaussian sigma of 150 nm (see Supplementary Methods and Supplementary Table 2 and Supplementary Fig. 3d). Nystatin treatment did not change domain size, while LatB increased the size of the confinement zone. Consistent with a reduction in confinement, LatB treatment also reduced the likelihood of dimer formation (Fig 5a). When analyzed based upon state, dimers are confirmed to be more mobile in LatB treated cells. Nystatin treatment had no effect on dimer diffusion (Fig 5b). Taken together, these data are consistent with a role for actin corrals in membrane compartmentalization and indicate that the cortical cytoskeleton has a larger influence on erbB1 than does cholesterol.
Figure 5.
Disruption of the actin cytoskeleton influences receptor dynamics. (a) Ratio of candidate pairs to dimers provides a measure of dimer frequency as a consequence of pharmacologic treatments. The drop in value for LatB-treated cells indicates that the disruption of cytoskeletal corrals reduces the likelihood for receptor encounters. (b) Summary of normalized diffusion coefficients for receptors bound to QD-EGF in the absence or presence of LatB or Nys treatment. States were identified by HMM and Viterbi analyses. Each group of coefficients is normalized to the free state component within that condition.
DISCUSSION
The technical advantages afforded by two-color QD tracking have permitted us to resolve several important and, as yet, unanswered questions related to erbB1 biology. First, we show that there is not a simple relationship between diffusion and dimer status. SPT using probes that distinguish between unoccupied and ligand-bound receptors revealed a broad range of diffusive behavior for monomers and dimers under all experimental conditions (Fig. 1b, Fig. 4d). In A431 cells, we calculate a diffusion coefficient for unoccupied erbB1 monomers of 0.05 μm2 s-1. There is only a slight change in diffusion for the short-lived dimers that form between two unoccupied receptors (0.038 μm2 s-1). Importantly, we are able to report that signaling competent, ligand-bound dimers slow dramatically (0.005 μm2 s-1) compared to ligand-bound dimers whose catalytic activity is inhibited by PD153035 (0.019 μm2 s-1). We conclude that reduced mobility is a complex reflection of the stability and size of the protein aggregate, as well as signaling-mediated changes in the local environment. Importantly, these data contradict the primary assumption of Chung, et al.4 that 2-fold changes in erbB1 diffusion can be used as the sole criteria for identifying dimers.
Second, we are now able to compare off rates for dimers formed on the surface of live cells. We show that dimers composed of 2EGF:2erbB1 are the most stable, confirming predictions based on a large prior body of evidence1. This result is inconsistent with at least one aspect of the negative cooperativity model that predicts EGF-bound monomers should have the lowest interaction stability8. Furthermore, with our long-term SPT, we consistently observed 2EGF:2erbB1 dimers formed from the collision of ligand-bound monomers, rather than binding of a second ligand to an existing 1EGF:2erbB1 complex 9, 21. While a functional role for the singly-liganded dimer may exist3, our results are contradictory to the idea that 1EGF:2erbB1 dimers are the predominant functional species 8. It is notable that dimers composed of 1EGF:2erbB1 are short lived (koff = 0.738 s-1 in A431 cells) while dimers between unoccupied receptors are even more transient (koff = 1.24 s-1). Considering the short lifetime, we predict that these complexes are markedly less efficient at initiating signal transduction than 2EGF:2erbB1 (koff = 0.271 s-1). Consistent with other reports7, 12, 40, we propose that dimers constitute the minimal signaling competent unit and that the strength of signal is likely to be directly related to the cumulative output from short and long-lived dimer events.
The dimer off rates in HeLa cells follow the same trends as those on A431 cells, but are overall slightly smaller (Fig 3c, Supplementary Table 2). This suggests that the cellular environment, such as the availability of binding partners, can influence dimer stability. These values will be useful for updating mathematical models of erbB1 signaling, which have typically used two-16 to 20-fold slower dissociation rates15, 17.
