Summary
Organism-wide RNA interference (RNAi) is due to the transport of mobile silencing RNA throughout the organism but the identities of these mobile RNA species in animals are unknown. Here we present genetic evidence that both the initial double-stranded RNA (dsRNA), which triggers RNAi, and at least one dsRNA intermediate produced during RNAi can act as or generate mobile silencing RNA in Caenorhabditis elegans. This dsRNA intermediate requires the long dsRNA-binding protein RDE-4, the endonuclease DCR-1, which cleaves long dsRNA into double-stranded short-interfering RNA (ds-siRNA), and the putative nucleotidyltransferase MUT-2 (RDE-3). However, single-stranded siRNA and downstream secondary siRNA produced upon amplification by the RNA-dependent RNA Polymerase RRF-1 do not generate mobile silencing RNA. Restricting inter-tissue transport to long dsRNA and directly processed siRNA intermediates rather than amplified siRNA may serve to modulate the extent of systemic silencing in proportion to available dsRNA.
Intercellular transport of RNA has been inferred in plants and animals undergoing gene silencing by RNAi1. In plants, short-interfering RNAs (siRNA) processed from long dsRNA move between cells through intercellular bridges called plasmodesmata and travel long distances via the phloem to convey gene-specific silencing information2,3,4. Although the nature of mobile silencing signals in animals is unknown, the conserved RNA transporter SID-1 is required for their import in Caenorhabditis elegans and has been implicated in RNA transport in other animals5,6,7. In addition, dsRNA expressed in multiple tissues can generate sid-1-dependent mobile silencing RNA through an as yet unknown pathway8. Since animals transcribe dsRNA from numerous loci9, understanding how mobile RNA is produced from dsRNA has broad implications for systemic control of gene expression.
Multiple distinct RNA species are produced during RNAi in C. elegans, but, it is unclear which of these are mobile (Fig. 1a)10,11,12,13. These RNA species include transcribed sense and anti-sense duplexes (dsRNA), double-stranded short-interfering RNA (ds-siRNA) generated upon cleavage of long dsRNA by the RDE-4–Dicer(DCR-1) complex, primary single-stranded siRNA generated upon cleavage of ds-siRNA by the Argonaute RDE-113, and the subsequent numerous secondary siRNAs generated by RNA-dependent RNA polymerases (RdRP) that are responsible for potent silencing of the target gene. In addition, enzymes that can modify RNA such as the putative nucleotidyltransferase MUT-214,15,16, which is required for efficient RNAi (Supplementary Fig. 1), may also generate RNA species that act as mobile RNA. Early studies using dsRNA injected into the cytoplasm of gut cells suggested that RNA silencing in gut cells is not required to transport a mobile silencing signal to the germline17,18. However, whether this signal is the injected exogenous dsRNA itself or a dsRNA-derived mobile RNA or both is unclear and how endogenously transcribed dsRNA leads to the production of mobile RNA is unknown.
Figure 1.
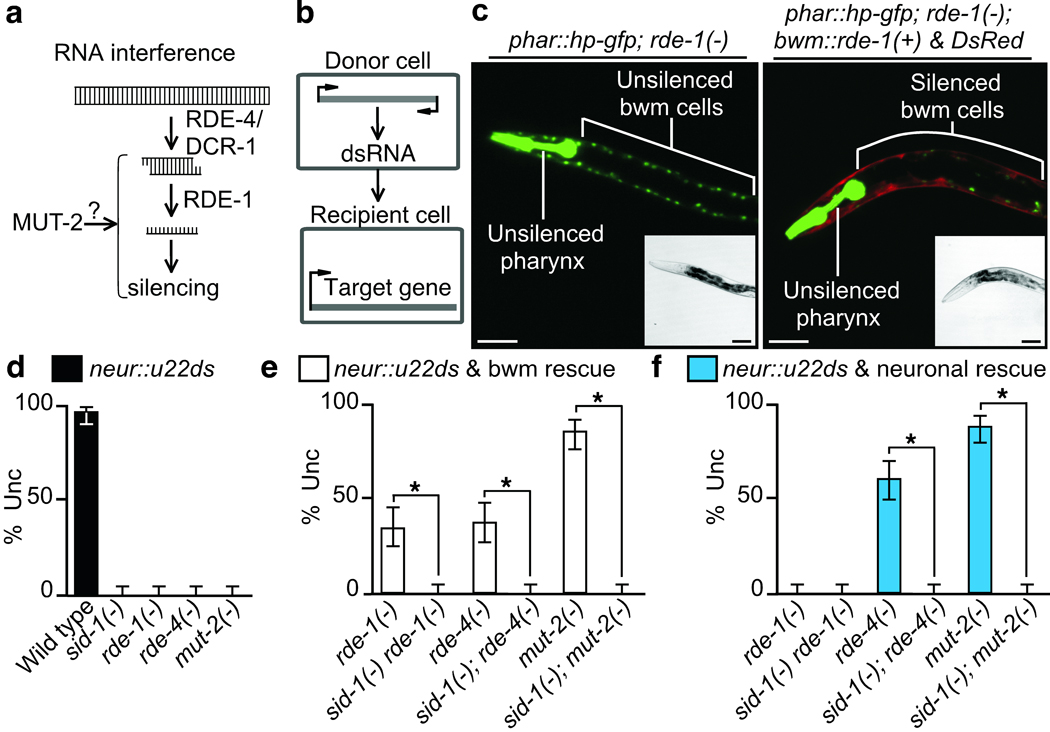
RNAi-independent biogenesis of mobile RNA from expressed dsRNA. (a) Schematic of RNAi within a cell. See text for details. (b) Schematic of assay to measure silencing due to mobile RNA. dsRNA that targets a gene in recipient cells are expressed only in other donor cells. (c) A representative rde-1(−) L4 animal that expresses gfp-hairpin RNA only in the pharynx (phar::hp-gfp) but gfp in pharynx and bwm cells (left panel) and one that in addition coexpress rde-1(+) and DsRed only in bwm cells (right panel). 