Abstract
Single polypyrrole (PPy) nanowire-based microfluidic aptasensors were fabricated using a one-step electrochemical deposition method. The successful incorporation of the aptamers into the PPy nanowire was confirmed by fluorescence microscopy image. The microfluidic aptasensor showed responses to IgE protein solutions in the range from 0.01 nM to 100 nM, and demonstrated excellent specificity and sensitivity with faster response and rapid stabilization times (~20 s). At the lowest examined IgE concentration of 0.01nM, the microfluidic aptasensor still exhibited ~0.32% change in the conductance. The functionality of this aptasensor was able to be regenerated using an acid treatment with no major change in sensitivity. In addition, the detection of cancer biomarker MUC1 was performed using another microfluidic aptasensor, which showed a very low detection limit of 2.66 nM MUC1 compared to commercially available MUC1 diagnosis assay (800 nM).
Keywords: Biosensors, microfluidics, conducting polymer, nanowire, aptamer
1. Introduction
Advanced biosensors based on nanostructured materials for point of care diagnosis are in high demand in recent years. Devices made from conducting polymer nanowires (Ramanathan et al. 2005; Yoon et al. 2009), semiconductor nanowires (Lee et al. 2009; Wang et al. 2005; Zheng et al. 2005), and carbon nanotubes (Chen et al. 2003; So et al. 2005; Star et al. 2003) have been used for the real-time detection of viruses, small molecules, and proteins because of their extraordinary electrical, mechanical, and thermal properties (Zheng et al. 2005). Although single-nanowire-based biosensors have achieved outstanding detection performances, there is still much room for further improvement in terms of the detection rate, sample volume consumption, specificity, fabrication complexity, and cost.
One strategy to significantly improve biosensors’ overall performances is to incorporate them into microfluidics. Microfluidic devices fabricated in poly(dimethylsiloxane) (PDMS) have several advantages such as the reduced consumption of samples and reagents, the comparatively fast and more sensitive reactions, and the good compatibility with biomolecules. (Kwakye and Baeumner 2003). So far, reversible sealing methods including mechanical fixation (Zaytseva et al. 2005) and adhesive tape sealing (McDonald et al. 2002) have been used to integrate pre-functionalized biosensor chips with microfluidic systems. Mechanical fixation usually requires screws, which complicates the device integration and miniaturization. In contrast, irreversible sealing provided by oxygen plasma treatment is a facile and easy method for microfluidic device fabrication. However pre-functionalized biosensors before microfluidic integration can possibly be deactivated by oxygen plasma treatment if they are directly exposed to the oxygen plasma. Hence, integrating with microfluidic devices with pre-functionalized single nanowire biosensors via irreversible sealing still poses a challenge.
Another strategy to improve the functionality of biosensors is to use aptamers as detection probes because they are highly specific to their targets. Antibodies have been extensively used as probes to detect targets such as prostate-specific antigen (PSA) and immunoglobulin (IgG) (Huang et al. 2002; Li et al. 2005; Zheng et al. 2005). Nevertheless, those probes can be easily denatured under severe conditions, and thus limit their use in practical applications. As an alternative probe, aptamers are more favorable because their target specificity and affinity are often superior to those of antibodies (So et al. 2005). Aptamers are artificial oligonucleotides (RNA or DNA) that can be isolated from combinatorial nucleic acid libraries using in vitro selection methods (Hermann and Patel 2000). As a result, aptamers can be synthesized at relatively low cost, and be easily engineered when necessary. When subjected to certain severe conditions, aptamers are stable or undergo reversible conformational change; under the same conditons proteins are irreversibly denatured. Because of these advantages, aptamers have been investigated and touted as novel probes for protein sensing with high specificity (Liao et al. 2008).
