Abstract
Since the isolation and characterization of dwarf1-1 (dwf1-1) from a T-DNA insertion mutant population, phenotypically similar mutants, including deetiolated2 (det2), constitutive photomorphogenesis and dwarfism (cpd), brassinosteroid insensitive1 (bri1), and dwf4, have been reported to be defective in either the biosynthesis or the perception of brassinosteroids. We present further characterization of dwf1-1 and additional dwf1 alleles. Feeding tests with brassinosteroid-biosynthetic intermediates revealed that dwf1 can be rescued by 22α-hydroxycampesterol and downstream intermediates in the brassinosteroid pathway. Analysis of the endogenous levels of brassinosteroid intermediates showed that 24-methylenecholesterol in dwf1 accumulates to 12 times the level of the wild type, whereas the level of campesterol is greatly diminished, indicating that the defective step is in C-24 reduction. Furthermore, the deduced amino acid sequence of DWF1 shows significant similarity to a flavin adenine dinucleotide-binding domain conserved in various oxidoreductases, suggesting an enzymatic role for DWF1. In support of this, 7 of 10 dwf1 mutations directly affected the flavin adenine dinucleotide-binding domain. Our molecular characterization of dwf1 alleles, together with our biochemical data, suggest that the biosynthetic defect in dwf1 results in reduced synthesis of bioactive brassinosteroids, causing dwarfism.
T-DNA-insertion mutagenesis has proven to be useful for the isolation of many important genes controlling plant growth and development (Choe and Feldmann, 1998). The Arabidopsis dwarf1 (dwf1) mutant was originally isolated from a T-DNA mutant population, and was the first mutant shown to cosegregate with the selectable marker in the T-DNA (Feldmann et al., 1989). The dwf1 mutant was identified because of its short stature, dark-green leaves, reduced fertility, and robust stems when grown in the light. Physiologically, dwf1 was not rescued by any of the known growth-promoting phytohormones such as GA3 or auxin (Feldmann et al., 1989). Using the plant DNA flanking the T-DNA as a probe, DWF1 was cloned and sequenced (accession no. U12400).
Independently, Takahashi et al. (1995) isolated a morphologically similar mutant, diminuto (dim), from a different T-DNA mutant collection. Cloning and sequencing revealed that dim is disrupted in the DWF1 sequence, indicating that it is an allele of dwf1. One year later, Kausch-mann et al. (1996) isolated another allele of DWF1 from a transposon-tagged population. They identified three tiny mutants named cabbage1, cabbage2, and cabbage3 (cbb1, cbb2, and cbb3). Altmann et al. (1995) found the sequence of genomic DNA flanking the transposon in cbb1 to be identical to that of DWF1. Kauschmann et al. (1996) originally found that cbb1 (dwf1-6) could be rescued by exogenous application of brassinosteroids, suggesting that cbb1 (dwf1-6) is defective in brassinosteroid biosynthesis. They also analyzed the expression of genes known to be involved in cell elongation, such as γ-tonoplast intrinsic protein (γ-TIP) (Höfte et al., 1992; Phillips and Huttly, 1994), and cell wall-modifying enzymes such as xyloglucan endotransglycosylases, including TOUCH4 (TCH4) (Xu et al., 1995) and MERI5 (Medford et al., 1991). The steady-state mRNA levels of TCH4 and MERI5 were lowered, whereas the expression of γ-TIP was increased in the cbb mutants. Based on this, they proposed that a defect in brassinosteroid biosynthesis in cbb1 (dwf1-6) leads to failure in cell elongation, which requires partial activity of the cell wall-modifying enzymes TCH4 and MERI5.
Brassinolide, the proposed end product of the brassinosteroid-biosynthetic pathway, is synthesized from sterol substrates. Therefore, plants defective in the biosynthetic steps leading from mevalonic acid to sterol, as well as in steps modifying sterols to brassinolide, could display the characteristic dwf phenotype. Currently, dwf7 is reported to be defective in a step of sterol biosynthesis (Choe et al., 1999), and three mutants, deetiolated2 (det2) (Li et al., 1996), dwf4 (Choe et al., 1998), and constitutive photomorphogenesis and dwarfism (cpd) (Szekeres et al., 1996), are defective in the brassinosteroid-specific biosynthetic steps, from campesterol to brassinolide. Specifically, dwf7 is blocked in the sterol C-5 desaturation step, which is the most upstream step identified in dwf mutants thus far (Choe et al., 1999). Fujioka et al. (1997) have shown det2 to be blocked in the 5α-reduction step converting campesterol to campestanol. Choe et al. (1998) have proposed that dwf4 is disrupted in the 22α-hydroxylation step, which is hypothesized to be the rate-limiting step in brassinosteroid biosynthesis. Finally, Szekeres et al. (1996) have found cpd to be defective in the 23α-hydroxylation step following dwf4. Both DWF4 and CPD have been assigned to the same group of Cyt P450 proteins (CYP90) because they share more than 40% identity. In addition to these biosynthetic mutants, Clouse et al. (1996) have also identified brassinosteroid insensitive1 (bri1), a mutant insensitive to brassinosteroids. Recently, BRI1 was cloned and was shown to encode a Leu-rich repeat receptor kinase, suggesting a role in brassinosteroid signal perception and transduction (Li and Chory, 1997). All of the brassinosteroid dwarf mutants share characteristic phenotypes in the light, as described above, as well as abnormal skotomorphogenesis in the dark, including short hypocotyls and expanded cotyledons.
Recent characterization of these dwf mutants provides compelling evidence that brassinosteroids are essential modulators for proper growth and development in plants. To understand all of the roles assigned to brassinosteroids in plants, the identification of the components of the brassinosteroid pathway and the regulation of endogenous brassinosteroid biosynthesis is critical. The proposed brassinosteroid-biosynthetic pathway predicts that there are at least 20 genes involved in brassinolide synthesis, which begins with squalene (Choe et al., 1999). To identify mutants in each biosynthetic step, we are characterizing a large collection of Arabidopsis dwarfs with the characteristic brassinosteroid dwarf phenotype. Currently, we have identified 12 different brassinosteroid loci. Six of these mutants, bri1 (dwf2) (Clouse et al., 1996; Li and Chory, 1997), cpd (dwf3) (Szekeres et al., 1996), dwf4 (Choe et al., 1998), det2 (dwf6) (Li et al., 1996), dwf7 (Choe et al., 1999), and dwf1 (Feldmann et al., 1989; Takahashi et al., 1995; Kauschmann et al., 1996), have been characterized.
Here we report further studies on dwf1. Because Kausch-mann et al. (1996) have shown that dwf1 can be rescued by exogenous application of brassinosteroids, we have used different methods to pinpoint the specific biosynthetic step that is defective in dwf1. First, we applied biosynthetic intermediates to dwf1 plants to identify compounds that rescued dwf1 phenotypes. In addition, we analyzed the endogenous brassinosteroid levels using GC-SIM to identify accumulated compounds. Based on this biochemical analysis, we found that a C-24 reduction step converting 24-methylenecholesterol to campesterol was blocked in dwf1. Coupled with sequence analyses of DWF1 and identification of the site of mutation in eight dwf1 alleles, we propose that DWF1 acts as a biosynthetic enzyme, catalyzing C-24 reduction in sterol biosynthesis.
