Abstract
We report methodological advances that extend the current capabilities of ion-abrasion scanning electron microscopy (IA–SEM), also known as focused ion beam scanning electron microscopy, a newly emerging technology for high resolution imaging of large biological specimens in 3D. We establish protocols that enable the routine generation of 3D image stacks of entire plastic-embedded mammalian cells by IA-SEM at resolutions of ~10 to 20 nm at high contrast and with minimal artifacts from the focused ion beam. We build on these advances by describing a detailed approach for carrying out correlative live confocal microscopy and IA–SEM on the same cells. Finally, we demonstrate that by combining correlative imaging with newly developed tools for automated image processing, small 100 nm-sized entities such as HIV-1 or gold beads can be localized in SEM image stacks of whole mammalian cells. We anticipate that these methods will add to the arsenal of tools available for investigating mechanisms underlying host-pathogen interactions, and more generally, the 3D subcellular architecture of mammalian cells and tissues.
Keywords: Ion-Abrasion Scanning Electron Microscopy, Focused Ion Beam Scanning Electron Microscopy, Correlative Light Electron Microscopy, T cells, HIV
INTRODUCTION
Ion-Abrasion Scanning Electron Microscopy (IA-SEM), also referred to as Focused Ion Beam Scanning Electron Microscopy, is a technology for 3D imaging that has been adapted recently to biology for the 3D imaging of large mammalian cells and tissues at nanoscale resolution (Heymann et al., 2009; Subramaniam, 2005). In this approach to 3D imaging, a gallium ion beam is used to abrade material progressively from the surface of the specimen and a scanning electron beam is used to generate an image from each newly exposed surface, producing a stack of images that provide a 3D representation of the imaged volume. With IA-SEM, we and others have previously reconstructed 3D images of yeast cells (Heymann et al., 2006), melanoma cells (Heymann et al., 2009), diatoms (Hildebrand et al., 2009), HIV-1-infected macrophages (Bennett et al., 2009), T cell-dendritic cell synapses (Felts et al., 2010), HeLa cells (Lucas et al., 2008), mouse neurons and arteries (Hekking et al., 2009; Knott et al., 2008; Merchan-Perez et al., 2009; Siskova et al., 2009), zebrafish vasculature (Armer et al., 2009) and diseased liver mitochondria (Murphy et al., 2010). Serial ultramicrotomy inside the chamber of the microscope in conjunction with block-face imaging has also been used as an alternative approach to generate 3D images of cells and tissues, although at lower resolutions in the direction orthogonal to the plane of sectioning (Denk and Horstmann, 2004; Shu et al., 2011). While the presently available methods have been useful to provide new, and sometimes unexpected insights into subcellular architecture in each of these cases, there is a need to develop additional tools to enhance the use of IA-SEM for investigating cellular mechanisms, especially in the context of correlative microscopy.
Recently, a variety of methods have been developed that combine TEM with light and fluorescence microscopy to allow correlative imaging using these two methods (Gaietta et al., 2002). These include approaches that involve hardware changes in the specimen chamber of the electron microscope to allow simultaneous fluorescence imaging (Agronskaia et al., 2008), as well as methods that are based on visualization of fluorescence in plunge-frozen samples with subsequent evaluation using cryo-electron microscopy (Sartori et al., 2007). Another approach used fluorescence en bloc light microscopy of thin-sectioned, high-pressure frozen and freeze-substituted eukaryotic cells and correlated it to the corresponding tomograms (Kukulski et al., 2011). One limitation of these methods is that since TEM imaging is restricted to regions of the sample that are less than 0.5 μm thick, correlative imaging is restricted to either a section from the cell, or to the thin edges of whole mammalian cells (Jimenez et al., 2010; Sartori et al., 2007). Since IA-SEM is capable of providing 3D images of whole mammalian cells, it is desirable to develop techniques that allow the imaging of entire cells first using live confocal microscopy and then by IA-SEM to obtain a correlated SEM image stack.
Once a SEM image stack is generated, methods that enable rapid feature identification in these large data sets must be developed. Image stacks with a lateral sampling of ~3 nm even from single mammalian cells can be very large (~1 gigabyte or larger) and pose significant technical challenges for mining information. While methods to carry out automated segmentation of these SEM image stacks involve procedures similar to those used in segmenting tomograms, locating small features of interest such as viruses poses an even greater challenge because they represent a much smaller fraction of the overall volume imaged. Feature identification using template-based approaches or machine learning (Narasimha et al., 2009) can provide a starting point for these analyses, but typically result in a very high frequency of detection of false positives because of the complexity and the low signal-to-noise ratios inherent to IA-SEM. One possible solution to high-fidelity, automated, feature recognition is to label the feature of interest with a fluorescent tag; then, the signal from the correlated confocal image volume could be used to identify the feature of interest while simultaneously excluding false positives generated by automatic segmentation.
Here, we present progress towards each of the problems identified above to further streamline the use of IA-SEM in biological imaging. We provide a perspective on the relative merits of different staining methods and identify conditions that produce the most useful image contrast. We present a method that enables regions identified in a fluorescence microscopic image to be easily identified and imaged with IA-SEM. We also present novel image processing tools for registration of light and electron microscopic images, and automated tools for segmentation to extract selected features such as 100 nm-sized viruses out of the large volume of mammalian cells. Together, these developments provide additional enhancements to tools for 3D imaging of mammalian cells at nanoscale resolution.
Materials and Methods
Cells, viruses and particles
For the staining screen, dendritic cells and CD4+ T cells were generated and purified from peripheral blood, and then incubated with HIV-1 BaL viruses as published previously (Felts et al., 2010). For the correlative experiment, the CD4+ T cell line H9 was obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. The cells were cultured in RPMI 1640 supplemented with 10% fetal bovine serum, 2 mM L-glutamine, and 100 U/mL Penicillin/Streptomycin and were >95% viable at the time of experiment. Infectious HIV-1 BaL viruses purified from supernatants of infected SupT1 CCR5 Cl.30 cells via sucrose gradients were treated with Alexa488-maleimide and Aldrithiol-2 (Sigma-Aldrich) to yield fluorescent viruses (kind gift from Dr. Jeff Lifson, SAIC Frederick). 100 nm-sized gold beads (BBI Inc, Cardiff, UK) were used as control particles because they are similar in size to HIV and are clearly visible by EM, and they were incubated with irrelevant fluorescent antibodies before the experiment.
High-Pressure Freezing and Freeze Substitution
H9 T cells mixed with 100 nm-sized gold beads and AT-2 treated HIV-1 BaL were placed into a solution of 4% paraformaldehyde (PFA) and 20% bovine serum albumin (BSA) for 20 minutes. The cells were spun down gently and then 0.7 μL of the pellet were added to a gold-covered planchette 0.1 μm deep and 1.5 μm wide (Leica Microsystems, Vienna, Austria). The planchette was then frozen in an EMPACT high-pressure freezer (Leica Microsystems, Vienna, Austria). Several staining solutions were tried. Those that produced poor staining or preservation lacked PFA and included conditions that used (i) the OTO method, (ii) BSA with only 0.5% uranyl acetate, (iii) osmium, uranyl acetate and glutaraldehyde with and without BSA and (iv) osmium, uranyl acetate, glutaraldehyde and water. The best staining solution, a mixture of 1% osmium tetroxide, 0.5% uranyl acetate and 0.5% glutaraldehyde in acetone, was prepared and dispensed into capsules (Leica Microsystems catalog #702738, Vienna, Austria) held in a metal block and then frozen in liquid nitrogen. The frozen staining solution was transferred to a styrofoam container filled with liquid nitrogen. The planchettes were then transferred to the styrofoam container and each planchette was placed face-up into a capsule. The capsule-containing block was then transferred to the chamber of a freeze substitution device (Leica Microsystems) maintained at −90 °C, and a warming protocol was initiated. The sample window was covered with aluminum foil to prevent destruction of the stain. The samples were maintained at −90 °C for 7 hours; warmed at a rate of 2 °C/hr for 32.5 hours to −25 °C; maintained at −25 °C for 12 hours, then warmed at a rate of 2 °C for 12.5 hours to 0 °C, and maintained at 0 °C for 3 hours. The block was then placed on ice and the staining solution was removed and washed with cold acetone every forty minutes (three times). After the last wash, the acetone was replaced with a solution containing 1:2 ratio of Embed-812 resin to acetone and stored at room temperature for 4 hours. The solution was finally replaced with a solution containing resin and acetone (2:1 ratio) for 2.5 hours. The planchettes were then removed from the capsules and placed face-up in inverted Beem capsules (EMS) whose pointed bottoms had been removed. Following the addition of 0.5 ml of pure resin, the Beem capsules were maintained at room temperature overnight, followed by baking in a 60 °C oven for 24 hours. The Beem capsules were subsequently removed from the hardened resin. Resin around the embedded planchette was removed with a Weck razor blade, and the block was dipped into liquid nitrogen, warmed and then dipped again, in order to dislodge the planchette gently from the resin. The planchette was removed and the sample-containing region of the resin block was shaped into a pyramid. The top was subsequently smoothed with an ultramicrotome and the top 2 mm of the block was then attached to a SEM stub. A comparison of data obtained from high pressure frozen, freeze substituted and corresponding chemically fixed T cells is shown in Figure S1. Because no substantive differences were observed in this preliminary comparison, chemically fixed cells were used in all of the studies reported here.
