Abstract
In contrast to elongation factor EF-Tu, which delivers aminoacyl-tRNAs to the ribosomal A-site, eukaryotic initiation factor eIF2 binds initiator Met-tRNAiMet to the P-site of the 40S ribosomal subunit. We used directed hydroxyl radical probing experiments to map the binding of Saccharomyces cerevisiae eIF2 on the ribosome and on Met-tRNAiMet. Our results identify a key binding-interface between domain III of eIF2γ and 18S rRNA helix h44 on the 40S subunit. Moreover, we showed that eIF2γ primarily contacts the acceptor stem of Met-tRNAiMet. Whereas the analogous domain III of EF-Tu contacts the T-stem of tRNAs, biochemical analyses demonstrated that eIF2γ domain III is important for ribosome, but not Met-tRNAiMet, binding. Thus despite their structural similarity, eIF2 and EF-Tu bind tRNAs in substantially different manners, and we propose that the tRNA-binding domain III of EF-Tu has acquired a new ribosome-binding function in eIF2γ.
Introduction
To facilitate translation elongation the GTPase EF-Tu in bacteria (eEF1A in eukaryotes) forms a ternary complex (TC) with GTP and an aminoacyl-tRNA, and then binds the aminoacyl-tRNA to the A-site of the ribosome in a codon-dependent manner. Recent x-ray crystallographic studies have provided a high resolution image of the EF-Tu–GTP–aminoacyl-tRNA–70S ribosome complex and revealed contacts between EF-Tu and rRNA elements near the A-site of the ribosome1,2. Functioning in an analogous manner to EF-Tu, the eukaryote-specific translation initiation factor eIF2 forms a TC with GTP and the specific initiator Met-tRNAiMet. However, in contrast to EF-Tu, which interacts with 70S elongating ribosomes, eIF2 binds Met-tRNAiMet to the small (40S) ribosomal subunit in eukaryotic cells to help form the 43S preinitiation complex (PIC) (reviewed in3). Binding of the factors eIF1 and eIF1A to the 40S subunit enhances TC binding, and creates an open, scanning-competent complex4–6. The resulting PIC binds an mRNA near the 5′ cap and then this 48S PIC scans the mRNA in search of a start codon. Base-pairing between the anticodon of the Met-tRNAiMet and a start codon in the mRNA triggers reconfiguration of the PIC from the open to a closed, scanning-arrested state. This open to closed transition is accompanied by movement of eIF1 away from eIF1A, completion of GTP hydrolysis, and release of Pi7. Joining of the 60S subunit yields an 80S ribosome with Met-tRNAiMet in the P-site base-paired with the start codon on the mRNA.
Previous x-ray crystallography and structural probing studies have provided insights into the ribosomal binding sites of several of the PIC factors. Directed hydroxyl radical mapping using factor tethered Fe(II) placed the core of eIF1A in the A-site of the 40S subunit8. This location is consistent with the x-ray structure of the analogous bacterial initiation complex which showed IF1 binding to the A-site of the 30S subunit9. Interestingly, the N-terminal and C-terminal tails of eIF1A extend from the A-site toward and under the P-site bound Met-tRNAiMet (ref. 8). Whereas the N-terminal tail does not contact the P-site, the C-terminal tail of eIF1A contacts the P-site and is proposed to interfere with Met-tRNAiMet fully entering the P-site (Pout state) in the open scanning complex8,10. Meanwhile, eIF1, which plays a critical role in start codon selection, binds near the P-site in a location where it might monitor codon-anticodon interactions that govern start site selection5. Upon start codon recognition and conversion of the PIC to the closed scanning arrested state, Met-tRNAiMet fully docks in the P-site (Pin state) displacing both the eIF1A C-terminal tail and eIF18,10. While eIF2 is known to bind Met-tRNAiMet, which is bound to the P-site of PICs, the binding site of eIF2 on the ribosome has not been determined.