MATERIALS AND METHODS
Mutant Isolation
The isolation of dwf1-1 and the cosegregation of the T-DNA with the dwarf phenotype are described by Feldmann et al. (1989). dwf1-3, dwf1-4, and dwf1-5 were isolated by screening dwarf mutants of the Enkheim-2 (En-2) ecotype obtained from the Nottingham Arabidopsis Stock Center (University of Nottingham, UK). These mutants were generously donated by Albert Kranz. Genetic-complementation tests were employed to determine allelism to dwf1-1. Three lines, 318, 355, and 356, were shown to be new alleles of dwf1. Moreover, it has been shown that dim (Takahashi et al., 1995) and cbb1 (Kauschmann et al., 1996) contain insertions in the DWF1 gene (Altmann et al., 1995). For consistency with Kauschmann et al. (1996), we will refer to dim and cbb1 as dwf1-2 and dwf1-6, respectively. In addition, we have isolated 43 new dwf mutants in a screen of approximately 50,000 M2 lines from an EMS-mutagenized population (ecotype Wassilewskija-2 [Ws-2]).
Dwarf mutants resembling dwf1 in both phenotype and brassinosteroid-feeding response were outcrossed to plants of the Columbia ecotype to test for linkage to markers near dwf1. We isolated DNA from individual F2 dwarfs, and tested the genetic linkage of the new mutants to dwf1, using SSLP markers as described by Bell and Ecker (1994). Previously, SSLP mapping of dwf1-1 showed linkage of dwf1 to nga162; the meiotic recombination ratio was 1 out of 40 chromosomes tested. Five mutants resembling dwf1 (WM1-7, WM3-1, WM5-5, WM9-3, and WM12-1) were also closely linked to nga162. Molecular characterization showed that these contained mutations in the DWF1 gene and, as such, were renamed dwf1-7 (WM1-7), dwf1-8 (WM3-1), dwf1-9 (WM5-5), dwf1-10 (WM9-3), and dwf1-11 (WM12-1) (see Table I).
Table I.
Alleles of the Arabidopsis dwf1 mutant
| Allele | Previous Name | Mutagen | Ecotype | Reference |
|---|---|---|---|---|
| dwf1-1 | T-31 | T-DNA | Ws-2 | Feldmann et al. (1989) |
| dwf1-2 | dim | T-DNA | Columbia | Takahashi et al. (1995) |
| dwf1-3 | 318 | Unknown | En-2 | NASCastock no. 318 |
| dwf1-4 | 355 | Unknown | En-2 | NASC stock no. 355 |
| dwf1-5 | 356 | Unknown | En-2 | NASC stock no. 356 |
| dwf1-6 | cbb1 | Ac/Dsb | C-24 | Kauschmann et al. (1996) |
| dwf1-7 | WM1-7 | EMS | Ws-2 | This paper |
| dwf1-8 | WM3-1 | EMS | Ws-2 | This paper |
| dwf1-9 | WM5-5 | EMS | Ws-2 | This paper |
| dwf1-10 | WM9-3 | EMS | Ws-2 | This paper |
| dwf1-11 | WM12-1 | EMS | Ws-2 | This paper |
Nottingham Arabidopsis Stock Center.
Ac/Ds, Activator/dissociation transposable element.
Growth Conditions and Feeding Tests
For plant growth in soil, seeds were sown in 5.5-cm pots filled with wet Metromix (Grace Sierra, Milpitas, CA). Pots were cold treated for 2 d before transfer to incubators (Percival, Boone, IA) set at 22°C with long-day conditions (16 h of light [240 μmol m−2 s−1] at 22°C and 75% RH; 8 h of dark at 21°C and 90% RH). Plants were subirrigated with a modified Hoagland solution (Feldmann and Marks, 1987) diluted 1:1 with deionized water as necessary. For brassinolide-dose experiments, seeds were surface-sterilized and sprinkled on agar-solidified medium containing Murashige-Skoog salts (GIBCO-BRL) supplemented with 0.5% Suc and 0.8% agar (Difco, Detroit, MI). Plates were cold treated for 2 d at 4°C before germination. After germination for 3 d, two to three seedlings at a similar growth stage were transferred to a single cell of a 24-well plate (Corning Inc., Corning, NY) prefilled with 1.5 mL of brassinosteroid-supplemented liquid Murashige-Skoog medium. Plates were sealed with porous tape (3M) and grown on a platform shaker (230 rpm) under continuous light (240 μmol m−2 s−1). As a control, an equivalent amount of ethanol (95%) was added to the medium, because ethanol was used as the solvent for brassinolide.
After 7 d of growth, seedlings from each well were placed in 0.05% toluidine blue in phosphate buffer, pH 4.4, and transferred to agar plates to measure the hypocotyl length using an ocular micrometer on a dissecting microscope. For inflorescence feeding experiments, dwf1-1 and Ws-2 wild-type plants were grown on soil until the inflorescence reached 1 cm in length. Inflorescence apices were marked by tying a string just below the unopened flowers to distinguish the portion of brassinosteroid-induced growth from untreated growth. Brassinosteroid-biosynthetic intermediates were diluted to the desired concentration with water containing 0.01% Tween 20, 10−6 m cathasterone, 6-deoxocathasterone, 22α-hydroxycampesterol, and 10−7 m brassinolide. Two microliters of each brassinosteroid solution was applied daily to the shoot tips of plants using a micropipette (Gilson P-20, Rainin Instrument, Emeryville, CA). After 1 week of treatment the pedicels and inflorescence above the string were measured to the nearest millimeter (n = 15). Choe et al. (1999) have described the methods used for the detection of endogenous brassinosteroid levels.
Molecular Cloning and Characterization of Mutations
Plant DNA flanking the T-DNA borders in dwf1-1 was isolated as described by Dilkes and Feldmann (1998). We used DNA from a right-border rescue as a probe to isolate corresponding cDNA clones from a λPRL-2 cDNA library constructed with λZipLox2 (GIBCO-BRL). The Arabidopsis Biological Resource Center (Ohio State University, Columbus) provided the library (stock no. CD4-7). In addition, genomic DNA clones were isolated from a λDASH-II (Stratagene) genomic library constructed using genomic DNA from the Ws-2 ecotype. To confirm the identity of the clone, a 7-kb restriction fragment from two genomic clones predicted to span the T-DNA insertion site was sequenced and compared with the sequences of the right-border-rescued fragment and cDNA (accession no. U12400). DNA sequencing was carried out using an automated sequencer (model ABI373, Perkin Elmer, Norwalk, CT) at Arizona Research Laboratories (University of Arizona, Tucson). We performed searches for similar sequences using the Basic Local Alignment Search Tool (BLAST) program (Altschul et al., 1997) at the National Center for Biotechnology Information. We aligned similar sequences identified from the BLAST search using the PileUp program of the software package from the Genetics Computer Group (GCG, Madison, WI). Boxing and highlighting of aligned sequences was performed using ALSCRIPT software provided by Barton (1993). We used the PSORT package (Nakai and Kanehisa, 1992; http://psort.nibb.ac.jp:8800/) to predict subcellular targeting.
