Abstract
One avenue for prevention and treatment of Alzheimer's disease involves inhibiting the aggregation of amyloid-β peptide (Aβ). Given the deleterious effects reported for Aβ dimers and trimers, it is important to investigate inhibition of the earliest association steps. We have employed quantized photobleaching of dye-labeled Aβ peptides to characterize four peptide-based inhibitors of fibrillogenesis and/or cytotoxicity, assessing their ability to inhibit association in the smallest oligomers (n = 2–5). Inhibitors were tested at acidic pH and in the presence of zinc, conditions that may promote oligomerization in vivo. Distributions of peptide species were constructed by examining dozens of surface-tethered monomers and oligomers, one at a time. Results show that all four inhibitors shift the distribution of Aβ species toward monomers; however, efficacies vary for each compound and sample environment. Collectively, these studies highlight promising design strategies for future oligomerization inhibitors, affording insight into oligomer structures and inhibition mechanisms in two physiologically significant environments.
Keywords: amyloid-beta peptide, soluble oligomers, aggregation inhibitor, acid-induced aggregation, metal-induced aggregation, single molecule spectroscopy
INTRODUCTION
Amyloid-β (Aβ) is a peptide of 39–43 amino acids that is formed through specific proteolytic cleavage of the transmembrane amyloid precursor protein (APP) [1] and normally circulates in brain plasma and cerebrospinal fluid at pico- to nanomolar concentrations [2]. Although Aβ monomers may be protective to neurons [3], numerous factors have been shown to promote the peptide's self-association into neurotoxic oligomers and aggregates implicated in Alzheimer's disease (AD; for a review, see ref. [4]). Among the multitude of aggregated species, Aβ fibrils exhibiting cross-β sheet structure [5, 6] have historically garnered the greatest attention, due to their prevalence in the senile plaques characteristic of AD brain [7–9]. Inhibiting Aβ fibrillogenesis has been a primary strategy in Alzheimer's drug development. A variety of small molecules [10, 11] and peptides [12] have been reported to inhibit fibril formation and/or reduce neurotoxicity of Aβ aggregates. Successful peptide inhibitors include block copolymers containing recognition and disrupting elements [13, 14], β-sheet breakers employing Pro residues [15, 16], N-methylated sequences [17–19], derivatives disubstituted at the α-carbon [20] and sequences containing D-amino acids [19, 21, 22].
Over the last decade, focus has shifted somewhat from the inhibition of fibrillogenesis, as several key results have linked small Aβ oligomers to the etiology of AD [23]. The accumulation of SDS-stable dimers and trimers [24–27] in the brain is strongly correlated with synapse loss, cognitive impairment and increased severity of end-stage AD [24, 25, 28]. Direct application of cell-derived dimers, trimers and tetramers results in impaired long-term potentiation (LTP) [29–31], short-term memory deficits [32] and synapse loss [33] in rodents. Additionally, Aβ dimers isolated from the cortex of human AD brain [34] and found in human cerebrospinal fluid [35] inhibit LTP in vivo. Collectively, these findings illustrate the crucial role of small Aβ oligomers in Alzheimer's progression and provide clear incentive to investigate inhibition of Aβ association in its earliest stages.
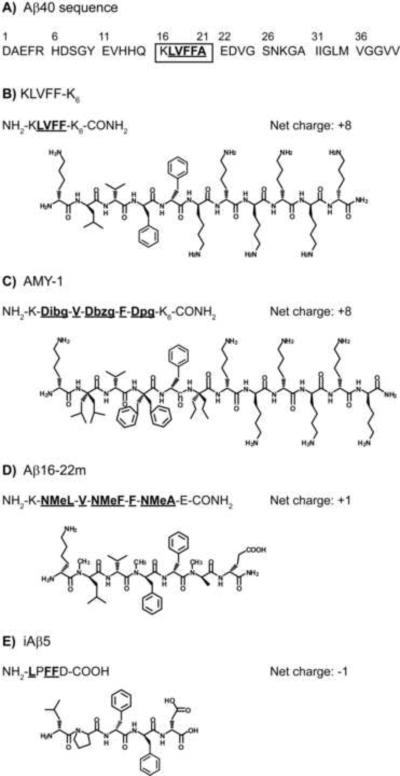
Toward that goal, we selected four successful inhibitors of fibrillogenesis and/or neurotoxicity, each with a distinct inhibition strategy (Figure 1) [14, 15, 17, 20], and examined their abilities to prevent or reverse association in the smallest oligomers (n = 2–5). Since oligomers and fibrils have been shown to possess different structures and form through distinct pathways [36], we were interested to determine which compound(s) would most successfully inhibit the earliest association steps. All four inhibitors employ recognition sequences similar to Aβ's central hydrophobic region (amino acids 16–21; see Figure 1A) and bind to the full-length peptide via a combination of hydrophobic side-chain interactions and backbone hydrogen bonds [37–40], the atomic-level details of which are not known. KLVFF-K6 (Figure 1B) contains residues 16–20 of Aβ with a lysine hexamer as a disrupting element. Murphy and coworkers reported that it significantly alters aggregation kinetics and aggregate morphology while reducing Aβ cytotoxicity [14, 41,42]; Moss et al. found that it inhibits monomer aggregation [43]. AMY-1 (Figure 1C) is a peptide analogue of KLVFF-K6 containing alternating α,α-disubstituted amino acids (ααAA). The Hammer group reported that equimolar concentrations of AMY-1 are highly effective in inhibiting Aβ fibrillogenesis: while the L-amino acids on one face of the inhibitor permit hydrogen-bonding to Aβ, steric effects of the ααAA on the other face effectively prevent continued association [20]. Aβ16–22m (Figure 1D) also functions by blocking binding of Aβ on one face of the Aβ-inhibitor complex, as N-methyl amino acids on one side of the inhibitor lack the amide hydrogens used in hydrogen bonding and β-sheet formation [17]. Meredith and colleagues reported that Aβ16–22m dramatically inhibits Aβ fibrillogenesis and successfully dissociates preformed fibrils [17]. Successful inhibition of fibril formation and dissociation of preformed fibrils have also been reported for iAβ5 (LPFFD, Figure 1E), developed by Soto and coworkers; in addition, this inhibitor was shown to prevent Aβ neurotoxicity [15]. iAβ5 is designed around residues 17–21 of Aβ: Asp is used in place of Ala-21 for improved solubility, and Pro is substituted in place of Val-18, harnessing the ability of proline to prevent and disrupt β-sheet formation [15, 38].
Figure 1.
Peptide inhibitors. A) Sequence of Aβ40 with residues numbered. (FAβB has a carboxyfluorescein (FAM) dye covalently bound to the N-terminus and a Lys-Biotin moiety attached to the C-terminus.) Amino acids 16–21 (boxed) comprise the central hydrophobic core that inhibitors are designed to recognize. The hydrophobic residues in this region are shown underlined in bold, both here and in each inhibitor sequence below. The net charge of each inhibitor at pH 7 is also listed for comparison. For these sequences, charges are largely unchanged in the range from pH 5.8 to 7.4. B) KLVFF-K6 employs a (primarily hydrophobic) KLVFF recognition element identical in sequence to Aβ16–20 and a hydrophilic disrupting element comprised of six Lys residues [14]. C) AMY-1 is an analogue of KLVFF-K6 that incorporates alternating α,α-disubstituted amino acids diisobutyl-, dibenzyl- and dipropyl-glycine (Dibg, Dbzg, and Dpg) on one face of the peptide [20]. D) Aβ16–22m is derived from Aβ16–22 and employs alternating N-methyl amino acids (NMeL, NMeF, NMeA) on one face of the peptide [17]. It has a hydrophobic core with charged terminal residues. E) iAβ5 is a pentapeptide designed around Aβ17–21 that uses Pro to disrupt β-structure. It is primarily hydrophobic with charged Asp incorporated to improve solubility [15].