The γ subunit of the heterotrimeric eIF2 complex shares significant amino acid sequence and structural similarity with EF-Tu (Fig. 1, and Supplementary Fig. 1 online). Whereas no structures have been obtained for eIF2γ, x-ray crystal structures have been obtained for the corresponding archaeal aIF2γ protein both free11,12 and in complex with full-length or truncated versions of aIF2α and aIF2β13–15,16 (Fig. 2b). Both aIF2γ and EF-Tu consist of three domains: an N-terminal GTP-binding domain, and β-barrel domains II and III (Fig. 1, and Supplementary Fig. 1 online). Despite the structural similarity, it is anticipated that eIF2γ and EF-Tu will have different docking arrangements on the A-versus P-sites of the ribosome, and this might lead to differences in how the two factors bind aminoacyl-tRNAs. Moreover, despite the wealth of structural information on aIF2 complexes, the ribosome-contacting surfaces aIF2 and eIF2 have not been identified. To gain insights into how eIF2 binds Met-tRNAiMet and then associates with the ribosome, we used directed hydroxyl radical probing to identify eIF2 contacts within the 48S PIC.
Figure 1.
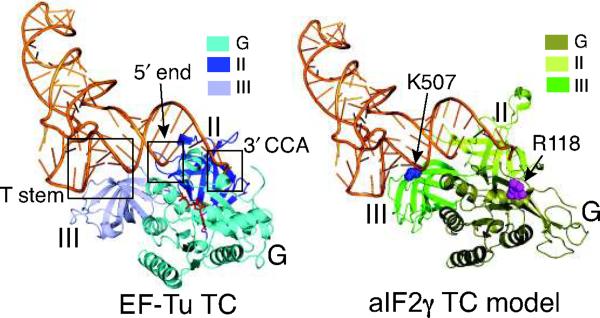
Structures of EF-Tu TC and aIF2γ TC model.
Ribbons representation of EF-Tu–GDPNP–Phe-tRNAPhe TC (pdb code: 1TTT18, left). The aIF2γ structure (pdb code: 2AHO15) was aligned to the EF-Tu structure using PyMOL software (DeLano Scientific) to make an aIF2γ–GDPNP–Phe-tRNAPhe TC model (right). Three interaction points between EF-Tu and Phe-tRNAPhe (T stem, 5′ end, and 3′CCA18) are boxed and labeled. The aIF2γ residues corresponding to residues K507 and R118 in S cerevisiae eIF2γ, which were used to tether Fe(II)-BABE for hydroxyl radical cleavage of tRNAiMet (see Fig. 3), are shown as blue and magenta spheres, respectively. GTP binding domain (G), domain II (II), and domain III (III) of both EF-Tu and aIF2γ are depicted using different shades of blue and green, respectively, as indicated in the inset beside each figure, and Phe-tRNAPhe is shown in orange. The same color schemes for EF-Tu, aIF2γ, and Phe-tRNAPhe are used in all figures.
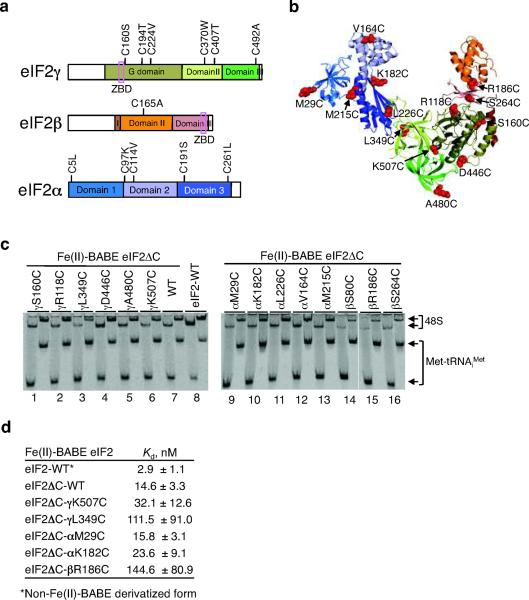
Figure 2.
Construction and analysis of eIF2 Cys mutants.