We identified the mutations in the dwf1 alleles by sequencing PCR-amplified DNA. Oligonucleotide sequences of the primers from 5′ to 3′ are: D1F1, GTTTGATGCAGTGAGGA; D1F2, TGAGGCCCAAGAGGAAGAAG; D1R5, ACGGCCCGAGAGAACATCAG; D1F3, ACAAGGAGAAGATGACTGC; D1R4, GGAACGCTGGTGCCCTAACG; D1F4, TACATTGATTCTTTTGCTCC; D1R3, GGAACACACGGACACCATCA; D1R2, TGGCGCATGACTCCGACCTT; D1F5, TGAATTGTATGAGGAGTGC; and D1R1, AAGTATCCGTTTAGGTTTTC. Genosys Biotechnologies (The Woodlands, TX) provided the PCR primers and Boehringer Mannheim supplied the Taq polymerase. We followed the manufacturer's method for PCR amplification. To obtain the longest PCR product spanning the entire coding region, we used the D1F1 and D1R1 primer pair. The PCR-amplified DNA fragments were gel purified (Prep-A-Gene DNA purification system, Bio-Rad) and subjected to sequencing using the primers described above. Using the BestFit program of the GCG software package, we compared DNA sequences from the mutant allele with the sequence of the wild type to identify base changes. Putative base changes in the mutant allele were confirmed by repeated sequencing of multiple PCR reactions (at least twice for each strand) to eliminate possible PCR artifacts.
RESULTS
Altered Development of Arabidopsis dwf1 Plants
dwf1-1 was originally isolated from the first population of 100 T-DNA lines generated by seed infection (Feldmann et al., 1989). To identify more alleles, 14,000 additional lines representing 21,000 insertion events were screened for dwarfs phenotypically similar to dwf1-1. Genetic-complementation tests showed that none of the 13 additional dwarfs isolated from the screen were allelic to dwf1-1. Next, we obtained all of the dwarf mutants maintained by the Nottingham Arabidopsis Stock Center and screened them for brassinosteroid-responsive dwarfs. Among 55 mutants analyzed, 3 were allelic to dwf1-1 (stock numbers 318, 355, and 356) and were therefore renamed dwf1-3, dwf1-4, and dwf1-5, respectively.
Figure 1 shows a comparison of the Ws-2 wild type with three dwf1 alleles at 35 d of age. The two En-2 alleles of dwf1 (dwf1-3 and dwf1-4) were morphologically distinct from the known null mutant dwf1-1 (described below). Unlike dwf1-1, they possess a shorter stature, a more tightly bunched rosette, and a more highly branched inflorescence (Fig. 1). These differences are probably due to ecotypic backgrounds. As evidence for this, when dwf1-5 plants were outcrossed twice into the Ws-2 ecotype, they looked more like dwf1-1 (data not shown). Finally, we isolated 43 dwf mutants from a screen of 50,000 M2 lines of an EMS-mutagenized population (ecotype Ws-2). Five mutants resembled dwf1 in phenotype and brassinosteroid-feeding response. Mapping using SSLP markers (Bell and Ecker, 1994) indicated that these five mutants are linked to dwf1-1. Allelism to dwf1 was further suggested by the identification of mutations in the DWF1 gene in these five lines (see below). Table I summarizes the dwf1 alleles and their origins.
Figure 1.
Comparison of 5-week-old Arabidopsis wild type, dwf1, and dwf4-1 plants. Bar = 2 cm. The characteristic phenotype of brassinosteroid dwarfs includes a short, robust inflorescence; round, dark-green leaves; and reduced fertility. dwf1-1 (Ws-2 background) is taller than dwf1-3 and dwf1-4 (En-2). Compared with dwf4, dwf1 alleles have a less-severe phenotype.
Morphological differences between dwf1-1 and wild-type plants are shown in Tables II and III. Measurements were taken from 35- and 111-d-old (mature) organs of dwf1-1 and Ws-2 wild-type plants. A comparison of their heights at 35 d indicates that dwf1-1 plants were 4-fold shorter than the wild type. The decrease in organ size caused by the dwf1-1 mutation was not uniform. Inflorescence length was the most affected. The internode distance was reduced to approximately 25% of that of the wild type (Table II; Fig. 1). Leaf length was one-half that of the wild type, although leaf width was not changed significantly (Table II), resulting in a round shape. The observation that leaf tissues between veins buckled suggests that the elongation of veins was more affected than the expansion of areoles (Fig. 1). Similarly, petiole length was 40% of that of the wild type. The total number of organs from the wild type and dwf1-1 was recorded. dwf1-1 plants produced more rosette leaves than did the wild type at 35 d (Table II). Furthermore, the total number of siliques, as well as the number of siliques on the primary inflorescence, was increased 4-fold compared with wild type at maturity. As previously described for the dwf7 mutant (Choe et al., 1999), dwf1 plants are not completely sterile. Although the defect in cell elongation affected the stamens more than the gynoecium, some stamens were long enough to reach the stigmatic surface for successful fertilization (data not shown; Choe et al., 1999).
Table II.
Morphometric analysis of Ws-2 wild-type and dwf1-1 plants
| Parameter | At 35
d
|
At Maturitya
|
||
|---|---|---|---|---|
| Ws-2 | dwf1-1 | Ws-2 | dwf1-1 | |
| mm | ||||
| Length | ||||
| Inflorescence | 25.80 ± 0.83 | 5.47 ± 0.40 | 26.99 ± 0.85 | 14.72 ± 0.88 |
| Internode | ndb | nd | 1.10 ± 0.03 | 0.27 ± 0.01 |
| Petiolec | 0.57 ± 0.03 | 0.22 ± 0.01 | nd | nd |
| Leaf bladec | 1.72 ± 0.02 | 0.80 ± 0.03 | nd | nd |
| Leaf blade widthc | 0.77 ± 0.02 | 0.84 ± 0.03 | nd | nd |
| no. | ||||
| Number of organs | ||||
| Rosette leaves | 7.1 ± 0.3 | 10.9 ± 0.6 | nd | nd |
| Rosette bolts | 3.6 ± 0.2 | 3.7 ± 0.3 | 6.4 ± 0.2 | 5.0 ± 0.2 |
| Branchesd | 1.8 ± 0.1 | 3.1 ± 0.2 | 2.8 ± 0.1 | 4.6 ± 0.2 |
| Siliquesd | 49.8 ± 4.0 | 38.0 ± 5.0 | 118.9 ± 12.1 | 492.6 ± 28.9 |
| Siliquese | 101.7 ± 11.3 | 72.4 ± 7.5 | 336.5 ± 28.6 | 1303.5 ± 52.1 |
| Seeds/silique | nd | nd | 37.7 ± 0.7 | 3.2 ± 1.0 |
The sizes of various organs were measured to the nearest millimeter, and the number of organs was counted in dwf1-1 and wild-type plants grown individually in 5.5-cm pots. Each value represents the mean of 10 or more plants ± se.
Maturity is considered to be the cessation of new flower development.
nd, Not determined.
Measured from the second leaf pair.
Number from the primary inflorescence.
Number from the entire plant.
Table III.
Timing of phase transitions
| Characteristic | Ws-2 | dwf1-1 |
|---|---|---|
| DAG | ||
| Bolt | 16.9 | 19.8 |
| Anthesis of first flower | 21.8 | 24.9 |
| First silique senescence | 34.6 | 44.6 |
| Termination of floweringa | 57 | 102 |
Anatomical characteristics associated with growth-phase changes were scored, and the days after germination (DAG) were recorded. Each number represents the mean of 10 or more plants.
Considered to be the senescence of the last flower.
Arabidopsis inflorescences are indeterminate (Bowman, 1994); however, wild-type plants typically cease flowering and senesce by 57 d after germination (Table III). All dwf1 mutants continued to grow and flower up to 110 d under controlled-growth conditions (Table III). Although dwf1-1 plants bolted and flowered later than wild-type plants, the primary cause of the expanded life cycle of dwf1 was due to a prolonged generative phase. dwf1-1 plants flowered for more than twice as long as wild-type plants. It is this extension of the generative phase that allows the dwf1-1 plants to overproduce siliques and eventually reach one-half the height of wild type at maturity (Table III).