100% of animals from three independent bwm::rde-1(+) & DsRed lines showed silencing as above. Scale bar, 50 µm. (d–f) A representative transgenic line that expresses unc-22-dsRNA under the control of the neuronal rgef-1 promoter (neur::u22ds) was generated in wild-type animals and crossed into the genetic backgrounds indicated. The uncoordinated twitching (Unc) due to the silencing of unc-22 was measured in these animals (black) and in animals that in addition had the corresponding RNAi gene rescued in body-wall muscles (white) or in neurons (blue). n=100 animals; error bars indicate 95% confidence intervals; and asterisks indicate significant differences (P<0.05). Partial silencing in rescued transgenic lines likely indicate that levels of the rescuing genes are inadequate for complete silencing in response to the low levels of neuronal unc-22 dsRNA. Consistently, feeding unc-22 dsRNA to animals with the same muscle-rescued transgenic lines above results in complete silencing (Fig. 3). See Supplementary Fig. 2 for details of constructs used.
Here we determine the genetic requirements for silencing due to mobile RNAs using well-characterized promoters to restrict the expression of dsRNA or RNAi pathway genes to specific tissues and examining target gene silencing in other tissues. In most experimental systems that use similar approaches, it is difficult to control for low levels of misexpression in the target tissues. Since SID-1 is strictly required for the import of mobile silencing RNAs8, SID-1-dependence of silencing serves to clearly distinguish silencing due to mobile RNA from silencing due to misexpression in the target tissues.
RESULTS
Long dsRNA is mobile in C. elegans
We examined how endogenously transcribed dsRNA produces mobile silencing RNA using mosaic animals (i.e. animals that have some mutant cells and some wild-type cells) in which a mutant donor tissue expresses dsRNA that targets a gene in a wild-type recipient tissue (Fig. 1b). To determine whether the activity of the primary Argonaute RDE-1 is required to produce a mobile silencing signal, we first expressed dsRNA targeting the green fluorescent protein (gfp) in the pharynx of rde-1(−) animals. We then coexpressed gfp and rde-1(+) in the body-wall muscle (bwm) cells, making bwm a wild-type recipient tissue (Fig. 1c and Supplementary Fig. 2). We observed gfp silencing in anterior rde-1(+) bwm cells. Thus, RNAi-mediated silencing in the pharynx is not required to produce and transport mobile RNA to the bwm cells. To determine whether RNAi pathway genes upstream of RDE-1 are required to produce a mobile silencing signal from expressed dsRNA, we developed a sensitive assay that measures silencing of an endogenous gene due to mobile RNA (Fig. 1d and Supplementary Fig. 2). Specifically, we introduced a neuronally expressed transgene that produces a ~560 bp dsRNA that targets the muscle gene unc-22 (neur::u22ds). All unc-22 silencing detected in animals with the neur::u22ds transgene required the RNAi pathway genes and the RNA transporter SID-1, showing that all silencing occurred through RNAi in these animals and was due to mobile RNA enabled RNAi (Fig. 1d). Using this source of mobile RNA, we detected unc-22 silencing in rde-4(−) animals that expressed rde-4(+) in bwm cells and in mut-2(−) animals that expressed mut-2(+) in bwm cells (Fig. 1e). Thus, neither dsRNA cleavage through RDE-4 recruitment of Dicer nor modification by the nucleotidyltransferase MUT-2 is required in neurons that express dsRNA for the generation and export of mobile RNA. Together, these results show that a mobile RNA is generated from transcribed long dsRNA, independent of processing by the canonical RNAi pathway, can generate a mobile silencing RNA.