(a) Removal of native Cys residues to generate Cys-lite eIF2ΔC. The designated Cys residues were mutated as indicated; note that Cys residues in the zinc binding domains (ZBD, pink box) of eIF2β and eIF2γ were left intact.
(b) Ribbon diagram of aIF2 showing sites of single Cys mutations (red spheres) in yeast eIF2ΔC. The aIF2 structure was assembled by aligning the structures of aIF2αγ (pdb code: 2AHO15) and aIF2γβ (pdb code: 2QMU16). The indicated mutations were introduced individually into S. cerevisiae eIF2 subunits; the S80C mutation in the eukaryote-specific N-terminal domain of eIF2β is not depicted because this element is not found in aIF2β. aIF2 subunit domains are colored as in panel a.
(c) Preformed TCs containing Fe(II)-BABE-derivatized (lanes 1–7, 9–16) or unmodified (lane 8) WT or the indicated mutant forms of eIF2 were assayed for 48S complex formation. The staggering of the bands is because the reactions were quenched by loading at 5 or 15 min onto a running native gel. The positions of 48S complexes and free Met-tRNAiMet are indicated. See also Supplementary Figure 2g online for rate constants obtained from the assay.
(d) Summary of Kd values for eIF2 TC binding to 40S subunits. Errors are s.d. from three independent measurements.
Results
Preparation of eIF2 Cys mutants
Yeast eIF2 contains 20 Cys residues among the three subunits (5 in eIF2α, 5 in eIF2β, and 10 in eIF2γ); however, 8 of these Cys residues (4 each in eIF2β and eIF2γ) are contained in essential zinc binding motifs. The 12 Cys residues not involved in coordinating Zn were mutated to either Ser or residues found in other species (Fig. 2a) to generate a Cys-lite version of eIF2 (eIF2ΔC). Next, single Cys residues were introduced at various surface exposed sites on all three subunits of eIF2 (see Fig. 2b). When expressed in a yeast strain in which the chromosomal genes encoding all three subunits, eIF2α (SUI2), eIF2β (SUI3), and eIF2γ (GCD11), were deleted, all of the mutants supported cell growth to the same extent as expression of WT eIF2 (Supplementary Fig. 2a–c online). Moreover, none of the mutants (except eIF2ΔC-γL349C) affected translational control of GCN4, a sensitive in vivo reporter of eIF2 function (Supplementary Fig. 2a–c online, and below). We conclude that the Cys mutations (except γL349C) have minimal effects on eIF2 function in vivo.
The various eIF2 mutants were purified, derivatized with Fe(II)-BABE, and then the activity of each modified protein was tested using a 48S complex formation assay. As shown in Figure 2c, and Supplementary Figure 2g online, Fe(II)-BABE modified WT eIF2ΔC was slightly less active than unmodified WT eIF2 (lane 7 versus 8) in binding [35S]Met-tRNAiMet to 40S ribosomes in the presence of eIF1, eIF1A and a model mRNA. Moreover, the eIF2ΔC complexes containing the βS80C, βR186C, βS264C, or γL349C mutant subunit displayed reduced 48S complex formation activity (Fig. 2c). Consistent with the results of the 48S complex formation assay, affinity measurements revealed that Fe(II)-BABE-eIF2ΔC bound 40S subunits with ~one-third the affinity of WT eIF2, and the βR186C and γL349C mutants bound 40S subunits with ~one-tenth the affinity of eIF2ΔC (Fig. 2d). In contrast, the γK507C mutant bound 40S subunits with a Kd of 32 nM, only twice the value obtained with eIF2ΔC (Fig. 2d). To avoid complications associated with altered 48S complex formation activity, 0.5 μM eIF2 was used in all cleavage assays. In preliminary experiments Fe(II)-BABE linked to the native Cys residues in yeast eIF1 produced 18S rRNA cleavages near the top of helix h44 (see Supplementary Note and Supplementary Fig. 2d–e online), consistent with the reported binding site for mammalian and Tetrahymena eIF117. Furthermore, toe-printing assays demonstrated that the 48S complexes were bound at the AUG start codon on the model mRNA (see Supplementary Note and Supplementary Fig. 2f online). We conclude that the yeast assay system produced bona fide 48S complexes.