Staining Screen
A flow chart depicting the various staining protocols is included (Figure S2). Briefly, dendritic and T cells isolated from peripheral blood mononuclear cells (PBMCs) were fixed with 2.5% glutaraldehyde in 100 mM sodium cacodylate buffer, pH 7.4, for 4 hours and stored at 4 °C overnight. Cells were washed three times for five minutes each in 100 mM sodium cacodylate buffer, pH 7.4. The sample was then fixed and stained with 2% osmium tetroxide, which itself was reduced with 1.5% potassium ferrocyanide, in cacodylate buffer for 1 hour. En bloc staining (i.e., additional staining using chemicals added post-fixation but pre-embedding) was tested using potassium permanganate, uranyl acetate, ammonium molybdate and phosphotungstic acid. For this purpose, the osmium-treated samples were divided into two aliquots, one with, and one without 1% potassium permanganate added for 4 hours in the dark at 4 °C. The two groups were then each divided into four additional groups to be stained with (i) saturated uranyl acetate in 70% methanol, (ii) 2% uranyl acetate in maleate buffer (pH 5.4), (iii) 1% phosphotungstic acid (pH 7.0), or (iv) ammonium molybdate (pH 6.2). Three separate staining times (1 hour, 4 hours or 12 hours) were evaluated to identify the condition that produced the best contrast and sample preservation. At the end of each staining time point, the pellet was rinsed for 5 minutes with deionized water three times. Each sample was then dehydrated by treatment with ethanol at progressive concentrations of 75%, 90%, 95%, and 100%. The samples were embedded in Eponate-12 (EMS, Hatfield, PA) using the following procedure: first, they were incubated in a mixture of resin and ethanol at the ratio 1:1 for 2 hours, followed by an overnight treatment with a mixture of resin and ethanol at the ratio 3:1, then in pure resin for 4 hours, and finally in fresh resin. The resin with the cell pellet was then polymerized in an oven at 70 °C for 24 or 48 hours.
In parallel, sample staining using the osmium-thiocarbohydrazide-osmium tetroxide (OTO) method was explored (Tanaka and Mitsushima, 1984). This method involves a specified iteration of thiocarbohydrazide and osmium tetroxide following the initial treatment with osmium. After fixation by 2.5% glutaraldehyde, the samples were rinsed in 100 mM sodium cacodylate buffer for three cycles of 5 minutes each, followed by a wash in deionized water for five cycles at 1 minute each. The pelleted cells were then treated with 1% osmium tetroxide in water for 30 minutes, and rinsed with water for three cycles of 5 minutes each. 1% thiocarbohydrazide in water was then added to the pelleted cells for 10 minutes, followed by another round of osmium treatment as before, to make an OTO sample. Additional rounds of thiocarbohydrazide and osmium were used to make an OTOTO and an OTOTOTO sample, though the latter was over-stained and therefore not pursued further. All the treated samples were finally washed in water for three cycles of 5 minutes each, and the pellets were then dehydrated in a graded ethanol series and embedded in Eponate-12 as described above. To expedite screening, 200 nm-thick sections from all the sample resin blocks were prepared using a Leica Ultracut ultramicrotome and placed on a carbon-coated silicon wafer, which was then attached to an SEM stub. An additional carbon layer was sputtered upon the sections for conductivity, and the samples were viewed en face with a scanning electron beam without focused ion beam milling. A comparison of images obtained after these staining protocols is shown in Figure S3. For the studies with higher contrast (Figure 1), cells from the human melanoma cell line MNT-1 were fixed on tissue culture plates using 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH7.2) and then treated with reduced, aqueous osmium tetroxide (2% osmium tetroxide, 1.5% potassium ferrocyanide) for one hour. Embedding was done using Embed-812 with acetone as the carrier, according to the manufacturer’s protocols.
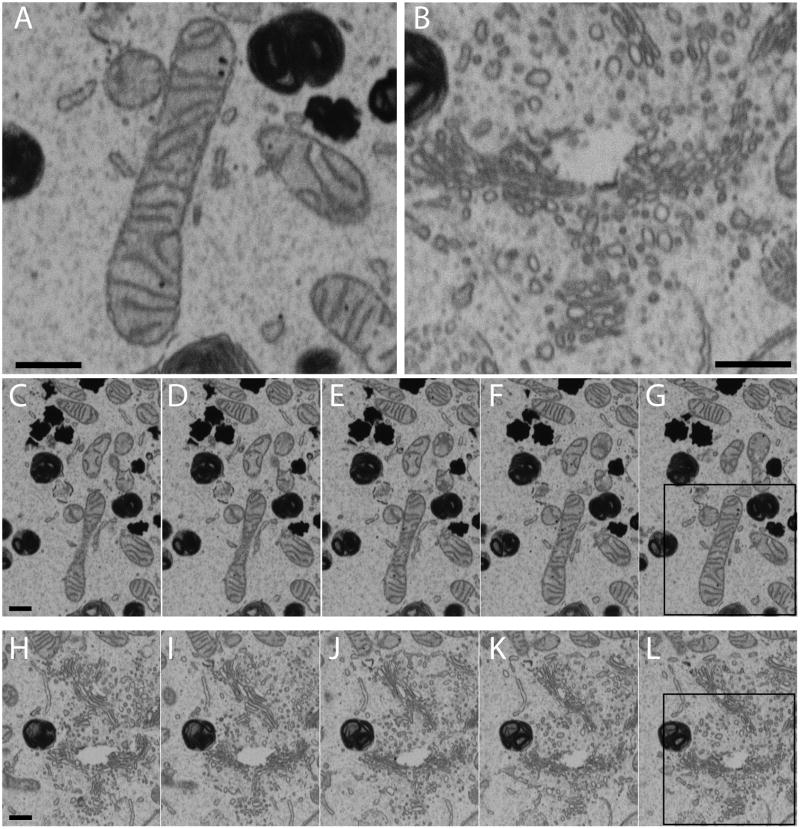
Figure 1. Representative images in a 3D images stack obtained by IA-SEM.
(A, B) Subregions highlighting the type of membrane contrast visible from mitochondria (A) and ER-like and Golgi-like structures (B) in an MNT-1 melanosome cell. (C–G) Successive 30 nm slices from a region of the cell which includes that shown in panel A. (H–L) Successive slices from a region of the cell which includes that shown in panel B. The boxed regions in panels (G) and (L) are the regions of the image that have been expanded in panels A and B respectively. The small heavily stained dark entities in all the images are melanosomes. Scale bars are 500 nm.