A processed dsRNA also moves between cells
To determine whether products of dsRNA processing by the canonical RNAi pathway are also mobile, we expressed dsRNA in a wild-type, RNAi proficient donor tissue and examined silencing in RNAi defective recipient tissues. If a processed RNA produced in the wild-type donor tissue can act as or generate a mobile silencing RNA, that RNA may bypass the lack of the earlier-acting RNAi pathway gene in the recipient tissue and cause silencing. Note that using this approach we cannot infer anything about RNAs that move between tissues but that fail to cause gene silencing.
To detect silencing triggered by mobile processed RNAs we rescued RNAi pathway mutants only in neurons of animals that contain the neur::u22ds transgene and measured silencing of the target gene unc-22 in mutant muscle cells. We detected unc-22 silencing in rde-4(−) animals that expressed rde-4(+) in neurons. Consistent with silencing due to mobile RNAs, SID-1 is required for the observed silencing (Fig. 1f). Since RDE-4 is required for DCR-1 cleavage of long dsRNA into ds-siRNA19, these mobile RNAs are either ds-siRNA or downstream RNAi products. To distinguish between these two possibilities, we similarly examined the role of the primary Argonaute RDE-1 in the production of mobile RNA. In contrast to the analogous experiment with RDE-4, we observed no detectable unc-22 silencing in rde-1(−) animals that express rde-1(+) in neurons. This observation suggests that primary siRNA and downstream RNAi products such as RdRP-dependent secondary siRNA are not mobile. Finally, we detected unc-22 silencing in mut-2(−) animals that expressed mut-2(+) in neurons (Fig. 1f) and this silencing was due to mobile RNA since it required SID-1 (Fig. 1d). Therefore, we infer that like RDE-4, MUT-2 functions upstream of RDE-1 to generate a species of mobile RNA that can bypass the need for MUT-2 activity in the recipient tissue.
Mobile RNAs are similarly made from other sources of dsRNA
We next tested whether other sources of silencing RNAs also rely on the same genes to produce mobile RNAs. Multicopy transgenes such as sur-5::gfp (which express nuclear-localized GFP in all somatic tissues) can generate mobile RNAs, presumably from trace amounts of dsRNA produced from the transgene8,20. We therefore generated rde-4(−);sur-5::gfp animals and moved a representative transgene that expresses rde-4 in bwm cells (bwm::rde-4(+)) into these animals. Significant silencing (P<0.05) was detected in non-muscle tissues in the resultant mosaic animals and was most easily observed in the prominent gut nuclei (Fig. 2a,b). Consistent with silencing due to mobile RNAs, SID-1 is required for the observed silencing of gut nuclei (Fig. 2c,d). However, when we moved a representative transgene that expressed rde-1(+) in bwm cells (bwm::rde-1(+)) into rde-1(−); sur-5::gfp animals, we observed no detectable silencing of GFP expression in the gut. In contrast, moving a representative transgene that expressed mut-2(+) in bwm cells (bwm::mut-2(+)) into mut-2(−); sur-5::gfp animals resulted in the silencing of GFP expression in the gut (Fig. 2d). The observed silencing was dependent on SID-1, showing that mobile RNA triggered the silencing in mut-2(−) gut cells (Fig. 2d). Therefore, as in the case of expressed dsRNA, multicopy transgenes also generate mobile RNAs that are upstream of RDE-1 and include those that are processed by RDE-4 and MUT-2.
Figure 2.
