Abstract
Aims/hypothesis
Intronic single nucleotide polymorphisms within the transcription factor 7-like 2 (TCF7L2) gene are associated with risk of type 2 diabetes. It is widely hypothesised that the predisposing variation is involved in cis-regulation of TCF7L2 activity. The aim of this study was to seek evidence for the existence of novel TCF7L2 isoforms encoded within the type 2 diabetes-associated genomic region.
Methods
We searched expressed sequence tag (EST) databases for novel TCF7L2 transcripts and sought to validate the function and integrity of any isoforms found using a combination of RT-PCR, western blotting and reporter gene techniques.
Results
Analysis of EST databases suggested the presence of an alternative polyadenylation site located in intron 4 of TCF7L2. We used 3′ rapid amplification of cDNA ends and real-time PCR to validate the integrity of this polyadenylation signal and show its wide use across human tissues. Western blotting results are consistent with the use of this polyadenylation signal to generate novel protein isoforms. The alternative polyadenylation signal results in the production of isoforms that retain the β-catenin binding domain but do not possess the high-mobility group box DNA-binding domain. Promoter–reporter gene assays suggest that these isoforms inhibit TCF7L2-dependent target genes by sequestering β-catenin.
Conclusions/interpretation
We have identified a novel polyadenylation signal within TCF7L2 that can result in the production of isoforms that act to repress TCF/LEF-dependent target genes. These findings may provide new insights into the association of TCF7L2 with susceptibility to type 2 diabetes.
Keywords: Alternative polyadenylation, TCF7L2, Type 2 diabetes
Introduction
Studies have shown single nucleotide polymorphism (SNPs) in TCF7L2 to be consistently associated with a risk of type 2 diabetes [1]. Despite evidence that attenuation of transcription factor 7-like 2 (TCF7L2) activity mimics the phenotypic effects seen in human carriers of alleles associated with risk [2–4], no protein-coding variation has been found to explain the association. Therefore it is likely that the association acts via an effect on TCF7L2 transcription, splicing or polyadenylation.
TCF7L2 belongs to the T cell factor/lymphoid-enhancer factor (TCF/LEF) family of transcription factors that use extensive alternative splicing to generate isoforms with unique temporal/spatial profiles and gene/context-dependent transactivation capabilities [5, 6]. Given that each TCF/LEF member binds to the same co-activator (β-catenin) and consensus DNA sequence, the molecular diversity generated by splicing may help to explain the lack of functional redundancy within this family.
Interestingly, most of the post-transcriptional events reported for TCF7L2 involve 3′ exons, which do not lie within the region associated with type 2 diabetes. To explain the link, we hypothesised that there might be as yet undiscovered isoforms transcribed within the associated linkage disequilibrium (LD) block.
Methods
Samples and mRNA analysis
Human tissue RNA from individual donors and pools of donors was purchased from Ambion (Austin, TX, USA) and AMS Biotechnology (Abingdon, UK). No individual’s RNA was present in more than one tissue sample. Snap-frozen islets were bought from the National Disease Research Interchange (Philadelphia, PA, USA), and RNA extracted (mirVana miRNA isolation kit; Ambion).
All RNA was DNase-treated (TURBO DNA-free kit; Ambion). For 3′ rapid amplification of cDNA ends (RACE), RNA was reverse-transcribed using an oligo-dT anchor primer (GCTGTCAACGATACGCTACGTAACGGCATGACAGTGT(15)) and amplified by nested PCR (Advantage 2 PCR Kit; Clontech, Paris, France) using gene-specific (GSP) and adaptor-specific (ASP) primers (GSP1_GCACACATTGTCGTAAGTAACCTCCCA, ASP1_GCTGTCAACGATACGCTACGTAAC, GSP2_GTAAGTAACCTCCCAGAGATGATGGCT, ASP2_CGCTACGTAACGGCATGACAGTG). PCR methods were as detailed in the manufacturers’ instructions. Second-round product was resolved by agarose gel electrophoresis and, owing to the presence of one band, sequenced using the ABI 3730 DNA sequencer (Applied Biosystems, Foster City, CA, USA).