Directed hydroxyl radical cleavage of Met-tRNAiMet by eIF2
In the crystal structure of the EF-Tu–GDPNP–Phe-tRNAPhe TC (pdb code: 1TTT18), three different regions of Phe-tRNAPhe interact with EF-Tu: the aminoacyl 3′-CCA-Phe, the 5′ end, and the T-stem (Fig. 1). Based on the structural similarity of aIF2γ and EF-Tu, we generated an aIF2γ TC model by aligning the structures of aIF2γ (pdb code: 2AHO15) and yeast initiator tRNAiMet (1YFG19) with EF-Tu and Phe-tRNAPhe, respectively, in the EF-Tu TC structure (Fig. 1, right panel). In this model the Met on Met-tRNAiMet binds in a groove between the G domain and domain II, and the body of the tRNA lies across domain III resulting in the anticodon portion of the L-shaped molecule projecting away from the factor (Fig. 1, right panel). Accordingly, the T-stem of Met-tRNAiMet is predicted to contact domain III of eIF2γ.
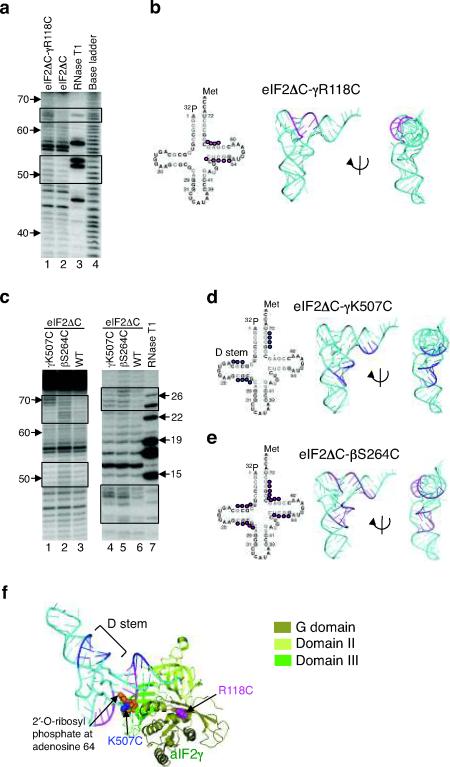
In order to probe the position of Met-tRNAiMet binding to eIF2, 48S complexes were formed using 5′ end-labeled Met-[32P]tRNAiMet and Fe(II)-BABE-modified forms of eIF2. Incorporation of the labeled Met-[32P]tRNAiMet into 48S complexes was confirmed using a native gel shift assay (see Supplementary Fig. 3 online). Cleavage of Met-[32P]tRNAiMet by hydroxyl radicals formed in the vicinity of the ferrous iron was analyzed by denaturing gel electrophoresis. In comparison to Fe(II)-BABE-treated eIF2ΔC, the modified eIF2ΔC-γR118C complex showed enhanced cleavage at bases C49 to G53 and G63 to C66, which are located in the T stem of tRNAiMet (Fig. 3a, lane 1 versus 2, and Fig. 3b). Hydroxyl radicals generated using the Fe(II)-modified eIF2ΔC-γK507C complex cleaved three different regions of tRNAiMet: C11 to C13 and C23 to G26, both in the D stem, and G68 to C71 in the 3′ side of the acceptor stem (Fig. 3c, lanes 1 and 4, and Fig. 3d). Interestingly, all of these γK507C cleavage sites lie on the inside face of the L-form structure of tRNAiMet (Fig. 3d). No significant tRNAiMet cleavages were observed when Fe(II)-BABE was tethered at other sites on eIF2α (M29C, K182C, or L226C), eIF2β (R186C) or eIF2γ (L349C, D446C [very weak cleavage at C25], or A480C), with the exception of the eIF2ΔC-βS264C complex which will be discussed below. The tRNAiMet cleavages obtained with the eIF2ΔC-γR118C and eIF2ΔC-γK507C complexes are inconsistent with the EF-Tu TC-like model (see Fig. 3f). Most strikingly, Fe(II) tethered to γK507C did not cleave the T stem, but instead resulted in cleavages in the D stem on the opposite face of the tRNA (Fig. 3f). Based on these results, we conclude that the structure of the eIF2 TC on the 48S complex differs significantly from the structure of the EF-Tu TC both free and on the ribosome.