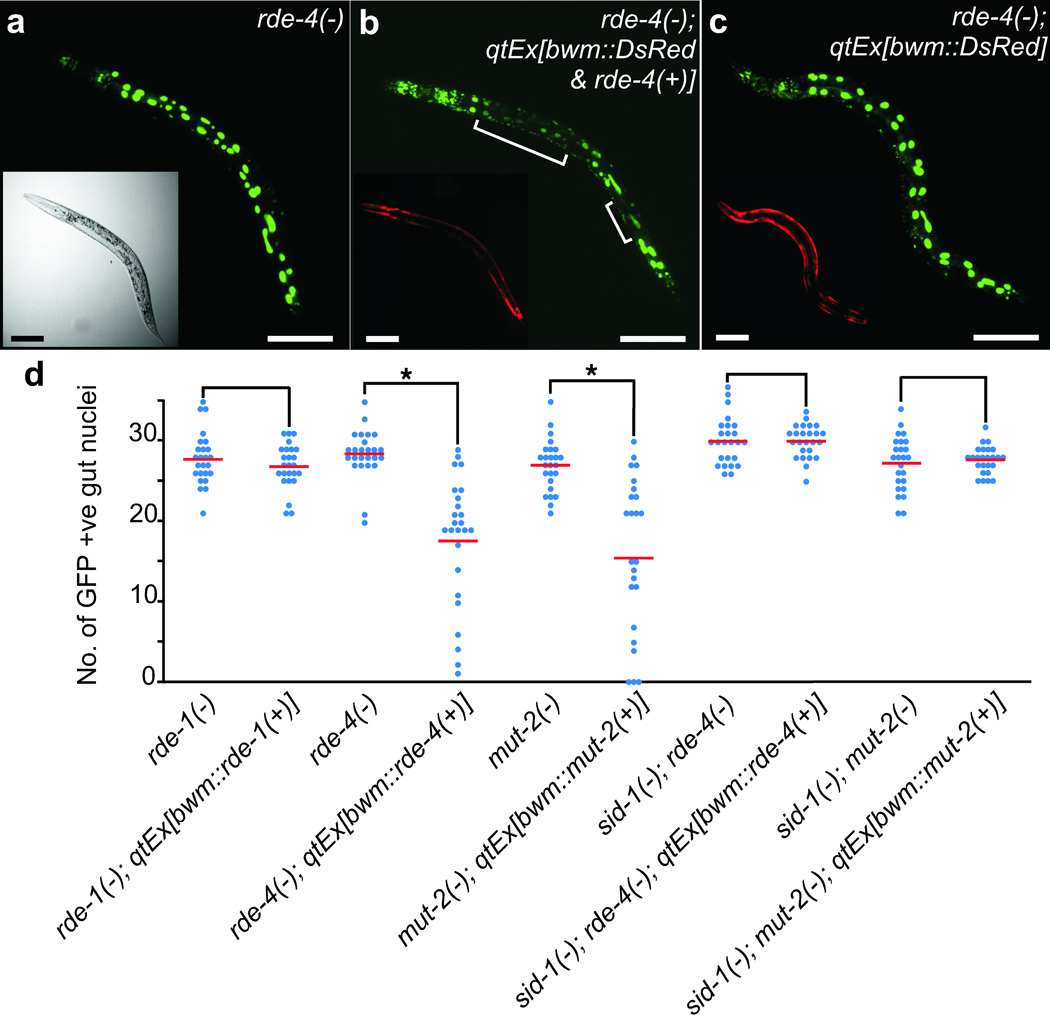
Restricted expression of RDE-4 and MUT-2 but not RDE-1 enables non-cell autonomous RNA silencing. (a–c) Representative animals that express nuclear-localized GFP in all cells (sur-5::gfp). (a) rde-4(−). (b) rde-4(−) animals with rde-4(+) and DsRed expressed in bwm cells (qtEx[bwm::rde-4(+) & DsRed]). (c) rde-4(−) animals that only express DsRed in bwm cells (qtEx[bwm::DsRed]). Brackets indicate silencing in gut nuclei. Insets are widefield (a) or red channel (b, c) images. Scale bars, 50 µm. Note that unlike overexpression of rde-4(+), overexpression of the coinjection marker DsRed did not result in any silencing of gfp expression [compare (b) and (c)]. When coexpressed with rde-4(+), DsRed expression was lower (enhanced in (b) inset to clearly indicate expression in the bwm), which likely reflects enhanced silencing of the DsRed transgene. (d) The number of brightly fluorescent gut nuclei that show sur-5::gfp expression were counted in rde-1(−), rde-4(−), and mut-2(−) mutant backgrounds as well as in mutant animals with a corresponding representative bwm rescue transgenes from Fig. 1e. Similar experiments done with rde-4(−); sid-1(−) and mut-2(−); sid-1(−) double mutant backgrounds are also shown. n=25 L4 animals. Averages (red bars), significant differences (brackets and *, P<0.05) and similar values (brackets) are indicated. Minor variations in the average number of nuclei (± 2 nuclei) observed between animals were not due to silencing of gfp expression but rather due to small changes in the number of intestinal nuclei (see Supplementary Fig. 4 and the discussion therein).
We next tested whether RDE-4 and MUT-2 but not RDE-1 can similarly process exogenously supplied dsRNA to produce mobile RNA. We fed bacteria that express gfp-dsRNA (feeding RNAi)21 to the above rde-1(−), rde-4(−), and mut-2(−) mutants that contain the sur-5::gfp transgene and that are rescued in muscle cells and examined silencing in the respective mutant gut cells. Consistent with our results with endogenously transcribed dsRNA, we found that gfp feeding RNAi increased silencing of GFP in the non-muscle cells of muscle-rescued rde-4(−) and mut-2(−) animals but not of muscle-rescued rde-1(−) animals (Fig. 3a). To assay silencing due to feeding RNAi targeting endogenous genes, we removed the sur-5::gfp transgenes from the transgenic bwm rescue lines and then fed these muscle-rescued animals bacteria that express dsRNA targeting the muscle gene unc-22 or that express dsRNA targeting the skin gene dpy-7 or that express dsRNA targeting the intestinal gene act-5. Silencing due to feeding RNAi of these endogenous genes was consistent with our results using gfp feeding RNAi and using endogenously transcribed dsRNA. Specifically, while we observed robust silencing of the muscle gene in all three strains of muscle-rescued animals, silencing of the skin and intestinal genes was detectable in muscle-rescued rde-4(−) and mut-2(−) animals (Fig. 3b) but not in muscle-rescued rde-1(−) animals (Fig. 3b, ref. 22). Thus, the silencing observed in these rde-4 and mut-2 mosaic animals by feeding RNAi is likely due to import of ingested long dsRNA into the rescued muscle cells followed by export of a processed mobile RNA that can silence the target genes in rde-4(−) and mut-2(−) cells.
