Abstract
The ability to visualize specific DNA sequences, on chromosomes and in nuclei, by fluorescence in situ hybridization (FISH) is fundamental to many aspects of genetics, genomics and cell biology. Probe selection is currently limited by the availability of DNA clones or the appropriate pool of DNA sequences for PCR amplification. Here, we show that liquid-phase probe pools from sequence capture technology can be adapted to generate fluorescently labelled pools of oligonucleotides that are very effective as repeat-free FISH probes in mammalian cells. As well as detection of small (15 kb) and larger (100 kb) specific loci in both cultured cells and tissue sections, we show that complex oligonucleotide pools can be used as probes to visualize features of nuclear organization. Using this approach, we dramatically reveal the disposition of exons around the outside of a chromosome territory core and away from the nuclear periphery.
Keywords: chromosome territories, exome, fluorescence in situ hybridization, nuclear organization, oligonucleotides, tissue sections
Introduction
Using standard fluorescence in situ hybridization (FISH) probes, specific loci, classes of DNA sequence, genomic regions and even whole chromosomes can be detected (Volpi and Bridger 2008; Levsky and Singer 2003). FISH probes can be labelled either directly with fluorescent nucleotides or indirectly and then detected with fluorescent antibodies or affinity molecules. DNA FISH probes to detect small genomic regions are typically generated from available cloned DNA resources, e.g. bacterial artificial chromosomes (BACs) or fosmids and so are constrained by the availability of such highly characterized resources.
DNA FISH probes for large regions of complex genomes, e.g. whole chromosomes or chromosomal regions, are typically generated by PCR amplification of all sequences from that region. Hybridization signals from repetitive sequences are then normally suppressed by annealing with unlabelled Cot1 DNA, whose composition is rather ill defined. Moreover, this is only effective for repeats of high enough copy number to be in the Cot1 fraction and is only practicable for organisms where there is a ready supply of CotI DNA. The high sequence complexity of these ‘chromosome paint’ probes means that the specific activity of label for any particular sequence is low. As a result, not all sequences encompassed by such chromosome paints can be detected effectively. This likely underlies the phenomenon of ‘looping out’ whereby hybridization signal for a specific region, detected with relatively low complexity probes (e.g. plasmid, fosmid, cosmid or BAC clones), can appear to be located outside of the host chromosome territory as detected by a paint (Mahy et al. 2002).
Sequence capture technologies were developed to enrich for designated regions of complex genomes with the aim of reducing complexity prior to sequencing (Albert et al. 2007; Okou et al. 2007; Hodges et al. 2007). This is revolutionizing the detection of sequence variants associated with Mendelian disease and with cancer (Meyerson et al. 2010). The oligonucleotides used are usually immobilized on a solid support, e.g. microarray, but more recently sequence capture in solution, using bead capture with biotinylated oligonucleotides, has been developed as an effective method for targeted sequencing (Bainbridge et al. 2010; Fairfield et al. 2011). Here, we have addressed whether the probe design principles used to select regions for targeted sequencing, can be developed to generate high-complexity pools of oligonucleotides that can act as improved repeat-free FISH probes. Indeed, we demonstrate the effectiveness of directly fluorescently labelled oligonucleotide pools as flexible, sensitive and specific probes for use in DNA FISH.
We show that labelled oligonucleotide pools robustly detect both small (15 kb) and larger (100 kb) genomic regions with high specificity and efficiency in both nuclei and chromosomes of cultured cells, as well as in the cell nuclei of tissue sections. Using an oligonucleotide pool that represents the exome from a single mouse chromosome, we reveal, for the first time en masse, that the exome (i.e. genic) portion of the genome is organized into decondensed chromatin that extends away from the core of chromosome territories and toward the nuclear interior.
Materials and methods
Oligonucleotide pools
Mouse oligonucleotide capture pools targeting various regions of the HoxD locus (Table 1) were produced at Roche NimbleGen (Madison, WI) using the proprietary method for sequence capture EZ probe pools (Parikh et al. 2010). Genome co-ordinates were taken from the USCS mouse genome browser (NCBI36/mm9). Hoxd probes were repeat masked and filtered for uniqueness using a cutoff of five close matches with a minimum match length of 38 nucleotides and allowing for up to five insertions or deletions. Mean oligo length was 75 bases but ranged from ~60–100 bases (Table 1). Each oligonucleotide synthesis culminated in the incorporation of a single fluorophore (6-FAM or Texas Red) on the 5′end, in lieu of the standard biotin used for sequence-capture applications.