We tested each of the four inhibitors in Figure 1 against samples of the earliest oligomers, prepared under acidic conditions (pH 5.8) and at physiological pH in the presence of Zn2+. These sample environments have been shown to promote Aβ aggregation [44–49] (and oligomerization [50]) in vitro and are also relevant to association in vivo. The acidic conditions mimic those of early endosomes where Aβ is produced [51, 52] and the zinc concentrations used are within the range observed in the brain [53–55], where zinc release during neurotransmission (at concentrations up to 300 μM [54, 55]) may cause Aβ oligomers to target synapses [56]. Single-molecule fluorescence spectroscopy was used to determine distributions of monomers and oligomers present under the various sample conditions, using a method we have reported previously [50] (which was further substantiated by the work of Steel and colleagues [57]). For these measurements, we used Aβ40 – the most prevalent form of Aβ in vivo [58] – that is covalently labeled at the N-terminus with carboxyfluorescein (FAM), and at the C-terminus with Lys-Biotin; this construct is abbreviated FAβB. The peptide is tethered to functionalized cover slips via biotin-streptavidin binding; a dense base layer of polyethylene glycol provides a non-fouling surface, preventing adsorption to the glass [50, 59]. Localized FAβB monomers and oligomers are interrogated, one at a time, through laser excitation. The quantized photobleaching of individual dye molecules permits quantification of the number of associated peptides [50, 57, 60]. By investigating dozens of peptide species, one by one, we have characterized the distributions of monomers and oligomers present under our conditions of interest. We report that all four inhibitors are successful in reducing association in the smallest oligomers, shifting the distribution of FAβB species toward monomers. The variations in efficacy observed for the different compounds in acidic versus zinc-containing samples suggest successful strategies for preventing or reversing the earliest Aβ association steps and afford new insight into oligomer structures and inhibition mechanisms in two physiologically significant environments.
EXPERIMENTAL METHODS
Materials
FAM-Aβ40-Lys-Biotin (FAβB) and iAβ5 (LPFFD) were purchased from AnaSpec, Inc. (Fremont, CA); the remaining three inhibitors (i.e., KLVFF-K6, AMY-1 and Aβ16–22m) were obtained from the Hammer research group, formerly of Louisiana State University. All peptides were stored at −20 °C upon receipt. Zinc(II) chloride (ZnCl2), sodium chloride (NaCl), 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) and phosphate buffered saline (PBS) powder were obtained from Fisher. Solutions were prepared in distilled, deionized water (18 MΩ) from a Millipore system. Functionalized cover slips (STREP-01) were purchased from MicroSurfaces, Inc. (Austin, TX). These are pre-coated with a dense layer of polyethylene glycol (PEG), a small percentage of which is functionalized with biotin (surface density ~1010/cm2); the biotin moieties are further bound to streptavidin, each of which has 2–3 accessible sites for binding of biotin-labeled FAβB [61]. Packaged cover slips were stored, vacuum-sealed, at ≤ −20 °C; after opening, unused cover slips were kept in a vacuum desiccator at room temperature, according to the manufacturer's instructions.
Sample Preparation
Phosphate buffered saline (PBS, 1×: 137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, and 1.47 mM KH2PO4) and HEPES/NaCl (100 mM HEPES, 50 mM NaCl) buffer solutions were prepared in distilled, deionized water; pH was adjusted as needed with HCl and/or NaOH. Lyophilized FAβB was added to prepared buffer after thawing for 30 minutes; these peptide stock solutions were made at ~1 μM concentrations, as determined from spectrophotometry (Beer's Law), using the molar absorptivity of the FAM dye. Additional solutes (e.g., ZnCl2, inhibitor compounds) were added at this point. Samples were then incubated at room temperature for one hour, after which they were diluted to final FAβB concentrations of approximately 60 pM.
Functionalized cover slips were prepared for use by dragging the bottom, uncoated sides across methanol-moistened lens tissues and edging the top, coated surfaces with Mylar frames (McMaster-Carr), to prevent sample loss. Samples (30–40 μL) were added to the centers of prepared cover slips already in place on the microscope, forming small pools of solution (area ~1 cm2), and left for 30 minutes to permit tethering of the biotin-labeled peptides to the streptavidin on the cover slips. At ~60 pM, assuming 100% binding of FAβB monomers to streptavidin, ~0.01% of the available streptavidin binding sites are occupied. Additional buffer solution (~20-μL aliquots) was added as needed (every ~45 minutes) to offset evaporation, which causes higher fluorescence signal and background.
Single-Molecule Fluorescence Spectroscopy
The apparatus used to detect surface-tethered FAβB monomers and oligomers (Figure 2A) was described previously [50]. Briefly, 488-nm light from a krypton-argon laser was focused to a diffraction-limited spot ~300 nm in diameter by the 100× objective (1.3 NA) of an inverted fluorescence microscope. A closed-loop nanopositioning stage raster-scanned samples over the laser spot, exciting surface-tethered peptide species. Fluorescence from excited dyes was collected by the objective, filtered to remove stray laser light (dichroic and holographic notch filters) and detected by an avalanche photodiode (APD) detector. (The 180-μm2 active area of the APD served as a pinhole to approximate confocal imaging.) TTL pulses generated by the APD were detected by a counter/timer board interfaced to custom LabVIEW software.
Figure 2.
Single-oligomer spectroscopy. A) Light (488 nm) from a Kr-Ar laser is focused to a diffraction-limited spot by the objective of an inverted fluorescence microscope. A nanopositioning stage scans the sample over the laser, which excites individual surface-tethered peptide species. Fluorescence is collected by the objective, filtered by dichroic (D) and notch (N) filters to remove laser light, and detected by an avalanche photodiode (APD) detector. B) During laser excitation, a monomer exhibits one characteristic fluorescence intensity (broadened by shot noise) until irreversible photobleaching occurs, returning the measured intensity to background levels. C) A dimer displays two distinct intensity levels above background. Initially, laser excitation results in fluorescence from both dyes, until one photobleaches, causing a discrete drop in measured intensity. Photobleaching of the second dye results in a return to background intensities.
Samples of surface-tethered FAβB peptides were imaged in 20×20-μm regions, generating plots of fluorescence intensity as a function of position; the laser power and scanning rate used (420–500 W/cm2 and 25–50 μm/s, respectively) were selected to minimize dye photobleaching during imaging. The bright spots detected in each image were examined, one by one, by directing the stage to “park” at each position; emitted photons were counted as a function of time (3- or 5-ms bins) to generate a fluorescence intensity profile for each spot. Excitation was continued until the detected intensity dropped to background levels, as determined from profiles of dark areas. Time-dependent intensity data were typically re-binned to plot counts per 30 ms, allowing for clearer discrimination of signal versus background levels. The number of dyes present in each profile was determined from the number of discrete photobleaching steps observed and the number of distinct intensity levels (at least 100 ms in duration) evident above background. Dozens of bright spots were interrogated for each sample, allowing for the construction of distributions (histograms) of the monomers and oligomers observed. Two-tailed t-tests were used to statistically compare oligomer distributions, assuming unequal variances with a confidence interval (α) equal to 0.05.