To quantify polyadenylation site usage in tissues, RNA was reverse-transcribed using the SuperScript III First-Strand Synthesis System (Invitrogen, Paisley, UK). TaqMan probes (Applied Biosystems) were designed to amplify the isoform-specific coding region and an intronic region downstream of the alternative cleavage site. Inventoried TaqMan probes (Applied Biosystems) were used to measure the expression of the housekeeping genes GAPDH, B2M and GUSB.
All quantitative PCR reactions were run on an ABI 7900HT using TaqMan Universal PCR Master Mix (Applied Biosystems). Relative expression was calculated using the comparative Ct method [7]. The GeNorm [8] algorithm was used to normalise expression of full-length TCF7L2.
Western blotting
Nuclear extracts from human adult pancreas, small intestine (AMS Biotechnology), HeLa cells (Santa Cruz Biotechnology, Santa Cruz, CA, USA), and MIN6 cells (lysed in RIPA buffer) were denatured and reduced in 2× Laemmli buffer. Extracts were separated on a 10% polyacrylamide gel and transferred to PVDF membrane before being blocked in 5% milk for 1 h at 4°C and incubated overnight at 4°C with primary antibody diluted to 1:1,000 (polyclonal antibody [pAb], sc-8631; Santa Cruz Biotechnology; monoclonal antibody [mAb], #2565; Cell Signaling Technology, Danvers, MA, USA). Horseradish peroxidase-conjugated secondary antibody (for pAb, sc-2020; for mAb, sc-2004; Santa Cruz Biotechnology) was added (1:20,000) for 1 h at room temperature before chemiluminescent detection using SuperSignal West Pico Substrate (Pierce, Rockford, IL, USA). Where appropriate, blots were stripped and reprobed for fibrillarin (sc-25397; Santa Cruz Biotechnology) or tubulin (T5168; Sigma Aldrich, St Louis, MO, USA). In the blocking peptide experiment, membrane was incubated overnight with blocking peptide (sc-8631P) and primary pAb.
Cloning and reporter gene assay
The coding sequence for isoforms generated using the alternative polyadenylation signal was amplified by PCR from human cDNA and cloned into pIRES2-EGFP (Clontech). This construct is called Poly(A). Subsequently, D16A Poly(A) was generated by site-directed mutagenesis (QuikChange II XL Site-Directed Mutagenesis Kit; Stratagene, La Jolla, CA, USA) to mutate amino acid residue 16, critical for β-catenin binding [9]. A vector encoding a constitutively active form of β-catenin [6] was a gift from A. Hecht (University of Freiburg, Germany). Mutations and correct insertion into vectors was verified by sequencing. Plasmids were introduced into MIN6 cells using Lipofectamine 2000 (Invitrogen). pTOPFLASH, a luciferase construct containing an ‘optimal’ TCF/LEF promoter with three copies of the TCF/LEF-binding motif, CCTTTGATC, was a gift from T. Dale (Cardiff University, UK). To control for differences in transfection efficiency, pRL-CMV (cytomegalovirus immediate early enhancer/promoter region) was co-transfected and Renilla luciferase measured. Cells were cultured for 48 h in complete MIN6 media prior to assay in a luminometer (Berthold Lumat LB 9507, Bad Wildbad, Germany) using the Dual-Glo luciferase system (Promega, Madison, WI, USA).
Results
An alternative polyadenylation signal within intron 4 of TCF7L2 is widely used in human tissues
Using expressed sequence tag (EST) databases, we searched within the type 2 diabetes-associated LD block for sequences that could be part of a novel TCF7L2 transcript. ECgene [10] (http://genome.ewha.ac.kr/ECgene/gbr/; accessed 18 July 2011) and AceView [11] (www.ncbi.nlm.nih.gov/IEB/Research/Acembly/; accessed 18 July 2011) provide evidence for spliced human and mouse transcripts, respectively, containing sequence extending into intron 4.