Table 1.
Details of the sequence capture probe pools used in this study. Genome co-ordinates are taken from the USCS mouse genome browser (NCBI36/mm9)
| Region | Fluorochrome | Genome co-ordinates | Genomic coverage (bp) | Number of probes | Probe length (bases) | ||||
|---|---|---|---|---|---|---|---|---|---|
| Start (mouse exome) | End | Mean | Range | Median | Mode | ||||
| MMU2 | Texas Red | Whole exome of MMU2 | 123,067 | 76.36 | 59–103 | 76.00 | 75.00 | ||
| HoxD locus | 6-FAM | 74,474,157 | 74,605,438 | 131,281 | 56,696 | 74.05 | 58–92 | 74.00 | 74.00 |
| Hoxd3-Hoxd1 | Texas Red | 74,575,438 | 74,605,438 | 30,000 | 12,991 | 74.57 | 61–89 | 75.00 | 74.00 |
| Hoxd1 | Texas Red | 74,590,438 | 74,605,438 | 15,000 | 5,962 | 74.74 | 62–87 | 75.00 | 74.00 |
| Hoxd1(b) | Texas Red | 74,590,438 | 74,597,938 | 7,500 | 3,310 | 74.96 | 62–87 | 75.00 | 74.00 |
To generate a Mus musculus chromosome 2 (MMU2) exome probe pool, a comprehensive mouse whole genome exome design was developed bioinformatically as a first step. This was generated by obtaining co-ordinates for CDS features from the non-redundant intersection of 21,770 unique ENSEMBL gene IDs, 14,479 unique VEGA gene IDs and 1,876 genes without ENSEMBL or VEGA gene IDs from UCSC Known Genes (Fairfield et al. 2011). All loci annotated as pseudogenes and those encoding the highly repetitive olfactory receptors were eliminated from the list. All mouse microRNA genes were included from mirDB (http://mirdb.org/miRDB/). The exons and mir were extracted as CDS features in the form of Chr, start, stop from the consolidated list using the UCSC genome browser (mouse build NCBI36/mm9). Exonic and or mir argets were padded to a minimum length of 100 bp, and overlapping regions were consolidated. Probes were repeat masked and filtered for uniqueness using a cutoff of five close matches with a minimum match length of 38 nucleotides, and allowing for up to five insertions or deletions. This genome-wide exome design consists of 205,360 consolidated regions and is comprised of a total real estate of 52,709,234 bp. The pool targeting the exome of the MMU2 was then designed by extracting all exons mapping to MMU2 from the genome-wide exome list and resulted in a new probe pool design consisting of 32,517 regions covering 5,405,862 bp.
Probe hybridization to cells
Four hundred nanogrammes (4 μl) of Texas Red or 6-carboxyfluorescein (6-FAM)-labelled sequence capture pools were added to (21 μl) hybridization mix (50% formamide, 2× SSC, 1% Tween20, 10% Dextran Sulphate), heated for 5 min at 70°C to ensure their single-stranded nature and then snap cooled on ice. To combine a standard chromosome paint with a sequence-capture probe pool, 15 μl of ready-to-use commercial MMU2 paint, already containing mouse CotI (Cambio) in hybridization mix was denatured for 5 min at 70°C, reannealled for 15 min at 37°C and briefly mixed with 400 ng of exome paint in hybridization mix before hybridizing to the slide.
Methanol/acetic acid (3:1) fixed mouse embryonic stem (ES) cells dropped onto glass slides were incubated in 100 μg/ml RNase A in 2× SSC for 1 h at 37°C, washed briefly in 2× SSC, put through an alcohol series and air dried. Slides were oven heated to 70°C for 5 min, denatured in 70% formamide/2× SSC (pH 7.5) for 1 min at 70°C, cooled in 70% ethanol on ice for 2 min and dehydrated through 90% and 100% ethanol for 2 min each before air drying.
Probe was hybridized to the slide under a sealed coverslip overnight at 37°C. Slides were washed the next day for 4 × 3 min in 2× SSC at 45°C, 4 × 3 min in 0.1× SSC at 60°C, mounted in Vectashield (Vector) with 0.5 μg/ml 4,6-diaminidino-2-phenylidole (DAPI) and sealed with rubber solution.