Herein, we present aptasensors based on aptamers incorporated polypyrrole nanowires (PPy-NWs) for rapid and label-free detection of proteins within microfluidic systems. Immunoglobulin E (IgE) is used in this work to demostrate the rapid response and high specificity of the aptasensor. According to this principle, another aptasenor is developed to detect Mucin 1 (MUC1), which is known to be overexpressed in almost of all human epithelial cell adenocarcinomas, including lung (Maeshima et al. 1997), prostate (Zhang et al. 1998), breast (Perey et al. 1992), and ovarian (Hough et al. 2000). The expression of MUC1 increases so much in many cases thus large amounts of the protein can be found in blood (Croce et al. 2003), thus makes the detection of MUC1 potentially useful for early cancers diagnosis.
2. Experimental
2.1 Chemicals
Pyrrole (≥99.5%), NaCl, and bovine serum albumin (BSA) were purchased from Sigma Aldrich. IgE protein was obtained from Athens Research & Technology, and MUC1 protein was purchased from peptide synthesis facility of the University of Pittsburgh. Aptamers for IgE and MUC1 were synthesized by Integrated DNA Technologies (IDT; Coralville, IA, USA). Phosphate buffer solution (PBS, pH 7.4) was used to prepare the BSA and IgE solutions with different concentrations.
2.2 Preparation of aptamer incorporated PPy-NW
The aptamers incorporated PPy-NW was electrochemically grown in a patterned polymethylmethacrylate (PMMA) nanochannel between two Au electrodes. An illustration of the preparation of aptamer incorporated PPy-NW is schematically shown in supplementary data Fig. S1A. A drop of electrolyte solution consisting of pyrrole monomer (5 mM), NaCl (10 mM), and aptamer (2 μM) was placed on the nanochannel between the electrodes. Then a constant current was applied between the Au electrode pair, and the corresponding potential across the electrodes was monitored during the NW growth. When the potential drops close to zero (normally around 500 μV), a NW is grown and connected between the electrodes (see Supplementary date Fig. S2). After the NWs preparation, the samples were rinsed with deionized water three times. The NWs were then examined using fluorescence microscopy.
2.3 Integration of microfluidic aptasensor
A PDMS microfluidic structure with six channels was fabricated through a replica molding process with a SU8 master. PDMS was selected for this work because it is a biocompatible material (McDonald and Whitesides 2002). The PDMS replica was treated with oxygen plasma for 1 minute, and irreversibly sealed on the chip containing the aptasensors. A hole was punched through the PDMS on both ends of each microchannel and tubings were inserted into these holes to complete the microfluidic aptasensor (see supplementary data Fig S3).
2.4 Conductance measurment
The real-time IgE detections in 10 mM phosphate salt buffer (PBS, pH=7.4) solution were performed at room temperature using the aptasensor with and without the microfluidic system. The PBS solution was placed on the aptasensor and then a constant current (100 uA) was applied across the NW. The conductance of the NW was monitored. When the conductance of the aptasensor stabilized in the PBS solution, the non-specific protein bovine serum albumin (BSA) solution was added, followed by several IgE solutions with different concentrations. The same procedures were performed for the IgE and MUC1 detections using the aptasensors within the microfluidic system in this work.
3. Results and disscusion
3.1 Fluorescence
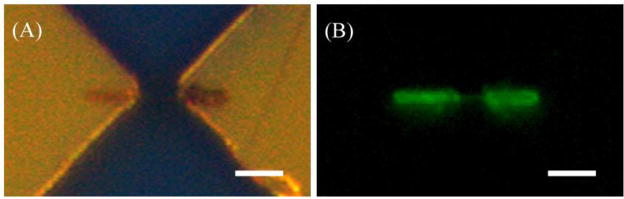
To confirm that the aptamers were successfully incorporated into the PPy-NWs, fluorescent-dye-labeled (Fluorescein amidite, FAM) aptamers were used during the electrochemical deposition. The physically adsorbed aptamers were removed by rinsing the NWs with deionized water for three times. As shown in Fig. 1A, the PPy-NW was connected with the gold electrodes by two thick ends that ensured a good contact. The fluorescence microscopy image (Fig. 1B) showed that the dye molecules were successfully entrapped in the PPy-NW. The NW exhibited green emission (bright region) with uniform distribution over its surface, which indicates that the aptamers were incorporated into the PPy-NW. Remarkably, clear fluorescence can be observed from most of the PPy-NWs incorporated with fluorescence-labeled aptamers, suggesting the excellent reproducibility of this aptamer incorporation approach.