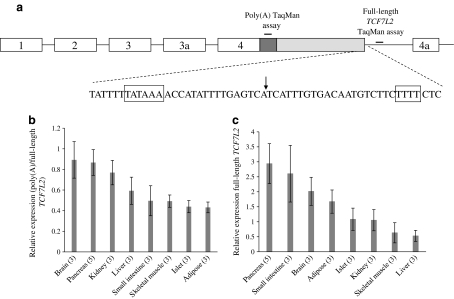
We hypothesised that there might be an alternative polyadenylation signal present, which would result in the use of an alternative translational stop codon and the production of isoforms possessing the β-catenin binding domain but not the HMG (high-mobility group) box DNA-binding domain. To determine whether an alternative polyadenylation signal is present in intron 4, we performed 3′ RACE on human pancreas cDNA and found a novel cleavage site at IVS4 + 1100. The 3′ end contains consensus sequences required for 3′ end formation in the correct spatial requirements (Fig. 1a). Real-time PCR analysis showed similar levels of truncated transcripts, relative to full-length TCF7L2 transcripts, across a number of tissues involved in the pathogenesis of type 2 diabetes (Fig. 1b). The production of full-length TCF7L2 transcripts was highest in pancreas, small intestine and brain, and lowest in kidney, skeletal muscle and liver (Fig. 1c). These results are consistent with a previous report examining TCF7L2 expression across human tissues [5].
Fig. 1.
Identification of an alternative polyadenylation signal within intron 4 of TCF7L2 widely used in human tissues. a Schematic depicting the location of the alternative polyadenylation signal within the human TCF7L2 gene. The dark grey rectangle represents the isoform-specific coding region, and the light grey rectangle the isoform-specific 3′ untranslated region, generated by the use of the novel polyadenylation signal. Cis sequences probably involved in 3′ end formation are detailed with boxes encompassing the polyadenylation sequence and U-rich element. The arrow indicates the cleavage site. Positions of the amplicons for TaqMan analysis are shown. b The relative use of the alternative polyadenylation site across a selection of human tissues involved in the pathogenesis of type 2 diabetes. Relative expression was determined using custom TaqMan assays that amplify the isoform-specific coding region and intronic sequences downstream of the alternative cleavage site. c The relative expression of full-length TCF7L2 expression across human tissues. Expression was normalised using the GeNorm algorithm, which selected B2M and GUSB as the two most stable housekeeping genes of those measured. All expression values are relative to that of one kidney sample. Numbers in brackets indicate the number of separate RNA samples on which expression was measured. Data are presented as the mean and standard error
Existence of isoforms utilising the alternative polyadenylation signal supported by western blotting
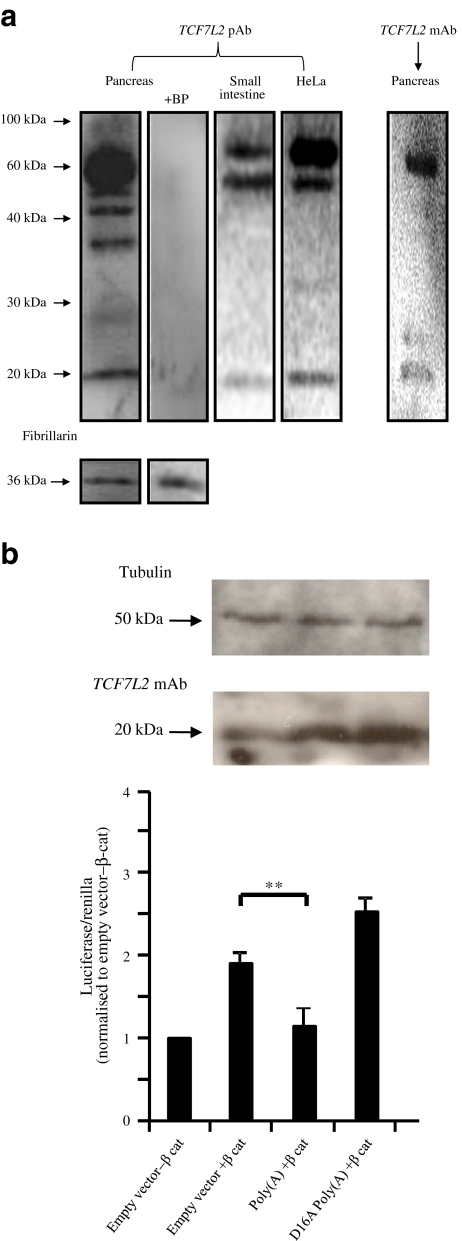
Western blot analysis using two antibodies targeting the N-terminus of TCF7L2, and nuclear extracts from human adult pancreas, small intestine and HeLa cells, shows we can detect full-length isoforms of the expected size (approximately 60 kDa) (Fig. 2a). The naive molecular mass of isoforms generated using the alternative polyadenylation signal would be approximately 20 kDa, and an interaction between the antibody and a protein of approximately 20 kDa was seen in all samples. Discrepancies in the number of high molecular mass bands observed across different samples may be explained by the presence of isoforms specifically produced in pancreas [5] and differences in antibody–antigen interactions between the two antibodies. A blocking peptide experiment confirmed the specificity of the bands observed with the pAb. Mass spectrometry could be used to unequivocally determine the identity of the proteins within the observed bands.