Probe hybridization to tissue sections
Paraffin embedded formalin-fixed mouse embryo sections were heated to 60°C for 20 min, xylene treated for 4 × 10 min, cleared with ethanol washes and microwaved in 0.1 M citrate buffer at pH 6.0 for 20 min, cooled for 20 min and then stored in water(Kerr et al. 2010). Slides were washed briefly in 2× SSC, incubated in 2× SSC at 75°C for 5 min before being denatured in 70% formamide/2× SSC at pH 7.5 for 3 min, put through an alcohol series and air dried. They were then hybridized to probe as above.
Image analysis
Slides were imaged as previously described (Kocanova et al. 2010). The proportion of nuclear area covered by hybridization signal was quantified from segmented chromosome territories. Domain areas were calculated using an in-house IPLab analysis script (Becton, Dickinson and Company, Franklin Lakes, NJ). In brief, The DAPI-stained nucleus was auto-segmented using the IPLab triangle algorithm. The FITC or Texas Red-labelled domains were then segmented by manually selecting the threshold value that included all the domain signal area. In practice, the two domains were clearly identifiable. In cases where the signal from one domain segmented into more than one area, a marquee was placed around the constituent segments and the total area calculated by the script. Only those segments lying inside the DAPI-stained nucleus were included in the analysis. The difference in the proportion of nuclear area covered by signals from chromosome paint probes was tested using the non-parametric Mann–Whitney U test.
To quantify the distribution of chromosome paint signals across the nucleus, DAPI-segmented nuclei were divided into five concentric shells of approximately equal area, eroded in from the edge of the nucleus (shell 1) to the nuclear centre (shell 5). The proportion of DAPI signal and chromosome paint hybridisation signals present in each of five shells was then calculated (Boyle et al. 2001). Differences in the proportion of signals from chromosome paint probes between erosion shells were tested using Student’s t test.
Results
Detection of small specific loci
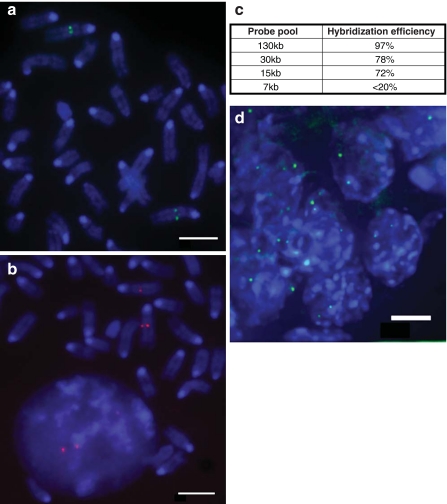
Previously, detection of specific loci by FISH has generally been limited by the availability of cloned resources such as plasmids, fosmid or BAC probes, but sequence capture pools are free from such constraints so potentially could be a route to a new generaration of flexible FISH probes. The design process for such oligonucleotides also means that repeat sequences can readily be excluded from the labelled probe pool. Previously, we have extensively analysed the nuclear organization of the murine HoxD locus on MMU2, both in ES cells and embryos (Morey et al. 2007). Therefore, to test the efficacy of such an approach, we first generated a 6-FAM directly labelled oligonucleotide pool representing a 130-kb domain encompassing the entire HoxD locus (Table 1). This represents the size of genomic region typically encompassed by a BAC-based probe. Hybridization signal from this probe clearly and efficiently (97%) detected the Hoxd locus on MMU2 in metaphase spreads (Fig. 1a) and in the nuclei of mouse ES cells, without the need for any suppression of repeats with Cot1 fraction.
Fig. 1.
Detection of specific loci with small sequence capture pools. a Metaphase chromosomes from mouse ES cells hybridized with 6FAM (green) 130 kb Hoxd sequence capture probe pool. DNA was stained with DAPI. Scale bar = 5 μm. b Mouse metaphase chromosomes hybridized with TexasRed 15 kb sequence capture probe for Hoxd1 (Table 1). Scale bar = 5 μm. c Efficiency with which probe pools of decreasing genomic target size detect loci on the chromatids of metaphase chromosomes (n > 160). d As in (a) but hybridization to nuclei in a mouse embryo section. Scale bar = 3 μm
To determine whether probe pools representing smaller genomic regions would also be effective we then analysed probe pools for progressively smaller sub-regions from the 3′end of Hoxd (Table 1). Hybridization efficiency for a 30-kb target pool was good—at almost 80%—and remained quite useable (>70%) down to a 15-kb pool (Fig. 1b, c). However, while functional below this size, with a 7.5-kb pool we found the hybridization efficiency (<20%) too low to be reliably used for the analysis of nuclear organization.