Fig. 1.
Optical (A) and fluorescence microscopy (B) images of an aptamer incorporated PPy-NW grown between the gold electrodes pair. The fluorescence image confirms that the aptamers have been successfully incorporated into the PPy-NW. Scale bars are 5.0 um.
3.2 IgE and MUC1 detection
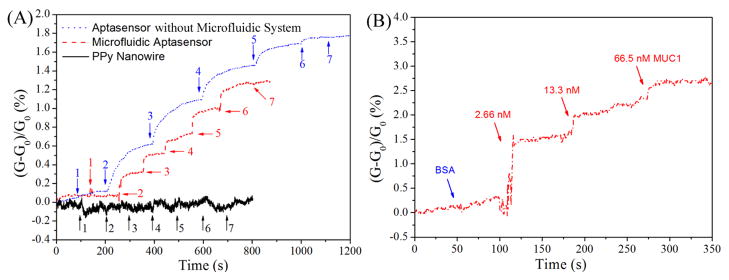
The normalized real-time response [(G-G0)/G0] of the aptasensors and the control sample are shown in Fig. 2A. For the IgE aptasensor (without microfludic system), non-appreciable changes in the curve were found upon the addition of BSA (1.5 uM) solution. In contrast, significant conductance increases were observed in the curve after the addition of IgE solutions. This observation indicates that the aptasensor has a good specificity to IgE protein. On the other hand, the negative control of PPy-NW (without aptamers) only showed negligible changes corresponding to BSA and IgE solutions, indicating the PPy-NWs have no significant interactions with the protiens in the absence of aptamers. Stepwise conductance increases were shown in the same figure upon the addition of IgE solutions on this aptasensor. After the addition of a 0.1 nM IgE solution, the conductance of this aptasensor increased rapidly and then stabilized slowly within 200 seconds. As the IgE concentration was increased to 1 nM, 10 nM, 100 nM, and 1 μM, consistent increases in the aptasensor conductance of 1.10%, 1.46%, 1.69%, and 1.77% were observed respectively, using the baseline as a reference. This response increase is attributed to an increase in the negative charge density on the PPy-NW resulting from the specific binding event (Yoon et al. 2009). The PBS solution of pH 7.4 caused the IgE protein to be negatively charged because it is higher than the isoelectric point of the IgE protein (pI=5.2–5.8). Therefore, the specific binding of the aptamer and the negatively charged IgE protein would result in an increased negative charge density on the PPy-NW, which could lead to the conductance increase.
Fig. 2.
(A) Normalized real-time response of aptasensor, microfluidic aptasensor, and PPy-NW to BSA and IgE solutions. Arrows indicate the points of injecting protein solution [Aptasebsor and PPy-NW: 1&7. 1.5 μM BSA; 2–6. 0.1, 1, 10, 100, 1000 nM IgE. Microfluidic aptasensor: 1&7. 1.5 μM BSA; 2–6. 0.01, 0.1, 1, 10, 100 nM IgE.]. (B) Normalized real-time detection of cancer biomarker MUC1 using MUC1 specific aptamer incorpoarted PPy-NW sensor within the microfluidic system.