Incubation for correlative imaging
LabTek chambered slides (Nalge Nunc, NY) were carefully separated and gridded cover slips (EMS Diasum, PA) were affixed to the bottom of the chambers in place of the slides. These coverslips were then Poly-L-Lysine coated at 4 °C overnight. H9 cells were incubated with excess fluorescent viruses and particles (particle:cell ratio of ~1000:1) for 30 minutes at 4 °C; the cells were then incubated for 5 minutes with the membrane dye DiD, washed three times, and allowed to adhere to the coverslips at 37 °C. Regions of interest were chosen and imaging was initiated immediately.
Live Fluorescence Imaging
Live cells were imaged using a 100× oil immersion lens in a Zeiss 510 Meta confocal laser scanning microscope (CLSM) equipped with lasers at 405, 488, 546 and 633 nm, motorized stage, and a heater that maintained stage temperature at 37 °C. Up to 5 regions were chosen and consecutive z stacks (z step size 1 micron or lower) were collected at low laser power. Cells were imaged almost continuously, resulting in a complete z stack in about 2 minutes at any given region. Cells did not show visible evidence of damage during image acquisition.
Transfer to microscope
At the end of live cell imaging, cells were fixed in 2.5% glutaraldehyde in a 0.1M sodium cacodylate buffer. After a 1 hour incubation period at room temperature, cells were washed gently with cacodylate buffer and imaged in a light microscope. A montage of low resolution images of the coverslip was generated for localization of regions of interest later. The cells were then fixed, stained and embedded according to standard protocols. Briefly, the cells were incubated for 1 hour with 1% osmium tetroxide in cacodylate buffer at room temperature, washed gently in water, and then incubated for 2 hours at room temperature with 1% uranyl acetate in maleate buffer. The sample was then dehydrated with ethanol in a step-wise manner (three five-minute incubations with 50%, 70%, and 90% ethanol followed by two 10 minute 100% ethanol incubations), and then embedded in Embed-812 (EMS, Hatfield, PA), by progressive incubations with ethanol and resin at ratios of 2:1, 1:1, and 1:2, followed by 100% resin. The chambers were heated at 60 °C for 24 hours, allowing the resin to polymerize. After resin polymerization, the cells were imaged by light microscopy again to ascertain location and preservation. The plastic of the chamber itself was removed using a Dremel tool (Bosch) and the gridded coverslip was removed by repeated heating in the oven followed by plunging in liquid nitrogen, revealing the embedded cell monolayer and the etched pattern of the coverslip that was transferred to the resin. The block was then fixed to an SEM stub using silver paint, sputter coated with gold and transferred to a Nova Nanolab 200 Dual-Beam Microscope (FEI Hillsboro, OR) for imaging. Representative images from the various stages of the transfer and localization process are shown in Figure S4.
Imaging with focused ion and electron beams
For most experiments, the resin blocks containing the cells were imaged in a Nova 200 Nanolab microscope (FEI, Hillsdale, OR). Using the gridded pattern from the coverslip and the light microscopic image as a guide, SEM images of the block face were taken at a high electron beam voltage (15kV) and low magnification (1000×). The higher accelerating voltage revealed details of the cells that allowed correlation with the light images (See Figure S4.) The cells that were imaged live by confocal microscopy were located, and the stub was re-oriented to allow for “slice and view” imaging by IA-SEM. A ~0.5 μm thick layer of platinum was deposited on the chosen region using a gas injector system coupled with the ion beam at 30 kV and 0.5 nA. A trench was created using the ion beam at 7 nA in front of the region, and the cliff face was smoothed with a 0.5 nA ion beam current. The sample was then sputter-coated with gold to reduce charging during image acquisition. 2D image stacks were collected using the slice-and-view’ imaging module (FEI, Hillsdale, OR) with a nominal sectioning thickness of 21 nm. Backscattered images were recorded at 3 kV with the electron beam set for a current of 0.27 nA, an extraction voltage of 50 V for the Through Lens Detector in BSE mode, and an effective magnification of 8931× (3.5 nm/pixel), corresponding to a horizontal field width of 14.3 μm (4096 × 3036 pixels). For selected experiments (Figure 1 and Supplementary Movie 1), the cells were imaged in a Zeiss NVision 40 (Carl Zeiss NTS, Germany). Here, the trench was created using a 6.5 nA ion beam current, and the sectioning of the resin was performed at a current of 1.5 nA. A series of 2D images were collected with a section thickness of 20 nm; images in this case were recorded at 4.8 kV using the 60 μm aperture and a suppressor voltage of 300 V for the energy selective backscatter electron detector. Images were recorded at a magnification of 16,400× and an image size of 2048 x 1536 pixels, resulting in a pixel size of 3.48 nm.
Image processing and segmentation
The stack of SEM images was first aligned using IMOD’s tiltxcorr program. The images were inverted in contrast and binned in the image plane to produce pixels of 21 nm, matching the 21 nm spacing of successive images in the stack, and resulting in isotropic voxels. To remove the vertical contrast gradient along the y-axis inherent to data collection by IA-SEM, the background value of each row was estimated by averaging a cell-free volume in x (horizontally) and in z (across the image stack). A polynomial equation was fit to the curve of contrast value versus image row using Prism (Graphpad Software, San Diego, CA). A gradient image was then generated by first creating a series of rows with an appropriate width whose values were calculated from the fit equation, and then the rows were concatenated to form an image. The average background value along the z-axis for each image was calculated, and to finally correct each image, the difference between the global and image background was subtracted from each gradient image before subtracting the gradient image from each corresponding sample image. Undesirable regions of some images, such as the top of the block, were deleted by masking. Finally, the images were denoised using anisotropic diffusion (Frangakis and Hegerl, 2001) with a lambda value of 0.05 over 7 iterations using procedures implemented in the IMOD image processing package (Kremer et al., 1996). See Figure S5 for a visualization of the individual processing steps. The plasma membrane, nucleus, mitochondria and vesicles were segmented using Amira (Amira, Visage Imaging, Andover, MA).
Cell membrane Alignment
For registration of light and electron microscopic data, the SEM image stack was aligned with the confocal data computationally. First, the cells from the binned 3D volume (21 nm/pixel) were automatically segmented in SimpleITK (the Insight Segmentation and Registration Toolkit (www.itk.org) (Ibanez et al., 2002)) with thresholding by Ostu’s methods (Otsu, 1979). The mask derived from this procedure was refined to remove holes and deformations with dimensions less than 100 nm. Then the mask was refined further with a binary opening filter to remove small features. The 1-pixel wide contour of this mask was extracted and convolved with a Gaussian kernel (0.45 micron sigma) to match the point spread function of the corresponding light microscopic image. The automatic registration procedure (implemented in ITK) was initialized with coarse manual alignment based on the observed orientation of cells and slicing arrangement of the confocal volume and SEM image stack. The ITK framework structures the registration process as an optimization problem with customizable options for transformations, optimizers, image-to-image metrics, and interpolators. Given the spatially smooth volumes resulting from the large kernel of the point-spread function, an iterative gradient descent optimizer was chosen. The quality metric used for the alignment was normalized correlation. No magnitude correction of the contour image was required because the metric is invariant to scalar multiplication of the image intensities. Because of the observed distortions in the volumes, an affine transformation that included shearing in addition to mere scaling, rotation and transformation, was required. The light microscopic volume was chosen as the fixed image, onto which the transform mapped the 3D image stack using a nearest neighbor interpolator. The optimization was carried out until transformation converged. The original solution yielded what appeared to be a qualitatively good solution. To evaluate the reproducibility of the registration, the parameters of the initial solution were randomly permuted by 33% of the total vector distance between the solution and the originally chosen initial condition to generate ten additional initial conditions. This random initialization can give implausible starting transformations, with many having a large amount of skew, yet nine of the ten conditions converged to the same final solution, validating its robustness.