Biochemical Characterization of the Brassinosteroid-Biosynthesis Defect in dwf1 Plants
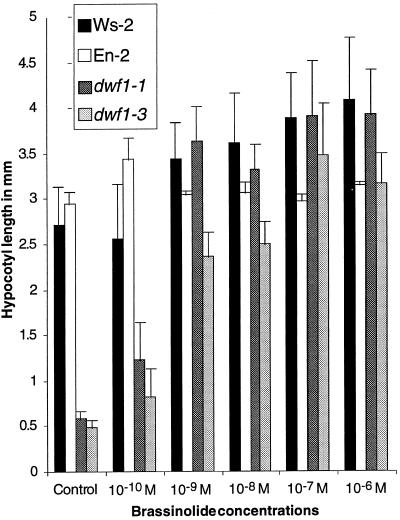
Clouse et al. (1996) have shown phenotypically similar dwarf mutants of Arabidopsis to be insensitive, whereas Li et al. (1996), Szekeres et al. (1996), and Choe et al. (1998) have reported others to be rescued by the exogenous application of brassinosteroids. Previously, Kauschmann et al. (1996) had shown that cbb1 (dwf1-6) was biochemically complemented by exogenous application of 24-epibrassinosteroids. To further examine this response in our dwf1 alleles, we applied different concentrations of brassinolide. Figure 2 summarizes the effect of different concentrations of brassinolide on the elongation of hypocotyls of Ws-2, En-2, dwf1-1, and dwf1-3 grown for 10 d in the light. The two ecotypes responded differently to increasing concentrations of brassinolide. Although En-2 showed a small increase after the application of 10−10 m brassinolide, elongation was not enhanced with higher concentrations. In contrast, Ws-2 hypocotyls displayed no increase at 10−10 m, but appeared to respond to higher concentrations of brassinolide. dwf1-1, in the Ws-2 background, showed an increase in length at 10−10 m and was comparable with the wild type at 10−9 m and higher (Fig. 2). dwf1-3, in the En-2 background, did not show a significant increase in length until the application of a 10−9 m concentration of brassinolide, and then continued to increase with concentrations up to 10−7 m. In addition to increasing the length of the hypocotyls, brassinolide application increased the size of dwf1-1 cotyledons and first leaves (data not shown). At concentrations of more than 10−9 m brassinolide, these organs were indistinguishable from those of the wild type, although cotyledon and hypocotyl morphology became distorted with higher brassinolide concentrations (data not shown).
Figure 2.
Response of Arabidopsis wild type (Ws-2 and En-2) and dwf1 alleles to different concentrations of exogenously applied brassinolide. The length of 15 or more hypocotyls grown for 10 d in liquid medium was measured. Bars = ±sd.
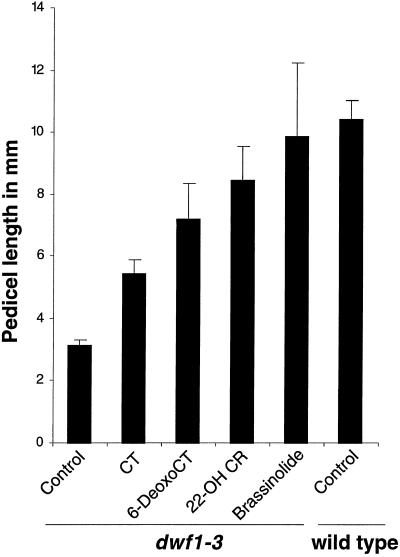
Rescue of dwf1-1 by exogenous application of brassinolide suggests that dwf1 is defective in brassinolide biosynthesis. Therefore, we wanted to pinpoint the exact step that is defective in dwf1. To this end, we examined the effectiveness of some of the brassinosteroid-biosynthetic intermediates in restoring the growth of dwf1 pedicels. dwf1 responds to cathasterone, 6-deoxocathasterone, 22α-hydroxycampesterol, and brassinolide with increased growth of pedicels (Fig. 3). The synthetic compound 22α-hydroxycampesterol has been used in brassinosteroid-feeding studies to overcome problems with the undetectable bioactivity of early biosynthetic intermediates in this bioassay, and rescue by 22α-hydroxycampesterol is interpreted as being complemented by campesterol (Choe et al., 1998). Therefore, complementation of dwf1 by 22α-hydroxycampesterol and downstream intermediates suggests that the biosynthetic defect in dwf1 resided before campesterol.
Figure 3.
Feeding tests with brassinosteroid-biosynthetic intermediates. Inflorescences of 4-week-old dwf1-3 plants were treated with specified biosynthetic intermediates for 1 week. Pedicel length (n = 15) was measured. All of the compounds tested induced a significant response from the pedicels compared with controls. In particular, 22α-hydroxycampesterol and brassinolide completely rescued the short pedicel length to the normal wild-type length. Bars = ±sd.
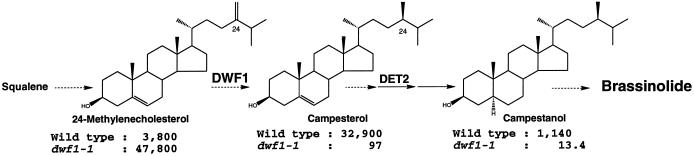
To further delimit the biosynthetic defect to a single step, we analyzed the endogenous levels of brassinosteroid-biosynthetic intermediates in dwf1-1 and the wild type using a GC-SIM assay. Figure 4 shows the endogenous amount of selected intermediates with their chemical structures. Compared with the wild type, 24-methylenecholesterol accumulates to a 12-fold higher level in dwf1-1, whereas the level of the downstream compound, campesterol, is decreased to 0.3% of the wild type. The level of the next compound, campestanol, is accordingly decreased (most likely due to a shortage of the substrate compound campesterol). Together, the accumulation of 24-methylenecholesterol with the simultaneous decrease of campesterol in dwf1-1 suggests that the C-24 reduction step is deficient in dwf1.
Figure 4.
Brassinosteroid-biosynthetic step found to be defective in dwf1 plants. The aerial parts of 5-week-old wild-type and dwf1 plants were subjected to analysis of endogenous brassinosteroid levels using GC-SIM. Accumulation of 24-methylenecholesterol with a simultaneous reduction in the campesterol level suggests that a specific step, C-24 reduction, was blocked in dwf1 mutants. Campesterol is known to serve as substrate for brassinolide biosynthesis.
Molecular Characterization of dwf1 Alleles
Feldmann et al. (1989) demonstrated that the dwf1-1 phenotype cosegregated with the kanamycin marker derived from an inserted T-DNA. Plant DNA flanking the T-DNA was isolated by plasmid rescue, as described by Dilkes and Feldmann (1998). The rescued clone was found to contain 300 bp of DNA flanking an intact right border. In Southern-blot analyses with genomic DNA from dwf1-1 and wild-type plants, restriction fragment length polymorphism was detected in the T-DNA line when the plant-flanking DNA was used as a probe. The plant-flanking DNA was used to isolate a cDNA clone. Plasmid-rescued DNA, the corresponding cDNA, and genomic clones spanning the entire coding region were sequenced. The cDNA sequence was deposited in GenBank (accession no. U12400). Takahashi et al. (1995) subsequently isolated the identical gene and named it DIMINUTO. As previously described by Takahashi et al. (1995), the DWF1 coding sequence is 1686 bp in length and is disrupted by one 92-bp intron 1282 bp downstream from the start codon.