Figure 3.
Directed hydroxyl radical cleavage of Met-[32P]tRNAiMet by Fe(II)-BABE-derivatized eIF2 in 48S complexes.
(a) eIF2ΔC-γR118C-Fe(II)-BABE cleavages. Hydroxyl radical cleavage products from 48S complexes were resolved on 10% (w/v) denatured polyacrylamide gels, and cleavage sites on [32P]tRNAiMet were determined by comparison with samples containing eIF2ΔC (WT) (lane 2). Sites of enhanced cleavage with eIF2ΔC-R118C are boxed. The tRNA ladders were prepared by digesting Met-[32P]tRNAiMet with RNase T1 (cleaves 3′ of G residue, lane 3) or by base cleavage (lane 4). The tRNA residue numbers are shown at the left.
(b) Sites (colored magenta) of eIF2ΔC-γR118C-Fe(II)-BABE directed hydroxyl radical cleavage on Met-[32P]tRNAiMet are shown on the secondary (left) and three-dimensional (pdb code: 1YFG19, middle and right) structures of tRNAiMet.
(c) Met-[32P]tRNAiMet cleavages in 48S complexes assembled with eIF2ΔC-γK507C-Fe(II)-BABE and eIF2ΔC-βS264C-Fe(II)-BABE, as in panel a. Cleavage products were resolved on 10% (left) or 20% (right) (w/v) denatured polyacrylamide gels.
(d,e) Summary of Met-[32P]tRNAiMet cleavages in panel c, depicted as in panel b.
(f) aIF2γ-GDPNP-tRNAiMet TC model showing the sites of tRNAiMet (cyan) cleavages by hydroxyl radicals generated at residues K507 (blue) and R118 (magenta). The model was generated by docking the structure of yeast tRNAiMet (pdb code: 1YFG19) on the aIF2γ TC structure from Fig. 1b. The RIT1 catalyzed 2′-O-ribosyl phosphate modification at residue 64 of tRNAiMet is shown as orange spheres.
Directed hydroxyl radical cleavage of 18S rRNA by eIF2
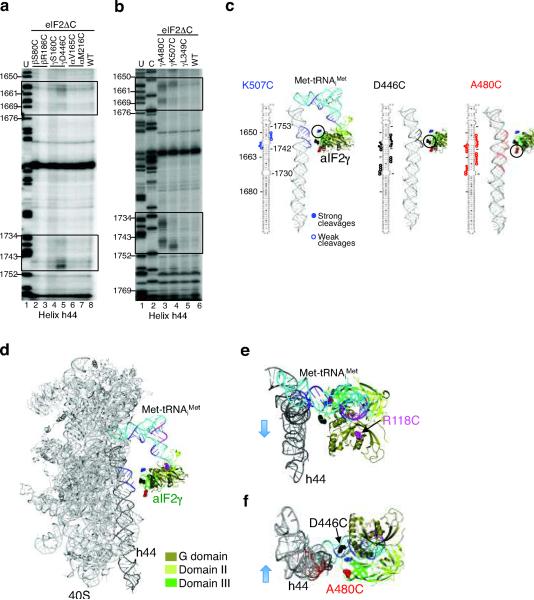
To determine how eIF2 contacts the 40S subunit, hydroxyl radicals were generated in 48S complexes containing Fe(II)-labeled eIF2, and 18S rRNA cleavage sites were identified by primer extension analyses using 32P-labeled primers. No cleavages beyond those obtained with eIF2ΔC were observed when eIF2ΔC complexes contained Fe(II) tethered to residues S80C or R186C in eIF2β, S160C in eIF2γ, or V165C or M216C in eIF2α (Fig. 4a and data not shown). In contrast, Fe(II)-modified eIF2ΔC-γD446C complexes produced cleavages at residues A1655 to A1659 and A1744 to A1749 and weaker, but not always reproducible, cleavages at residues A1667 to A1671 and G1736 to U1738 (Fig. 4a, lane 5 versus 8). These cleavages map to both strands of the upper and middle portions of 18S rRNA helix h44, a prominent landmark that extends from the bottom of the head to the bottom of the body on the subunit interface side of the 40S subunit (Fig. 4c, middle panel, and Fig. 4d).
