Abstract
DNA ligases finalize DNA replication and repair through DNA nick-sealing reactions that can abort to generate cytotoxic 5′-adenylation DNA damage (5′-AMP). Aprataxin (Aptx) catalyses direct reversal of 5′-AMP adducts to protect genome integrity. Here, the structure of an Aptx-DNA-AMP-Zn complex reveals active site and DNA interaction clefts formed by fusing a HIT (histidine triad) nucleotide hydrolase with an unprecedented DNA minor groove binding C2HE Zn-finger (Znf). An Aptx helical wedge interrogates the base stack for DNA end/nick sensing. HIT-Znf, the wedge, and an "[F/Y]PK" pivot motif cooperate to distort terminal DNA base-pairing and direct 5′-AMP into the active site pocket. Structural and mutational data support a wedge-pivot-cut HIT-Znf catalytic mechanism for 5′-AMP adduct recognition and removal, and suggest mutations impacting protein folding, the active site pocket, and the pivot underlie Aptx dysfunction in the neurodegenerative disorder Ataxia Oculomotor Apraxia 1 (AOA1).
Maintenance of genomic integrity is completed in the ultimate step of DNA replication and repair transactions when eukaryotic ATP-dependant DNA ligases seal DNA nicks. Ligation is a three step reaction involving (1) adenylation of a ligase active site lysine, (2) transadenylation of DNA 5′-phosphate, and (3) phosphodiester bond formation with AMP release 1,2. When ligases engage nicks harboring DNA-distorting adducts and DNA repair metabolites, including common products of abundant cellular oxidative DNA damage3, ligation can fail at the last step, leaving 5′-adenylated DNA termini. DNA 5′-adenylation must then be reversed to prevent persistent DNA single strand breaks (SSBs) and genome instability3,4,5. Aprataxin (Aptx) proofreads DNA ligase errors to restore ligatable DNA 5′-phosphates via a poorly understood DNA 5′-AMP hydrolase activity3,4,5 (Fig. 1a).
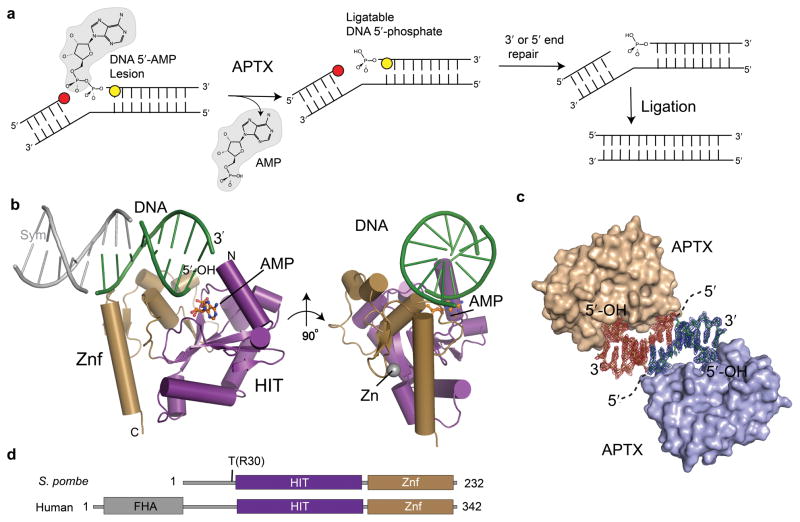
Figure 1. X-ray crystal structure of the Aptx–DNA–AMP–Zn quaternary complex.
(a) To restore ligatable DNA 5′-phosphates, Aptx reverses DNA 5′-adenylation from abortive ligation reactions created when DNA ligases engage nicks with 3′ (red circles) or 5′ (yellow circles) nick distorting lesions. (b) Overall Aptx–DNA–AMP–Zn complex architecture. Orthogonal views of the fused Aptx HIT-Znf assembly with the HIT domain (purple) and Znf (gold) collaborating to assemble the structure specific DNA end (green) binding surface. A DNA pseudo-duplex axis is formed by pairing of 3-bases across a crystallographic 2-fold axis. Symmetry related DNA is shown in grey. (c) DNA protein complex formation in the crystal. σ-A weighted Fo-Fc positive difference density map calculated before building the DNA model is displayed for symmetry related molecules (red and blue density, contoured at 2.4σ). (d) Domain architectures of S. pombe and human Aprataxin. S. pombe Aptx lacks an N-terminal FHA domain, but retains the HIT-Znf catalytic core. "T" marks site of tryptic proteolysis delineating a minimal conserved catalytic core encompassing residues 30-232 of S. pombe Aptx.
The importance of Aptx DNA processing functions in mammals is underscored by the fact that mutations in human Aprataxin (APTX) are linked to the autosomal recessive neurological disorders Ataxia with Oculomoter Apraxia 1 (AOA1) 6,7, Ataxia with coenzyme Q10 (coQ10) deficiency 8, and additional syndromes clinically overlapping with multiple system atrophy (MSA)9. AOA1 cells amass DNA damage with exposure to oxidative stress5,10 or treatment with the topoismerase poison and anti-cancer drug camptothecin11. Further evidence that budding12 and fission yeast13 Aptx homolog mutants confer clastogen sensitivity underlines conserved ancient Aptx DNA damage repair functions. It is hypothesized that Aptx dysfunction causes accumulation of DNA-adenylates in AOA1 patient cells, results in persistent DNA single strand breaks, and impacts the transcription apparatus3,14,15. Both missense and truncating Aptx substitutions are linked to neurodegeneration, but how Aptx is inactivated by mutation remains unclear.
The Aptx catalytic domain is homologous to the histidine triad (HIT) family of nucleotide hydrolases and transferases16,17. Biochemical studies support a 2-step Aptx catalytic mechanism with formation of a covalent AMP-enzyme intermediate, and disruption of DNA base-pairing proximal to the 5′-adenylate18. Mammalian Aptx FHA domain phosphorylation-dependent protein-protein interactions with Xrcc119, Xrcc4 19 and MDC1 20 implicate the Aptx DNA-deadenylase action in multiple DNA repair pathways, including base excision repair (BER), single strand break repair (SSBR) and DNA double strand break repair (DSBR). Thus, Aptx repairs abortive ligation products in the context of varied DNA architectures. Aptx processes 5′-adenylated blunt and nicked DNA substrates with comparable efficiency15 and DNaseI protection mapping of Aptx bound to adenylated and non-adenylated gapped substrates show very similar footprints, and are characterized by marked DNA binding asymmetry about the 5′-adenylation site18. In these studies, DNA contacts are mediated almost entirely to the duplex DNA downstream of the 5′-adenylate (10-11bp of duplex downstream of a 1-base gap bearing a 5′-adenylate, and 2-3 bases of ssDNA upstream of the gap. So, critical determinants of DNA binding and catalysis involve a DNA-end interaction. A putative Aptx Zn-binding domain confers substrate interaction specificity for 5′-adenylated nicked or DNA end polynucleotide substrates18. However, how Aptx sees adenylated nicks or DNA ends through a common dsDNA binding mode, accesses the 5′-AMP lesion, and drives direct enzymatic reversal of ligation errors remains ill-defined due to the absence of protein or protein-nucleic acid complex structural information for any Aptx homolog.