Fig. 2.
The use of the alternative polyadenylation signal generates protein isoforms with the ability to inhibit TCF/LEF-dependent transcription. a TCF7L2 protein production in human adult pancreas, small intestine and HeLa cells using a pAb. The specificity of the antibody–protein interaction at 20 kDa is proven by a blocking peptide (+BP) experiment and use of a monoclonal TCF7L2 antibody (mAb) predicted to bind to the truncated isoform. Fibrillarin was used as a loading control. b An isoform generated by the use of the alternative polyadenylation signal inhibits TCF/LEF-dependent target genes by sequestering β-catenin. MIN6 cells were co-transfected with firefly and Renilla luciferase reporter genes and expression vectors as indicated. The β-catenin (β-cat) construct encodes a constitutively active form of this protein. Firefly luciferase production was driven by a promoter containing three copies of the consensus TCF/LEF-binding motif, CCTTTGATC. Renilla luciferase was measured to correct for differences in transfection efficiency. Reporter gene activities were determined 48 h post-transfection. Bars represent relative luciferase activities compared with control transfected with empty vector and β-catenin. Mean values and standard deviations from five independent experiments are given. Western blotting was performed to determine TCF7L2 and tubulin protein production in transfected samples. **p < 0.01
Effect of overexpression of truncated isoform on transcriptional activation
We next tested the prediction that these novel isoforms bind to β-catenin but not to DNA, and hence might act to repress the activity of other TCF7L2 isoforms on their target promoters. In clonal MIN6 beta cells, transfection of a vector encoding a constitutively active form of β-catenin was able to transactivate a luciferase reporter gene downstream of a TCF/LEF promoter. Co-transfection of β-catenin and the truncated isoform significantly reduced luciferase production (empty vector + β-catenin vs Poly(A) + β-catenin; p = 0.003). Co-transfection of β-catenin and a mutated version of the truncated isoform (D16A Poly(A)), with reduced affinity for β-catenin, did not inhibit luciferase production. Western blotting confirms that both versions of the truncated isoform were produced, and at a higher level than that seen in control cells (Fig. 2b). These results suggest that isoforms generated by the use of the alternative polyadenylation signal inhibit TCF/LEF-dependent transcription by a mechanism involving β-catenin sequestration.
Discussion
In this paper, we have identified a novel polyadenylation signal within TCF7L2 that results in the production of isoforms that may inhibit the activity of full-length TCF7L2 isoforms. Given the context-dependent nature of TCF7L2 isoforms, it would be unwise to extrapolate this effect to all TCF/LEF-dependent genes in vivo, or to exclude alternative roles for this isoform in regulating gene expression. Indeed, a similarly truncated TCF7L2 isoform in mouse has been shown to inhibit TCF/LEF-dependent target genes but potentiate the co-activation of non-TCF/LEF-dependent target genes by β-catenin [12].
Phenotypic characterisation suggests that SNPs in TCF7L2 modulate the risk of type 2 diabetes, primarily through an effect on insulin secretion [3]. It has been shown, specifically in islets, that the risk-associated T allele of the diabetes-associated SNP, rs7903146, exists in a more open chromatin state than the opposing C allele, and that the T allele possesses greater enhancer activity selectively in islet beta cell lines [13, 14]. This increased enhancer effect for the risk-associated allele is somewhat at odds with the results from RNA interference experiments conducted in beta cells, which suggest that a loss of TCF7L2 function causes a predisposition to diabetes [2, 4]. To reconcile the differences in experimental findings, it is possible that the T allele enhances the production of a repressive TCF7L2 isoform, such as the one described in this study. A large number of human islet samples would likely be needed to test this hypothesis.