The obtained detection results (Fig. 2A dot-line; without microfluidic system) indicate that the aptasensor was almost saturated at 1 μM. Therefore, the IgE concentration was optimized from 0.01 to 100 nM for the microfluidic aptasensor. As shown in Fig. 2A, stepwise responses were also observed. The conductance of this microfluidic aptasensor changed rapidly and stabilized quickly (~20 s) after the IgE solutions were injected. A similarly enhanced response was reported previously and attributed to the enhanced effects of diffusion and mass transport of the biomarkers within the microchannel (Kwakye and Baeumner 2003). From baseline, the conductance of this microfluidic aptasensor increased consistently from 0.32% to 1.26% as the IgE concentration increased from 0.01 to 100 nM. The signal-to-noise ratio was 3.6:1 at the lowest IgE concentration. So far, the reported detection limits for IgE are about 0.01 nM and 10 nM using conventional surface plasma resonance (SPR) measurements (Kim et al. 2009) and quartz crystal microbalance (QCM) biosensors (Yao et al. 2009) respectively, which are the same or higher than the value achieved in this work. Rather than optical methods such as SPR technique, electrical detection possesses advantages including not only the portability, but also the independence from the optical path length, sample turbidity, extremely low cost, and low power requirements.
Another microfluidic aptasensor was then fabricated for the detection of cancer biomarker MUC1 using MUC1-specific-aptamer as the detection probe. As shown in Fig. 2B, time-dependent conducatance measurements recorded on the microfluidic aptasensor exhibited no change in the conductance upon addition of 1.5 uM BSA. However, a significant increase in the conductance was observed after the injection of 2.66 nM MUC1 solution, demostrating the high specificity of this microfluidic aptasensor. As the concentrations of MUC1 were subsequently increased to 13.3 nM and 66.5 nM, the condunctance of the microfluidic aptasensor was increased to 2.25% and 2.8% respectively. As far as we are aware, the lowest detection concentration for the most popularly used CA 15-3 assay, which is a currently commercially available ELISA kit for the detection of MUC1, is estimated to be 800 nM of MUC1 (Cheng et al. 2009). The results showed here indicates that the aptamer-based detection method within the microfluidic system is not only rapid but also sensitive, with a very low detection concentration of 2.66 nM MUC1.
3.3 Sensitivity
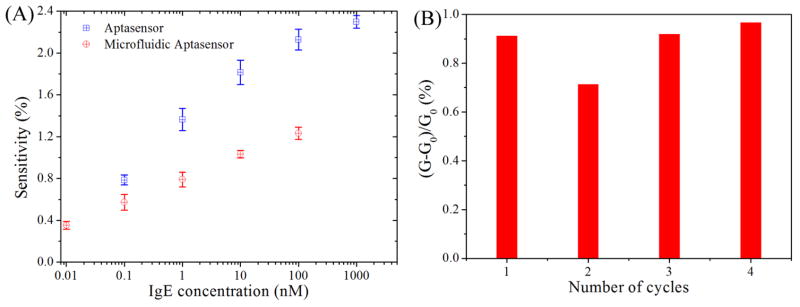
Fig. 3A shows the sensitivities of the aptasensor and microfluidics aptasensor obtained from three NWs for each case. The aptasensor was very sensitive exhibiting 0.79±0.04% change in its conductance even at the IgE concentration of 0.1 nM (20 ng/ml), which is about 2 times greater than that obtained from an aptamer-doped PPy film (Liao et al. 2008). On the other hand, the microfluidic aptasensor showed responses to IgE solutions within the range from 0.01 nM (2 ng/ml) to 100 nM (20 μg/ml), showing a 0.57±0.07% change in the conductance at the IgE concentration of 0.1 nM. Despite the small decrease in sensitivity of the microfluidic aptasensor, the reproducible responses of the microfluidic aptasensor remain within distinguishable level. Clearly, both high sensitivity and fast response are important for biosensors. For this microfluidic aptasensor, the sensitivity was decreased probably because of the degradation the biomolecules during the microfluidic device integration process (i.e. the sealing of PDMS replica on the chip). However, using microfluidic system in this experiment showed great improvement on the response time. Optimizing the microfluidic device integration process could increase the sensitivity of the microfluidic aptasensor in future.
Fig. 3.