To clarify Aptx mechanism and provide insights into how Aptx is inactivated in disease, we report structural and biochemical characterizations of a Schizosaccharomyces Pombe (S. pombe) Aptx–DNA–AMP–Zn complex. Our results capture a molecular snapshot defining the salient features of the Aptx DNA-processing mechanism, provide a structural paradigm for DNA damage sensing and processing by DNA-nick and -end cleansing enzymes in the DNA damage response, and establish a molecular platform for understanding Aptx mutations in neurodegenerative disease.
RESULTS
Aptx domain mapping and structure determination
A catalytically active and structurally ordered domain was defined in S. pombe Aptx by proteolysis (Supplementary Fig.1a). The trypsin-stable Aptx core encompasses the predicted HIT and Znf domains (residues 30–232, Aptxcat hereafter, Fig. 1a-c). Full-length Aptx (AptxFL) and Aptxcat both display comparable 5′ DNA-deadenylation activity on a nicked 5′-adenylated plasmid substrate (Supplementary Fig.1b).
To characterize Aptx DNA binding and deadenylation of DNA substrates, we crystallized a DNA end-bound Aptx–DNA–AMP–Zn quaternary complex. Unlike vertebrate Aptx orthologs with proposed C2H2 Zn-binding motifs, the second histidine of this motif is replaced with a glutamate in fungal Aprataxins6,7 (Supplementary Fig. 2). Structural C2HE Zn-binding cores have not been reported. Further, it has not been demonstrated directly that Aprataxins bind Zn2+, so we assessed and confirmed Aptx Zn-binding by X-ray anomalous absorption of Aptx–DNA–AMP–Zn crystals, and then exploited the Zn anomalous properties to phase the complex structure to 2.35 A resolution (see Methods). Although 3 strands designed to anneal and form a 1 bp gap with a 5′ phosphorylated terminus at the gap were included in the crystallization mix, the crystallized complex contained a duplex with two strands, with Aptx bound at the DNA-end bearing a 5′ OH (see Methods, Figs.1b,1c, Supplementary Fig.4a).
Aptx–DNA–AMP–Zn Complex Architecture
The structure unveils the union of the histidine triad (HIT) nucleoside hydrolase related fold 21 (aa 33-153) with a Znf DNA binding domain (aa 154-232) that together comprise Aptxcat (Fig. 1b). The Aptx HIT domain has a central five-stranded anti-parallel β-sheet (β1-β5) that is cradled on three sides by eight helical elements (α1-α6, 3101 and 3102). The Znf and HIT domains assemble a positively charged dsDNA interaction scaffold (Fig. 2a). A network of 20 highly conserved, and primarily hydrophobic residues reinforce the HIT-Znf assembly (Supplementary Fig. 3), indicating that the relative alignment and connectivity of the domains is crucial for function. The DNA is an end-bound duplex in the crystals, with symmetry contacts across a crystallographic two-fold axis extending a DNA pseudo-helix via interactions involving a 3-base bridge (Figs. 1b,c and Supplementary Fig. 4a). The footprint of Aptx on the DNA end is consistent wtih DNAseI mapping studies defining Aptx binding to the duplex region on the 5′ end of an adenylated substrate (Supplementary Fig. 4c).
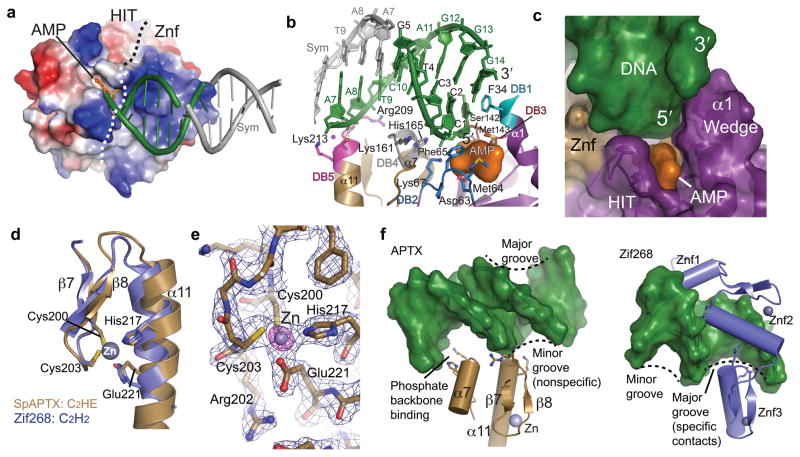
Figure 2. Aptx structure-specific DNA binding.
(a) The Aptx HIT and Znf surfaces assemble an electropositive DNA binding platform. AMP (orange) binds in a recessed hydrophobic pocket. A dotted line demarcates surface boundaries of the HIT and Znf domains. (b) Five conserved regions (DB1-5) mediate Aptx DNA binding contacts. DNA base stacking from DB1 (turquoise), DB3 (red) probe and interrogate the dsDNA end base stack for end recognition. DB2 (blue) and DB4 (grey) bind the incoming 5′ strand, whereas DB5 engages the complementary "non-adenylated" strand. (c) Surface representation of the DNA base-stacking wedge. The AMP product is sandwiched between the wedge, 5′ strand, and the HIT-Znf active site. (d) The Aptx C2HE zinc finger is structurally related to C2H2 zinc fingers. Aptx E221 replaces the second histidine of a canonical C2H2. (e) A model-phased Zn anomalous difference Fourier map (purple, displayed at 14σ) reveals the position of the single bound Zn. Blue electron density is the final σ-A weighted 2Fo-Fc map is contoured at 1.4 σ. (f) The Aptx C2HE Znf domain engages the DNA minor groove. Left: the Aptxββα-fold and α7 bind the phosphate backbone in a novel minor groove DNA binding mode. Right: canonical major groove interaction by the Zif268 C2H2 domains.
The Aptx–DNA–AMP–Zn quaternary complex shows how Aptx has evolved a protein surface suited to specifically bind and direct the hydrolysis of 5′-adenylated DNA substrates, a property that is unique to the Aptx HIT. Other HIT superfamily nucleotide hydrolases including FHIT and PKCI 16,17,21 are dimeric molecules, with extended 10-stranded inter-dimer β-sheets 17. For instance, in the protein Kinase C interacting protein (PKCI) that hydrolyzes ADP, dimerization completes assembly of the active site by contributing residues in trans across the dimer interface. In contrast, Aptx is monomeric, and a Znf domain β-strand (β6) caps the Aptx HIT β-sheet, blocking formation of a canonical HIT dimer. In Aptx, the Znf further unexpectedly contributes residues to a composite HIT-Znf active site, and decorates the HIT catalytic machinery core with an extensive electropositive dsDNA-interaction surface (Fig. 2a).