Figure 3.
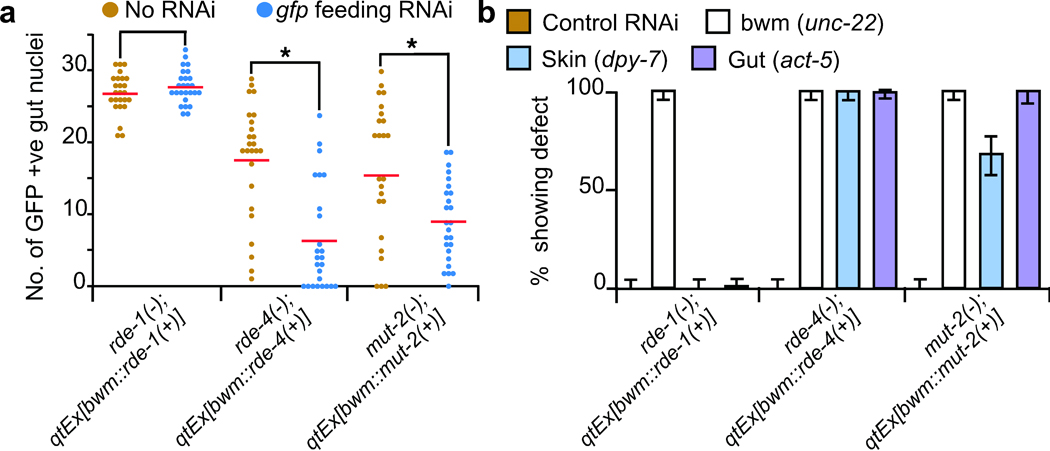
RDE-4 and MUT-2 but not RDE-1 processed ingested dsRNA is mobile. (a) Feeding RNAi of rde-1(−), rde-4(−), and mut-2(−) animals with sur-5::gfp and their corresponding representative bwm rescue transgenic lines used in Fig. 2d. The number of brightly fluorescent gut nuclei that show sur-5::gfp expression were counted in L4 animals that were fed either control bacteria (brown, re-plotted from Fig. 2d) or bacteria expressing gfp-dsRNA (blue). n=25 animals. Averages (red bars), significant differences (brackets and *, P<0.05) and similar values (brackets) are indicated. (b) Feeding RNAi of strains in (a) after removal of sur-5::gfp. L4 animals were fed L4440 (control) or dsRNA targeting the muscle gene unc-22 (bwm) or the skin gene dpy-7 (skin) or the gut gene act-5 (gut) and the percentage of L4 progeny that showed the corresponding defects were determined. n=100 L4 animals; error bars indicate 95% confidence intervals.
Therefore, both multicopy transgenes and ingested dsRNA use the same genetic pathway to produce short mobile silencing RNA.
Two classes of upstream dsRNAs are mobile RNAs
Taken together, our results suggest a model whereby upstream dsRNA species such as long dsRNA and ds-siRNA act as or generate mobile RNA, while all silencing RNAs produced after cleavage of ds-siRNA by RDE-1 cannot cause silencing in rde-1(−) cells (Fig. 4a). Since Dicer can cleave long dsRNA in the absence of MUT-212 and since MUT-2 acts upstream of RDE-1 to generate mobile RNA, one possible role for MUT-2 in RNAi is to modify ds-siRNA. Despite MUT-2 having the required catalytic residues, a systematic test of putative nucleotidyltransferases using in vitro assays failed to reveal how MUT-2 might modify RNA23. Nevertheless, consistent with our model, overexpression of neither mut-2(+) nor rde-4(+) in bwm cells of rde-1(−); sur-5::gfp animals resulted in detectable silencing (Supplementary Table 1). Further, neither overexpression of mut-2(+) in bwm cells of rde-4(−); sur-5::gfp animals nor overexpression of rde-4(+) in the bwm cells of mut-2(−);sur-5::gfp animals resulted in detectable silencing (Supplementary Table 1), suggesting that RDE-4 and MUT-2 act in the same pathway to generate mobile RNA.
Figure 4.