RESULTS
Single-molecule fluorescence measurements were utilized to assess the efficacy of each inhibitor in preventing or reversing acid- and zinc-promoted oligomerization. For each sample environment, all four peptides were evaluated at 10:1 inhibitor:Aβ ratios (~600 pM inhibitor concentrations); the more promising compounds were further studied at equimolar concentrations (~60 pM). The distribution of FAβB monomers and small oligomers in a particular sample was constructed by interrogating dozens of spatially resolved peptide species (observed as bright spots in fluorescent images), one at a time, as described previously [50]. Each mono- or oligomer was continuously excited until its emission intensity dropped to a background level; the number of associated peptides was determined from the discrete photobleaching steps observed in the time-dependent fluorescence profile (see Figure 2).Note 1 A monomer exhibits one characteristic fluorescence signal (broadened by shot noise) from its single dye; the signal drops sharply to background upon dye photobleaching (Figure 2B) [62, 63]. (Brief visits to background intensities may occur prior to irreversible photobleaching, due to intersystem crossing [62, 63], but these “blinks” are rare at our low excitation rates.) A dimer undergoes two photobleaching steps, one for each dye, and therefore exhibits two discrete intensity levels above background (Figure 2C); a trimer shows three intensity levels, and so on. FAβB monomers through pentamers are reliably identified on this basis, as we have demonstrated [50]. Larger oligomers often exhibit smooth, quasi-exponential decays in their intensity profiles, due partly to rapid dye photobleaching, and are more difficult to quantify. Consistent with our earlier findings [50], we observed only a small number of oligomers with n > 5, ~2 % of all species for metal-free samples at pH 7.4, ~5 % in the presence of zinc, and ~7 % at pH 5.8. Based on their fluorescence intensities, we estimate that the largest of these oligomers may contain tens of peptides.
Multiple samples were investigated to characterize each set of conditions. Oligomer distributions for replicate samples were found to be highly reproducible (as noted previously [50]); as such, their statistically indistinguishable data sets (p >> 0.05, typically 0.3 < p < 0.9) were combined to generate composite distributions containing at least 100 individual peptide species. To facilitate comparison across different inhibitors and environments, the composite histograms are depicted below in terms of percentages of monomers and small oligomers.
Inhibiting Association at Acidic pH
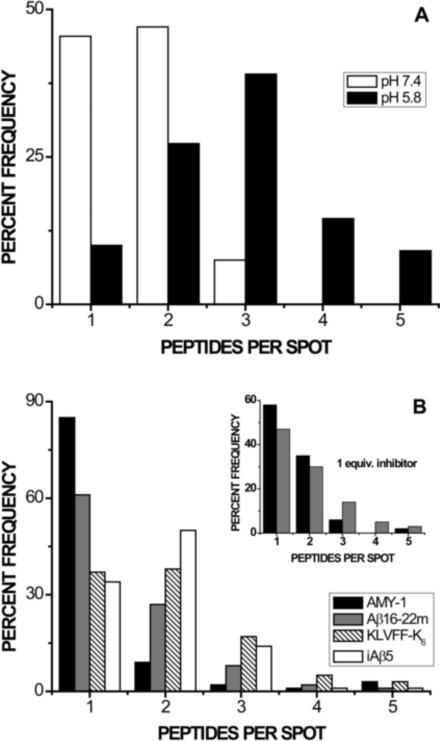
Figure 3 depicts distributions of FAβB monomers and small oligomers acquired in PBS buffer at pH 5.8. In the absence of inhibitor, acidic conditions promote increased association over that observed at physiological pH, as shown in panel 3A (and as reported previously [50]). Incubation with 10 molar equivalents of any of the four peptides (Figure 3B) results in a statistically significant shift toward FAβB monomers, as compared to inhibitor-free samples at pH 5.8 (p << 0.01); however, none of the inhibitors significantly affect the number of unquantifiable larger oligomers, which remains consistent in the absence or presence of inhibitor at ~7 %. Samples containing KLVFF-K6 and iAβ5 are statistically indistinguishable and exhibit the lowest degree of inhibition. Samples containing AMY-1 reproducibly show the greatest percentage of monomers (85 %); Aβ16–22m ranks second in efficacy, producing 61 % monomers.
Figure 3.
Inhibition of acid-promoted oligomerization (pH 5.8). Each distribution represents at least 100 small FAβB species from multiple samples, interrogated one at a time. The total percentage of monomer and each oligomer observed (up to n = 5) is plotted in order to facilitate comparison between different sample environments; each sample additionally contains ~2–7% larger oligomers, not shown. A) As we have reported previously [50], acidic pH (black bars) significantly shifts the distribution of FAβB monomers and oligomers toward larger species, as compared to samples at pH 7.4 (white bars). B) The presence of ten molar equivalents of any inhibitor at pH 5.8 significantly shifts the distribution of FAβB species toward monomers, as compared to inhibitor-free samples (pH 5.8, Panel A). AMY-1 is most successful in producing monomers of FAβB. Inset: The most successful inhibitors AMY-1 and Aβ16–22m were tested at concentrations equimolar with FAβB. Under these conditions, the resulting distributions are statistically indistinguishable.
Composite distributions for AMY-1 and Aβ16–22m present at 1 molar equivalent (Figure 3B, inset) are statistically indistinguishable from each other; however, even at this reduced concentration, both inhibitors successfully shift the distribution of observed FAβB species toward monomers (p << 0.01 compared to inhibitor-free samples). Inhibition by Aβ16–22m is apparently unaffected by concentration changes in this range, as the distributions for 1 and 10 equivalents of this inhibitor are statistically indistinguishable (p = 0.37). AMY-1 is significantly less effective when present at one versus ten molar equivalents (p < 0.01).
Inhibiting Association in the Presence of Zinc
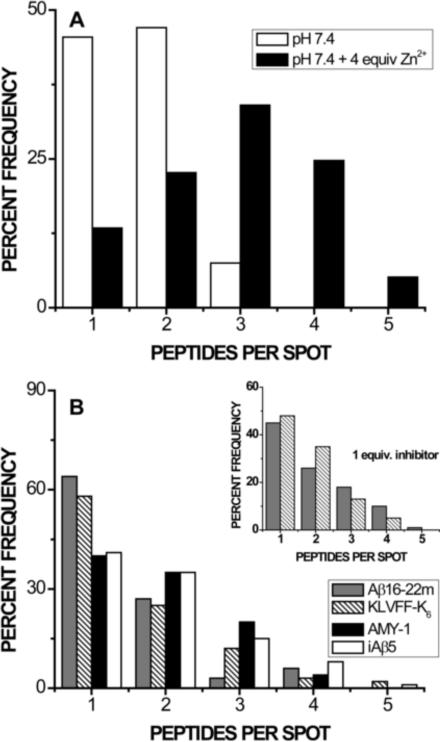
The addition of 4 molar equivalents of Zn2+ at pH 7.4 promotes increased oligomerization over that observed for metal-free samples, as shown in Figure 4A (and reported previously [50]). As observed under acidic conditions, incubation with 10 molar equivalents of any of the four peptides results in a statistically significant shift toward FAβB monomers (Figure 4B), as compared to inhibitor-free samples in the presence of zinc (p << 0.01). Under these conditions, Aβ16–22m and KLVFF-K6 are most successful in reducing oligomerization; distributions for these two inhibitors are statistically indistinguishable from each other, but show significantly improved inhibition over AMY-1 or iAβ5. In fact, inhibition by Aβ16–22m or KLVFF-K6 in the presence of zinc is comparable to that afforded by the zinc chelator clioquinol, reported previously [50]: FAβB oligomer distributions are statistically indistinguishable, with 64, 58, and 54 % monomers for samples containing ten equivalents of Aβ16–22m, KLVFF-K6, and clioquinol, respectively. The presence of inhibitor in zinc-containing samples also seems to reduce the percentage of larger FAβB oligomers (n > 5), from ~5 % in inhibitor-free samples to ~4 % (iAβ5), ~3 % (AMY-1, KLVFF-K6), or ~1 % (Aβ16–22m).