The primary structure of the DWF1 protein (561 amino acids) was subject to analyses using PSORT software (Nakai and Kanehisa, 1992), which revealed that the protein is likely to be targeted to endomembrane systems such as the ER, Golgi apparatus, and mitochondria as an integral protein, with amino acids 27 to 43 serving as the membrane-spanning domain. Based on the criteria set by Hicks and Raikhel (1995) and Robbins et al. (1991), the NUCDISC (discrimination of nuclear-localization signals) subprogram of the PSORT package did not find any nuclear-localization signals. To test the expression of the gene, Arabidopsis EST database searches for sequences identical to DWF1 were performed. Fourteen EST clones were identified (K3F6TP, 106C3T7, 137P3T7, E4F9T7, 128K1T7, G10C7T7, E1D10T7, G9E12T7, 94O7XP, 176H17T7, 94O7T7, VBV10-12592, G5D11T7, and VCVDH10). As predicted by the abundance of EST clones, the steady-state level of the DWF1 transcript was present in all organs and at all developmental stages tested (data not shown; Takahashi et al., 1995).
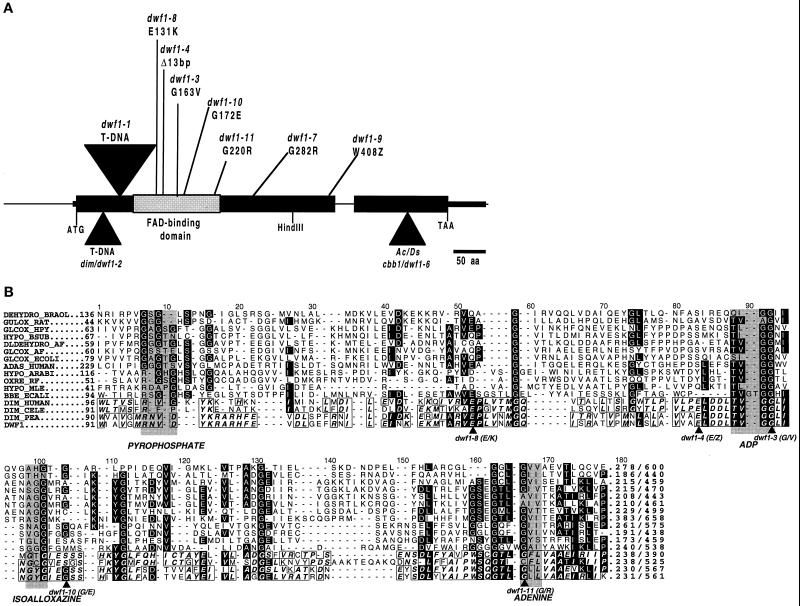
Sequencing the genomic DNA from the various mutants and comparing it with the wild type showed that there were single mutations in all of the mutant alleles. Figure 5A schematically represents DWF1, including the positions of various mutations. Most mutations (7 of 10) were located in the amino-terminal half of the gene. The T-DNA is inserted 200 bp downstream from the start codon in dwf1-1. To determine whether the T-DNA insertion disrupted DWF1 gene expression, we performed northern-blot analysis with total RNA from dwf1-1 and from the wild type. When the blot was probed with DWF1 cDNA, we detected no DWF1 RNA in the dwf1-1 sample, although this RNA was easily detected in the wild-type sample (data not shown). This indicated that steady-state RNA levels in dwf1-1 were below the threshold for detection. Along with the position of the T-DNA, this suggests that dwf1-1 is a null mutation.
Figure 5.
Schematic representation of Arabidopsis DWF1 locus with characterized mutations (A) and aligned amino acid sequences of proposed FAD-binding domains from various oxidoreductases (B). Elucidation of DWF1 organization resulted from comparison of cDNA with genomic DNA. Sites of the FAD-binding domain (Mushegian and Koonin, 1996; Fraaije et al., 1998) and mutations identified from dwf1 alleles, including dim (dwf1-2; Takahashi et al., 1995) and cbb1 (dwf1-6; Kauschmann et al., 1996), are indicated. Sequences corresponding to the region of the FAD-binding domain depicted in A are aligned to show conserved residues and relative positions of mutations. Accession numbers for the following are given in parentheses: DEHYD_BRAOL (2760543), GULOX_RAT (625202), GLCOX_HPY (2313619), HYPO_BSUB (1770026), DLDEHYD_AF (2650235), GLCOX_AF (2649802), GLCOX_ECOLI (1707917), ADAS_HUMAN (2498106), HYPO_ARABI (2618686), OXRE_RF (1169648), HYPO_MLE (3150105), BBE_ECALI (400972), DIM_HUMAN (3182980), DIM_CELE (3182979), DIM_PEA (3182981), DWF1 (U12400). Similar sequences were identified using gapped BLAST (Altschul et al., 1997), followed by alignment using PileUp software (GCG). Box shading was carried out using the ALSCRIPT package developed by Barton (1993).
dwf1-4 carries a 13-bp deletion from bases 437 to 449, causing a frame shift that generates a stop codon eight amino acids downstream of the deletion, indicating that it is also a null allele. Sequencing of dwf1-5 revealed that it contained the same mutation as dwf1-4, suggesting that these two alleles arose from the same mutational event. Moreover, the EMS-induced allele dwf1-9 contains a transition mutation from guanine to adenine at 1279, changing Trp into a stop codon. Four of the five remaining alleles contained substitution mutations that changed Gly into a charged amino acid (dwf1-7, dwf1-10, and dwf1-11) or Val (dwf1-3). Finally, dwf1-8 caused a change from a negatively charged Glu to a positively charged Lys.
To better understand DWF1 function, we performed database searches with the DWF1 cDNA and its deduced protein sequence. BLAST searches (Altschul et al., 1997) produced two categories of proteins; one displayed global similarity with DWF1. This group included DIM-like proteins isolated from pea (88% similarity; Shimizu and Mori, 1996; accession no. 3182981), Caenorhabditis elegans (53%; accession no. 3182979), and human (60%; Nomura et al., 1994; accession no. 3182980). The other group of proteins showed similarities to only a part of the DWF1 protein from amino acids 91 to 231. Figure 5B illustrates the results of the local sequence alignment. The second group of proteins is represented by sequences from bacteria through plants and animals, and the functions of many of these proteins are unknown.
Proteins with known functions include l-gulonolactone oxidase of rat (GULOX_RAT; Koshizaka et al., 1988), l-galactono-1,4-lactone dehydrogenase of Brassica oleracea (DEHYD_BRAOL; Ostergaard et al., 1997), alkyldihydroxyacetonephosphate synthase of human (ADAS_HUMAN; de Vet et al., 1997), berberine-bridge-forming enzyme of Eschscholzia californica (BBE_ECALI; Dittrich and Kutchan, 1991), glycolate oxidase subunit of Helicobacter pylori (GLCOX_HPY; Tomb et al., 1997), and d-lactate dehydrogenase of Archaeoglobus fulgidus (DLDEHYD_AF; Klenk et al., 1997). Mushegian and Koonin (1995) initially proposed that the domain illustrated in Figure 5B is conserved among a broad spectrum of oxidoreductases requiring FAD as a prosthetic group. In the FAD-binding domain shown in Figure 5B, 46 of 140 residues (91–231) in DWF1 are conserved. Ten of the 49 conserved residues are Gly. In support of the importance of the conserved domain, mutations in our dwf1 alleles correspond well to the conserved residues. Among eight dwf1 mutations, six are located in or before this FAD-binding domain. Four mutations are located in highly conserved residues (dwf1-2, dwf1-8, dwf1-10, and dwf1-11), and one that creates a premature stop codon (dwf1-3) is predicted to delete the FAD-binding domain.