Figure 4.
Directed hydroxyl radical cleavage of 18S rRNA by Fe(II)-BABE-derivatized eIF2 in 48S complexes.
(a,b) Primer extension analysis of 18S rRNA helix h44 cleavages by Fe(II)-BABE linked to the indicated positions in eIF2α, eIF2β or eIF2γ. Lanes marked “U” and “C” present 18S rRNA sequencing reactions using reverse transcriptase and the indicated dideoxynucleotide. Positions of cleaved nucleotides are boxed and the numbering of helix h44 residues is shown on the left.
(c) Sites of 18S rRNA helix h44 cleavages by eIF2ΔC-γK507C-Fe(II)-BABE (left, blue), eIF2ΔC-γD446C-Fe(II)-BABE (middle, black), and eIF2ΔC-γA480C-Fe(II)-BABE (right, red) are displayed on secondary and tertiary, taken from the crystal structure of the yeast ribosome (pdb code: 3O3020), structure models of helix h44. aIF2γ and Met-tRNAiMet are docked on helix h44 as described in the text.
(d–f) 40S–aIF2γ–Met-tRNAiMet complex model viewed from the A-site, d; from above, e; and from below, f. Met-tRNAiMet was docked in the P-site of the yeast 40S ribosome (pdb code: 3O3020) as observed in the structure of the bacterial 70S ribosome (pdb code: 2J0021). aIF2γ was docked on the acceptor stem of Met-tRNAiMet as in the EF-Tu TC (Fig. 1b); however, domain III of aIF2γ was positioned toward helix h44 and aligned consistent with the cleavage data. aIF2γ residues corresponding to K507 (blue), D446 (black), A480 (red) and R118 (magenta) are shown as spheres and the helix h44 and Met-tRNAiMet cleavage sites are shown in matching colors for the three sites of Fe(II)-BABE modification.
To help orient the docking of eIF2γ on the 40S subunit, three additional eIF2γ mutants were examined: L349C, A480C and K507C. As shown in Figure 2b, the Ala480 and Lys507 residues lie on opposite sides of Asp446 with all three residues on the same face of domain III of eIF2γ. The Leu349 residue is in domain II of eIF2γ near the eIF2α binding site and remote from Asp446. Consistent with the results obtained with the D446C mutant, tethering Fe(II) to A480C yielded cleavages at four different positions in helix h44: U1656 to A1660, A1667 to A1671, U1735 to A1740, and A1744 to A1749 (Fig. 4b–c). Likewise, hydroxyl radicals generated at γK507C cleaved A1655 to G1658 and G1747 to A1750 in helix h44 (Fig. 4b–c). When Fe(II) was tethered to γL349C no specific cleavages above background (eIF2ΔC) were observed in the 18S rRNA (Fig. 4b). This lack of cleavage could reflect the distance of γL349 from the ribosome or the poor binding of this mutant to the ribosome. As shown on the secondary and tertiary structure models of the 18S rRNA (see Fig. 4c and Supplementary Fig. 4b online), the three eIF2γ domain III mutants, D446C, A480C, and K507C, produced cleavages in overlapping regions of helix h44, yet each mutant yielded a distinct pattern of cleavages. Based on the location of the cleavages, K507 lies closest to the top of helix h44, followed by D446, and then A480 (Fig. 4c). The distances between the centers of the cleavages observed with each mutant, as mapped on the structure of helix h44, are comparable to the distances between the three residues in aIF2γ (Fig. 4c–f), consistent with the notion that domain III of eIF2γ docks very close to the 40S subunit.