Automatic Virus Locator
A graphical user interface (GUI) program called itkISE was created to automatically locate HIV-1 virions in the cellular reconstructions. A Canny edge detector (Canny, 1986) was utilized to locate potential image contours of the particles. The Hough transform was initially developed to detect lines (Hart, 2009), and has been generalized to detect arbitrary analytical shapes. A variation of this classic Hough transform algorithm was implemented for spherical objects, with the transformed space consisting of the center and the average radius of the spheres. This method allows specification of the radius range, potential eccentricity and estimated numbers of the particles. Spheres with diameters from 90 nm to 230 nm were sought in 4 batches of 35 nm widths – i.e., 90–125, 125–160, 160–195 and 195–230 nm. All spheres outside or on the plasma membrane were accepted, while those inside the cell were rejected.
RESULTS AND DISCUSSION
Establishment of sample processing protocols for IA-SEM
To explore the staining protocols that generated the highest useful contrast in conjunction with chemical fixation, we screened a variety of conditions (see Materials and Methods and Figure S2 for more details). Our strategy was to test various well-established reagents for TEM such as uranyl acetate, osmium tetroxide, potassium permanganate, phosphotungstic acid and ammonium molybdate, and to evaluate how they performed in terms of obtaining maximal contrast in IA-SEM imaging. Overall, we tested a set of 24 different conditions expected to span a wide range in terms of contrast generation and preservation of cellular structure. We recorded images from each of these conditions to identify those that provided the best balance between high contrast and minimal damage to subcellular structure (Figure S3). From our analysis, we concluded that staining with osmium (reduced or non-reduced), followed by en bloc staining with uranium in maleate buffer, appeared to be the best compromise to obtain the best contrast with maximal preservation of membrane structure, and we therefore chose non-reduced osmium plus uranyl acetate (UA) for the correlative imaging studies (Figure S3H).
In Figure 1, we provide an example of the type of contrast that can be typically achieved using our preferred specimen preparation procedures. Despite being an image recorded from the surface of a plastic-embedded specimen using a scanning electron beam, key organelles such as mitochondria and intracellular membrane networks such as the endoplasmic reticulum and the Golgi are readily identified. The overall image quality in a single SEM image is still slightly inferior to an image that can be recorded from a thin section using transmission electron microscopy (See Heymann et al., 2009 for a detailed comparison.) Nevertheless, the images are clear enough to allow visualization of organelles and allow for rapid determination of the 3D membrane architecture of large mammalian cells without the need for the more time-consuming procedure of generating serial sections by ultramicrotomy followed by transmission electron microscopic imaging and image stack registration. Additionally, with FIB milling, SEM stacks can now be reliably obtained with ~20 nm spacing between successive images (Figure 1C–G, H–L), yielding data with significantly better resolution along the z-axis (perpendicular to the plane of the SEM image) than stacks obtained using mechanical sectioning with a microtome.
Correlative confocal and IA-SEM imaging
To obtain correlative images using fluorescence and IA-SEM imaging, CD4+ T cells were mixed with fluorescent antibody-coated, 100 nm-diameter gold spheres and HIV-1-Ax488, a fluorescently labeled preparation of HIV-1 BaL. The top of a LabTek chambered slide (Nalge Nunc, NY) was removed and affixed to photo-etched, square-gridded cover slips (EMS Diasum, PA) coated with Poly-L-Lysine. The cells were washed briefly, allowed to adhere to the slide and equilibrate to 37 °C, and then imaged with a confocal laser scanning microscope. Sequential, time-lapse, multichannel confocal images were collected for several regions of interest every 8 minutes over a 32 minute period, revealing particle trafficking in these cells (Figs. 2A–E). While the gold spheres (blue) remained relatively static at the surface of the cells, the virions (green) showed slow to moderate lateral motion on the cell membrane, averaging approximately 0.2 μm/minute. We did not observe internalization of fluorescent particles, and all the fluorescently labeled entities were found to be associated with the plasma membrane or in extracellular regions. The cells were fixed with 2.5% glutaraldehyde within 6 minutes of live cell imaging, followed by staining and embedding in the chamber using the osmium/UA procedure discussed above. The resin block was separated from the plastic and glass and attached to a stub, and the region of interest was located for IA-SEM imaging using the coded grid pattern as described in the Methods section.
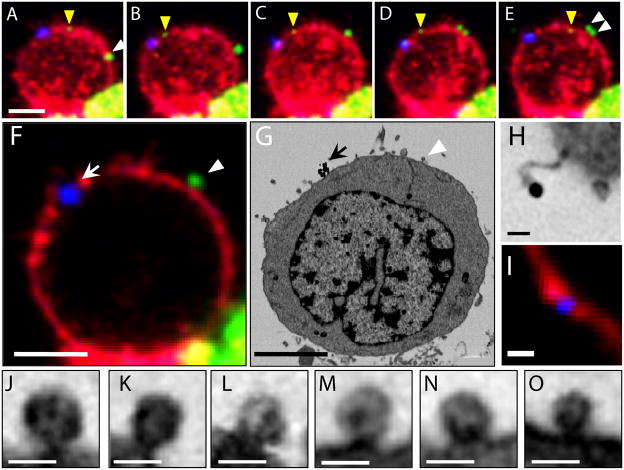
Figure 2. Localization of HIV-1 and gold particles in correlated light and electron microscopic volumes.
(A–E) Fluorescent, time-lapse, projection images of a T cell bound to HIV-1. Virions are labeled green, T cell membrane labeled red and 100 nm colloidal gold labeled blue. The yellow arrowhead points to a single HIV-1 virion. The white arrowheads point to a pair of HIV-1 particles. (F) 800 nm-thick confocal slice through same T cell at the last time point, as in panel E. (G) A manually chosen image from the SEM image stack corresponding to Panel F. Corresponding fluorescent gold particles (arrow) and HIV-1 (arrowhead) are indicated. (H, I) Single fluorescent gold bead as imaged by SEM and light microscopy. (J–O) Putative virions from other areas in the SEM image stack. The scale bar is 5 μm for panels A–G, 1 μm for panel I, and 200 nm for panels H and J–O.
As an illustration of the utility of this type of correlative imaging, we show a single slice from the confocal image (Figure 2F) alongside an SEM image (Figure 2G) from the same cell. The fluorescent gold particles, which are large and relatively static on the membrane, appear as a small cluster (arrows in Figs. 2F and 2G) and provide a quick marker to roughly register the two images by visual examination. Inspection of the SEM image stack also reveals the presence of a particle with dimensions and appearance consistent with that of HIV-1, and at roughly the location expected from the confocal imaging (Figs. 2F and 2G; arrow heads). Although definitive registration requires the more rigorous automated procedures described in the next section, the visual inspection is sufficient to locate both clusters and individual gold particles (Figs. 2F, G, and H, I, respectively). Several virions could also be identified in the SEM image stack with no corresponding fluorescence signal, consistent with the expectation that chemical fluorescence labeling of HIV-1 is likely to be partial. The imaging parameters we selected were optimized to provide enough signal and resolution to identify putative, individual virions in the SEM image stacks as illustrated by the slices from selected sub-regions shown in Figs. 2J–O.
3D registration of light and electron microscopic images
While manual correlation between light and electron microscopic images provides an approximate registration, it is not amenable to high-throughput imaging or for rapid location of multiple regions of interest. Additionally, automated registration methods are objective, reproducible, and do not depend on the user’s ability to manually align the images. Feature-based registration methods typically begin by extracting critical points or landmarks with methods such as SIFT (Cheung and Hamarneh, 2009) or other multi-scale approaches (Lindeberg, 1998), before finding correspondences. However, several issues complicate feature-based registration of light and electron microscopic volumes: the vastly different magnifications of the two imaging modalities, the different underlying physics involved in generating the images, and further, the possibility of particle (virion or gold bead) movement and incomplete fluorescent labeling in these experiments. Given the lack of any kind of statistical correspondence between regions or edges, direct image-to-image intensity registration does not produce adequate results, and other image-to-image metrics (mutual information, Kullback-Leibler distance) (Narasimha et al., 2008) fail to converge to an optimal alignment. We therefore developed a computational strategy that allowed for the rapid and unbiased 3D registration of the confocal and IA-SEM images via extraction of the cell membrane from the SEM image stack via segmentation. We reasoned that the cell membrane, which was brightly labeled in the confocal images and easily identified by thresholding in the SEM image stack, was an appropriate feature for registration.