DISCUSSION
Comparison of Different dwf Mutants
Brassinosteroids have long been known to be involved in many different developmental events throughout the life cycle of plants (Mandava, 1988). However, definitive evidence for the role of brassinosteroids during plant growth has remained unclear until the recent characterization of mutants defective in brassinosteroid biosynthesis or perception. Brassinosteroid dwarf mutants, including dwf1, are characterized by multiple phenotypes: reduced height, robust stems, reduced fertility, prolonged life cycle, dark-green color, round and curled leaves, and, when grown in the dark, short hypocotyls and expanded cotyledons. This phenotype has been reported for several brassinosteroid dwarfs, including bri1 (dwf2) (Clouse et al., 1996), cpd (dwf3) (Szekeres et al., 1996), dwf4 (Choe et al., 1998), det2 (dwf6) (Li et al., 1996), and dwf7 (Choe et al., 1999). Therefore, it is worth comparing dwf1 with other dwarf loci.
At 35 d of age, dwf1-1 was 5.47 cm tall (Table I), whereas dwf4-1 grew to only half of this height, or 2.8 cm (Azpiroz et al., 1998). In addition, the heights of dwf2-1 (bri1), dwf3-1 (cpd), and dwf6-1 (det2) were 1.4, 1.92, and 5.1 cm, respectively (B.P. Dilkes, B. Schulz, S. Choe, R. Azpiroz, and K.A. Feldmann, unpublished data). The length of dark-grown hypocotyls for these mutants was proportional to the severity of their phenotype in the light (data not shown). Thus, based on their severity, the dwf loci can be divided into two groups: standard and small dwarfs. The standard dwarfs include dwf1 and dwf6 (det2), whereas the small dwarfs include dwf2 (bri1), dwf3 (cpd), and dwf4. The earlier the biosynthetic defect, the less severe the phenotype. In other words, biosynthetic defects in the standard dwarfs reside before the DWF4 step, the putative rate-limiting step in brassinosteroid biosynthesis (Fujioka et al., 1995; Choe et al., 1998), whereas small dwarfs correspond to the DWF4-mediated steps and steps following them.
This relationship is reversed in GA-biosynthetic dwarfs such as ga1 to ga5 (Ross et al., 1997). Mutants blocked in the early reactions of GA biosynthesis, such as ga1, ga2, and ga3 (defective in steps before GA12), display extreme dwarfism compared with ga4 and ga5, which are defective in later biosynthetic steps (Koornneef and van der Veen, 1980; Finkelstein and Zeevaart, 1994). It is thought that later biosynthetic steps in GA biosynthesis are mainly modification reactions conferring increased bioactivity to GAs through network reactions of many redundant isozymes (Finkelstein and Zeevaart, 1994). Thus, GA-biosynthetic mutants could still synthesize a limited number of active GAs (Finkelstein and Zeevaart, 1994). It is also possible that brassinosteroid-biosynthetic reactions downstream of the DWF4-mediated step are more critical to conferring functionality to brassinosteroids than upstream reactions.
One difference between the GA- and brassinosteroid-biosynthetic pathways may be that the genes involved in the reactions upstream of the DWF4 step are genetically redundant. In support of this, we have detected lightly hybridizing bands with DWF1 and DWF7 probes in northern and Southern blots (A. Tanaka, B.P. Dilkes, S. Choe, F.E. Tax, and K.A. Feldmann, unpublished data). A DWF7 homolog has been isolated and is being characterized (A. Tanaka, S. Choe, F.E. Tax, and K.A. Feldmann, unpublished data). In addition, the possibility of a second gene for DET2 was reported by Fujioka et al. (1997). Further analysis of these cross-hybridizing genes in the upstream reactions through cloning, sequencing, and expression studies, including temporal and spatial localization studies, will provide us with in-depth knowledge about the regulation of brassinosteroid biosynthesis in relation to the phenotypic severity of these dwarf mutants.
Delayed Senescence in dwf Mutants
dwf1 mutants and other brassinosteroid dwarf mutants show an expanded life span. Dwarf plants both maintain green leaves and produce flowers for a longer period of time than the wild type. Bolting, first flower anthesis, first silique senescence, and termination of flowering are all delayed in dwf1 (Table III). Similarly, Azpiroz et al. (1998) reported that completion of one generation takes 98 d for dwf4-1, compared with 57 d for the wild type. The cause of the extended life span for brassinosteroid dwf mutants is not known. However, one possible explanation is that a long life cycle is associated with reduced fertility. Guarente et al. (1998) and Nooden (1988) have shown that developing seeds in need of mobilized nutrients trigger the onset of senescence as part of a global nutrient-recycling program.
Nooden (1988) showed that removal of the developing seed pods of soybean greatly delayed senescence of leaves and whole plants because of an increased quantity of cytokinins transported from the roots. Similarly, it is possible that the failure of seed development in dwarf mutants to produce signals for the onset of senescence could be the reason that dwf1 displayed an expanded life span. In support of this, the generative phase was significantly expanded in dwf1. It took 82 d for dwf1-1 to progress from bolting to the termination of flowering, whereas 40 d were required for the wild type. However, the vegetative phase of dwf1 was not delayed significantly (Table III). Furthermore, there are data (B. Dilkes, R. Azpiroz, S. Choe, and K. Feldmann, unpublished data) suggesting that the generation time was proportional to the fertility of brassinosteroid dwf mutants: the less fertile (i.e. dwf4 and bri1 [dwf2]) the longer the generation time. Despite the delayed senescence, we do not know whether intrinsic, age-dependent senescence was also delayed or if it was unaffected. Hensel et al. (1993) have suggested that age-dependent senescence can be uncoupled from reproductive development. Recently, Park et al. (1998) and Weaver et al. (1998) have cloned and tested many senescence-related genes in relation to various internal and external signals. Exploring the expression of the senescence-related genes in dwf1 plants could provide more detailed information about the mechanism of delayed senescence in dwf1 and about the role of brassinosteroids in the senescence process.
Possible Function of DWF1 Protein
In addition to the morphological analysis of dwf1, we characterized the molecular basis of several dwf1 alleles, and showed that dwf1 was defective in the conversion of 24-methylenecholesterol to campesterol in the sterol-biosynthetic pathway leading to brassinosteroid biosynthesis. Analysis of the amino acid sequence of DWF1 indicated that it probably resides in the endomembrane system, most likely in the ER. It has been shown that most of the steroid-biosynthetic enzymes need to be located in membranes for proper functioning (for review, see Bach and Benveniste, 1997). In addition, part of the DWF1 protein sequence showed significant similarity to oxidases from many different organisms, including bacteria and humans (Fig. 5B). Mushegian and Koonin (1995) initially found that this putative domain was shared by many oxidases that require FAD as a coenzyme.
More recently, based on the crystal structure of an 8α-(N3-histidyl)-FAD-containing flavoprotein, vanillyl-alcohol oxidase, Fraaije et al. (1998) reported that there were two major domains: a FAD-binding domain and a substrate-binding domain. The FAD-binding domain was characterized by subdomains for the binding of pyrophosphate, ADP, isoalloxazine, and adenine (Fraaije et al., 1998; Fig. 5B). According to their findings, the residues in the FAD-binding domain were highly conserved among enzymes involved in a diverse range of redox reactions. The absence of residues for isoalloxazine-ring linkages such as His, Cys, or Tyr in the pyrophosphate subdomain in DWF1 (undernoted as pyrophosphate in Fig. 5B) suggests that DWF1 binds to its coenzyme, FAD, not by a covalent bond but by a dissociable bond, which is common for many FAD-dependent enzymes (Mewies et al., 1998). Furthermore, conservation of the subdomains in DWF1 indicated that it belonged to this novel group of oxidases. Six of eight dwf1 mutations reported in this paper interfered with this domain.