Figure 4.
Inhibition of zinc-promoted oligomerization at pH 7.4. Each distribution represents at least 100 small FAβB species from multiple samples, interrogated one at a time. The total percentage of monomer and each oligomer observed (up to n = 5) is plotted in order to facilitate comparison between different sample environments; each sample additionally contains ~1–5% larger oligomers, not shown. A) As we have reported previously [50], the presence of 4 equivalents of Zn2+ (black bars) significantly shifts the distribution of FAβB monomers and oligomers toward larger species, as compared to nominally metal-free samples at pH 7.4 (white bars). B) The addition of ten molar equivalents of any inhibitor to samples containing 4 equivalents of Zn2+ (pH 7.4) significantly shifts the distribution of FAβB species toward monomers, as compared to inhibitor-free samples (Panel A). Aβ16–22m and KLVFF-K6 are most successful in producing monomers of FAβB. Inset: Aβ16–22m and KLVFF-K6 were tested at concentrations equimolar with FAβB. Each successfully shifts the distribution of FAβB species toward monomers at this reduced concentration; as was observed for 10 equivalents of inhibitor, the two distributions are statistically indistinguishable.
At one molar equivalent (Figure 4B, inset), both Aβ16–22m and KLVFF-K6 again cause a significant shift of FAβB species toward monomers (p << 0.01 compared to the inhibitor-free distribution in Figure 4A); however, the two compounds cannot be distinguished in terms of efficacy. The effectiveness of Aβ16–22m is somewhat decreased from that observed with 10 equivalents of the peptide (p < 0.01), demonstrating a concentration-dependent response, whereas the distribution for KLVFF-K6 at one equivalent cannot be distinguished from that observed when the inhibitor is present in excess (p = 0.28).
Evaluating Inhibitor Consistency in Different Sample Environments
Statistical comparison of the histograms in Figures 3B and 4B confirms that oligomer distributions for Aβ16–22m-containing samples at pH 5.8 and in the presence of zinc are indistinguishable (p = 0.77 for 10 equivalents of inhibitor); the same is true for distributions obtained in the presence of iAβ5 (p = 0.68). In contrast, comparison of oligomer distributions for AMY-1- or KLVFF-K6-containing samples verifies statistically significant differences for acidic versus zinc-containing environments (p < 0.01).
DISCUSSION
A number of recent studies have linked Aβ dimers and trimers to symptoms of Alzheimer's disease, including synapse loss, short-term memory deficits and LTP impairment [29–35], providing clear incentive to identify inhibitors of the earliest Aβ association steps. Further, since the earliest Aβ oligomers may be found in acidic endosomes where Aβ is produced [51, 52] and recruited to synapses by zinc release during neurotransmission [56], inhibiting oligomerization in acidic and zinc-containing environments is of particular interest. Through single-oligomer spectroscopy, we have successfully characterized the efficacies of four peptides with distinct inhibition strategies, determining distributions of monomers and small oligomers in each of these environments. To our knowledge, these are some of the first empirical studies to investigate inhibition of the earliest oligomerization steps in the presence of acid or zinc.
The inhibition of FAβB oligomerization exhibited by all four peptides, particularly when present at equimolar concentrations (also observed for iAβ5 under at pH 5.8; data not shown) reveals that these molecules successfully bind to small oligomers and/or monomers and shift the equilibrium [64] of oligomeric species toward smaller assemblies. Recognition may be facilitated by β-character in the central hydrophobic region of the full-length peptides and the presence of a turn and/or salt bridge around residues 23–28, common features of the fibrils these compounds are designed to bind [65] which have recently been discovered in simulations of Aβ monomers and small oligomers [66–69].
Although the most effective inhibitors in each sample environment afford a 5- to 8-fold increase in the percentage of FAβB monomers over that observed for inhibitor-free samples, the efficacies of individual compounds vary significantly in acidic versus metal-containing environments (Figures 3 and 4). These variations highlight the most successful strategies for inhibition of acid- versus zinc-promoted oligomerization and inform on the nature of acidic versus zinc-containing oligomers. We recognize that our results for FAβB do not specifically inform on association of native Aβ40. However, they should afford a few insights regarding the inhibition of oligomerization in these important sample environments, given that fluorophore attachment at the N-terminus has been shown to preserve the biological activity of Aβ, including fibril morphology [57], binding to senile plaques [70], neurotoxicity [71] and membrane permeability [57].
Under acidic conditions, the two most successful oligomerization inhibitors possess similar structural motifs and inhibition strategies. AMY-1 (Figure 1C) and Aβ16–22m (Figure 1D) differ significantly in terms of net charge and hydrophobic/hydrophilic character, as the latter lacks the hexalysine disrupting element; however, both peptides adopt β-strand secondary structure [17, 20] (confirmed at pH 5.8; data not shown), unlike the less effective iAβ5 and KLVFF-K6 [42] compounds. In addition, once bound, AMY-1 and Aβ16–22m both use substituents on one face of the peptide to block further association: the bulky α,α-groups in AMY-1 sterically prevent the close approach of neighboring chains, while the N-methyl groups in Aβ16–22m prevent hydrogen bonding. These commonalities in structure and design may be key factors for successful inhibition of the earliest association steps under acidic conditions. Interestingly, molecular dynamics simulations on Aβ peptides at pH 6 showed that acidic conditions cause increased β-character and increased solvent accessibility in the central hydrophobic region [72], which may lead to improved Aβ recognition by β-structured inhibitors such as AMY-1 and Aβ16–22m.
In the presence of zinc at physiological pH, the two most effective peptide inhibitors, Aβ16–22m and KLVFF-K6, exhibit equal capability to prevent or reverse the earliest Aβ association steps (Figure 4B), but share few other commonalities. Aβ16–22m is β-structured [17] and primarily hydrophobic, with a net charge of +1; in contrast, KLVFF-K6 is unstructured [42] and highly charged (net +8), with distinct hydrophobic and hydrophilic regions (Figure 1).Note 2 In terms of inhibition strategy, while Aβ16–22m utilizes its N-methyl groups to prevent association on one face of the peptide, as discussed above, KLVFF-K6 uses a lysine hexamer to disrupt association electrostatically. Cairo et al. proposed that electrostatic interactions between these lysine residues and the Glu-22 and Asp-23 residues of Aβ also stabilize inhibitor binding, permitting superior recognition of small Aβ oligomers at physiological pH [73]. In the current studies, the increased positive charge on oligomers containing bound zinc ions may further improve the disruption capability of the oligolysine unit.