Probe pools were also very effective at detecting a specific locus with high efficiency and low background in the nuclei of mouse embryo sections (Fig. 1d).
Complex oligonucleotide probe pools can paint specific chromosomes
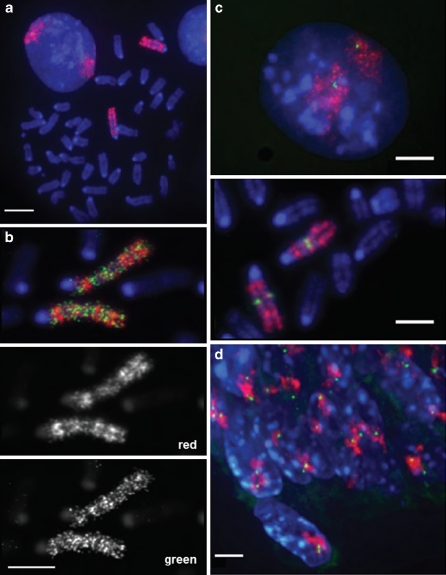
FISH probes for detecting whole chromosomes or chromosomal regions are complex and typically are generated by PCR amplification of all sequences from that region. To test the effectiveness of sequence capture oligonucleotides for the detection of specific chromosomes, we generated a Texas Red-labelled probe pool representing the whole exome of MMU2 (Table 1). Since repeat sequences were eliminated from the probe pool at the design stage, we hybridized this probe to mouse metaphase chromosomes and nuclei without any repeat suppression by added mouse CotI DNA. Bright hybridization signal was confined to the chromosomes two with, remarkably, little to no background elsewhere in the genome (Fig. 2a). The distribution of hybridization signals gave a distinct R-band pattern along the chromosome length, consistent with the observation that CpG islands (generally marking gene promoters) are concentrated in R-bands regions of the mouse genome (Cross et al. 1997). Dual hybridization of this probe set together with an FITC-labelled commercially available paint for MMU2 also revealed this banding pattern and the better quality hybridization signal from the sequence capture probe (Fig. 2b).
Fig. 2.
Detection of specific loci and whole chromosomes with sequence capture pools. a Metaphase chromosome spread and interphase nucleus from mouse ES cells hybridized with TexasRed MMU2 exome sequence-capture probe pool without the addition of any mouse CotI DNA for repeat suppression. DNA was stained with DAPI. Scale bar = 5 μm. b Metaphase chromosomes from mouse ES cells hybridized with TexasRed MMU2 exome sequence capture probe pool and FITC-labelled (green) standard MMU2 chromosome paint (Cambio). Scale bar = 5 μm. Grey-scale images of the red exome paint signal and the green chromosome paint are shown below. c Mouse ES cell nucleus (top) and metaphase chromosomes (bottom) co-hybridized with TexasRed MMU2 exome and 6FAM (green) 130 kb Hoxd sequence capture pools. Scale bar = 3 μm. d As in (c) but hybridization to mouse embryo section. Scale bar = 3 μm
We combined the Texas Red MMU2 exome sequence capture pool with the 6-FAM labelled 130-kb probe pool specific for the Hoxd locus to demonstrate the efficient detection of large complex and smaller specific genomic targets together on chromosomes and nuclei from cultured cells (Fig. 2c) and on nuclei of tissue sections (Fig. 2d).
Complex oligonucleotide probe pools can reveal chromosome organization
By combining traditional chromosome paint probes (labelled by PCR amplification of catch-linkered chromosomal DNA or purchased commercially) with probes for specific loci, we have previously demonstrated by FISH that gene-rich transcriptionally active loci are decondensed in the nucleus (Gilbert et al. 2004) and appear to lie outside of the chromosome territory as defined by chromosome paint signal (Mahy et al. 2002; Chambeyron and Bickmore 2004). These regions were interpreted as being in decondensed and mobile chromatin that can dynamically loop out from the main mass of the chromosome territory (Morey et al. 2007, 2009; Muller et al. 2010). It is the open chromatin structure of such regions that likely reduces their effective detection by low specific activity chromosome paints.