Finally, this work directly addresses the role for membrane organization in promoting signal initiation34. We show that the same two receptors can repeatedly encounter each other within membrane domains, with sequential periods of dimerization. Since encounters between proteins are diffusion-limited, stochastic processes, this cannot be explained without evoking contribution of membrane compartmentalization. Results in LatB-treated cells indicate a prominent role for actin corrals in establishing confinement zones that promote dimerization (Fig. 5a). Remarkably, the only change in Nys-treated cells was a slower diffusion rate (Supplementary Fig. 5), consistent with a major role for cholesterol in influencing membrane fluidity, but suggesting that cholesterol is not the primary factor in erbB1 compartmentalization.
An important feature of this study is the ability to quantify diffusion within confinement zones. We confirm that diffusion becomes more restricted within microdomains26, 41, 42, but we also have the ability to specifically identify pairs of receptors that are co-confined. Ligand-bound but PD153035-inhibited receptors do not demonstrate as dramatic a change in diffusion when co-confined. We speculate that repeated dimer events can propagate signals that alter the local environment, which in turn slows receptors and promotes rebinding. These signals could influence diffusion by promoting the formation of protein scaffolds or inducing cytoskeletal assembly. Notably, there is not a marked reduction in mobility when the candidate pairs in a domain are composed of one unoccupied and one ligand-bound receptor (EGF&VHH, Figure 4d). This supports the model that the less stable dimers formed by these receptors cannot sustain signals that lead to receptor immobilization and membrane rearrangement.
The new quantitative methods described here capture dynamic receptor interactions at the single molecule level, providing detail that is obscured in traditional methods. Since dimerization is a common mechanism for signal initiation, our approach can be applied across many receptor systems to further our understanding of dimerization kinetics, receptor mobility and the influences of membrane structure on regulating signal transduction.
Materials and Methods
Reagents
Biotinylated EGF was purchased at a 1:1 stoichiometry (Invitrogen, Carlsbad, CA) and singly biotinylated VHH fragment EGb4 was produced using the BirA expression system23. Biotin-EGF or biotin-VHH were conjugated to Qdot® 655 or Qdot® 585-streptavidin conjugate (Invitrogen, Carlsbad, CA) in PBS + 1% (w/v) BSA to generate stock solutions of 30 nM 1:1 monovalent QD-conjugates19. Stock solutions were stored at 4°C and used for up to two weeks. Biotin-EGF was conjugated to Qdot® 625-streptavidin for bleedthrough experiments.
Cell culture
A431 Human Epithelial carcinoma cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) without phenol red (Sigma-Aldrich, St. Louis, MO), with 10% (v/v) fetal bovine serum (FBS; Invitrogen, Carlsbad, CA), penicillin and streptomycin. HeLa cells were cultured in DMEM, 10% (v/v) FBS, antibiotics. 1% Hepes (v/v), and 1% (v/v) sodium pyruvate. For live cell imaging, cells were plated in 8-well Lab-Tek chambers (Nunc, Rochester, NY) to achieve <50% confluency.
Cell treatment for SPT experiments
Cells in Lab-Tek chambers were imaged in Tyrode’s with 0.1% (w/v) BSA and 20 mM glucose. Prior to imaging, cells were serum starved in DMEM lacking FBS for 4 hr. QD-EGF was added at a concentration of 2 pM and QD-VHH at 20 pM to the appropriate imaging chambers and monitored for up to 7 minutes following labeling. Experiments with PD153035 (Tocris Bioscience, Ellisville, MO) were performed after serum starvation in the presence of 1 μM inhibitor; drug was also present during live cell imaging buffer. Where applicable, 10 nM non-fluorescent EGF was added as a dark ligand. Experiments with Latrunculin B (Sigma-Aldrich, St. Louis, MO) or Nystatin (Sigma-Aldrich, St. Louis, MO) were performed following 10 minutes of pre-treatment at 500 nM and 5 μg mL-1, respectively.