The observation that three very different inhibitors – Aβ16–22m, KLVFF-K6, and clioquinol (studied previously [50]) – are equally successful in combating the first stages of zinc-promoted association suggests that zinc-containing oligomers may be more weakly associated and/or more structurally diverse than those formed at pH 5.8. The same is apparently true for assemblies containing tens of associated peptides: whereas none of the four inhibitors successfully reduce the percentage of larger oligomers (n > 5) at pH 5.8, all of them apparently decrease the prevalence of these species in zinc-containing samples. These findings are also consistent with reports from several groups, which suggest that Aβ oligomers containing zinc appear looser and are more easily dissociated than those without coordinated metal ions [47, 74, 75].Note 3
Despite these apparent variations in oligomer structure, the efficacies of two of the four inhibitors are basically unaffected by the different environments. We attribute the limited efficacy of iAβ5 to less successful recognition of FAβB oligomers and suggest that its “β-sheet-breaker” inhibition strategy is better suited to targeting fibrils with well-defined cross-β structure, as opposed to the early oligomers investigated here. This inference is qualitatively consistent with previous in vitro studies of Aβ oligomers: Hoshi et al. reported that iAβ5 does not inhibit formation of spherical oligomers [76], while Cairo et al. found that it does not bind to the small Aβ(10–35) assemblies used in their SPR studies [73].
In contrast to iAβ5, Aβ16–22m exhibits superior efficacy against the earliest assemblies in both acidic and zinc-containing samples, due to highly successful recognition of full-length peptides in both environments. (Affinity is somewhat greater for acidic versus zinc-containing species, as evidenced by the concentration-independent inhibition observed at pH 5.8.) Molecular dynamics simulations of Aβ16–22m and Aβ16–20m (in which Leu-17 and Phe-19 are N-methylated) revealed that the extended, β-strand conformations of the inhibitors greatly facilitated binding to small, β-structured Aβ16–22 assemblies, due to reduced entropic costs [77, 78]. The same reasoning likely applies here, since acidic conditions (pH 6) [72] and zinc binding [79] are reported to increase the β-character in the central hydrophobic (recognition) region of the full-length peptide. Simulations also suggest that the inhibitor's rather rigid, extended structure permits it to intercalate between peptide strands (in addition to binding modes preventing lateral and longitudinal association), facilitating the removal of associated peptides [77, 78].
CONCLUSIONS
In summary, we have evaluated four peptide inhibitors of Aβ fibrillogenesis based on their abilities to inhibit acid- and zinc-promoted oligomerization physiologically relevant conditions. Single-oligomer fluorescence measurements on FAβB species revealed that all four inhibitors successfully combat the earliest association steps in both acidic and zinc-containing environments. The most successful inhibitors at pH 5.8, AMY-1 and Aβ16–22m, are β-structured, with blocking substituents on one face of the peptide. In the presence of zinc, the disparate inhibition strategies employed by Aβ16–22m, KLVFF-K6 and the zinc chelator clioquinol are equally effective, suggesting that zinc-containing oligomers may be looser and/or more structurally diverse. The N-methylated inhibitor Aβ16–22m is most successful across both sample environments, likely due to its rather rigid, extended conformation, which is expected to facilitate recognition and permit intercalation between associated peptides.
ACKNOWLEDGEMENTS
We gratefully acknowledge support from the National Science Foundation (NSF CHE-0848824) and the National Center for Research Resources (NIH P20 RR-016461), in addition to the Winthrop University Research Council. We also thank Dr. Robert Hammer for providing three of the inhibitors studied.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The number of observed dyes corresponds directly to the number of singly labeled Aβ peptides in the excited region. As we discussed in ref. 49, under our conditions (i.e., ~1010 streptavidin per square centimeter of coverslip surface, ~0.01% occupation of streptavidin binding sites by FAβB), we can reasonably assert that the observation of multiple dye-labeled peptides at a single bright spot in a fluorescent image is indicative of oligomerization, rather than binding of several monomers within the focal volume.
Circular dichroism spectra of AMY-1 and KLVFF-K6 in acidic and zinc-containing environments match those reported for metal-free samples at physiological pH (data not shown).
In light of the rather broad success for inhibition of zinc-promoted oligomerization, it is perhaps surprising that AMY-1 demonstrates only mild efficacy in this environment, significantly lower than that exhibited at pH 5.8. Given the superior effectiveness of Aβ16–22m, which has similar secondary structure and inhibition strategy, and KLVFF-K6, which contains the same hexalysine disrupting element, we speculate that AMY-1's bulky α-substituents hinder the approach of AMY-1 to FAβB oligomers in the presence of zinc more so than they do at acidic pH.
REFERENCES
- 1.Kang J, Lemaire HG, Unterbeck A, Salbaum JM, Masters CL, Grzeschik KH, Multhaup G, Beyreuther K, Muller-Hill B. The precursor of Alzheimer's disease amyloid A4 protein resembles a cell-surface receptor. Nature. 1987;325:733–736. doi: 10.1038/325733a0. [DOI] [PubMed] [Google Scholar]
- 2.Seubert P, Vigo-Pelfrey C, Esch F, Lee M, Dovey H, Davis D, Sinha S, Schlossmacher MG, Whaley J, Swindlehurst C, McCormack R, Wolfert R, Selkoe DJ, Lieberburg I, Schenk D. Isolation and quantitation of soluble Alzheimer's β-peptide from biological fluids. Nature. 1992;359:325–327. doi: 10.1038/359325a0. [DOI] [PubMed] [Google Scholar]
- 3.Giuffrida ML, Caraci F, Pignataro B, Cataldo S, De Bona P, Bruno V, Molinaro G, Pappalardo G, Messina A, Palmigiano A, Garozzo D, Nicoletti F, Rizzarelli E, Copani A. The monomer state of beta-amyloid: where the Alzhiemer's disease protein meets physiology. J. Neurosci. 2009;29:10582–10587. doi: 10.1523/JNEUROSCI.1736-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lazo ND, Maji SK, Fradinger EA, Bitan G, Teplow DB. In: Amyloid proteins: The beta-sheet conformation and disease. Sipe J, editor. vol. 2. Wiley-VCH; Weinheim, Germany: 2005. pp. 385–491. [Google Scholar]