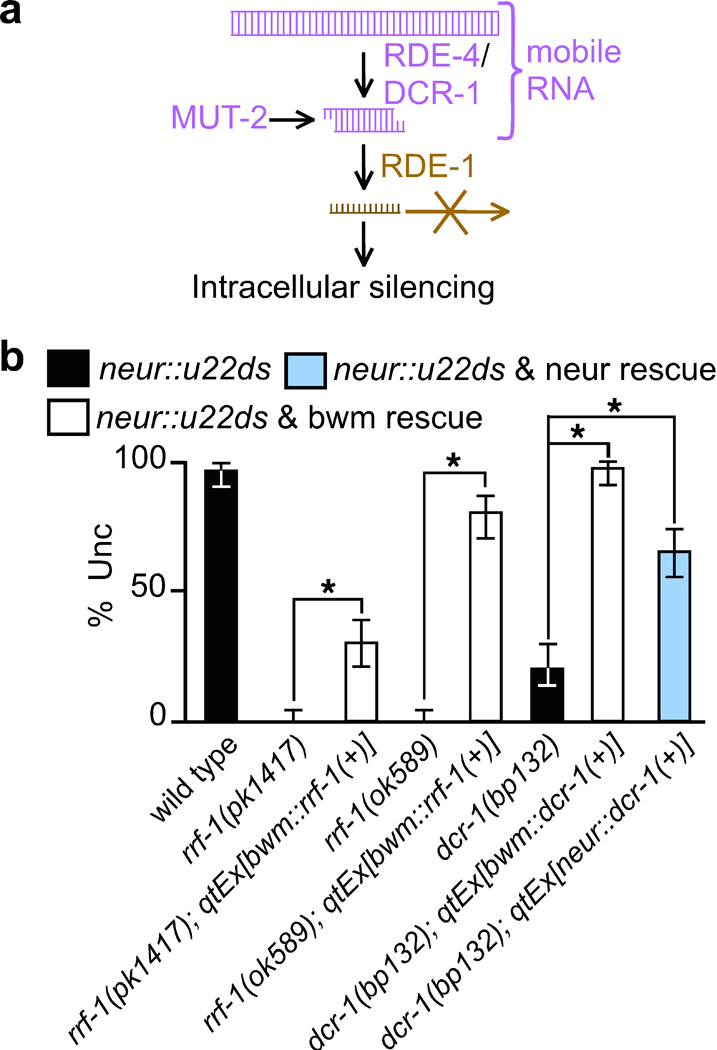
Biogenesis of mobile RNA in C. elegans. (a) Schematic of the biogenesis pathway. Double-stranded forms of RNA produced during the early steps of RNA interference act as or generate mobile RNAs. RNAs produced after the Argonaute RDE-1 cleaves ds-siRNA to release single stranded RNA13 are restricted to intracellular silencing. MUT-2 expression enables the generation and export of mobile RNA possibly through enzymatic modification of dsRNA. Since expressed as well as ingested dsRNA generate mobile RNA, additional regulation in response to the environment and selection of specific endogenous loci to make mobile RNAs is likely. (b) RdRP activity is not required for mobile RNA production and dsRNAs both upstream and downstream of Dicer generate mobile RNAs. The representative transgene used in Fig. 1 to express unc-22-dsRNA under the control of the neuronal rgef-1 promoter (neur::u22ds) was crossed into the genetic backgrounds indicated. Two deletion alleles of rrf-1 (pk1417 & ok589) were rescued with rrf-1(+) in bwm and the missense allele dcr-1(bp132) was rescued with dcr-1(+) in bwm and in neurons. Silencing of unc-22 was measured (% Unc) in the mutant animals (black) and in animals with the corresponding RNAi gene rescued in bwm (white) and in neurons (blue). n=100 animals. 95% confidence intervals (error bars) and significant differences (brackets and *, P<0.05) are indicated. See Supplementary Fig. 2 for details of constructs used.
The following tests provide additional support for the model that long and short dsRNAs, but not single-stranded siRNA act as mobile silencing RNA: (1) The RdRP RRF-1, which makes the numerous downstream secondary siRNAs, is not required for the generation of mobile RNAs (Fig. 4b); (2) Rescuing a partial loss-of-function dcr-1 mutant24 in the recipient tissue (which increases processing of imported Dicer precursors [long dsRNA]) improves silencing, presumably of imported long dsRNA (Fig. 4b); (3) Rescuing the dcr-1 mutant in donor tissues also increased silencing in recipient cells, presumably by increased transport of ds-siRNA (Fig. 4b); and (4) expression of Inhibitors of RNAi in recipient cells, including the conserved exonuclease ERI-1 that can degrade ds-siRNA25, inhibited silencing (Supplementary Fig. 3).
DISCUSSION
We provide evidence for the existence of at least two distinct species of mobile RNA in C. elegans: one that is produced from long dsRNA independent of RNAi genes in donor tissues but requires all tested RNAi genes for silencing in recipient tissues and one that requires RDE-4, DCR-1, and MUT-2 for production in donor tissues but not for silencing in recipient tissues.