Fluorescence Microscopy
Wide field imaging for SPT was performed using an Olympus IX71 inverted microscope equipped with a 60× 1.2 N.A. water objective; an objective heater (Bioptechs, Butler, PA) maintained temperature at (34-36°C). Wide field excitation was provided by a mercury lamp with either a 436/10 nm BP excitation filter and a 50/50 neutral density filter. Emission was collected by an electron multiplying CCD camera (Andor iXon 887) using a QuadView image splitter (Optical Insights) to simultaneously image two spectrally distinct QDs. QD emission was collected using a 600 nm dichroic and the appropriate filters, 655/40 nm and 585/20 BP (Chroma, Rockingham, VT). A single pixel is equivalent to 267 nm.
Image Processing
All image processing was performed using Matlab (The MathWorks, Inc., Natick, MA) in conjunction with the image processing library DIPImage (Delft University of Technology). For descriptions of specific analysis routines see Supplementary Information.
GPU Single Particle Tracking and Track Elongation
Images were acquired at 20 frames per s for a total of 1,000 frames. Single molecule localization and trajectory connection were carried out as previously described29. Complete derivation of the track elongation algorithm is provided in Supporting Online Text. To elongate short tracks, the positions (pi and pi+j) with the smallest Δt for temporally independent tracks are compared using equation 6. Temporally independent tracks with the best p-value that pass a user-defined cutoff are augmented for form an elongated trajectory. Due to temporal independence of short trajectories, the cutoff and Δt for track elongation can be relaxed slightly from those used in the initial connection algorithm without introducing artifacts. This process is repeated until no temporally independent tracks that pass the cutoff remain. An example comparison of short and elongated trajectories is shown in Supplementary Fig 1.
Squared Displacement Analysis
Diffusion analysis of trajectories was performed by square displacement analysis and two-component fitting, as previously described30.
Correlated Motion Analysis
The degree of correlation between pairwise trajectories was determined as previously described25.
Fiducial Data Acquisition and Image Registration
See supplemental text for mathematical details. Images were registered using a calibration image of multi–fluorophore fluorescent beads (0.2 μm Tetraspeck, Invitrogen, Carlsbad, CA) that have an emission spectrum covering the two spectral windows. The camera was sampled by moving a single bead across the field of view using a mechanical stage (Prior Pro Scan II, Rockland, MA) to obtain good sampling over the field of view. A fiducial data set was acquired at the beginning and end of each chamber tested. Corresponding beads are identified in each channel and fit to a polynomial calibration model28.
Hidden Markov Model
See supplemental text for mathematical details. We use a three state model: 1) Dimer; 2) Co-confined; 3) Free. The observed parameter in the Hidden Markov Model is the separation. For the dimer and domain model, the distribution of the displacements between the QDs is modeled by a zero mean Gaussian distribution in each (x,y) dimension using σdimer and, σdomain respectively. The resulting observed distribution of displacements is a convolution of the actual displacements and the errors in measurement, which are also assumed to contribute as unbiased Gaussian distributions. The value σdimer is taken as that expected from combining information from crystal structure measurements and the size of QDs, whereas σdomain is varied to find the best fit across all data sets of a specific condition. For the Free model, the probability density is calculated as a function of the observed distance in the previous frame and a characteristic diffusion constant.
The set of rate constants is found by maximizing the likelihood over all interactions of two QDs for a specific condition, The errors on each parameter are given as standard errors and are calculated as (Hi,i-1)-0.5 where H is the Hessian matrix of the negative log-likelihood and i denotes one of the estimated rate constants. Given the set of rate parameters found in the estimation step, the Viterbi algorithm36 is used to identify the most likely state within individual QD interactions.
Statistical Analysis
Errors in two-component fits for square displacement are reported as 95% confidence intervals.