We first extracted the contour of the cell from a thresholded segmentation of the 3-fold binned SEM image stack, and transformed it by convolving with a Gaussian kernel (Figure 3A) so that it emulated the lower resolution point spread function of the confocal image (Figure 3B). The two volumes were registered (as described in the Methods section) and the voxels corresponding to the volume derived from IA-SEM imaging were mapped by the inverse of the registration solution onto an evaluated, super-sampled envelope of the cell boundary in the confocal image. A correlated set of volumes was generated by transforming the coordinates of the confocal image onto the SEM image stack and visualized after blending of the two volumes (Figure 3C). To evaluate whether this approach could generate an accurate and reproducible registration of the two image volumes, multiple optimizations of the transformation parameters were performed on the dataset, starting from randomized positions offset by 33% from the first, original solution. Nine out of ten randomly generated conditions converged with a mean normalized cross-correlation value of 0.61 and standard deviation of 0.001 suggesting that our approach resulted in overlaid volumes that likely represented a true registration of the light microscopic and SEM datasets. The average distance between the transformed points and the corresponding point of original solution is 0.33 μm with a variance of 0.24 μm. The resolution of the light microscope volume is ~300 nm, so the error in the reproducibility of the registration is likely limited by the resolution of the confocal images. This extent of variation in the solution may also be attributed to the inexact alignment, resulting from deformation of the cells during handling not captured by the affine transformation. The converging paths of the registration optimizations starting from the random initial locations are shown plotted on derived axes from the twelve-dimensional parameter space after normalization and distance preserving multidimensional scaling (Figure 3D).
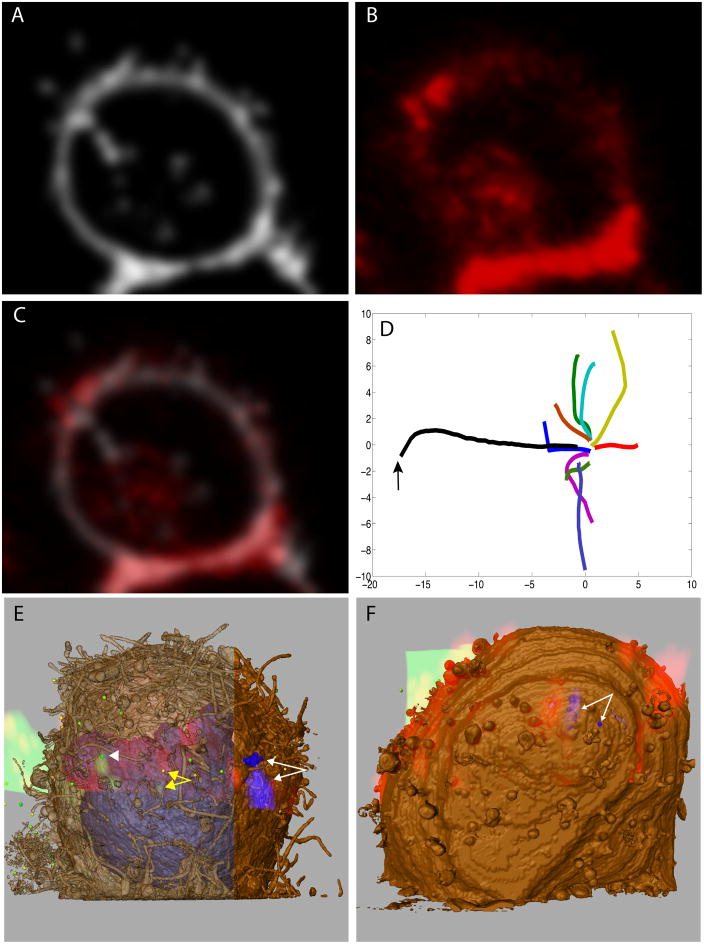
Figure 3. Registration of the live confocal volume to the SEM image stack.
(A) An optical point spread function-convolved image of the segmented T cell plasma membrane of the SEM image stack. (B) A 1 μm-thick slice from the confocal channel corresponding to the labeled T cell membrane, which alone was used for light and electron microscopic registration. (C) Overlay of the images from panels A & B. (D) Convergent paths of multiple optimizations from random initial parameters displayed on two axes derived from the twelve-dimensional parameter space. The arrow points to the initial, manual alignment. (E) A volume rendering of the confocal data for a T cell over the segmented SEM image stack. The plasma membrane from the SEM data is colored brown (either opaque or transparent to allow clear visualization of fluorescence), and gold particles blue, and these are colored red and blue, respectively, for the confocal volume. The white arrowhead points to a green, 125 nm-diameter, virion-sized, segmented sphere and the overlapping green fluorescent signal. The yellow double-headed arrow points to a 160 nm-diameter sphere which is 1.0 μm from the green virion density, the same density indicated by a yellow arrowhead in 2A–E. (F) A volume rendering of the confocal volume for another T cell overlaid with the data derived by IA-SEM.
The outcome of this type of automated, quantitative correlative 3D imaging is illustrated in Figs. 3E and 3F. Two different T cells are shown where the confocal image volume, including all fluorescence channels, has been overlaid with the corresponding segmented SEM image stack to identify the locations of the fluorescent virions and gold beads on the cell membrane. The overlay shows that the location of the closest virion candidate either overlapped with, or was <1 μm away from, the HIV fluorescent signal (white arrowhead and yellow arrows in 3E). The center-to-center distance between the density in the optical image and the SEM image of the virion on the left is ~ 0.5 μm. The virion apparent by the visual inspection in Fig. 2G is within 1.0 μm (center-to-center distance) from the fluorescent signal from HIV-1 (yellow arrow in Figure 3E). The lack of perfect overlap between the optical and electron microscopic images very likely reflects the movements of the virion relative to the rest of the cell in the time period between the recording of the last live confocal image and the time when fixation prevents further movements. In Figs. 3E–F, though the center-to-center distances between the gold in the two volumes are estimated to be as much as 1.6 μm and 0.93 μm respectively, the fluorescent signal partially overlaps the location of the gold particles in the image stacks in each case. This corresponds to an area of uncertainty of ~ 0.1% of the total surface area of the cell, rendering it an effective method to at least locate particles with reasonable precision, perhaps to be followed by a brief manual confirmation when necessary.
Algorithm for automated identification of viruses in SEM image stacks
The automated membrane registration described above allows the localization of specific components such as fluorescently labeled virions guided by the confocal image, but it is also useful more generally to independently locate virions by automated segmentation of the SEM data stack, and confirm registration by the overlap of fluorescent virion density with the segmented HIV-1-sized spheres. To this end, we implemented a computational method to automatically locate and segment spheres of a given diameter. A graphical user interface was created that allowed the user to interactively select the number and diameter range of the spheres (Figure 4A–B). Edges in the reconstruction were located using a Canny edge detection method, and candidate spheres were identified with a Hough transform voting algorithm (Figure 4C; detected edges are displayed in red and a candidate sphere on the membrane is shown in green). Because no virions were seen inside cells in the confocal reconstruction, all candidate spheres within the cell were rejected, leaving behind only those on the membrane surface or present in the extracellular milieu. Figure 4D shows a representative slice through the automated segmentation of a T cell image stack, where the membranes are colored gray and one of numerous candidate spheres in black (boxed). The inset shows an unbinned, full-resolution digital slice through the same candidate particle, establishing that this feature has dimensions expected from a virus particle. The segmentation of the entire cell shown both transparent (Figure 4E) and shaded (Figure 4F) shows that the overlay of the confocal image with the set of all putative hits obtained by the sphere finder algorithm permits localization of the virions identified using automated image segmentation. The algorithm also validates the registration, that is, the virion candidates lie amid the aggregated, fluorescent, virion density from the confocal volume that could not be resolved from each other at optical resolutions. Additionally, a single isolated virion identified by the automated sphere finder was not only visually apparent in the SEM image stack (Figure 2G, arrowhead), but was also within 1μm of a fluorescent signal corresponding to HIV (yellow arrows in Figure 3E). A movie of the entire SEM image stack at various stages of image processing is included in Supplementary movie M2, and the same segmentation and overlay process from two other T cells is presented in Figure 5.