(A) The sensitivity of the aptasensor with and without microfluidic system as a function of IgE concentration, data were collected from three aptasensors for each case. (B) Regeneration of the aptasensor upon treatment with 0.01 M HCl. The data are collected with IgE concentration of 0.1 nM.
3.4 Reusablity
The possibility of reusing a sensor several times with or without a regeneration step is highly desirable, but typically difficult to achieve. For example, the dissociation of antibody-antigen complexes can usually be achieved by treating with acid solution. However, the antibodies are irreversibly damaged to some extent (Liss et al. 2002). In contrast, the aptamers are almost capable of reversible regeneration. The reversible conformational change of the anti-IgE aptamer was investigated with acid treatment. The used aptasensor was incubated in 1 μL of 0.01 M hydrochloric acid for 15 min and followed by thorough washing with 10 mM PBS buffer. As shown in Fig. 3B, the as-prepared aptasensor showed 0.91% response to the 0.1 nM IgE solution and the incubation restored the aptasensor functionality for up to three repeated measurements with no significant loss of sensitivity. This result indicates the good reusability of this aptasensor.
4. Conclusions
In conclusion, we have successfully demonstrated the Ppy-NW-based microfluidic aptasensor for rapid and real-time detection of IgE and MUC1 with high sensitivity and specificity. The one-step incorporation of biomolecules into the dimension-controllable conducting polymer nanowire eliminates post-synthesis surface modification and simplifies the device fabrication process. The fluorescence microscopic image confirmed the successful incorporation of the aptamers. This microfluidic aptasensor provided fast response and stabilization times. Furthermore, the functionality of this aptasensor can be simply restored by acid treatment with no major change in sensitivity. Importantly, the proposed method for aptasensor fabrication can be easily adapted for the development of various aptasensors based on different conducting polymers and different aptamers for specific targets. A microfluidic aptasensor for the detection of MUC1 was accordingly developed. This aptasensor acheived a very low detection concentration of 2.66 nM MUC1 in this research. These results indicates the possibility for developing rapid and sensitive biosensor devices for the detection of cancer biomarkers.
Supplementary Material
Acknowledgments
This project is supported by National Science Foundation (NSF) ECCS 0824035 and National Institute of Health (NIH) 1R21B008825 and Mascaro Center of Sustainable Innovation (MCSI). Minhee Yun acknowledges the Korea Brain Pool.
Appendix A. Supplementary data
Supplementary data associated with this article can be found in the online version.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Chen RJ, Bangsaruntip S, Drouvalakis KA, Kam NWS, Shim M, Li YM, Kim W, Utz PJ, Dai HJ. Proc Natl Acad Sci U S A. 2003;100(9):4984–4989. doi: 10.1073/pnas.0837064100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AKH, Su HP, Wang A, Yu HZ. Anal Chem. 2009;81(15):6130–6139. doi: 10.1021/ac901223q. [DOI] [PubMed] [Google Scholar]