Supplementary Material
Acknowledgments
This work was supported by National Science Foundation CAREER MCB-0845062 and the Oxnard Foundation (D.S.L.), by National Institutes of Health R21RR024438 (K.A.L.), by National Institutes of Health R01CA119232 (B.S.W.) and by NIH P50GM085273. S.T.L-N. and P.J.C. were supported by National Science Foundation Integrative Graduate Education and Research Traineeships. We are grateful for the contributions of Geoffrey Graff (ideocraft) in composing images for figures. We thank colleagues in the New Mexico Spatiotemporal Modeling Center for valuable input as well as technical support in the University of New Mexico Cell Pathology Laboratory. Images in this paper were generated in the University of New Mexico & Cancer Center Fluorescence Microscopy Shared Resource.
Abbreviations
- A431
epidermoid carcinoma cell line
- EGF
epidermal growth factor
- erbB1
erythroblastic leukemia viral oncogene homolog 1 or epidermal growth factor receptor
- HMM
Hidden Markov Model
- LatB
Latrunculin B
- Nys
Nystatin
- QD
quantum dot
- SD
square displacement
- SPT
single particle tracking
- VHH
variable fragment of heavy chain antibody
Footnotes
Author Contributions S.T.L. performed experiments. S.T.L. and K.A.L. developed the HMM analysis. P.J.C. and K.A.L. implemented the overlay algorithm. D.S.L., B.S.W., S.T.L. and K.A.L. designed and interpreted experiments. R.R. and P.M.v.B. provided essential camelid antibodies. All authors contributed to preparing the manuscript. D.S.L. directed the project.
No conflicts of interest are declared.
References
- 1.Schlessinger J. Ligand-induced, receptor-mediated dimerization and activation of EGF receptor. Cell. 2002;110:669–672. doi: 10.1016/s0092-8674(02)00966-2. [DOI] [PubMed] [Google Scholar]
- 2.Ferguson KM, et al. EGF activates its receptor by removing interactions that autoinhibit ectodomain dimerization. Mol Cell. 2003;11:507–517. doi: 10.1016/s1097-2765(03)00047-9. [DOI] [PubMed] [Google Scholar]
- 3.Alvarado D, Klein DE, Lemmon MA. Structural basis for negative cooperativity in growth factor binding to an EGF receptor. Cell. 2010;142:568–579. doi: 10.1016/j.cell.2010.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chung I, et al. Spatial control of EGF receptor activation by reversible dimerization on living cells. Nature. 2010;464:783–787. doi: 10.1038/nature08827. [DOI] [PubMed] [Google Scholar]
- 5.Clayton AH, et al. Ligand-induced dimer-tetramer transition during the activation of the cell surface epidermal growth factor receptor-A multidimensional microscopy analysis. J Biol Chem. 2005;280:30392–30399. doi: 10.1074/jbc.M504770200. [DOI] [PubMed] [Google Scholar]
- 6.Hofman EG, et al. Ligand-induced epidermal growth factor receptor (EGFR) oligomerization is kinase-dependent and enhances internalization. J Biol Chem. 2010 doi: 10.1074/jbc.M110.164731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lidke DS, Lidke KA, Rieger B, Jovin TM, Arndt-Jovin DJ. Reaching out for signals: filopodia sense EGF and respond by directed retrograde transport of activated receptors. J Cell Biol. 2005;170:619–626. doi: 10.1083/jcb.200503140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Macdonald JL, Pike LJ. Heterogeneity in EGF-binding affinities arises from negative cooperativity in an aggregating system. Proc Natl Acad Sci U S A. 2008;105:112–117. doi: 10.1073/pnas.0707080105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sako Y, Minoghchi S, Yanagida T. Single-molecule imaging of EGFR signalling on the surface of living cells. Nat Cell Biol. 2000;2:168–172. doi: 10.1038/35004044. [DOI] [PubMed] [Google Scholar]