Aptx utilizes an unprecedented sequence-independent Znf DNA interaction mechanism for its action as a general DNA processing enzyme. Dali protein structural similarity searches reveal the Aptx C2HE Zn-binding core is closely related to the classical C2H2 Zn-finger ββα-fold found in Zif268, with a short anti-parallel β-sheet and α-helix 22,23 (Fig. 2d). Four residues in two Zn-binding motifs (Cys200 and Cys203 from ZB1 in the Znfβ7-β8 sheet, and His217, Glu221 from ZB2 of helix α11) tetrahedrally coordinate bound Zn2+ (Fig. 2d and 2e, Supplementary Fig.5). In Aptx, a key additional helical element (α7) and DNA binding recognition loops (DB4 and DB5) assemble around the C2HE ββα core (Fig. 2b and 2f). The Aptx Znf binds exclusively via non-specific DNA minor groove sugar-phosphate contacts, whereas canonical C2H2 Zn-fingers are typically tandem repeats that specifically recognize the DNA major groove with ββα helix side chains 22,23. Hence, two critical observations from the present structure revise our fundamental understanding of the C2H2 zinc finger fold, which is one of the largest DNA binding superfamilies known 23. First, a C2HE motif can structurally and functionally replace a C2H2 for zinc coordination, and second, in contrast with canonical major groove sequence-specific binding C2H2 binding architectures, the Aptx C2HE fold assembles a structure-specific interaction scaffold for binding damaged DNA.
Aptx DNA binding
DNA-protein complementarity explains how Aptx achieves structure specific DNA binding and deadenylation activities on 5′-adenylated nicks and DNA ends. Three key interactions define Aptx structure-specific DNA binding: (1) 5′-adenylated strand sugar-phosphate binding by both the HIT and Znf, (2) Znf non-adenylated strand phosphate backbone binding, and (3) DNA-end base stacking and interrogation by a helical hydrophobic "wedge" (Fig. 2b, 2c).
A contiguous HIT-Znf basic surface binds the dsDNA minor groove and DNA end, with ~990 A2 of solvent accessible surface buried at the Aptx–DNA–AMP interface. Ten residues from five DNA binding (DB) recognition loops, including DB1-DB3 of the HIT, and DB4-DB5 of the Znf, comprise the DNA binding surface (Fig. 2b). HIT domain residues Phe65 and Lys67 of DB2, and Ser142-Met143 of DB3 combine with Lys161 and His165 of Znf DB4 to engage and direct the 5′-adenylated strand into the HIT active site. Intriguingly, the structure uncovers a [F/Y]PK motif in the Aptx DB2 loop which is related in structure to a 5′ adenylated strand binding FPR motif of eukaryotic DNA Ligase I 1. We hypothesize convergent evolution may have selected these structurally analogous motifs for 5′-adenylated DNA binding and processing.
Through-water contacts from Lys213 (loop DB5) combine with Arg209 phosphate backbone binding to engage the complementary, "non-adenylated" strand (Fig. 2b). The DB1-DB4 DNA binding residues of S. pombe Aptx are strictly conserved with vertebrate Aptx homologs, but DB5 is not. S. pombe Aptx DB5 contacts are replaced by a conserved Znf DB4 Lys-Lys pair at the N-terminus of α7 of vertebrate Aptx (Supplementary Fig.2, Supplementary Fig.5) which, based on the positioning of this loop, we predict would interact with the non- adenylated strand. Hydrogen bonding to the DNA phosphate backbone mediated by the main chain amide of the C2HE-ββα core at the electropositive end of the helix α11 dipole also likely contributes significantly to the DNA-protein interaction binding energy.
The third prominent and defining feature of the protein-DNA interaction is a helical hydrophobic "wedge" formed by HIT domain amino-terminal helicies (3101-α1) that penetrates into the dsDNA-end base-pair stack (Fig. 2b and 2c). Interactions of the HIT domain wedge with the DNA end, plus the AMP contacts, constitute 40 % of the DNA–adduct binding surface. The conserved aromatic residue Phe34 (Tyr195 in hAptx) of loop DB1 π-stacks with and distorts the terminal C G base pair. In the case of nicked or DNA-end substrate binding, α1 base-stack capping is also likely a critical determinant of structure-specific DNA binding.
Aptx DNA interaction motifs
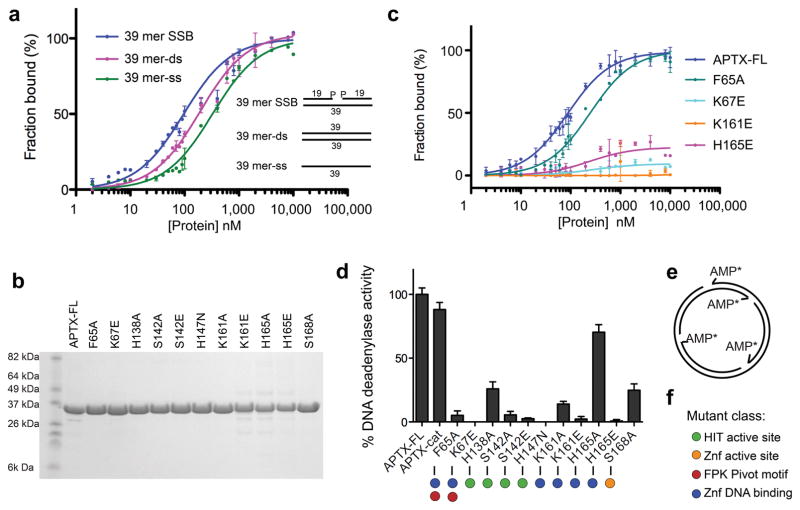
To evaluate functional roles for DNA binding contacts observed in our structures, we measured AptxFL equilibrium binding affinities on FITC-conjugated DNA substrates for Aptx mutants (Fig.3a-c). AptxFL displays a binding preference for a gapped substrate with a 3′-phosphate (a mimic of an oxidative single strand break (SSB, Kd=80 nM) over dsDNA (Kd=190 nM) or ssDNA (Kd=330 nM) (Fig. 3a). Additional DNA contacts to 2-3bp upstream on gap substrate defined by footprinting, but not observed in our structure (Supplementary Fig. 4c) may explain the slight preference for binding of a gap over blunt ended DNA. An F65A (FPK motif) mutant has lower affinity binding on the SSB substrate (Kd=250 nM), and mutations K67E, K161E and H165E impaired DNA binding to a greater extent. These Znf domain and FPK motif mutants also severely impaired plasmid DNA-deadenylation activity (Figs 3d-f), supporting roles in 5′-adenylated strand binding for DNA deadenylation repair. Thus our mutagenesis results, the observed approach of the 5′ DNA end into the Aptx HIT active site, and the close juxtaposition of the 5′ end to the bound AMP product in the active site are consistent with Aptx mediating 5′-deadenylation repair 3,4,5, but not with the proposed DNA 3′-phosphatase and 3′-phosphoglycolate processing activities 24.