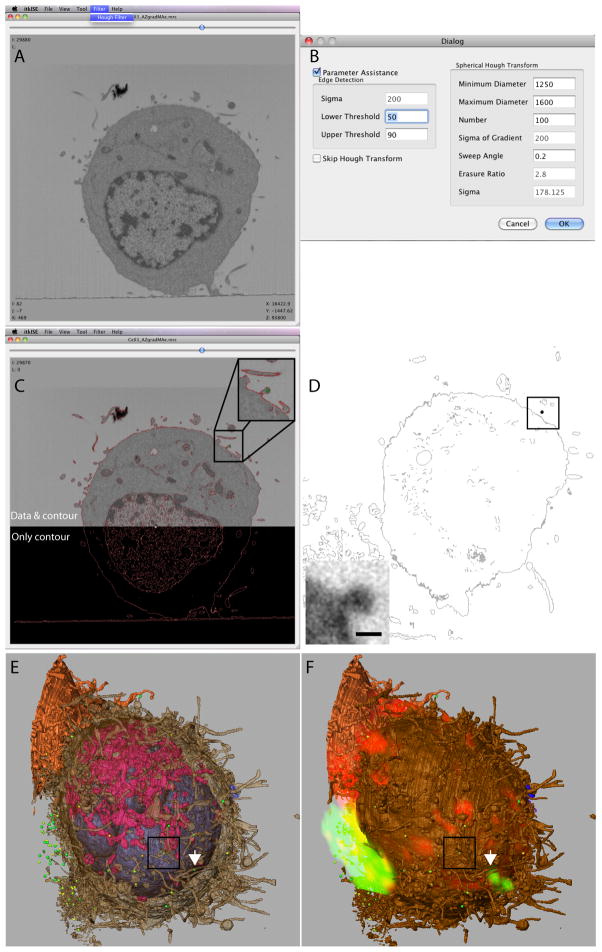
Figure 4. Automated sphere finding and evaluation with confocal images.
(A) Screen capture image of the user interface for the itkISE automated sphere finder algorithm. (B) Typical search criteria for HIV-1 virions are shown. (C) Example of a successful identification of a membrane-bound virion (green, inset) using itkISE. The edges detected are shown as red contours and is overlaid with the corresponding EM image (top). (D) A slice through an automatically segmented T cell volume. Cellular membranes are in grey and a candidate sphere 90–125 nm in diameter is in black. (Inset) A sub-image from the SEM image stack at its original resolution corresponding to the same sphere. The scale bar is 100 nm. (E) Segmented T cell showing semi-transparent plasma membrane in brown, nucleus in blue, mitochondria in pink and automatically segmented spheres 90 to 160 nm in diameter in green. The white arrow points to the segmented virion highlighted by the white arrowhead in 3E. The box encloses the same candidate sphere displayed in 4C and 4D. (F) The same segmentation as in D, superimposed with the registered confocal volume rendering of the same T cell. The intracellular organelles have not been highlighted for the sake of clarity.
Figure 5.
Automatic sphere finding algorithm and its use in correlative imaging. (A, B) A slice through two automatically segmented T cells. Edges (membranes) are in grey and candidate spheres 90–125 nm in diameter are in black. Scale bar 100 nm. (C,D) T cells showing semi-transparent plasma membrane in brown, nucleus in blue and automatically segmented spheres 90 to 160 nm in diameter in green. (E,F) The same segmentation as in C and D, superimposed with the registered volume rendering of the confocal data from the same T cells.
CONCLUSION
The use of IA-SEM for imaging cells and tissues is a relatively recent development, and many future advances can be anticipated that increase throughput, image contrast and specimen preservation. Nevertheless, even with currently available technology, the use of IA-SEM in conjunction with confocal microscopy can provide valuable mechanistic information. A schematic summary of the general strategy we have developed for merging experimentally imaged 3D volumes from confocal microscopy and IA-SEM from the same cell is shown in Figure 6. Representative slices from the confocal volume and the SEM image stack reveal the scope of information contained in these correlated datasets. The confocal images show the dynamics of fluorescently labeled entities at light microscopic resolution, while the data collected using IA-SEM allow localization of the labeled entities at resolutions that are 1 to 2 orders of magnitude higher than that achieved using light microscopy. Correlative imaging in the context of light and electron microscopy has received much interest lately (Brown et al., 2009; Hanson et al., 2010; Mironov and Beznoussenko, 2009; Watanabe et al., 2011), and the studies we present here show that the idea of correlative imaging can be extended to imaging whole cells using IA-SEM and live confocal microscopy. The development of procedures for automated registration of confocal and electron microscopic images and the application of methods for the detection of specific subcellular features such as viruses further enhance the value of correlative imaging for the study of a variety of cellular processes, including those associated with virus-cell and cell-cell interactions.
Figure 6.
Correlative 3D imaging of an entire eukaryotic cell by fluorescence and electron microscopy reveals complementary information from the two imaging methods. 3D volumes of a CD4+ T cell exposed to HIV-1 and gold particles generated by confocal microscopy (A) and IA-SEM (B) were registered and correlated. The volumes were generated from a stack of 2D images in orthogonal axes. An arbitrary slice from both stacks is also shown. Fluorescently labeled entities such as HIV-1 and cell membranes are imaged by confocal microscopy; registering these with corresponding densities in the SEM image stack reveals the correlated ultrastructure of the imaged cell.
Supplementary Material
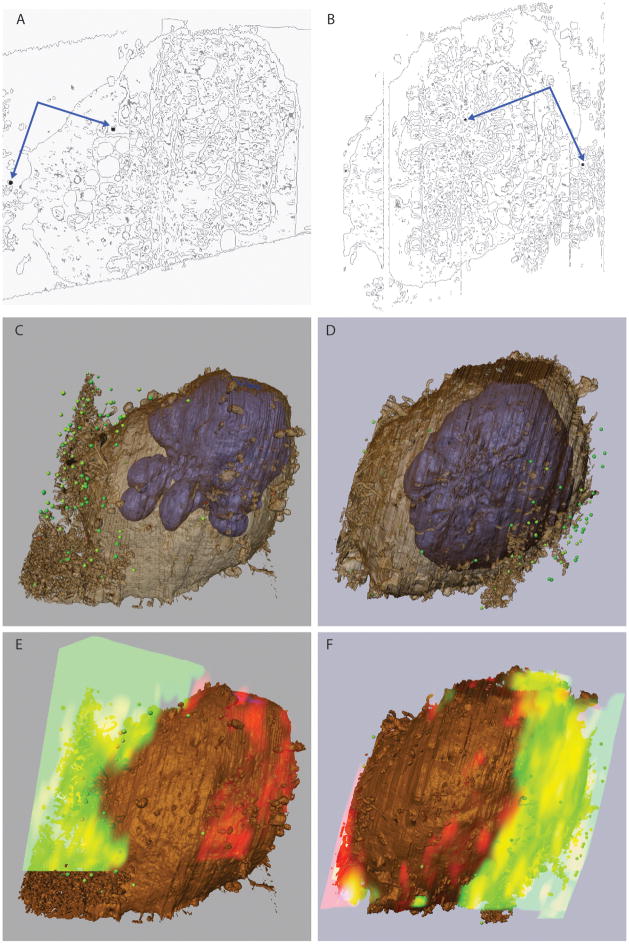
(A) Slice through a SEM image stack of a room temperature-stained T cell. (B–E) Images of subcellular organelles from the cell shown in (A): mitochondria (B, C), putative Golgi (D) and endoplasmic reticulum (E). (F) Slice through a SEM image stack of a HPF/FS T cell. (G–J) Images of subcellular organelles from the cell shown in (F): mitochondria (G, H), putative Golgi (I) and endoplasmic reticulum (J). Scale bars in panels B, C, E are the same as in panel D and scale bars in G, H, I are the same as panel J. All scale bars are 1 micron.