- 10.Chen L, Merzlyakov M, Cohen T, Shai Y, Hristova K. Energetics of ErbB1 transmembrane domain dimerization in lipid bilayers. Biophys J. 2009;96:4622–4630. doi: 10.1016/j.bpj.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dawson JP, et al. Epidermal growth factor receptor dimerization and activation require ligand-induced conformational changes in the dimer interface. Mol Cell Biol. 2005;25:7734–7742. doi: 10.1128/MCB.25.17.7734-7742.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lemmon MA, et al. Two EGF molecules contribute additively to stabilization of the EGFR dimer. EMBO J. 1997;16:281–294. doi: 10.1093/emboj/16.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sherrill JM, Kyte J. Activation of Epidermal Growth Factor Receptor by Epidermal Growth Factor. Biochemistry. 1996;35:5705–5718. doi: 10.1021/bi9602268. [DOI] [PubMed] [Google Scholar]
- 14.Sturani E, et al. Kinetics and regulation of the tyrosine phosphorylation of epidermal growth factor receptor in intact A431 cells. Mol Cell Biol. 1988;8:1345–1351. doi: 10.1128/mcb.8.3.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kholodenko BN, Demin OV, Moehren G, Hoek JB. Quantification of Short Term Signaling by the Epidermal Growth Factor Receptor. Journal of Biological Chemistry. 1999;274:30169–30181. doi: 10.1074/jbc.274.42.30169. [DOI] [PubMed] [Google Scholar]
- 16.Hsieh MY, et al. Stochastic simulations of ErbB homo and heterodimerisation: potential impacts of receptor conformational state and spatial segregation. IET Syst Biol. 2008;2:256–272. doi: 10.1049/iet-syb:20070073. [DOI] [PubMed] [Google Scholar]
- 17.Shankaran H, Wiley HS, Resat H. Modeling the effects of HER/ErbB1-3 coexpression on receptor dimerization and biological response. Biophys J. 2006;90:3993–4009. doi: 10.1529/biophysj.105.080580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Treanor B, Batista FD. Organisation and dynamics of antigen receptors: implications for lymphocyte signalling. Curr Opin Immunol. 2010;22:299–307. doi: 10.1016/j.coi.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 19.Kusumi A, Sako Y, Yamamoto M. Confined Lateral Diffusion of Membrane Receptors as Studied by Single Particle Tracking (Nanovid Microscopy). Effects of Calcium-Induced Differentiation in Cultured Epithelial Cells. Biophysical Journal. 1993;65:2021–2040. doi: 10.1016/S0006-3495(93)81253-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simson R, Sheets ED, Jacobson K. Detection of temporary lateral confinement of membrane proteins using single-particle tracking analysis. Biophys J. 1995;69:989–993. doi: 10.1016/S0006-3495(95)79972-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ichinose J, Murata M, Yanagida T, Sako Y. EGF signalling amplification induced by dynamic clustering of EGFR. Biochem Biophys Res Commun. 2004;324:1143–1149. doi: 10.1016/j.bbrc.2004.09.173. [DOI] [PubMed] [Google Scholar]
- 22.Nagy P, Claus J, Jovin TM, Arndt-Jovin DJ. Distribution of resting and ligand-bound ErbB1 and ErbB2 receptor tyrosine kinases in living cells using number and brightness analysis. Proc Natl Acad Sci U S A. 2010;107:16524–16529. doi: 10.1073/pnas.1002642107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orr G, et al. Cholesterol dictates the freedom of EGF receptors and HER2 in the plane of the membrane. Biophys J. 2005;89:1362–1373. doi: 10.1529/biophysj.104.056192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lidke DS, et al. Quantum dot ligands provide new insights into erbB/HER receptor-mediated signal transduction. Nat Biotechnol. 2004;22:198–203. doi: 10.1038/nbt929. [DOI] [PubMed] [Google Scholar]