Animal mobile silencing RNAs differ from plant mobile RNAs
In plants, mobile RNAs move between cells through relatively non-selective intercellular bridges called plasmodesmata2,3. In the plant Arabidopsis, grafting experiments between genetically distinct source and target tissues have enabled the molecular identification of mobile RNAs. These studies have identified both single-stranded siRNA and ds-siRNA whose movement to distant tissues correlates with mobile RNAs2,3. In addition, accumulating evidence supports the intercellular movement of microRNAs, tasiRNAs, and mRNAs26.
Our results indicate that in C. elegans, long dsRNA and a form of ds-siRNA can move between cells (Fig. 4a). Unexpectedly, and in contrast to what is observed in plants, single-stranded siRNAs produced by RdRP amplification are either not mobile, or if mobile, are incapable of causing detectable silencing in recipient cells. Consistent with mobile silencing signals being restricted to double-stranded forms of RNA, most systemic RNAi silencing observed in C. elegans is dependent on SID-15,8, which is exquisitely selective for dsRNA27. This restriction couples the extent of RNAi spreading to the amount of primary dsRNA produced within cells or imported from the environment.
A conserved pathway to make animal mobile RNAs
Since a mammalian SID-1 homolog can transport ds-siRNAs into mammalian cells7, ds-siRNA, perhaps modified by a nucleotidyltransferase, may move between mammalian cells. Importantly, short dsRNAs can escape the interferon response that results in non-specific effects in differentiated mammalian cells28, thus their transport between differentiated tissues should be tolerated. In contrast, the transport of long dsRNA will result in specific gene silencing only in undifferentiated mammalian cells. Furthermore, the protein activities required to make short mobile RNA in worms are found in most animals: dsRNA-binding proteins, such as RDE-4, that act with Dicer (e.g. PACT and TRBP with human Dicer29) and β-nucleotidyltransferases, such as MUT-2, that play a role in RNA silencing14,15,16. Modulation of such conserved biochemical pathways may contribute to the tissue- and environment-dependent differences in silencing due to mobile RNA that are observed in C. elegans30,8. Regulated transport of mobile RNA is evident in plants, where mobile RNA produced in metabolic source tissues control gene expression in distant metabolic sink tissues31. Similarly, C. elegans mobile RNAs are preferentially imported into cells that express SID-1 at high levels32,8, suggesting that SID-1 expression produces a sink for mobile RNA. Therefore, short dsRNAs produced from endogenous loci in a mammalian cell may control gene expression in another cell type that expresses a SID-1 homolog.
Supplementary Material
Acknowledgements
We thank Katerina Ragkousi, Susan Mango, and members of the Hunter lab, particularly Kenneth Pang, Jacqueline Brooks, and Daniel Schott for comments on the manuscript; the C. elegans Genetics Center for some strains; Dr. Hong Zhang, National Institute of Biological Sciences, Beijing, China for HZ202; Steven Ekman for two constructs; and the NIH (K99 to A.M.J) and NSF (to C.P.H) for funding.
Footnotes
Note: Supplementary Information is available on the Nature Structural & Molecular Biology website.
Author contributions A.M.J. performed the experiments and G.A.G. generated most of the DNA constructs; A.M.J. and C.P.H. designed the study, analyzed the data, and wrote the paper. All authors discussed the results and commented on the manuscript.