Figure 3. Aptx DNA binding and deadenylation activity.
(a) Substrate specific DNA binding by AptxFL. DNA binding to FITC-conjugated 39-mer SSB (1bp gap with 3′- and 5′-phosphates), 39-mer ssDNA and 39-mer dsDNA substrates was monitored by fluorescence anisotropy. Binding isotherms are expressed as fraction bound. Error bars reflect standard deviation of three measurements. Inset: schematics of DNA binding substrates. (b) Commassie stained gel of purified AptxFL structure-based mutants. (c) DNA binding activity of Aptx 5′ adenylated strand binding Znf and FPK motif mutants. Binding to the SSB substrate (as in panel a) shown. (d) DNA deadenylase activity of structure-specific Aptx mutants. Measured release of α-P32-AMP (relative to WT) from an abortively ligated nicked (form II) φX174 is shown. Error bars are standard deviation of two measurements.
Despite maintaining HIT reaction chemistry, Aptx is a comparatively poor AMP-lysine, Ap4A and ATP nucleotide hydrolase relative to other HIT family enzymes25. Based on structural observations, we hypothesize that Aptx specificity for DNA substrate is dictated by the HIT-Znf DNA contact surface. To test this, we monitored Ap4A hydrolysis of Aptx DNA binding variants (Supplementary Fig. 6). Whereas an active site nucleophile mutant (H147N) displays no Ap4A hydrolase activity, mutants ablating the high affinity DNA interaction and DNA deadenylation activities (H165E and K161E, Fig. 3c,d) have a limited impact on Ap4A hydrolysis (Supplementary Fig.6). Thus, augmentation of the HIT architecture with a novel HIT-Znf DNA interaction scaffold defined here illustrates the versatile evolutionary integration of HIT reaction chemistry for enzymatic deadenylation of structurally distinct DNA substrates by Aptx. Observation of DNA-protein contacts proximal to the active site (Fig. 2b) (to Ser142, Met143 and Phe34) further suggests that DNA driven induced-fit assembly of the active site may guard against inappropriate non-specific nucleotide metabolism (eg. ATP or ADP hydrolysis) by Aptx.
Accessing DNA 5′-adenylate
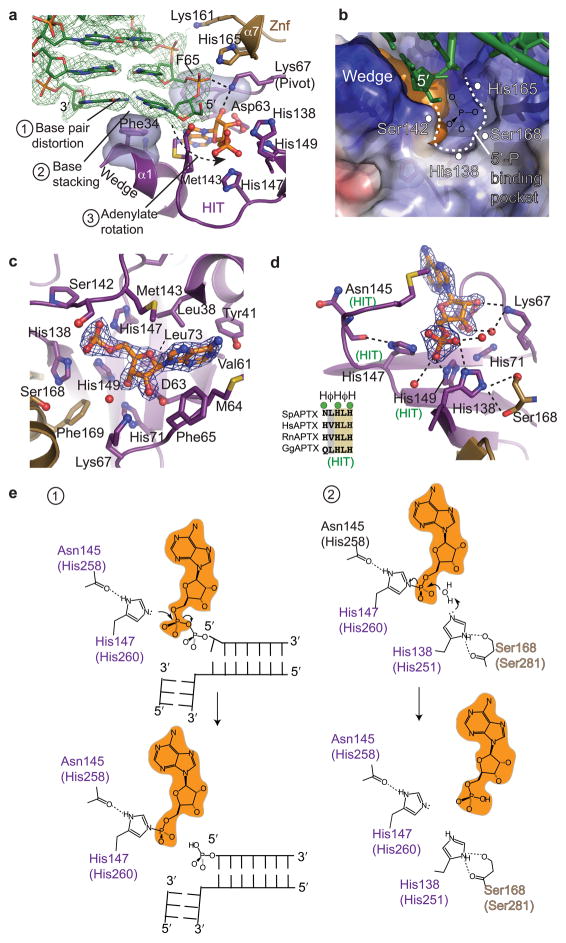
Unlike the nucleotide-hydrolyzing HIT superfamily members, Aptx must overcome steric impediments of the DNA duplex for access to the 5′-adenylate pyrophosphate linkage during catalysis. The DNA is slightly under-wound and bent relative to B-DNA. The terminal C G base pair is distorted by base stacking with Phe34 (wedge) and sugar phosphate binding by the Znf and FPK motifs (Fig. 4a). Acting as a central pivot point for adenylate rotation, the FPK motif (referred here to as the "pivot") binds both to the approaching penultimate 5′-phosphate and to the AMP adduct 2′-hydroxyl. A fused HIT-Znf adenylate binding pocket recognizes AMP, which is sandwiched between the α1-wedge, the HIT active site, and the 5′ strand (Fig. 4b, 2c). Six residues, (Leu38, Tyr41, Met64, Phe65, Lys73, and Met143) bind the adenosine base (Fig. 4c). van Der Waals interactions and hydrogen bonding by the FPK pivot (Lys67, Phe65), combined with the Asp63 side chain hydrogen bonds to the ribose 2′ and 3′ hydroxyls, precisely orient AMP in the active site, orthogonal to the 5′-strand.
Figure 4. Aptx adenylate access and catalytic mechanism.
(a) Aptx DNA end binding and adenylate rotation is mediated by the HIT domain wedge and pivot (FPK) elements that extract and align 5′-adenylate into the HIT-Znf active site. Unbiased σ-A weighted Fo-Fc positive difference density map (green, contoured at 2.4σ) calculated before building the DNA model is displayed. Phe65 and Lys67 of the pivot directly bind the sugar-phosphate backbone of the incoming 5′-strand, and AMP. (b) DNA 5′-phosphate binding pocket. An electrostatic surface potential with electropositive surface (blue) and electronegative surface (red) is displayed with a orange surface representation for the AMP. The DNA 5′-phosphate binding pocket is indicated as a dotted line. (c) The AMP binding pocket is displayed with bound AMP. Blue electron density is finalσ-A weighted 2Fo-Fc map, contoured at 1.4 σ. (d) The Aptx active site catalytic residues. Canonical HIT motif residues are marked (green "HIT"). Residues His138 and His168 are conserved in the Aprataxins, and assemble to from the Aptx HIT-Znf composite active site. (e) Structure based mechanism for 2-step direct reversal of DNA 5′-adenylation.