The null allele, dwf1-1, contained an insertion upstream of the FAD-binding domain, and dwf1-4 contained a stop codon in the middle of the domain. In addition, in dwf1-3 and dwf1-11, residues belonging to the ADP- and the adenine-contacting regions, respectively, were altered (Fig. 5B). The severity of dwf1 alleles was correlated with the location of the mutations relative to the FAD-binding domain. The mutants in the Ws-2 background, dwf1-1, dwf1-8, dwf1-10, and dwf1-11, which are located at or before the FAD-binding domain, displayed more severe phenotypes than dwf1-7 and dwf1-9, in which the mutations were found after the FAD-binding domain (data not shown). With the addition of dwf1-2 (dim) (T-DNA insertion upstream of FAD-binding domain [Takahashi et al., 1995]) and dwf1-6 (cbb1) (transposon insertion 515 amino acids downstream of the start codon [Kauschmann et al., 1996]), 7 of 10 mutations were localized in or before the FAD-binding domain. This suggests that the conserved residues in the FAD-binding domain are critical for DWF1 function, perhaps through binding of the coenzyme FAD to the DWF1 apoenzyme leading to redox reactions catalyzed by the flavoenzyme.
Aside from the putative FAD-binding domains, we can speculate on the role assigned to the rest of the protein. First, as Fraaije et al. (1998) have pointed out, one part of the protein may play the role of the substrate-binding domain, and this binding domain may be variable in sequence between diverse redox enzymes. Second, the rest of the protein sequence may confer an enzymatic function other than that of an oxidase. For instance, it has been proposed that campesterol is produced via isomerization of Δ24(28) to Δ24(25) as an intermediate before the double bond is saturated by the reductase activity. Thus, the isomerization and reduction of the Δ24(25) double bond could be performed by the rest of the protein in this multifunctional enzyme.
In conclusion, loss of the enzyme that converts 24-methylenecholesterol to campesterol in dwf1 plants caused a dramatic reduction in campesterol biosynthesis (Fig. 4) to approximately 0.3% of wild-type levels. The reduction of campesterol resulted in reduced biosynthesis of brassinolide, which seemed to be the direct cause of altered development in dwf1 plants. In addition to dwf7, dwf1 is now a second sterol-specific biosynthetic mutant showing a characteristic dwarf phenotype that can be rescued to wild type by the exogenous application of brassinosteroids.
NOTE ADDED IN PROOF
While this manuscript was in the review process, Klahre et al. (1998) also showed that diminuto (dwf1-2) was defective in the conversion of 24-methylenecholesterol to campesterol in brassinosteroid biosynthesis. In addition, the authors provided evidence that DIM/DWF1 is associated with the endomembrane system rather than with the nucleus.
Abbreviations:
- EMS
ethyl methanesulfate
- EST
expressed sequence tag
- SIM
selective ion monitoring
- SSLP
simple sequence length polymorphism
Footnotes
This research was supported by the National Science Foundation (grant no. 9604439 to K.A.F.) and by a Grant-in-Aid for Scientific Research (B) from the Ministry of Education, Science, Sports, and Culture of Japan (grant no. 10460050 to S.F.).
LITERATURE CITED
- Altmann T, Felix G, Jessop A, Kauschmann A, Uwer U, Pena-Cortes H, Willmitzer L. Ac/Ds transposon mutagenesis in Arabidopsis thaliana: mutant spectrum and frequency of Dsinsertion mutants. Mol Gen Genet. 1995;247:646–652. doi: 10.1007/BF00290357. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azpiroz R, Wu Y, LoCascio JC, Feldmann KA. An Arabidopsis brassinosteroid-dependent mutant is blocked in cell elongation. Plant Cell. 1998;10:219–230. doi: 10.1105/tpc.10.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach TJ, Benveniste P. Cloning of cDNAs or genes encoding enzymes of sterol biosynthesis from plants and other eukaryotes: heterologous expression and complementation analysis of mutations for functional characterization. Prog Lipid Res. 1997;36:197–226. doi: 10.1016/s0163-7827(97)00009-x. [DOI] [PubMed] [Google Scholar]
- Barton GJ. ALSCRIPT: a tool to format multiple sequence alignments. Protein Eng. 1993;6:37–40. doi: 10.1093/protein/6.1.37. [DOI] [PubMed] [Google Scholar]
- Bell CJ, Ecker JR. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- Bowman J (1994) Arabidopsis: An Atlas of Morphology and Development. Springer-Verlag, New York, pp 3–11
- Choe S, Dilkes BP, Fujioka S, Takatsuto S, Sakurai A, Feldmann KA. The DWF4gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22α-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell. 1998;10:231–243. doi: 10.1105/tpc.10.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Feldmann KA (1998) T-DNA mediated gene tagging. In K Lindsey, ed, Transgenic Plant Research. Harwood Academic Publishers, Amsterdam, The Netherlands, pp 57–73
- Choe S, Noguchi T, Fujioka S, Takatsuto S, Tanaka A, Yoshida S, Ross AS, Tax FE, Feldmann KA. Arabidopsis dwf7ste1–2 is defective in the Δ7sterol C5 desaturation step leading to brassinosteroid biosynthesis. Plant Cell. 1999;11:207–222. [PMC free article] [PubMed] [Google Scholar]
- Clouse SD, Langford M, McMorris TC. A brassinosteroid-insensitive mutant in Arabidopsis thalianaexhibits multiple defects in growth and development. Plant Physiol. 1996;111:671–678. doi: 10.1104/pp.111.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vet EC, van den Broek BT, van den Bosch H. Nucleotide sequence of human alkyl-dihydroxyacetonephosphate synthase cDNA reveals the presence of a peroxisomal targeting signal 2. Biochim Biophys Acta. 1997;1346:25–29. doi: 10.1016/s0005-2760(97)00014-3. [DOI] [PubMed] [Google Scholar]
- Dilkes BP, Feldmann KA (1998) Cloning genes from T-DNA tagged mutants. In J Martinez-Zapater, J Salinas, eds, Methods in Molecular Biology: Arabidopsis Protocol. Humana Press, Totowa, NJ, pp 339–351 [DOI] [PubMed]
- Dittrich H, Kutchan TM. Molecular cloning, expression, and induction of berberine bridge enzyme, an enzyme essential to the formation of benzophenanthridine alkaloids in the response of plants to pathogenic attack. Proc Natl Acad Sci USA. 1991;88:9969–9973. doi: 10.1073/pnas.88.22.9969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann KA, Marks MD. Agrobacterium-mediated transformation of germinating seeds of Arabidopsis thaliana: a non-tissue culture approach. Mol Gen Genet. 1987;208:1–9. [Google Scholar]
- Feldmann KA, Marks MD, Christianson ML, Quatrano RS. A dwarf mutant of Arabidopsisgenerated by T-DNA insertion mutagenesis. Science. 1989;243:1351–1354. doi: 10.1126/science.243.4896.1351. [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Zeevaart JAD. Gibberellin and abscisic acid biosynthesis and response. In: Meyerowitz E, Somerville C, editors. Arabidopsis. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. pp. 523–554. [Google Scholar]