- 5.Eanes ED, Glenner GG. X-ray diffraction studies on amyloid filaments. J. Histochem. Cytochem. 1968;16:673–677. doi: 10.1177/16.11.673. [DOI] [PubMed] [Google Scholar]
- 6.Bonar L, Cohen AS, Skinner MM. Characterization of the amyloid fibril as a cross-β protein. Proc. Soc. Exp. Biol. Med. 1969;131:1373–1375. doi: 10.3181/00379727-131-34110. [DOI] [PubMed] [Google Scholar]
- 7.Glenner GG, Wong CW. Alzheimer disease and Down's syndrome: sharing of a unique cerebrovascular amyloid fibril protein. Biochem. Biophys. Res. Commun. 1984;122:1131–1135. doi: 10.1016/0006-291x(84)91209-9. [DOI] [PubMed] [Google Scholar]
- 8.Masters CL, Simms G, Weiman NA, Multhaup G, McDonald BL, Beyreuther K. Amyloid plaque core protein in Alzheimer disease and Down syndrome. Proc. Natl. Acad. Sci. USA. 1985;82:4245–4249. doi: 10.1073/pnas.82.12.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hardy JA, Higgins GA. Alzheimer's disease: the amyloid cascade hypothesis. Science. 1992;256:184–185. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- 10.DaSilva KA, Shaw JE, McLaurin J. Amyloid-β fibrillogenesis: structural insight and therapeutic intervention. Exp. Neurol. 2010;223:311–321. doi: 10.1016/j.expneurol.2009.08.032. [DOI] [PubMed] [Google Scholar]
- 11.LeVine H., III Small molecule inhibitors of Abeta assembly. Amyloid. 2007;14:185–197. doi: 10.1080/13506120701461020. [DOI] [PubMed] [Google Scholar]
- 12.Sciarretta KL, Gordon DJ, Meredith SC. Amyloid, prions, and other protein aggregates, Part C. In: Kheterpal I, Wetzel R, editors. Methods in enzymology. vol. 413. Academic; New York: 2006. pp. 273–312. [Google Scholar]
- 13.Burkoth TS, Benzinger TLS, Jones DNM, Hallenga K, Meredith SC, Lynn DG. C-terminal PEG blocks the irreversible step in β-amyloid(10–35) fibrillogenesis. J. Am. Chem. Soc. 1998;120:7655–7656. [Google Scholar]
- 14.Pallitto MM, Ghanta J, Heinzelmann P, Kiessling LL, Murphy RM. Recognition sequence design for peptidyl modulators of β-amyloid aggregation and toxicity. Biochemistry. 1999;38:3570–3578. doi: 10.1021/bi982119e. [DOI] [PubMed] [Google Scholar]
- 15.Soto C, Sigurdsson EM, Morelli L, Kumar RA, Castano EM, Frangione B. β-sheet breaker peptides inhibit fibrillogenesis in a rat brain model of amyloidosis: implications for Alzheimer's therapy. Nat. Med. 1998;4:822–826. doi: 10.1038/nm0798-822. [DOI] [PubMed] [Google Scholar]
- 16.Permanne B, Adessi C, Fraga S, Frossard MJ, Saborio GP, Soto CJ. Are β-sheet breaker peptides dissolving the therapeutic problem of Alzheimer's disease? J. Neural Transm. Suppl. 2002:293–301. doi: 10.1007/978-3-7091-6139-5_27. [DOI] [PubMed] [Google Scholar]
- 17.Gordon DJ, Sciarretta KL, Meredith SC. Inhibition of β-amyloid(40) fibrillogenesis and disassembly of β-amyloid(40) fibrils by short β-amyloid congeners containing N-methyl amino acids at alternate residues. Biochemistry. 2001;40:8237–8245. doi: 10.1021/bi002416v. [DOI] [PubMed] [Google Scholar]
- 18.Gordon DJ, Tappe R, Meredith SC. Design and characterization of a membrane permeable N-methyl amino acid-containing peptide that inhibits Aβ1–40 fibrillogenesis. J. Pept. Res. 2002;60:37–55. doi: 10.1034/j.1399-3011.2002.11002.x. [DOI] [PubMed] [Google Scholar]
- 19.Kokkoni N, Stott K, Amijee H, Mason JM, Doig AJ. N-Methylated peptide inhibitors of β-amyloid aggregation and toxicity: optimization of the inhibitor structure. Biochemistry. 2006;45:9906–9918. doi: 10.1021/bi060837s. [DOI] [PubMed] [Google Scholar]
- 20.Etienne MA, Aucoin JP, Fu Y, McCarley RL, Hammer RP. Stoichiometric inhibition of amyloid β-protein aggregation with peptides containing alternating α,α-disubstituted amino acids. J. Am. Chem. Soc. 2006;128:3522–3523. doi: 10.1021/ja0600678. [DOI] [PubMed] [Google Scholar]
- 21.Tjernberg LO, Lilliiehook C, Callaway DJE, Naslund J, Hahne S, Thyberg J, Terenius L, Nordstedt C. Controlling amyloid β-peptide fibril formation with protease-stable ligands. J. Biol. Chem. 1997;272:12601–12605. doi: 10.1074/jbc.272.19.12601. [DOI] [PubMed] [Google Scholar]
- 22.Chalifour RJ, McLaughlin RW, Lavoie L, Morissette C, Tremblay N, Boule M, Sarazin P, Stea D, Lacombe D, Tremblay P, Gervais FJ. Stereoselective interactions of peptide inhibitors with the β-amyloid peptide. J. Biol. Chem. 2003;278:34874–34881. doi: 10.1074/jbc.M212694200. [DOI] [PubMed] [Google Scholar]
- 23.Walsh DM, Selkoe DJ. Aβ oligomers – a decade of discovery. J. Neurochem. 2007;101:1172–1184. doi: 10.1111/j.1471-4159.2006.04426.x. [DOI] [PubMed] [Google Scholar]
- 24.McLean CA, Cherny RA, Fraser FW, Fuller SJ, Smith MJ, Beyreuther K, Bush AI, Masters CL. Soluble pool of Aβ amyloid as a determinant of severity of neurodegeneration in Alzheimer's disease. Ann. Neurol. 1999;46:860–866. doi: 10.1002/1531-8249(199912)46:6<860::aid-ana8>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 25.Lue LF, Kuo YM, Roher AE, Brachova L, Shen Y, Sue L, Beach T, Kurth JH, Rydel RE, Rogers J. Soluble amyloid β peptide concentration as a predictor of synaptic change in Alzheimer's disease. Am. J. Pathol. 1999;155:853–862. doi: 10.1016/s0002-9440(10)65184-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roher AE, Chaney MO, Kuo Y-M, Webster SD, Stine WB, Haverkamp LJ, Woods AS, Cotter RJ, Tuohy JM, Krafft GA, Bonnell BS, Emmerling MR. Morphology and toxicity of Abeta(1–42) dimer derived from neuritic and vascular amyloid deposits of Alzheimer's disease. J. Biol. Chem. 1996;271:20631–20635. doi: 10.1074/jbc.271.34.20631. [DOI] [PubMed] [Google Scholar]
- 27.Enya M, Morishima-Kawashima M, Yoshimura M, Shinkai Y, Kusui K, Khan K, Games D, Schenk D, Sugihara S, Yamaguchi H, Ihara Y. Appearance of sodium dodecyl sulfate-stable amyloid beta-protein (Abeta) dimer in the cortex during aging. Am. J. Pathol. 1999;154:271–279. doi: 10.1016/s0002-9440(10)65273-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Terry RD, Masliah E, Salmon DP, Butters N, DeTeresa R, Hill R, Hansen LA, Katzman R. Physical basis of cognitive alterations in Alzheimer's disease: synapse loss is the major correlate of cognitive impairment. Ann. Neurol. 1991;30:572–580. doi: 10.1002/ana.410300410. [DOI] [PubMed] [Google Scholar]
- 29.Walsh D, Klyubin I, Fadeeva J, William K, Cullen W, Anwyl R, Wolfe M, Rowan M, Selkoe D. Naturally secreted oligomers of the Alzheimer amyloid beta-protein potently inhibit hippocampal long-term potentiation in vivo. Nature. 2002;416:535–539. doi: 10.1038/416535a. [DOI] [PubMed] [Google Scholar]