A single cell pellet was divided to assess each of the conditions noted in the grid.
(A) 1-hour incubation with 2% osmium tetroxide reduced with 1.5% potassium ferrocyanide. (B) 4-hour incubation with 1% uranyl acetate in maleate buffer. (C) 4-hour incubation with 1% phosphotungstic acid. (D) 1-hour incubation with 1% ammonium molybdate. (E–G) Evaluation of OTO staining protocols for IA-SEM. (E) 30-minute incubation with 1% osmium tetroxide alone. (F) 30 minute incubation with 1% osmium tetroxide, followed by 10 minute incubation with thiocarbohydrazide, followed by another incubation with 1% osmium tetroxide. (G) OTO followed by another cycle of thiocarbohydrazide and osmium tetroxide. (H) 1-hour incubation with 1% osmium tetroxide followed by 1 hour with 1% uranyl acetate. Scale bars are 1 micron.
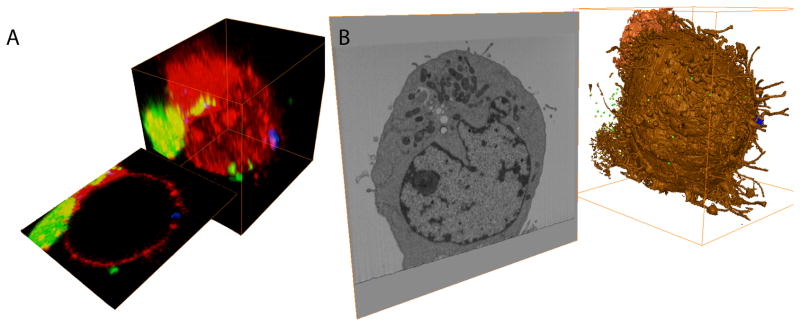
(A) A phase contrast image of the region of interest (ROI). (B) A confocal section through the three cells of interest. (C) A phase contrast image of the ROI embedded in resin. (D) A 3 keV scanning secondary electron image of the block surface, with the ROI boxed in red. The inset depicts how deep the 3 keV beam would penetrate, about 20 nm. (E) A 15 keV, backscattered image of the ROI showing the cells of interest and the milling and viewing direction. The inset depicts how deep the 15 keV beam would penetrate, about 1 μm. (F) An image of the trench and side walls milled in front of and around the ROI.
(1) Raw image. (2) Image binned 5-fold and inverted after alignment. (3) Computed gradient image. (4) Image in (2) after subtraction by image in (3). (5) Thresholded mask generated from image in (2). (6) Masked image obtained by multiplying image in (4) by image in (5). (7) Final 3D denoised image.
Representative image stack from a MNT-1 melanosome cell, representing 150 slices.
Representative 3D image stack from a T cell exposed to fluorescent HIV-1 and gold beads. Section 1: 3D processed image stack through T cell. Section 2: Image stack after applying Automatic Virus Locator algorithm (all edges corresponding to 90–160 nm spheres in the volume appear as bright specks). Section3: Segmented SEM image stack of T cell. The cell membrane is colored brown; the nucleus is blue; the mitochondria are pink and putative virions picked up by the Automatic Virus Locator are displayed as green spheres
Acknowledgments
This work was supported by funds from the Center for Cancer Research at the National Cancer Institute, NIH, Bethesda, MD. We thank Dr. Jeff Lifson and Julian Bess for providing the fluorescently labeled HIV-1 samples.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agronskaia AV, Valentijn JA, van Driel LF, Schneijdenberg CT, Humbel BM, et al. Integrated fluorescence and transmission electron microscopy. J Struct Biol. 2008;164:183–9. doi: 10.1016/j.jsb.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Armer HE, Mariggi G, Png KM, Genoud C, Monteith AG, et al. Imaging transient blood vessel fusion events in zebrafish by correlative volume electron microscopy. PLoS One. 2009;4:e7716. doi: 10.1371/journal.pone.0007716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett AE, Narayan K, Shi D, Hartnell LM, Gousset K, et al. Ion-abrasion scanning electron microscopy reveals surface-connected tubular conduits in HIV-infected macrophages. PLoS Pathog. 2009;5:e1000591. doi: 10.1371/journal.ppat.1000591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown E, Mantell J, Carter D, Tilly G, Verkade P. Studying intracellular transport using high-pressure freezing and Correlative Light Electron Microscopy. Semin Cell Dev Biol. 2009;20:910–9. doi: 10.1016/j.semcdb.2009.07.006. [DOI] [PubMed] [Google Scholar]
- Canny J. A Computational Approach to Edge-Detection. Ieee Transactions on Pattern Analysis and Machine Intelligence. 1986;8:679–698. [PubMed] [Google Scholar]
- Cheung W, Hamarneh G. n-SIFT: n-Dimensional Scale Invariant Feature Transform. Ieee Transactions on Image Processing. 2009;18:2012–2021. doi: 10.1109/TIP.2009.2024578. [DOI] [PubMed] [Google Scholar]
- Denk W, Horstmann H. Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS Biol. 2004;2:e329. doi: 10.1371/journal.pbio.0020329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felts RL, Narayan K, Estes JD, Shi D, Trubey CM, et al. 3D visualization of HIV transfer at the virological synapse between dendritic cells and T cells. Proc Natl Acad Sci U S A. 2010;107:13336–41. doi: 10.1073/pnas.1003040107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangakis AS, Hegerl R. Noise reduction in electron tomographic reconstructions using nonlinear anisotropic diffusion. J Struct Biol. 2001;135:239–50. doi: 10.1006/jsbi.2001.4406. [DOI] [PubMed] [Google Scholar]
- Gaietta G, Deerinck TJ, Adams SR, Bouwer J, Tour O, et al. Multicolor and electron microscopic imaging of connexin trafficking. Science. 2002;296:503–7. doi: 10.1126/science.1068793. [DOI] [PubMed] [Google Scholar]
- Graham LL, Beveridge TJ. Evaluation of freeze-substitution and conventional embedding protocols for routine electron microscopic processing of eubacteria. J Bacteriol. 1990;172:2141–9. doi: 10.1128/jb.172.4.2141-2149.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson HH, Reilly JE, Lee R, Janssen WG, Phillips GR. Streamlined embedding of cell monolayers on gridded glass-bottom imaging dishes for correlative light and electron microscopy. Microsc Microanal. 2010;16:747–54. doi: 10.1017/S1431927610094092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart PE. How the Hough Transform Was Invented. Ieee Signal Processing Magazine. 2009;26:18–22. [Google Scholar]
- Hekking LH, Lebbink MN, De Winter DA, Schneijdenberg CT, Brand CM, et al. Focused ion beam-scanning electron microscope: exploring large volumes of atherosclerotic tissue. J Microsc. 2009;235:336–47. doi: 10.1111/j.1365-2818.2009.03274.x. [DOI] [PubMed] [Google Scholar]
- Heymann JA, Hayles M, Gestmann I, Giannuzzi LA, Lich B, et al. Site-specific 3D imaging of cells and tissues with a dual beam microscope. J Struct Biol. 2006;155:63–73. doi: 10.1016/j.jsb.2006.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymann JA, Shi D, Kim S, Bliss D, Milne JL, et al. 3D imaging of mammalian cells with ion-abrasion scanning electron microscopy. J Struct Biol. 2009;166:1–7. doi: 10.1016/j.jsb.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand M, Kim S, Shi D, Scott K, Subramaniam S. 3D imaging of diatoms with ion-abrasion scanning electron microscopy. J Struct Biol. 2009;166:316–28. doi: 10.1016/j.jsb.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibanez L, Ng L, Gee J, Aylward S. Registration patterns: the generic framework for image registration of the insight toolkit. Biomedical Imaging; 2002; Proceedings. 2002 IEEE International Symposium on.2002. pp. 345–348. [Google Scholar]
- Jimenez N, Van Donselaar EG, De Winter DA, Vocking K, Verkleij AJ, et al. Gridded Aclar: preparation methods and use for correlative light and electron microscopy of cell monolayers, by TEM and FIB-SEM. J Microsc. 2010;237:208–20. doi: 10.1111/j.1365-2818.2009.03329.x. [DOI] [PubMed] [Google Scholar]