- Croce MV, Isla-Larrain MT, Demichelis SO, Gori JR, Price MR, Segal-Eiras A. Breast Cancer Res Treat. 2003;81(3):195–207. doi: 10.1023/A:1026110417294. [DOI] [PubMed] [Google Scholar]
- Hermann T, Patel DJ. Science. 2000;287(5454):820–825. doi: 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]
- Hough CD, Sherman-Baust CA, Pizer ES, Montz FJ, Im DD, Rosenshein NB, Cho KR, Riggins GJ, Morin PJ. Cancer Res. 2000;60(22):6281–6287. [PubMed] [Google Scholar]
- Huang NP, Voros J, De Paul SM, Textor M, Spencer ND. Langmuir. 2002;18(1):220–230. [Google Scholar]
- Kim YH, Kim JP, Han SJ, Sim SJ. Sens Actuator B-Chem. 2009;139(2):471–475. [Google Scholar]
- Kwakye S, Baeumner A. Anal Bioanal Chem. 2003;376(7):1062–1068. doi: 10.1007/s00216-003-2063-2. [DOI] [PubMed] [Google Scholar]
- Lee HS, Kim KS, Kim CJ, Hahn SK, Jo MH. Biosens Bioelectron. 2009;24(6):1801–1805. doi: 10.1016/j.bios.2008.08.036. [DOI] [PubMed] [Google Scholar]
- Li C, Curreli M, Lin H, Lei B, Ishikawa FN, Datar R, Cote RJ, Thompson ME, Zhou CW. J Am Chem Soc. 2005;127(36):12484–12485. doi: 10.1021/ja053761g. [DOI] [PubMed] [Google Scholar]
- Liao W, Randall BA, Alba NA, Cui XT. Anal Bioanal Chem. 2008;392(5):861–864. doi: 10.1007/s00216-008-2354-8. [DOI] [PubMed] [Google Scholar]
- Liss M, Petersen B, Wolf H, Prohaska E. Anal Chem. 2002;74(17):4488–4495. doi: 10.1021/ac011294p. [DOI] [PubMed] [Google Scholar]
- Maeshima A, Miyagi A, Hirai T, Nakajima T. Pathol Int. 1997;47(7):454–460. doi: 10.1111/j.1440-1827.1997.tb04524.x. [DOI] [PubMed] [Google Scholar]
- McDonald JC, Chabinyc ML, Metallo SJ, Anderson JR, Stroock AD, Whitesides GM. Anal Chem. 2002;74:1537–1545. doi: 10.1021/ac010938q. [DOI] [PubMed] [Google Scholar]
- McDonald JC, Whitesides GM. Accounts Chem Res. 2002;35(7):491–499. doi: 10.1021/ar010110q. [DOI] [PubMed] [Google Scholar]
- Perey L, Hayes DF, Maimonis P, Abe M, Ohara C, Kufe DW. Cancer Res. 1992;52(9):2563–2568. [PubMed] [Google Scholar]
- Ramanathan K, Bangar MA, Yun M, Chen W, Myung NV, Mulchandani A. J Am Chem Soc. 2005;127(2):496–497. doi: 10.1021/ja044486l. [DOI] [PubMed] [Google Scholar]
- So HM, Won K, Kim YH, Kim BK, Ryu BH, Na PS, Kim H, Lee JO. J Am Chem Soc. 2005;127(34):11906–11907. doi: 10.1021/ja053094r. [DOI] [PubMed] [Google Scholar]
- Star A, Gabriel JCP, Bradley K, Gruner G. Nano Lett. 2003;3(4):459–463. [Google Scholar]
- Wang WU, Chen C, Lin KH, Fang Y, Lieber CM. Proc Natl Acad Sci U S A. 2005;102(9):3208–3212. doi: 10.1073/pnas.0406368102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao CY, Qi YZ, Zhao YH, Xiang Y, Chen QH, Fu WL. Biosens Bioelectron. 2009;24(8):2499–2503. doi: 10.1016/j.bios.2008.12.036. [DOI] [PubMed] [Google Scholar]
- Yoon H, Lee SH, Kwon OS, Song HS, Oh EH, Park TH, Jang J. Angew Chem-Int Edit. 2009;48(15):2755–2758. doi: 10.1002/anie.200805171. [DOI] [PubMed] [Google Scholar]
- Zaytseva NV, Goral VN, Montagna RA, Baeumner AJ. Lab Chip. 2005;5:805–811. doi: 10.1039/b503856a. [DOI] [PubMed] [Google Scholar]
- Zhang SL, Zhang HS, Reuter VE, Slovin SF, Scher HI, Livingston PO. Clin Cancer Res. 1998;4(2):295–302. [PubMed] [Google Scholar]
- Zheng GF, Patolsky F, Cui Y, Wang WU, Lieber CM. Nat Biotechnol. 2005;23(10):1294–1301. doi: 10.1038/nbt1138. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.