This level of chromosome organization is not an artefact of FISH since it can be seen in living cells by combining photoactivatable histone marking of a specific chromosome territory with lacO-lacI-GFP marking of a specific locus (Muller et al. 2010). However, from these studies of a handful of loci, it is hard to know the extent of this level of nuclear organization. Because looped out loci are often gene rich, we reasoned that the probe pool representing all the exons from a specific chromosome (MMU2) might be able to visualize these regions en masse.
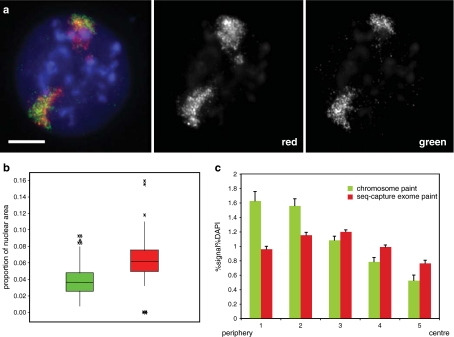
Indeed, there were striking differences in the hybridization signal from the exome probe compared with that from the commercial whole MMU2 paint in interphase nuclei of mouse ES cells (Fig. 3a). Firstly, hybridization signal from the sequence capture MMU2 exome pool covered a larger proportion of the nuclear area (0.063 ± 0.002) compared with that from the conventional chromosome paint (0.039 ± 0.001, p = 0.000) (Fig. 3b). This indicates the better ability of the exome probe pool to detect the more decondensed gene-rich regions of the chromosome territory.
Fig. 3.
Analysing the nuclear organization of chromosomes. a Left, interphase nucleus from mouse ES cells hybridized with TexasRed MMU2 exome sequence capture probe pool and FITC-labelled (green) standard MMU2 chromosome paint (Cambio). Scale bar = 5 μm. Grey-scale images of the red exome paint signal and the green chromosome paint are shown to the right. b Box plots showing the proportion of nuclear area covered by hybridization signal from the standard MMU2 chromosome paint (green) and the MMU2 exome oligonucleotide pool (red). Boxed area shows the median and interquartile range; asterisks indicate outliers. n > 198 chromosome territories. c Erosion analysis to assess the distributions of hybridization signals from TexasRed MMU2 exome sequence capture probe pool (red) and FITC-labelled MMU2 chromosome paint (green), relative to that of DNA (DAPI), across five concentric shells from the edge (shell 1) to the centre (shell 5) of the nucleus. Graph shows mean (+/– SEM) percent hybridization signal in a particular erosion shell, normalized to the percentage of DAPI in that shell, averaged from 68 ES cell nuclei
We also noted that the exome paint hybridization signal appeared to extend much further into the centre of the nucleus than the signal from the conventional chromosome paint (Fig. 3a). To quantify this we compared the relative proportion of hybridization signals from each paint across five shells of equal area eroded from the periphery (shell 1) through to the centre (shell 5) of >60 nuclei (Fig. 3c) (Boyle et al. 2001). This confirmed the relative depletion from the nuclear periphery of signal from the exome probe pool, compared with that from the conventional paint (mean signal/DAPI ratio in erosion shell 1 = 0.95 ± 0.04 for exome paint, 1.61 ± 0.13 for conventional paint, p < 0.001). Conversely, a higher proportion of exome paint signal (signal/DAPI = 0.76 ± 0.04) was in the most internal nuclear erosion shell 5 compared with signal from the conventional paint (0.52 ± 0.07, p = 0.006). Finally, there is also a significant rise in the proportion of exome paint signal in going from shell 1 (0.95 ± 0.04) to 2 (1.14 ± 0.04, p = 0.01) which is not seen for the conventional MMU2 chromosome paint (shell 1, 1.61 ± 0.13; shell 2, 1.54 ± 0.10, p = 0.67). The rather peripheral localization of hybridization signal from the conventional MMU2 paint in mouse ES cell nuclei is consistent with a previous analysis (Mayer et al. 2005).
The relative enrichment of exome probe signal away from the edge of the nucleus, compared with the bulk MMU2 chromosome territory, is consistent with the radial nuclear organization reported in many cell types, including mouse ES cells (Gilbert et al. 2005; Mayer et al. 2005; Peric-Hupkes et al. 2010; Joffe et al. 2010).