- Knott G, Marchman H, Wall D, Lich B. Serial section scanning electron microscopy of adult brain tissue using focused ion beam milling. J Neurosci. 2008;28:2959–64. doi: 10.1523/JNEUROSCI.3189-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–6. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Kukulski W, Schorb M, Welsch S, Picco A, Kaksonen M, et al. Correlated fluorescence and 3D electron microscopy with high sensitivity and spatial precision. J Cell Biol. 2011;192:111–9. doi: 10.1083/jcb.201009037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeberg T. Feature Detection with Automatic Scale Selection. International Journal of Computer Vision. 1998;30:79–116. [Google Scholar]
- Lucas MS, Gasser P, Günthert M, Mercer J, Helenius A, et al. In: Aretz A, et al., editors. Correlative 3D microscopy: CLSM and FIB/SEM tomography used to study cellular entry of vaccinia virus; EMC 2008 14th European Microscopy Congress; 1–5 September 2008; Berlin Heidelberg, Aachen, Germany: Springer; 2008. pp. 361–362. [Google Scholar]
- Merchan-Perez A, Rodriguez JR, Alonso-Nanclares L, Schertel A, Defelipe J. Counting Synapses Using FIB/SEM Microscopy: A True Revolution for Ultrastructural Volume Reconstruction. Front Neuroanat. 2009;3:18. doi: 10.3389/neuro.05.018.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironov AA, Beznoussenko GV. Correlative microscopy: a potent tool for the study of rare or unique cellular and tissue events. J Microsc. 2009;235:308–21. doi: 10.1111/j.1365-2818.2009.03222.x. [DOI] [PubMed] [Google Scholar]
- Murk JL, Posthuma G, Koster AJ, Geuze HJ, Verkleij AJ, et al. Influence of aldehyde fixation on the morphology of endosomes and lysosomes: quantitative analysis and electron tomography. J Microsc. 2003;212:81–90. doi: 10.1046/j.1365-2818.2003.01238.x. [DOI] [PubMed] [Google Scholar]
- Murphy GE, Lowekamp BC, Zerfas PM, Chandler RJ, Narasimha R, et al. Ion-abrasion scanning electron microscopy reveals distorted liver mitochondrial morphology in murine methylmalonic acidemia. J Struct Biol. 2010;171:125–32. doi: 10.1016/j.jsb.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimha R, Ouyang H, Gray A, McLaughlin S, Subramaniam S. Automatic joint classification and segmentation of whole cell 3D images. Pattern Recognition. 2009;42:1067–1079. [Google Scholar]
- Narasimha R, Aganj I, Bennett AE, Borgnia MJ, Zabransky D, et al. Evaluation of denoising algorithms for biological electron tomography. J Struct Biol. 2008;164:7–17. doi: 10.1016/j.jsb.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsu N. A Threshold Selection Method from Gray-Level Histograms. IEEE T Syst Man Cyb. 1979;9:62–66. [Google Scholar]
- Sartori A, Gatz R, Beck F, Rigort A, Baumeister W, et al. Correlative microscopy: bridging the gap between fluorescence light microscopy and cryo-electron tomography. J Struct Biol. 2007;160:135–45. doi: 10.1016/j.jsb.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Shu X, Lev-Ram V, Deerinck TJ, Qi Y, Ramko EB, et al. A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biol. 2011;9:e1001041. doi: 10.1371/journal.pbio.1001041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siskova Z, Page A, O'Connor V, Perry VH. Degenerating synaptic boutons in prion disease: microglia activation without synaptic stripping. Am J Pathol. 2009;175:1610–21. doi: 10.2353/ajpath.2009.090372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam S. Bridging the imaging gap: visualizing subcellular architecture with electron tomography. Curr Opin Microbiol. 2005;8:316–22. doi: 10.1016/j.mib.2005.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Mitsushima A. A preparation method for observing intracellular structures by scanning electron microscopy. J Microsc. 1984;133:213–22. doi: 10.1111/j.1365-2818.1984.tb00487.x. [DOI] [PubMed] [Google Scholar]
- Walther P, Ziegler A. Freeze substitution of high-pressure frozen samples: the visibility of biological membranes is improved when the substitution medium contains water. J Microsc. 2002;208:3–10. doi: 10.1046/j.1365-2818.2002.01064.x. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Punge A, Hollopeter G, Willig KI, Hobson RJ, et al. Protein localization in electron micrographs using fluorescence nanoscopy. Nat Methods. 2011;8:80–4. doi: 10.1038/nmeth.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Slice through a SEM image stack of a room temperature-stained T cell. (B–E) Images of subcellular organelles from the cell shown in (A): mitochondria (B, C), putative Golgi (D) and endoplasmic reticulum (E). (F) Slice through a SEM image stack of a HPF/FS T cell. (G–J) Images of subcellular organelles from the cell shown in (F): mitochondria (G, H), putative Golgi (I) and endoplasmic reticulum (J). Scale bars in panels B, C, E are the same as in panel D and scale bars in G, H, I are the same as panel J. All scale bars are 1 micron.
A single cell pellet was divided to assess each of the conditions noted in the grid.
(A) 1-hour incubation with 2% osmium tetroxide reduced with 1.5% potassium ferrocyanide. (B) 4-hour incubation with 1% uranyl acetate in maleate buffer. (C) 4-hour incubation with 1% phosphotungstic acid. (D) 1-hour incubation with 1% ammonium molybdate. (E–G) Evaluation of OTO staining protocols for IA-SEM. (E) 30-minute incubation with 1% osmium tetroxide alone. (F) 30 minute incubation with 1% osmium tetroxide, followed by 10 minute incubation with thiocarbohydrazide, followed by another incubation with 1% osmium tetroxide. (G) OTO followed by another cycle of thiocarbohydrazide and osmium tetroxide. (H) 1-hour incubation with 1% osmium tetroxide followed by 1 hour with 1% uranyl acetate. Scale bars are 1 micron.
(A) A phase contrast image of the region of interest (ROI). (B) A confocal section through the three cells of interest. (C) A phase contrast image of the ROI embedded in resin. (D) A 3 keV scanning secondary electron image of the block surface, with the ROI boxed in red. The inset depicts how deep the 3 keV beam would penetrate, about 20 nm. (E) A 15 keV, backscattered image of the ROI showing the cells of interest and the milling and viewing direction. The inset depicts how deep the 15 keV beam would penetrate, about 1 μm. (F) An image of the trench and side walls milled in front of and around the ROI.
(1) Raw image. (2) Image binned 5-fold and inverted after alignment. (3) Computed gradient image. (4) Image in (2) after subtraction by image in (3). (5) Thresholded mask generated from image in (2). (6) Masked image obtained by multiplying image in (4) by image in (5). (7) Final 3D denoised image.
Representative image stack from a MNT-1 melanosome cell, representing 150 slices.
Representative 3D image stack from a T cell exposed to fluorescent HIV-1 and gold beads. Section 1: 3D processed image stack through T cell. Section 2: Image stack after applying Automatic Virus Locator algorithm (all edges corresponding to 90–160 nm spheres in the volume appear as bright specks). Section3: Segmented SEM image stack of T cell. The cell membrane is colored brown; the nucleus is blue; the mitochondria are pink and putative virions picked up by the Automatic Virus Locator are displayed as green spheres