- Fraaije MW, Van Berkel WJ, Benen JA, Visser J, Mattevi A. A novel oxidoreductase family sharing a conserved FAD-binding domain. Trends Biochem Sci. 1998;23:206–207. doi: 10.1016/s0968-0004(98)01210-9. [DOI] [PubMed] [Google Scholar]
- Fujioka S, Inoue T, Takatsuto S, Yanagisawa T, Yokota T, Sakurai A. Biological activities of biosynthetically-related congeners of brassinolide. Biosci Biotechnol Biochem. 1995;59:1973–1975. [Google Scholar]
- Fujioka S, Li J, Choi YH, Seto H, Takatsuto S, Noguchi T, Watanabe T, Kuriyama H, Yokota T, Chory J and others. The Arabidopsis deetiolated2mutant is blocked early in brassinosteroid biosynthesis. Plant Cell. 1997;9:1951–1962. doi: 10.1105/tpc.9.11.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L, Ruvkun G, Amasino R. Aging, life span, and senescence. Proc Natl Acad Sci USA. 1998;95:11034–11036. doi: 10.1073/pnas.95.19.11034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensel LL, Grbic V, Baumgarten DA, Bleecker AB. Developmental and age-related processes that influence the longevity and senescence of photosynthetic tissues in Arabidopsis. Plant Cell. 1993;5:553–564. doi: 10.1105/tpc.5.5.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks GR, Raikhel NV. Protein import into the nucleus: an integrated view. Annu Rev Cell Dev Biol. 1995;11:155–188. doi: 10.1146/annurev.cb.11.110195.001103. [DOI] [PubMed] [Google Scholar]
- Höfte H, Hubbard L, Reizer J, Ludevid D, Herman EM, Chrispeels MJ. Vegetative and seed-specific forms of tonoplast intrinsic protein in the vacuolar membrane of Arabidopsis thaliana. Plant Physiol. 1992;99:561–570. doi: 10.1104/pp.99.2.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauschmann A, Jessop A, Koncz C, Szekeres M, Willmitzer L, Altmann T. Genetic evidence for an essential role of brassinosteroids in plant development. Plant J. 1996;9:701–713. [Google Scholar]
- Klahre U, Noguchi T, Fujioka S, Takatsuto S, Yokota T, Nomura T, Yoshida S, Chua NH. The Arabidopsis DIMINUTO/DWARF1gene encodes a protein involved in steroid synthesis. Plant Cell. 1998;10:1677–1690. doi: 10.1105/tpc.10.10.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenk HP, Clayton RA, Tomb JF, White O, Nelson KE, Ketchum KA, Dodson RJ, Gwinn M, Hickey EK, Peterson JD and others. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- Koornneef M, van der Veen JH. Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana(L.) Heynh. Theor Appl Genet. 1980;58:257–263. doi: 10.1007/BF00265176. [DOI] [PubMed] [Google Scholar]
- Koshizaka T, Nishikimi M, Ozawa T, Yagi K. Isolation and sequence analysis of a complementary DNA encoding rat liver l-gulono-gamma-lactone oxidase, a key enzyme for l-ascorbic acid biosynthesis. J Biol Chem. 1988;263:1619–1621. [PubMed] [Google Scholar]
- Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- Li J, Nagpal P, Vitart V, McMorris TC, Chory J. A role for brassinosteroids in light-dependent development of Arabidopsis. Science. 1996;272:398–401. doi: 10.1126/science.272.5260.398. [DOI] [PubMed] [Google Scholar]
- Mandava NB. Plant growth-promoting brassinosteroids. Annu Rev Plant Physiol Plant Mol Biol. 1988;39:23–52. [Google Scholar]
- Medford JI, Elmer JS, Klee HJ. Molecular cloning and characterization of genes expressed in shoot apical meristems. Plant Cell. 1991;3:359–370. doi: 10.1105/tpc.3.4.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mewies M, McIntire WS, Scrutton NS. Covalent attachment of flavin adenine dinucleotide (FAD) and flavin mononucleotide (FMN) to enzymes: the current state of affairs. Protein Sci. 1998;7:7–20. doi: 10.1002/pro.5560070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushegian AR, Koonin EV. A putative FAD-binding domain in a distinct group of oxidases including a protein involved in plant development. Protein Sci. 1995;4:1243–1244. doi: 10.1002/pro.5560040623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura N, Miyajima N, Sazuka T, Tanaka A, Kawarabayasi Y, Sato S, Nagase T, Seki N, Ishikawa K, Tabata S. Prediction of the coding sequences of unidentified human genes. I. The coding sequences of 40 new genes (KIAA0001-KIAA0040) deduced by analysis of randomly sampled cDNA clones from human immature myeloid cell line KG-1. DNA Res. 1994;1:27–35. doi: 10.1093/dnares/1.1.27. [DOI] [PubMed] [Google Scholar]
- Nooden LD. Whole plant senescence. In: Nooden LD, Leopold AC, editors. Senescence and Aging in Plants. San Diego, CA: Academic Press; 1988. pp. 391–439. [Google Scholar]
- Ostergaard J, Persiau G, Davey MW, Bauw G, Van Montagu M. Isolation of a cDNA coding for l-galactono-gamma-lactone dehydrogenase, an enzyme involved in the biosynthesis of ascorbic acid in plants: purification, characterization, cDNA cloning, and expression in yeast. J Biol Chem. 1997;272:30009–30016. doi: 10.1074/jbc.272.48.30009. [DOI] [PubMed] [Google Scholar]
- Park JH, Oh SA, Kim YH, Woo HR, Nam HG. Differential expression of senescence-associated mRNAs during leaf senescence induced by different senescence-inducing factors in Arabidopsis. Plant Mol Biol. 1998;37:445–454. doi: 10.1023/a:1005958300951. [DOI] [PubMed] [Google Scholar]
- Phillips LP, Huttly AK. Cloning of two gibberellin-regulated cDNAs from Arabidopsis thaliana by subtractive hybridization: expression of the tonoplast water channel, γ-TIP is increased by GA3. Plant Mol Biol. 1994;24:603–615. doi: 10.1007/BF00023557. [DOI] [PubMed] [Google Scholar]
- Robbins J, Dilworth SM, Laskey RA, Dingwall C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell. 1991;64:615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- Ross JJ, Murfet IC, Reid JB. Gibberellin mutants. Physiol Plant. 1997;100:550–560. [Google Scholar]
- Shimizu S, Mori H. A cDNA from Pisum sativum encoding the DIMINUTO homolog (accession no. D86494) (PGR 96-079) Plant Physiol. 1996;112:862. [Google Scholar]
- Szekeres M, Nemeth K, Koncz-Kalman Z, Mathur J, Kausch-mann A, Altmann T, Redei GP, Nagy F, Schell J, Koncz C. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;85:171–182. doi: 10.1016/s0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Gasch A, Nishizawa N, Chua N-H. The DIMINUTO gene of Arabidopsisis involved in regulating cell elongation. Genes Dev. 1995;9:97–107. doi: 10.1101/gad.9.1.97. [DOI] [PubMed] [Google Scholar]
- Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA and others. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- Weaver LM, Gan S, Quirino B, Amasino RM. A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatment. Plant Mol Biol. 1998;37:455–469. doi: 10.1023/a:1005934428906. [DOI] [PubMed] [Google Scholar]
- Xu W, Purugganan MM, Polisensky DH, Antosiewicz DM, Fry SC, Braam J. Arabidopsis TCH4, regulated by hormones and the environment, encodes a xyloglucan endotransglycosylase. Plant Cell. 1995;7:1555–1567. doi: 10.1105/tpc.7.10.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]