- 25.Andrews NL, et al. Actin restricts FcepsilonRI diffusion and facilitates antigen-induced receptor immobilization. Nat Cell Biol. 2008;10:955–963. doi: 10.1038/ncb1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roullier High-Affinity Labeling and Tracking of Individual Histidine-Tagged Proteins in Live Cells Using Ni2+ Tris-nitrilotriacetic Acid Quantum Dot Conjugates. NanoLetters. 2009;9:1228–1234. doi: 10.1021/nl9001298. [DOI] [PubMed] [Google Scholar]
- 27.Hofman EG, et al. EGF induces coalescence of different lipid rafts. J Cell Sci. 2008;121:2519–2528. doi: 10.1242/jcs.028753. [DOI] [PubMed] [Google Scholar]
- 28.Churchman LS, Okten Z, Rock RS, Dawson JF, Spudich JA. Single molecule high-resolution colocalization of Cy3 and Cy5 attached to macromolecules measures intramolecular distances through time. Proc Natl Acad Sci U S A. 2005;102:1419–1423. doi: 10.1073/pnas.0409487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith CS, Joseph N, Rieger B, Lidke KA. Fast, single-molecule localization that achieves theoretically minimum uncertainty. Nat Methods. 2010;7:373–375. doi: 10.1038/nmeth.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Keijzer S, et al. A spatially restricted increase in receptor mobility is involved in directional sensing during Dictyostelium discoideum chemotaxis. J Cell Sci. 2008;121:1750–1757. doi: 10.1242/jcs.030692. [DOI] [PubMed] [Google Scholar]
- 31.Fry DW, et al. A specific inhibitor of the epidermal growth factor receptor tyrosine kinase. Science. 1994;265:1093–1095. doi: 10.1126/science.8066447. [DOI] [PubMed] [Google Scholar]
- 32.Egeblad M, Mortensen OH, van Kempen LC, Jaattela M. BIBX1382BS, but not AG1478 or PD153035, inhibits the ErbB kinases at different concentrations in intact cells. Biochem Biophys Res Commun. 2001;281:25–31. doi: 10.1006/bbrc.2001.4302. [DOI] [PubMed] [Google Scholar]
- 33.Rabiner LR. A Tutorial on Hidden Markov Models and Selected Applications in Speech Recognition. Proceedings of the IEEE. 1989;77:257–286. [Google Scholar]
- 34.Chen Y, Yang B, Jacobson K. Transient confinement zones: a type of lipid raft? Lipids. 2004;39:1115–1119. doi: 10.1007/s11745-004-1337-9. [DOI] [PubMed] [Google Scholar]
- 35.Lillemeier BF, Pfeiffer JR, Surviladze Z, Wilson BS, Davis MM. Plasma membrane-associated proteins are clustered into islands attached to the cytoskeleton. Proc Natl Acad Sci U S A. 2006;103:18992–18997. doi: 10.1073/pnas.0609009103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Forney GD. Viterbi algorithm. Proceedings of the IEEE. 1973;61:268–278. [Google Scholar]
- 37.Hillman GM, Schlessinger J. Lateral diffusion of epidermal growth factor complexed to its surface receptors does not account for the thermal sensitivity of patch formation and endocytosis. Biochemistry. 1982;21:1667–1672. doi: 10.1021/bi00536a030. [DOI] [PubMed] [Google Scholar]
- 38.Livneh E, et al. Large Deletions in the Cytoplasmic Kinase Domain of the Epidermal Growth Factor Receptor Do Not Affect Its Lateral Mobility. Journal of Cell Biology. 1986;103:327–331. doi: 10.1083/jcb.103.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao Z, Zhang W, Yang Y, Xu L, Fang X. Single-molecule diffusion study of activated EGFR implicates its endocytic pathway. Biochem Biophys Res Commun. 2008;369:730–734. doi: 10.1016/j.bbrc.2008.02.084. [DOI] [PubMed] [Google Scholar]
- 40.Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 41.Daumas F, et al. Confined diffusion without fences of a g-protein-coupled receptor as revealed by single particle tracking. Biophys J. 2003;84:356–366. doi: 10.1016/S0006-3495(03)74856-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Douglass AD, Vale RD. Single-molecule microscopy reveals plasma membrane microdomains created by protein-protein networks that exclude or trap signaling molecules in T cells. Cell. 2005;121:937–950. doi: 10.1016/j.cell.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.