Discussion
Here, we have shown that the flexible design, high specificity and efficiency of sequence capture oligonucleotide probe pools, which obviate the need for repeat suppression, makes them a powerful alternative to conventional probes for analysis of genomes and nuclear organization in multiple organisms by FISH. Yamada et al. (2011) have similarly recently demonstrated the effectiveness of using indirectly labelled oligonucleotide pools for detecting specific sequences in the human genome by FISH. In that case, the oligonucleotide pools were amplified by PCR and then the amplified DNA was labelled with fluorochromes using a commercial labelling kit. This may give higher detection efficiency for very small probes, but the workflow means more processing time is required for labelled probe generation and sequence biases may be introduced during the PCR amplification steps.
Using a probe pool that targets the exome of a specific chromosome, we provide a graphic demonstration that chromosome paints normally only reveal the most compact core of chromosome territories and are not able to detect the extensive amount of gene-rich sequence located external to this. Whilst we have used an exome focused probe pool to reveal a particular aspect of chromosome organization in the nucleus, the flexibility of sequence capture pool design opens up possibility of analysing the nuclear distribution of other classes of DNA sequence en masse.
Acknowledgements
This work was supported by the Medical Research Council, UK and by an ERC advanced grant 249956 to WAB. We thank Paul Perry (MRC HGU) for help with image analysis algorithms.
Competing interest
The authors from Roche NimbleGen Inc, recognize a competing interest in the publication of this manuscript as employees of the company.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- 6-FAM
6-Carboxyfluorescein
- BACs
Bacterial artificial chromosomes
- CDS
coding sequence
- Cot
Concentration x time
- DAPI
4,6-Diaminidino-2-phenylidole
- ES cells
embryonic stem cells
- FISH
Fluorescence in situ hybridization
- FITC
Fluorescein isothiocyanate
- ID
identifier
- MMU
Mus musculus
- PCR
Polymerase chain reaction
- SSC
saline-sodium citrate
- UCSC
University of California Santa Cruz Genome Browser
References
- Albert TJ, Molla MN, Muzny DM, Nazareth L, Wheeler D, Song X, Richmond TA, Middle CM, Rodesch MJ, Packard CJ, Weinstock GM, Gibbs RA. Direct selection of human genomic loci by microarray hybridization. Nat Methods. 2007;4:903–905. doi: 10.1038/nmeth1111. [DOI] [PubMed] [Google Scholar]
- Bainbridge MN, Wang M, Burgess DL, Kovar C, Rodesch MJ, D’Ascenzo M, Kitzman J, Wu YQ, Newsham I, Richmond TA, Jeddeloh JA, Muzny D, Albert TJ, Gibbs RA. Whole exome capture in solution with 3 Gbp of data. Genome Biol. 2010;11:R62. doi: 10.1186/gb-2010-11-6-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle S, Gilchrist S, Bridger JM, Mahy NL, Ellis JA, Bickmore WA. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum Mol Genet. 2001;10:211–219. doi: 10.1093/hmg/10.3.211. [DOI] [PubMed] [Google Scholar]
- Chambeyron S, Bickmore WA. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 2004;18:1119–1130. doi: 10.1101/gad.292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross SH, Lee M, Clark VH, Craig JM, Bird AP, Bickmore WA. The chromosomal distribution of CpG islands in the mouse: evidence for genome scrambling in the rodent lineage. Genomics. 1997;40:454–461. doi: 10.1006/geno.1996.4598. [DOI] [PubMed] [Google Scholar]
- Fairfield H, Gilbert J, Barter M, et al. (2011) Mutation discovery in mice by whole exome sequencing. Genome Biology 12(9): R86 [DOI] [PMC free article] [PubMed]
- Gilbert N, Boyle S, Fiegler H, Woodfine K, Carter NP, Bickmore WA. Chromatin architecture of the human genome: gene-rich domains are enriched in open chromatin fibres. Cell. 2004;118:555–566. doi: 10.1016/j.cell.2004.08.011. [DOI] [PubMed] [Google Scholar]
- Gilbert N, Gilchrist S, Bickmore WA. Chromatin organization in the mammalian nucleus. Int Rev Cytol. 2005;242:283–336. doi: 10.1016/S0074-7696(04)42007-5. [DOI] [PubMed] [Google Scholar]