A critical role of Phe65 in stabilizing the DNA 5′-adenylate for catalysis is highlighted by a large reduction in DNA deadenylation activity (5% activity) in the F65A mutant (Fig. 3c, 3d). Our crystals contain an AMP product and a DNA end bearing a 5′OH in the active site. A positively charged pocket formed by HIT and Znf domain residues His138, Ser168, His165, and Ser142 is positioned to directly bind the DNA 5′-phosphate in a 5′-adenylated substrate complex (Fig. 4b). Consistent with this proposal, mutation of any of these residues impairs DNA deadenylation activity (Fig. 3d). Thus, structures and mutagenesis support a general mechanism for accessing DNA-5′ adenylates involving wedge-mediated DNA end or nicked DNA base stacking, FPK pivot and wedge driven adenylate rotation, and terminal base pair distortion.
Aptx catalytic mechanism
The present structural data provides the basis for evaluating and revising catalytic mechanism proposals made in the absence of Aptx structures. A proposed 2-step mechanism for HIT hydroylases16 involves first, the transfer of the AMP nucleotide to the active site of the enzyme through nucleophilic attack, forming a covalent enzyme-AMP intermediate, and second, hydrolysis of the intermediate with AMP release. Unexpectedly however, like the DNA interaction platform, both the HIT and Znf domains assemble the Aptx active site. The first position of the HIT HφHφH motif (Asn145 of S. pombe Aptx) is not conserved in aprataxins, but this side chain projects away from the active site, and contributes a main chain carbonyl to the active site (Fig. 4d). Three additional histidines conserved across the HIT protein superfamily (His147, His149, His71), and a His/Ser pair unique to aprataxins (HIT domain His138 and Znf Ser168), are also found in close proximity to the AMP product (Fig. 4d).
Our structure supports a mechanism wherein step 1 involves nucleophilic attack of DNA 5′ adenylate by His147 to generate a covalent enzyme-AMP intermediate (Fig. 4e). His147 is stabilized by a close hydrogen bond with the Asn145 main chain carbonyl. Complete loss of activity for a H147N mutant (Fig. 3d, Supplementary Fig. 6), the orientation of the bound AMP product phosphate in the active site, and previous observation of a transient covalent intermediate with the equivalent histidine of hAptx (His260) 18 are consistent with His147 (hAptx H260) identity as the step 1 nucleophile.
For the second step, the third HIT motif histidine is suggested to direct general acid base catalysis (enzyme-AMP hydrolysis) in FHIT by activating a water nucleophile 17. However, the equivalent histidine of Aptx (His149) is poorly oriented to act in this capacity. A HIT superfamily conserved histidine outside the core "HIT" motif (His71) positions His149, which in turn binds the product α-phosphate. Based on the observed alignment of the AMP product, His149 more likely acts in stabilizing the step 1 intermediate. From the structure, Ser168 of Znf-α7 aligns His168, which in turn hydrogen bonds to a product AMP phosphate oxygen found inline with H147-Nε and the AMP α-phosphate (the bridging oxygen in a 5′ adenylated DNA substrate). Based on this geometry, the Aptx specific His168-Ser168 pair appears better positioned to activate solvent for hydrolysis of the enzyme-AMP intermediate in Aprataxins (Fig. 4e). His168 may also act as a proton donor to the bridging oxygen in the first step. In support of catalytic roles, H138A) and S168A mutants significantly impair plasmid deadenylation activity (~25% activity for both) (Fig. 3d).
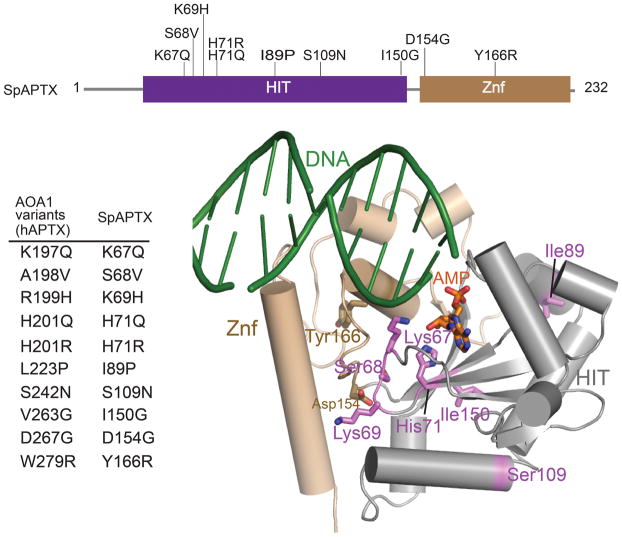
AOA1 mutations in neurodegenerative disease
To understand heritable hAptx mutations causing neurodegenerative disease, hAptx AOA1-linked substitutions were mapped onto the S. pombe Aptx structure (Fig. 5). R247X and W279X are nonsense mutants that would delete large portions of the Znf domain, likely causing loss of DNA binding and folding defects (Supplementary Fig. 5). Of 13 recorded AOA1 missense substitutions (reviewed in 14), 10 occur at conserved positions in the S. pombe structure. Mutants A198V, R199H W297R and D267G are near the HIT-Znf interface and may therefore impair interdomain interactions. V263G and L223P map to conserved buried HIT hydrophobic residues. Thus AOA1 mutations A198V, R199H, L223P, V263G, D267G and W279R mutations all change residues with apparent critical protein folding roles, predicting that these substitutions destabilize hAptx.
Figure 5. AOA1 mutations.
hAptx AOA1 associated single amino acid substitutions. Corresponding S. pombe Aptx variants for are shown in tabular format, and are mapped onto the S. pombe Aptx structure.
Three mutations (K197Q, H201Q and H201R) directly impact DNA binding and active site chemistry. Based on our structural data, H201Q and H201R mutants are predicted to occlude or distort the AMP binding pocket, preventing efficient substrate interaction. The K197Q variant is linked to mild form of Ataxia which does not present with oculomotor apraxia26, suggesting this mutation retains partial biological function. hAptx Lys197 is the adenylate-pivot lysine (S. pombe Aptx Lys67, Figs. 4a and 4c), such that substitution with a Gln may impair DNA binding and/or pivot-driven orientation of the 5′ adenylate for catalysis.
Adenylate processing of nicks and gaps
DNA ligase chemistry is sensitive to a variety of distorting forms of DNA damage and DNA mismatches flanking a nick (Fig. 1a). Aptx repairs 5′-adenylate abortive ligation products in the context of DNA repair intermediates, including 3′ abasic sites, 3′-8-oxo-dG5, 5′-abasic sites (5′deoxyribose phosphate, 5′-dRP) arising from metabolism of damaged DNA bases during BER15, and oxidative single-strand breaks bearing a 3′-phosphate and 1-bp nick3. Also, a 5′-adenylate may have the propensity to base stack with a DNA end, nick, or gap, requiring Aptx to actively extrude or alternatively stabilize extrahelical 5′-adenylate conformations in a manner analogous to DNA glycosylases27.