- 30.Walsh DM, Townsend M, Podlisny MB, Shankar GM, Fadeeva JV, Agnaf OE, Hartley DM, Selkoe DJ. Certain inhibitors of synthetic amyloid beta-peptide (Abeta) fibrillogenesis block oligomerization of natural Abeta and thereby rescue long-term potentiation. J. Neurosci. 2005;25:2455–2462. doi: 10.1523/JNEUROSCI.4391-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Townsend M, Shankar GM, Mehta T, Walsh DM, Selkoe DJ. Effects of secreted oligomers of amyloid β-protein on hippocampal synaptic plasticity: a potent role for trimers. J. Physiol. 2006;572:477–492. doi: 10.1113/jphysiol.2005.103754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cleary JP, Walsh DM, Hofmeister JJ, Shankar GM, Kuskowski MA, Selkoe DJ, Ashe KH. Natural oligomers of the amyloid-beta protein specifically disrupt cognitive function. Nat. Neurosci. 2005;8:79–84. doi: 10.1038/nn1372. [DOI] [PubMed] [Google Scholar]
- 33.Shankar GM, Bloodgood BL, Townsend M, Walsh DM, Selkoe DJ, Sabatini BL. Natural oligomers of the Alzheimer amyloid beta-protein induce reversible synapse loss by modulating an NMDA-type dlutamate receptor-dependent signaling pathway. J. Neurosci. 2007;27:2866–2875. doi: 10.1523/JNEUROSCI.4970-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shankar GM, Li S, Mehta TH, Garcia-Munoz A, Shepardson NE, Smith I, Brett FM, Farrell MA, Rowan MJ, Lemere CA, Regan CM, Walsh DM, Sabatini BL, Selkoe DJ. Amyloid-β protein dimers isolated directly from Alzheimer's brains impair synaptic plasticity and memory. Nat. Med. 2008;14:837–842. doi: 10.1038/nm1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klyubin I, Betts V, Welzel AT, Blennow K, Zetterberg H, Wallin A, Lemere CA, Cullen WK, Peng Y, Wisniewski T, Selkoe DJ, Anwyl R, Walsh DM, Rowan MJ. Amyloid β protein dimer-containing human CSF disrupts synaptic plasticity: prevention by systemic passive immunization. J. Neurosci. 2008;28:4231–4237. doi: 10.1523/JNEUROSCI.5161-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Necula M, Kayed R, Milton S, Glabe CG. Small molecule inhibitors of aggregation indicate that amyloid β oligomerization and fibrillization pathways are independent and distinct. J. Biol. Chem. 2007;282:10311–10324. doi: 10.1074/jbc.M608207200. [DOI] [PubMed] [Google Scholar]
- 37.Hilbich C, Kisters-Woike B, Masters CL, Beyreuther K. Substitutions of hydrophobic amino acids reduce the amyloidogenicity of Alzheimer's disease βA4 peptides. J. Mol. Biol. 1992;228:460–473. doi: 10.1016/0022-2836(92)90835-8. [DOI] [PubMed] [Google Scholar]
- 38.Wood SJ, Wetzel R, Martin JD, Hurle MR. Prolines and amyloidogenicity in fragments of the Alzheimer's peptide β/A4. Biochemistry. 1995;34:724–730. doi: 10.1021/bi00003a003. [DOI] [PubMed] [Google Scholar]
- 39.Esler WP, Stimson ER, Ghilardi JR, Lu YA, Felix AM, Vinters HV, Mantyh PW, Lee JP, Maggio JE. Point substitution in the central hydrophobic cluster of a human beta-amyloid congener disrupts peptide folding and abolishes plaque competence. Biochemistry. 1996;35:13914–13921. doi: 10.1021/bi961302+. [DOI] [PubMed] [Google Scholar]
- 40.Tjernberg LO, Naslund J, Lindquist F, Johansson J, Karlstrom AR, Thyberg J, Terenius L, Nordstedt C. Arrest of β-amyloid fibril formation by a pentapeptide ligand. J. Biol. Chem. 1996;271:8545–8549. doi: 10.1074/jbc.271.15.8545. [DOI] [PubMed] [Google Scholar]
- 41.Lowe TL, Strzelec A, Kiessling LL, Murphy RM. Structure-function relationships for inhibitors of β-amyloid toxicity containing the recognition sequence KLVFF. Biochemistry. 2001;40:7882–7889. doi: 10.1021/bi002734u. [DOI] [PubMed] [Google Scholar]
- 42.Kim JR, Murphy RM. Mechanism of accelerated assembly of β-amyloid filaments into fibrils by KLVFFK6. Biophys. J. 2004;86:3194–3203. doi: 10.1016/S0006-3495(04)74367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moss MA, Nichols MR, Reed DK, Hoh JH, Rosenberry TL. The peptide KLVFF-K6 promotes β-amyloid(1–40) protofibril growth by association but does not alter protofibril effects on cellular reduction of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) Mol. Pharmacol. 2003;64:1160–1168. doi: 10.1124/mol.64.5.1160. [DOI] [PubMed] [Google Scholar]
- 44.Barrow CJ, Zagorski MG. Solution structures of β peptide and its constituent fragments: relation to amyloid deposition. Science. 1991;253:179–182. doi: 10.1126/science.1853202. [DOI] [PubMed] [Google Scholar]
- 45.Snyder SW, Ladror US, Wade WS, Wang GT, Barrett LW, Matayoshi ED, Huffaker HJ, Krafft GA, Holzman TF. Amyloid-β aggregation: selective inhibition of aggregation in mixtures of amyloid with different chain lengths. Biophys. J. 1994;67:1216–1228. doi: 10.1016/S0006-3495(94)80591-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wood SJ, Maleeff B, Hart T, Wetzel R. Physical, morphological, and functional differences between pH 5.8 and 7.4 aggregates of the Alzheimer's amyloid peptide Aβ. J. Mol. Biol. 1996;256:870–877. doi: 10.1006/jmbi.1996.0133. [DOI] [PubMed] [Google Scholar]
- 47.Klug GMJA, Losic D, Subasinghe SS, Aguilar M-I, Martin LL, Small DH. β-amyloid protein oligomers induced by metal ions and acid pH are distinct from those generated by slow spontaneous aging at neutral pH. Eur. J. Biochem. 2003;270:4282–4293. doi: 10.1046/j.1432-1033.2003.03815.x. [DOI] [PubMed] [Google Scholar]
- 48.Mantyh PW, Ghilardi JR, Rogers S, DeMaster E, Allen CJ, Stimson ER, Maggio JE. Aluminum, iron, and zinc ions promote aggregation of physiological concentrations of β-amyloid peptide. J. Neurochem. 1993;61:1171–1174. doi: 10.1111/j.1471-4159.1993.tb03639.x. [DOI] [PubMed] [Google Scholar]
- 49.Bush AI, Pettingell WH, Multhaup G, d'Paradis M, Vonsattel JP, Gusella JF, Beyreuther K, Masters CL, Tanzi RE. Rapid induction of Alzheimer A beta amyloid formation by zinc. Science. 1994;265:1464–1467. doi: 10.1126/science.8073293. [DOI] [PubMed] [Google Scholar]
- 50.Dukes KD, Rodenberg CF, Lammi RK. Monitoring the earliest amyloid-β oligomers via quantized photobleaching of dye-labeled peptides. Anal. Biochem. 2008;382:29–34. doi: 10.1016/j.ab.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 51.Kaether C, Schmitt S, Willem M, Haass C. Amyloid precursor protein and notch intracellular domains are generated after transport of their precursors to the cell surface. Traffic. 2006;7:408–415. doi: 10.1111/j.1600-0854.2006.00396.x. [DOI] [PubMed] [Google Scholar]