- Hodges E, Xuan Z, Balija V, Kramer M, Molla MN, Smith SW, Middle CM, Rodesch MJ, Albert TJ, Hannon GJ, McCombie WR. Genome-wide in situ exon capture for selective resequencing. Nat Genet. 2007;39:1522–1527. doi: 10.1038/ng.2007.42. [DOI] [PubMed] [Google Scholar]
- Joffe B, Leonhardt H, Solovei I. Differentiation and large scale spatial organization of the genome. Curr Opin Genet Dev. 2010;20:562–569. doi: 10.1016/j.gde.2010.05.009. [DOI] [PubMed] [Google Scholar]
- Kerr E, Kiyuna T, Boyle S, Saito A, Thomas JS, Bickmore WA. Changes in chromatin structure during processing of wax-embedded tissue sections. Chromosome Res. 2010;18:677–688. doi: 10.1007/s10577-010-9147-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocanova S, Kerr EA, Rafique S, Boyle S, Katz E, Caze-Subra S, Bickmore WA, Bystricky K. Activation of estrogen-responsive genes does not require their nuclear co-localization. PLoS Genet. 2010;6:e1000922. doi: 10.1371/journal.pgen.1000922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levsky JM, Singer RH. Fluorescence in situ hybridization: past, present and future. J Cell Sci. 2003;116:2833–2838. doi: 10.1242/jcs.00633. [DOI] [PubMed] [Google Scholar]
- Mahy NL, Perry PE, Bickmore WA. Gene density and transcription influence the localization of chromatin outside of chromosome territories detectable by FISH. J Cell Biol. 2002;159:753–763. doi: 10.1083/jcb.200207115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer R, Brero A, von Hase J, Schroeder T, Cremer T, Dietzel S. Common themes and cell type specific variations of higher order chromatin arrangements in the mouse. BMC Cell Biol. 2005;6:44. doi: 10.1186/1471-2121-6-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet. 2010;11:685–696. doi: 10.1038/nrg2841. [DOI] [PubMed] [Google Scholar]
- Morey C, Da Silva NR, Perry P, Bickmore WA. Nuclear reorganisation and chromatin decondensation are conserved, but distinct, mechanisms linked to Hox gene activation. Development. 2007;134:909–919. doi: 10.1242/dev.02779. [DOI] [PubMed] [Google Scholar]
- Morey C, Kress C, Bickmore WA. Lack of bystander activation shows that localization exterior to chromosome territories is not sufficient to up-regulate gene expression. Genome Res. 2009;19:1184–1194. doi: 10.1101/gr.089045.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller I, Boyle S, Singer RH, Bickmore WA, Chubb JR. Stable morphology, but dynamic internal reorganisation, of interphase human chromosomes in living cells. PLoS One. 2010;5:e11560. doi: 10.1371/journal.pone.0011560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okou DT, Steinberg KM, Middle C, Cutler DJ, Albert TJ, Zwick ME. Microarray-based genomic selection for high-throughput resequencing. Nat Methods. 2007;4:907–909. doi: 10.1038/nmeth1109. [DOI] [PubMed] [Google Scholar]
- Parikh H, Deng Z, Yeager M, Boland J, Matthews C, Jia J, Collins I, White A, Burdett L, Hutchinson A, Qi L, Bacior JA, Lonsberry V, Rodesch MJ, Jeddeloh JA, Albert TJ, Halvensleben HA, Harkins TT, Ahn J, Berndt SI, Chatterjee N, Hoover R, Thomas G, Hunter DJ, Hayes RB, Chanock SJ, Amundadottir L. A comprehensive resequence analysis of the KLK15-KLK3-KLK2 locus on chromosome 19q13.33. Hum Genet. 2010;127:91–99. doi: 10.1007/s00439-009-0751-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, Graf S, Flicek P, Kerkhoven RM, van Lohuizen M, Reinders M, Wessels L, van Steensel B. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol. Cell. 2010;38:603–613. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpi EV, Bridger JM. FISH glossary: an overview of the fluorescence in situ hybridization technique. Biotechniques. 2008;45:385–386. doi: 10.2144/000112811. [DOI] [PubMed] [Google Scholar]
- Yamada NA, Rector LS, Tsang P, Carr E, Scheffer A, Sederberg MC, Aston ME, Ach RA, Tsalenko A, Sampas N, Peter B, Bruhn L, Brothman AR. Visualization of fine-scale genomic structure by oligonucleotide-based high-resolution FISH. Cytogenet Genome Res. 2011;132:248–254. doi: 10.1159/000322717. [DOI] [PubMed] [Google Scholar]