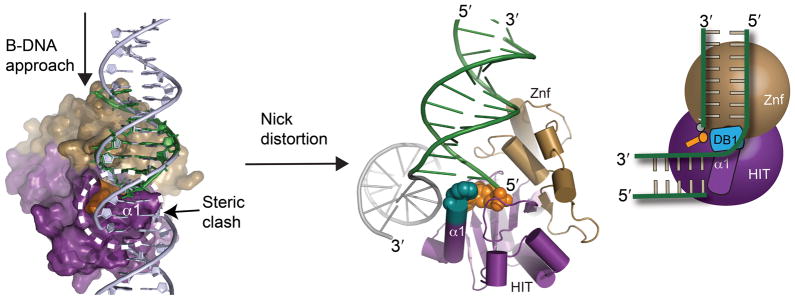
To understand how a nicked or gap DNA substrate might interact with Aptx, we modeled a B-DNA approach to the Aptx active site based on the end-bound structure. Our structures predict the α1-wedge would clash with the upstream region of a bound B-form DNA nick or gap, so we speculate DNA distortion is required to accommodate binding (Fig. 6). The upstream region of the nick may interact with a positively charged Aptx surface not defined by our end bound structure. To circumvent variable DNA access obstacles, the collective results suggest that
Figure 6. Nicked and gap DNA binding by Aptx.
Molecular model for Aptx interaction with a nicked or gapped 5′-adenylated substrate is shown (center). A steric clash of the wedge (α1) with the upstream region of a nick implies nicked or gapped DNA must be bent for binding and access to the 5′-adenylate lesion.
DNA base stack interrogation by the α1 hydrophobic wedge provides a mechanism to bind, distort and gain access to a 5′-adenylated terminus, regardless of the DNA end or nick context encountered by Aptx (Supplementary Movie 1). DNA bending of nicked or gap DNA SSBR intermediates may have important roles in facilitating control and transfer of cytotoxic DNA damage intermediate through substrate handoffs 28 to polynucleotide kinase for repair of 3′ phosphate12, 29, or to DNA polymerase for gap filing and 5′-deoxyribose phosphate lyase processing 15. An alternative mechanism, which is supported by the observation that AMP competes with binding to a 5′-phosphorylated substrate, is that in step 1 of the Aptx reaction, 5′-deadenylation promotes release of the 5′ phosphorylated DNA product from Aptx for subsequent processing by downstream enzymes (Fig. 1a).
DISCUSSION
Our results reveal an elegant "wedge-pivot-cut" mechanism for DNA 5′ adenylate access and processing. Structure specific DNA binding by the HIT-Znf composite surface for engagement of DNA end or nicked substrates illuminates impressive and diverse augmentation of the core nucleotide hydrolase/transferase chemistry of the HIT superfamily. The HIT domain and Znf are evidently evolutionarily separable elements, imparting alternative scaffolding for accommodating distinct nucleotide and polynucleotide substrates.
For sequence independent DNA engagement and genome maintenance, structures and functional data unexpectedly unveil a previously unrecognized C2HE structural Zinc coordinating element that is related to the classical C2H2 Zn-finger fold. Vertebrate Aptx homologs bear a C2H2 Zn-binding core, underscoring structural and functional interchangeability of C2H2 and C2HE Zn-scaffolding in the Aprataxins. Thus, the Aptx structure expands the repertoire of C2H2-like Zn-fingers to include sequence-independent DNA binding architectures.
The ubiquity of the C2H2 DNA binding class, which is widespread throughout nature and predicted to be found in ~3.0% of human proteins 23,30, may be much larger than previously estimated, given expansion to include C2HE. In general, pharmacological inhibition of DNA repair pathways may sensitize cancer cells to current radiation therapies and chemotherapeutic regimens. Cellular Aptx levels are a marker for therapeutic efficacy for the colorectal cancer treatment topoisomerase poison irinotecan 31, and targeted knockdown of Aprataxin DNA binding and/or catalytic activity may therefore increase efficacy of treatment of cancer. Conservation of the Aptx active site suggests that the crystal structure defined here provides molecular templates for development of human Aptx inhibitors.
ONLINE METHODS
Protein Expression and Purification
Full-length recombinant S. pombe AptxFL and Quickchange (Stratagene) generated mutants (F65A, K67E, H138A, S142A, S142E, H147N, K161A, K161E, H165A, H165E, and S168A) were expressed from pET29b (Novagen) as C-terminal 6x-His tagged proteins in E. coli BL21 Gold(DE3) cells (Stratagene) grown at 16 C in Terrific Broth. Following Ni-NTA affinity chromatography, proteins were purified by Superdex 75 (GE Healthcare) gel filtration, and anion exchange chromatography. For crystallization, the trypsin stable catalytic core Aptxcat construct was expressed from pET15b (Novagen) as an N-terminally hexa-histidine tagged protein and purified similar to AptxFL, except the 6xHis tag and linker peptide were removed by thrombin cleavage after the Ni-NTA affinity step.
Crystallization
Aptx–DNA–AMP–Zn quaternary complex crystals were grown by sitting drop vapor diffusion by mixing 200 nL ternary complex solution (10 mg mL-1 Aptxcat, with 1 mM AMP, DNA substrate at a 1.5:1 molar excess DNA substrate) 150 mM NaCl, 20 mM Tris-HCl pH 7.5, 0.1% β-mercaptoethanol, and 200 nL of crystallization solution 1 (0.2 M sodium-potassium tartrate, 20% w/v polyethylene glycol 3350). We performed co-crystallization screens with a variety nicked and blunt DNAs bearing 5′ phosphates at the nick or ends. However, we trapped a 5′-hydroxyl DNA-end bound structure, which is also bound to an AMP product nucleotide. A "dual-product" state (with both AMP and 5′phosphate) likely does not occur stably during the two-step deadenylation reaction, as there is not enough space in the active site to accommodate both. Further, AMP competes with binding to a 5′-phosphorylated gapped DNA substrate (Supplementary Fig. 4b). This explains why our crystallization experiments containing 1 mM AMP selected for a 5′-OH DNA end-bound conformation. Crystals appeared following 2 months incubation at 4 oC. For data collection, crystals were transferred to a cryoprotectant solution (0.2 M sodium-potassium tartrate, 20% w/v polyethylene glycol 3350, 26% ethylene glycol) and flash cooled in liquid nitrogen.
For the DNA crystallization substrate, three DNA oligonucleotides (A: 5′(OH)-CCCTG-3′(OH) , B : 5′(PO4)-TTCCGATA-3′(OH), and C: 5′(OH)-TATCGGAATCAGGG-3′(OH), designed to form a 1 bp gap) were included in the crystallization mix. However, the crystallized complex contained only a duplex with two strands (strands A and C), with Aptx bound at a DNA-end, probably in part due to sub-stoichiometric quantities of strand B. Crystallographic symmetry contacts forming a pseudo-duplex axis across a crystallographic 2-fold are mediated by partial pairing of 5′ overhangs of strand C, which extends the 5 bp duplex formed with annealing of strands A and C (Supplementary Fig. 4).