Acknowledgements
This study was funded by Wellcome Trust grants (WT081278MA to L.W. Harries and 081958/Z/07/Z to G.A. Rutter) and grants to G.A. Rutter from the MRC (GO401641) and the European Community’s Seventh Framework Programme (FP7/2007-2013) for the Innovative Medicine Initiative under grant agreement no. 115005.
Contribution statement
All authors were involved in drafting and revising the article, and in the conception, design, analysis and interpretation of the data. All the authors approved the final version of the manuscript to be published.
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- ASP
Adaptor-specific primer
- EST
Expressed sequence tag
- GSP
Gene-specific primer
- LD
Linkage disequilibrium
- mAb
Monoclonal antibody
- pAb
Polyclonal antibody
- RACE
Rapid amplification of cDNA ends
- SNP
Single nucleotide polymorphism
- TCF/LEF
T cell factor/lymphoid-enhancer factor
Footnotes
An erratum to this article can be found at http://dx.doi.org/10.1007/s00125-011-2327-x
References
- 1.Cauchi S, El Achhab Y, Choquet H, et al. TCF7L2 is reproducibly associated with type 2 diabetes in various ethnic groups: a global meta-analysis. J Mol Med. 2007;85:777–782. doi: 10.1007/s00109-007-0203-4. [DOI] [PubMed] [Google Scholar]
- 2.da Silva Xavier G, Loder MK, McDonald A, et al. TCF7L2 regulates late events in insulin secretion from pancreatic islet beta-cells. Diabetes. 2009;58:894–905. doi: 10.2337/db08-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lyssenko V, Lupi R, Marchetti P, et al. Mechanisms by which common variants in the TCF7L2 gene increase risk of type 2 diabetes. J Clin Invest. 2007;117:2155–2163. doi: 10.1172/JCI30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shu L, Matveyenko AV, Kerr-Conte J, Cho JH, McIntosh CH, Maedler K. Decreased TCF7L2 protein levels in type 2 diabetes mellitus correlate with downregulation of GIP- and GLP-1 receptors and impaired beta-cell function. Hum Mol Genet. 2009;18:2388–2399. doi: 10.1093/hmg/ddp178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prokunina-Olsson L, Welch C, Hansson O, et al. Tissue-specific alternative splicing of TCF7L2. Hum Mol Genet. 2009;18:3795–3804. doi: 10.1093/hmg/ddp321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weise A, Bruser K, Elfert S, et al. Alternative splicing of Tcf7l2 transcripts generates protein variants with differential promoter-binding and transcriptional activation properties at Wnt/beta-catenin targets. Nucleic Acids Res. 2010;38:1964–1981. doi: 10.1093/nar/gkp1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vandesompele J, de Preter K, Pattyn F, et al. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:RESEARCH0034 [DOI] [PMC free article] [PubMed]
- 9.Fasolini M, Wu X, Flocco M, Trosset JY, Oppermann U, Knapp S. Hot spots in Tcf4 for the interaction with beta-catenin. J Biol Chem. 2003;278:21092–21098. doi: 10.1074/jbc.M301781200. [DOI] [PubMed] [Google Scholar]
- 10.Kim N, Shin S, Lee S. ECgene: genome-based EST clustering and gene modeling for alternative splicing. Genome Res. 2005;15:566–576. doi: 10.1101/gr.3030405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thierry-Mieg D, Thierry-Mieg J. AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006;7(Suppl 1):S12 11–14. doi: 10.1186/gb-2006-7-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kennell JA, O’Leary EE, Gummow BM, Hammer GD, MacDougald OA. T cell factor 4N (TCF-4N), a novel isoform of mouse TCF-4, synergizes with beta-catenin to coactivate C/EBPalpha and steroidogenic factor 1 transcription factors. Mol Cell Biol. 2003;23:5366–5375. doi: 10.1128/MCB.23.15.5366-5375.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaulton KJ, Nammo T, Pasquali L, et al. A map of open chromatin in human pancreatic islets. Nat Genet. 2010;42:255–259. doi: 10.1038/ng.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stitzel ML, Sethupathy P, Pearson DS, et al. Global epigenomic analysis of primary human pancreatic islets provides insights into type 2 diabetes susceptibility loci. Cell Metab. 2010;12:443–455. doi: 10.1016/j.cmet.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]