- 52.Rajendran L, Honsho M, Zahn TR, Keller P, Geiger KD, Verkade P, Simons K. Alzheimer's disease β-amyloid peptides are released in association with exosomes. Proc. Natl. Acad. Sci. USA. 2006;103:11172–11177. doi: 10.1073/pnas.0603838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Molina JA, Jimenez-Jimenez FJ, Aguilar MV, Meseguer I, Matos-Vega CJ, Gonzalez-Munoz MJ, de Bustos F, Porta J, Orti-Pareja M, Zurdo M, Barrios E, Martinez-Para MC. Cerebrospinal fluid levels of transition metals in patients with Alzheimer's disease. J. Neural Transm. 1998;105:479–488. doi: 10.1007/s007020050071. [DOI] [PubMed] [Google Scholar]
- 54.Assaf SY, Chung SH. Release of endogenous zinc(2+) from brain tissue during activity. Nature. 1984;308:734–736. doi: 10.1038/308734a0. [DOI] [PubMed] [Google Scholar]
- 55.Howell GA, Welch M, Frederickson CJ. Stimulation-induced uptake and release of zinc in hippocampal slices. Nature. 1984;308:736–738. doi: 10.1038/308736a0. [DOI] [PubMed] [Google Scholar]
- 56.Deshpande A, Kawai H, Metherate R, Glabe CG, Busciglio J. A role for synaptic zinc in activity-dependent Aβ oligomer formation and accumulation at excitatory synapses. J. Neurosci. 2009;29:4004–4015. doi: 10.1523/JNEUROSCI.5980-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ding H, Wong PT, Lee EL, Gafni A, Steel DG. Determination of the oligomer size of amyloidogenic protein β-amyloid(1–40) by single-molecule spectroscopy. Biophys. J. 2009;97:912–921. doi: 10.1016/j.bpj.2009.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shoji M, Gold TE, Ghiso J, Cheung TT, Estus S, Shaffer LM, Cai XD, McKay DM, Tintner R, Frangione B, Younkin SG. Production of the Alzheimer amyloid β protein by normal proteolytic processing. Science. 1992;258:126–129. doi: 10.1126/science.1439760. [DOI] [PubMed] [Google Scholar]
- 59.Guo A, Zhu X-Y. Functional protein microarrays in drug discovery. In: Predki P, editor. Drug discovery series. vol. 8. CRC Press; Boca Raton, FL: 2007. pp. 53–71. [Google Scholar]
- 60.Abe K, Kaya S, Hayashi Y, Imagawa T, Kikumoto M, Oiwa K, Katoh T, Yazawa M, Taniguchi K. Correlation between the activities and the oligomeric forms of pig gastric H/K-ATPase. Biochemistry. 2003;42:15132–15138. doi: 10.1021/bi035686x. [DOI] [PubMed] [Google Scholar]
- 61.Guo A, Microsurfaces, Inc. Personal communication. [Google Scholar]
- 62.Ha T, Enderle T, Chemla DS, Selvin PR, Weiss S. Quantum jumps of single molecules at room temperature. Chem. Phys. Lett. 1997;271:1–5. [Google Scholar]
- 63.Xie SX, Trautman JK. Optical studies of single molecules at room temperature. Annu. Rev. Phys. Chem. 1998;49:441–480. doi: 10.1146/annurev.physchem.49.1.441. [DOI] [PubMed] [Google Scholar]
- 64.Bitan G, Kirkitadze MD, Lomakin A, Vollers SS, Benedek GB, Teplow DB. Amyloid β-protein (Aβ) assembly: Aβ40 and Aβ42 oligomerize through distinct pathways. Proc. Natl. Acad. Sci. USA. 2003;100:330–335. doi: 10.1073/pnas.222681699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Petkova AT, Ishii Y, Balbach JJ, Antzutkin ON, Leapman RD, Delaglio F, Tycko R. A structural model for Alzheimer's β-amyloid fibrils based on experimental constraints from solid state NMR. Proc. Natl. Acad. Sci. USA. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kent A, Jha AK, Fitzgerald JE, Freed KF. Benchmarking implicit solvent folding simulations of the amyloid β(10–35) fragment. J. Phys. Chem. B. 2008;112:6175–6186. doi: 10.1021/jp077099h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Urbanc B, Cruz L, Ding F, Sammond D, Khare S, Buldyrev SV, Stanley HE, Dokholyan NV. Molecular dynamics simulation of amyloid-β dimer formation. Biophys. J. 2004;87:2310–2321. doi: 10.1529/biophysj.104.040980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jang S, Shin S. Computational study on the structural diversity of amyloid beta peptide (Aβ10–35) oligomers. J. Phys. Chem. B. 2008;112:3479–3484. doi: 10.1021/jp076450w. [DOI] [PubMed] [Google Scholar]
- 69.Dulin F, Callebaut I, Colloc'h N, Mornon J-P. Sequence-based modeling of Aβ42 soluble oligomers. Biopolymers. 2007;85:422–437. doi: 10.1002/bip.20675. [DOI] [PubMed] [Google Scholar]
- 70.Prior R, D'Urso D, Frank R, Prikulis I, Cleven S, Ihl R, Pavlakovic G. Selective binding of soluble Abeta1–40 and Abeta1–42 to a subset of senile plaques. Am. J. Pathol. 1996;148:1749–1756. [PMC free article] [PubMed] [Google Scholar]
- 71.Fulop L, Penke B, Zarandi M. Synthesis and fluorescent labeling of beta-amyloid peptides. J. Pept. Sci. 2001;7:397–401. doi: 10.1002/psc.346. [DOI] [PubMed] [Google Scholar]
- 72.Khandogin J, Brooks CL., III Linking folding with aggregation in Alzheimer's β-amyloid peptides. Proc. Natl. Acad. Sci. USA. 2007;104:16880–16885. doi: 10.1073/pnas.0703832104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cairo CW, Strzelec A, Murphy RM, Kiessling LK. Affinity-based inhibition of β-amyloid toxicity. Biochemistry. 2002;41:8620–8629. doi: 10.1021/bi0156254. [DOI] [PubMed] [Google Scholar]
- 74.Hu W-P, Chang G-L, Chen S-J, Kuo Y-M. Kinetic analysis of β-amyloid peptide aggregation induced by metal ions based on surface plasmon resonance biosensing. J. Neurosci. Methods. 2006;154:190–197. doi: 10.1016/j.jneumeth.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 75.Garai K, Sahoo B, Kaushalya K, Desai R, Maiti S. Zinc lowers amyloid-β toxicity by selectively precipitating aggregation intermediates. Biochemistry. 2007;46:10655–10663. doi: 10.1021/bi700798b. [DOI] [PubMed] [Google Scholar]
- 76.Hoshi M, Sato M, Matsumoto S, Noguchi A, Yasutake K, Yoshida N, Sato K. Spheroidal aggregates of β-amyloid (amylospheroid) show high neurotoxicity and activate tau protein kinase I/glycogen synthase kinase-3β. Proc. Natl. Acad. Sci. USA. 2003;100:6370–6375. doi: 10.1073/pnas.1237107100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li W, Zhang J, Su Y, Wang J, Qin M, Wang W. Effects of zinc binding on the conformational distribution of the amyloid-β peptide based on molecular dynamics simulations. J. Phys. Chem. B. 2007;111:13814–13821. doi: 10.1021/jp076213t. [DOI] [PubMed] [Google Scholar]
- 78.Chebaro Y, Derreumaux P. Targeting the early steps of Aβ16–22 protofibril disassembly by N-methylated inhibitors: A numerical study. Proteins. 2009;75:442–452. doi: 10.1002/prot.22254. [DOI] [PubMed] [Google Scholar]
- 79.Soto P, Griffin MA, Shea J-E. New insights into the mechanism of Alzheimer amyloid-β fibrillogenesis inhibition by N-methylated peptides. Biophys. J. 2007;93:3015–3025. doi: 10.1529/biophysj.107.112086. [DOI] [PMC free article] [PubMed] [Google Scholar]