X-ray Diffraction Data Collection, Phasing and Refinement
X-ray data was collected at 105K on beamline 22-ID of the Advanced Photon Source. Data reduction and scaling was performed with the HKL2000 suite 32. The Aptx–DNA–AMP–Zn structure was solved using a SAD (single-wavelength anomalous dispersion) data set collected at the Zn K-edge. A single Zn position in the crystallographic asymmetric unit was located and refined with SOLVE35. Phase ambiguity in initial single wavelength anomalous dispersion (SAD) phases (overall figure of merit of phasing, FOM =0.20) were resolved by density modification in DM (FOM=0.74). A combination of automated fitting in RESOLVE35 and Manual fitting in O (v10.0) 33 produced a model that was refined to 2.35 A in REFMAC34 and PHENIX35. The final model (R=16.5, Rfree=21.1) displays good geometry (MOLPROBITY35 ramachandran statistics, 97.0%, favored, 0.5% outliers).
Fluorescence DNA Binding Assays
The DNA binding was monitored using the change in fluorescence polarization (FP) of 5′-FITC-labeled gapped duplex DNA. A 39 nucleotide oligomer (FITC-5′-CAGAGTCATAATATGCAGGTGCAGGTTATTCATGCTCGG-3′) was used as single-stranded DNA substrate; the 5′ FITC-labeled 39mer was annealed to a complementary strand (5′-CCGAGCATGAATAACCTGCACCTGCATATTATGACTCTG-3′) to form the double-stranded DNA substrate; and a 39 nucleotide oligomer (5′-CCGAGCATGAATAACCTGCACCTGCATATTATGACTCTG-3′) was annealed to two complementary strands, a 5′ FITC-labeled 19-mer (FITC-5′-CAGAGTCATAATATGCAGG-3′-(P)), and a 19-mer (5′-(P) GCAGGTTATTCATGCTCGG-3′) to form the single-nucleotide gapped duplex single-strand break (SSB) DNA substrate. DNA substrates (1 nM) were mixed with Aptx protein at concentrations ranging from 2 nM to 10 μM, in binding buffer (20 mM Tris, pH 7.5, containing 30 mM NaCl, 5 mM EDTA, 1 mM DTT, 1μl ml−1β-mercaptoenthanol, 5% sucrose) in a total reaction volume of 50 μl. Binding reactions were incubated in black flat bottom 96 well plates for 30 min at 4°C, and FP measurements were collected at room temperature with the POLARstar Omega microplate reader (BMG Labtech) using excitation and emission wavelengths of 485 and 520 nm, respectively. Kd values for the DNA interactions were calculated by fitting data expressed as a fraction bound ratio from FP measurements, with a 1-site binding model in GraphPad Prism.
AP4A Nucleotide Hydrolysis Assays
AMP release from hydrolysis of Ap4A was monitored using the Transcreener AMP FP Assay (BellBrook Labs). Ap4A substrates (1μM) were mixed with protein (16 nM) in reaction buffer (20 mM Tris, pH 7.5, containing 30 mM NaCl, 1μl mL−1 β-mercaptoenthanol), 25 μl of detection and stop buffer containing 4 nM AMP Alexa633 Tracer and 14.8 μg mL−1 AMP antibody, in a total volume of 50 μl. The reactions were incubated in black flat bottom 96 well plates for 60 minutes at 20 oC, and FP data was acquired on a POLARstar Omega microplate reader (BMG Labtech) using excitation and emission wavelengths of 630 and 670nm, respectively.
DNA Deadenylation reactions
DNA-adenylates were formed by incubation of double-stranded, nicked, circular form of ΦX174 DNA (RF II; from NEB) with 100 nM T4 DNA ligase. Abortive ligation reactions were stopped by addition of EDTA (40 mM), and unincorporated radioactivity removed by passage through a G25 spin column. Plasmid deadenylation reactions were performed essentially as described18, using Aptx proteins at 0.4 nM.
Supplementary Material
Table 1.
Data collection and refinement statistics
| Aptx–DNA–AMP–Zn complex | |
|---|---|
| Data collection | |
| Space group | I432 |
| Cell dimensions | |
| a, b, c (A) | 157.05 |
| α, β γ (°) | 90.00 |
| Zn-SAD
|
|
| Wavelength | 1.2830 |
| Resolution (A) | 50–2.35 (2.43–2.35) |
| Rmerge* | 10.2 (46.2) |
| I / σI | 20.9 (4.7) |
| Completeness (%) | 100.0 (100.0) |
| Redundancy | 7.7 (7.6) |
| Refinement | |
| Resolution (A) | 50–2.35 |
| No. reflections | 25992 |
| Rwork / Rfree | 16.5/21.1 |
| No. atoms | 2175 |
| Protein | 1645 |
| DNA | 287 |
| AMP | 23 |
| Zn | 1 |
| Solvent | 219 |
| B-factors | |
| Protein | 23.9 |
| DNA | 50.6 |
| AMP | 23.7 |
| Zn | 41.6 |
| Solvent | 35.1 |
| R.m.s deviations | |
| Bond lengths (A) | 0.009 |
| Bond angles (°) | 1.062 |
One crystal was used for each dataset.
Values in parentheses are for highest-resolution shell.
Acknowledgments
This research was supported by the intramural research program of the US National Institutes of Health (NIH), National Institute of Environmental Health Sciences (NIEHS) (1Z01ES102765-01 to R.S.W). We thank Lars Pedersen, Tom Kunkel, and Sam Wilson for discussions and critical reading of the manuscript, the Advanced Photon Source (APS) Southeast Regional Collaborative Access Team (SER-CAT) staff for assistance with crystallographic data collection, and Jason Williams of the NIEHS Protein Microcharacterization Core Facility for mass spectrometry analysis.
Footnotes
ACCESSION CODES
Coordinates for the Aptx–DNA–AMP–Zn complex are in the RCSB protein data bank (ID code 3SZQ).