References
- 1.Jose AM, Hunter CP. Transport of sequence-specific RNA interference information between cells. Annu. Rev. Genet. 2007;41:305–330. doi: 10.1146/annurev.genet.41.110306.130216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Molnar A, et al. Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science. 2010;328:872–875. doi: 10.1126/science.1187959. [DOI] [PubMed] [Google Scholar]
- 3.Dunoyer P, et al. Small RNA duplexes function as mobile silencing signals between plant cells. Science. 2010;328:912–916. doi: 10.1126/science.1185880. [DOI] [PubMed] [Google Scholar]
- 4.Dunoyer P, et al. An endogenous, systemic RNAi pathway in plants. EMBO J. 2010;29:1699–1712. doi: 10.1038/emboj.2010.65. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Winston WM, Molodowitch C, Hunter CP. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science. 2002;295:2456–2459. doi: 10.1126/science.1068836. [DOI] [PubMed] [Google Scholar]
- 6.Feinberg EH, Hunter CP. Transport of dsRNA into cells by the transmembrane protein SID-1. Science. 2003;301:1545–1547. doi: 10.1126/science.1087117. [DOI] [PubMed] [Google Scholar]
- 7.Wolfrum C, et al. Mechanisms and optimization of in vivo delivery of lipophlic siRNAs. Nat. Biotechnol. 2007;25:1149–1157. doi: 10.1038/nbt1339. [DOI] [PubMed] [Google Scholar]
- 8.Jose AM, Smith JJ, Hunter CP. Export of RNA silencing from C. elegans tissues does not require the RNA channel SID-1. Proc. Natl. Acad. Sci. USA. 2009;106:2283–2288. doi: 10.1073/pnas.0809760106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat. Rev. Mol. Cell Biol. 2009;10:637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 11.Grishok A. RNAi mechanisms in Caenorhabditis elegans. FEBS Lett. 2005;579:5932–5939. doi: 10.1016/j.febslet.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 12.Aoki K, Moriguchi H, Yoshioka T, Okawa K, Tabara H. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J. 2007;26:5007–5019. doi: 10.1038/sj.emboj.7601910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steiner FA, Okihara KL, Hoogstrate SW, Sijen T, Ketting RF. RDE-1 slicer activity is required only for passenger-strand cleavage during RNAi in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 2009;16:207–211. doi: 10.1038/nsmb.1541. [DOI] [PubMed] [Google Scholar]
- 14.Chen CC, et al. A member of the polymerase beta nucleotidyltransferase superfamily is required for RNA interference in C. elegans. Curr. Biol. 2005;15:378–383. doi: 10.1016/j.cub.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 15.Moazed D, et al. Studies on the mechanism of RNAi-dependent heterochromatin assembly. Cold Spring Harb. Symp. Quant. Biol. 2006;71:461–471. doi: 10.1101/sqb.2006.71.044. [DOI] [PubMed] [Google Scholar]
- 16.van Wolfswinkel JC, et al. CDE-1 affects chromosome segregation through uridylation of CSR-1-bound siRNAs. Cell. 2009;139:135–148. doi: 10.1016/j.cell.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 17.Tabara H, et al. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- 18.Grishok A, Tabara H, Mello CC. Genetic requirements for inheritance of RNAi in C. elegans. Science. 2000;287:2494–2497. doi: 10.1126/science.287.5462.2494. [DOI] [PubMed] [Google Scholar]
- 19.Parker GS, Eckert DM, Bass BL. RDE-4 preferentially binds long dsRNA and its dimerization is necessary for cleavage of dsRNA to siRNA. RNA. 2006;12:807–818. doi: 10.1261/rna.2338706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Habig JW, Aruscavage PJ, Bass BL. In C. elegans, high levels of dsRNA allow RNAi in the absence of RDE-4. PLoS One. 2008;3:e4052. doi: 10.1371/journal.pone.0004052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- 22.Qadota H, et al. Establishment of a tissue-specific RNAi system in C. elegans. Gene. 2007;400:166–173. doi: 10.1016/j.gene.2007.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwak JE, Wickens M. A family of poly(U) polymerases. RNA. 2007;13:860–867. doi: 10.1261/rna.514007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ren H, Zhang H. Wnt signaling controls temporal identities of seam cells in Caenorhabditis elegans. Dev. Biol. 2010;345:144–155. doi: 10.1016/j.ydbio.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 25.Kennedy S, Wang D, Ruvkun G. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature. 2004;427:645–649. doi: 10.1038/nature02302. [DOI] [PubMed] [Google Scholar]
- 26.Hyun TK, Uddin MN, Rim Y, Kim JY. Cell-to-cell trafficking of RNA and RNA silencing through plasmodesmata. Protoplasma. 2011;248:101–116. doi: 10.1007/s00709-010-0225-6. [DOI] [PubMed] [Google Scholar]
- 27.Shih JD, Hunter CP. SID-1 is a ds-RNA selective ds-RNA gated channel. RNA. 2011;17:1057–1065. doi: 10.1261/rna.2596511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elbashir SM, et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 29.Kok KH, Ng M-HJ, Ching Y-P, Jin D-Y. Human TRBP and PACT directly interact with each other and associate with Dicer to facilitate the production of small interfering RNA. J. Biol. Chem. 2007;282:17649–17657. doi: 10.1074/jbc.M611768200. [DOI] [PubMed] [Google Scholar]
- 30.Timmons L, Tabara H, Mello CC, Fire AZ. Inducible systemic RNA silencing in Caenorhabditis elegans. Mol. Biol. Cell. 2003;14:2972–2983. doi: 10.1091/mbc.E03-01-0858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tournier B, Tabler M, Kalantidis K. Phloem flow strongly influences the systemic spread of silencing in GFP Nicotiana benthamiana plants. Plant J. 2006;47:383–394. doi: 10.1111/j.1365-313X.2006.02796.x. [DOI] [PubMed] [Google Scholar]
- 32.Calixto A, Chelur D, Topalidou I, Chen X, Chalfie M. Enhanced neuronal RNAi in C. elegans using SID-1. Nat. Methods. 2010;7:554–559. doi: 10.1038/nmeth.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.