AUTHOR CONTRIBUTIONS
P.T. performed and analyzed experiments, and helped write the manuscript. C.D.A., R.K.,J.S.W. performed experiments. P.D.R. and J.K. analyzed results. I.A. designed experiments and analyzed results. R.S.W designed research, performed experiments, analyzed results and wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests
References
- 1.Pascal JM, O’Brien PJ, Tomkinson AE, Ellenberger T. Human DNA ligase I completely encircles and partially unwinds nicked DNA. Nature. 2004;432:473–478. doi: 10.1038/nature03082. [DOI] [PubMed] [Google Scholar]
- 2.Ellenberger T, Tomkinson AE. Eukaryotic DNA ligases: structural and functional insights. Annu Rev Biochem. 2008;77:313–338. doi: 10.1146/annurev.biochem.77.061306.123941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahel I, et al. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature. 2006;443:713–716. doi: 10.1038/nature05164. [DOI] [PubMed] [Google Scholar]
- 4.Reynolds JJ, et al. Defective DNA ligation during short-patch single-strand break repair in ataxia oculomotor apraxia 1. Mol Cell Biol. 2009;29:1354–1362. doi: 10.1128/MCB.01471-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harris JL, et al. Aprataxin, poly-ADP ribose polymerase 1 (PARP-1) and apurinic endonuclease 1 (APE1) function together to protect the genome against oxidative damage. Hum Mol Genet. 2009;18:4102–4117. doi: 10.1093/hmg/ddp359. [DOI] [PubMed] [Google Scholar]
- 6.Date H, et al. Early-onset ataxia with ocular motor apraxia and hypoalbuminemia is caused by mutations in a new HIT superfamily gene. Nat Genet. 2001;29:184–188. doi: 10.1038/ng1001-184. [DOI] [PubMed] [Google Scholar]
- 7.Moreira MC, et al. The gene mutated in ataxia-ocular apraxia 1 encodes the new HIT/Zn-finger protein aprataxin. Nat Genet. 2001;29:189–193. doi: 10.1038/ng1001-189. [DOI] [PubMed] [Google Scholar]
- 8.Quinzii CM, et al. Coenzyme Q deficiency and cerebellar ataxia associated with an aprataxin mutation. Neurology. 2005;64:539–54. doi: 10.1212/01.WNL.0000150588.75281.58. [DOI] [PubMed] [Google Scholar]
- 9.Baba Y, et al. Aprataxin (APTX) gene mutations resembling multiple system atrophy. Parkinsonism Relat Disord. 2007;13:139–142. doi: 10.1016/j.parkreldis.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 10.Hirano M, et al. DNA single-strand break repair is impaired in aprataxin-related ataxia. Ann Neurol. 2007;61:162–174. doi: 10.1002/ana.21078. [DOI] [PubMed] [Google Scholar]
- 11.Mosesso P, et al. The novel human gene aprataxin is directly involved in DNA single-strand-break repair. Cell Mol Life Sci. 2005;62:485–491. doi: 10.1007/s00018-004-4441-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daley JM, Wilson TE, Ramotar D. Genetic interactions between HNT3/Aprataxin and RAD27/FEN1 suggest parallel pathways for 5′ end processing during base excision repair. DNA Repair (Amst) 2010;9:690–699. doi: 10.1016/j.dnarep.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deshpande GP, et al. Screening a genome-wide S. pombe deletion library identifies novel genes and pathways involved in genome stability maintenance. DNA Repair (Amst) 2009;8:672–679. doi: 10.1016/j.dnarep.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet. 2008;9:619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- 15.Rass U, Ahel I, West SC. Actions of aprataxin in multiple DNA repair pathways. J Biol Chem. 2007;282:9469–9474. doi: 10.1074/jbc.M611489200. [DOI] [PubMed] [Google Scholar]
- 16.Lima CD, Klein MG, Hendrickson WA. Structure-based analysis of catalysis and substrate definition in the HIT protein family. Science. 1997;278:286–290. doi: 10.1126/science.278.5336.286. [DOI] [PubMed] [Google Scholar]
- 17.Brenner C. Hint, Fhit, and GalT: function, structure, evolution, and mechanism of three branches of the histidine triad superfamily of nucleotide hydrolases and transferases. Biochemistry. 2002;41:9003–9014. doi: 10.1021/bi025942q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rass U, Ahel I, West SC. Molecular mechanism of DNA deadenylation by the neurological disease protein aprataxin. J Biol Chem. 2008;283:33994–34001. doi: 10.1074/jbc.M807124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clements PM, et al. The ataxia-oculomotor apraxia 1 gene product has a role distinct from ATM and interacts with the DNA strand break repair proteins XRCC1 and XRCC4. DNA Repair (Amst) 2004;3:1493–1502. doi: 10.1016/j.dnarep.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 20.Becherel OJ, et al. CK2 phosphorylation-dependent interaction between aprataxin and MDC1 in the DNA damage response. Nucleic Acids Res. 2010;38:1489–1503. doi: 10.1093/nar/gkp1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lima CD, et al. MAD analysis of FHIT, a putative human tumor suppressor from the HIT protein family. Structure. 1997;5:763–774. doi: 10.1016/s0969-2126(97)00231-1. [DOI] [PubMed] [Google Scholar]
- 22.Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 23.Klug A. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu Rev Biochem. 2010;79:213–231. doi: 10.1146/annurev-biochem-010909-095056. [DOI] [PubMed] [Google Scholar]
- 24.Takahashi T, et al. Aprataxin, causative gene product for EAOH/AOA1, repairs DNA single-strand breaks with damaged 3′-phosphate and 3′-phosphoglycolate ends. Nucleic Acids Res. 2007;35:3797–3809. doi: 10.1093/nar/gkm158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kijas AW, Harris JL, Harris JM, Lavin MF. Aprataxin forms a discrete branch in the HIT (histidine triad) superfamily of proteins with both DNA/RNA binding and nucleotide hydrolase activities. J Biol Chem. 2006;281:13939–13948. doi: 10.1074/jbc.M507946200. [DOI] [PubMed] [Google Scholar]
- 26.Tranchant C, Fleury M, Moreira MC, Koenig M, Warter JM. Phenotypic variability of aprataxin gene mutations. Neurology. 2003;60:868–870. doi: 10.1212/01.wnl.0000048562.88536.a4. [DOI] [PubMed] [Google Scholar]
- 27.Huffman JL, Sundheim O, Tainer JA. DNA base damage recognition and removal: new twists and grooves. Mutat Res. 2005;577:55–76. doi: 10.1016/j.mrfmmm.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 28.Wilson SH, Kunkel TA. Passing the baton in base excision repair. Nat Struct Biol. 2000;7:176–178. doi: 10.1038/73260. [DOI] [PubMed] [Google Scholar]
- 29.Bernstein NK, et al. The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase. Mol Cell. 2005;17:657–670. doi: 10.1016/j.molcel.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 30.Klug A. The discovery of zinc fingers and their development for practical applications in gene regulation and genome manipulation. Q Rev Biophys. 2010;43:1–21. doi: 10.1017/S0033583510000089. [DOI] [PubMed] [Google Scholar]
- 31.Dopeso H, et al. Aprataxin tumor levels predict response of colorectal cancer patients to irinotecan-based treatment. Clin Cancer Res. 2010;16:2375–2382. doi: 10.1158/1078-0432.CCR-09-3275. [DOI] [PubMed] [Google Scholar]
- 32.Otwinowski Z, Minor W. Methods in Enzymology. In: Carter CW Jr, Sweets RM, editors. Macromolecular Crystallography, part A. 276 . Academic Press; 1997. pp. 307–326. [Google Scholar]
- 33.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47 ( Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 34.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 35.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.