Model of the 40S–aIF2γ–Met-tRNAiMet complex
In the GDPNP-bound structure of the archaeal aIF2αγ complex from Sulfolobus solfataricus (pdb code: 2AHO)15, the aIF2γ subunit adopts an active conformation, similar to EF-Tu–GTP, so this aIF2γ structure was chosen for modeling studies of the 40S–aIF2–Met-tRNAiMet complex. The 40S subunit structure was obtained from the recent crystal structure of the yeast ribosome (pdb code: 3O3020), and the orientation of the P-site tRNA was modeled based on the structure of the Thermus thermophilus 70S ribosome containing bound P- and E-site tRNAs (pdb code: 2J0021). Docking of the yeast tRNAiMet structure (pdb code: 1YFG19) in the P-site of the 40S subunit placed the codon-anticodon interaction in the P-site at the top of helix h44 (see Fig. 4d). The aminoacyl end of the L-shaped Met-tRNAiMet projects down toward the body of the 40S subunit (Figs. 4c–d). Interestingly, the distance between helix h44 and the aminoacyl end of the Met-tRNAiMet in this model (43 Å) is consistent with the distance between Lys507 (Gly403 in S. solfataricus aIF2γ) and the proposed aIF2γ amino acid binding site located between the G domain and domain II in analogy with EF-Tu (32 Å).
To fine tune the docking of aIF2γ between the aminoacyl-tRNA and helix h44, we first focused on the helix h44 cleavages observed when hydroxyl radicals were generated at residues Lys507, Asp446 and Ala480. Domain III of aIF2γ was oriented on helix h44 such that Lys507 (Ss Gly403) lay above Asp446 (Ss Met345), which in turn was above Ala480 (Ss Lys375), consistent with the partially overlapping cleavage patterns observed by hydroxyl radicals generated at these sites (Fig. 4c). The absence of rRNA cleavages flanking helix h44 limits the ability to precisely position eIF2γ on helix h44. However, it is notable that when viewed from below, residues D446 and A480 lie on opposite sides of helix h44, consistent with the bias of the cleavages to opposite sides of the helix (Fig. 4f). A modest reconfiguration of the 3′ CCA end of the tRNAiMet enabled its docking on aIF2γ. Interestingly, in this model the elbow of the L-shaped tRNAiMet is rotated nearly 180° from its position in the EF-Tu–GTP–Phe-tRNAPhe complex (see Figs. 1, 4d, and 5a–b). In support of this dramatic reconfiguration of the eIF2 versus EF-Tu TC model, docking of aIF2γ in the EF-Tu mode of binding on the P-site bound Met-tRNAiMet places domain III of aIF2γ remote from helix h44 (see Supplementary Fig. 4a online).
Figure 5.
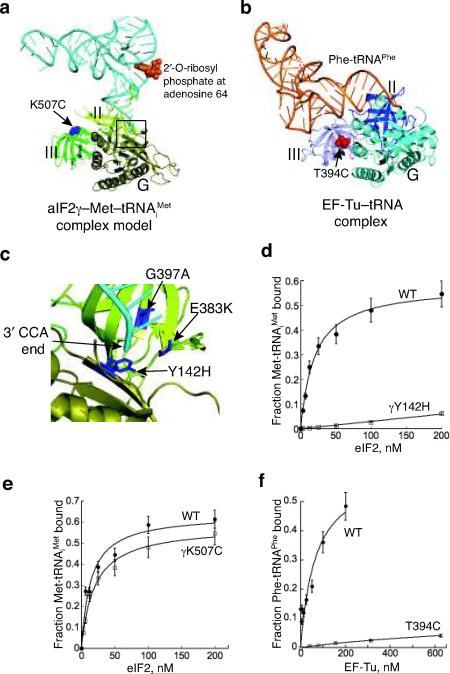
eIF2γ binding site for 3′ end of Met-tRNAiMet.
(a) aIF2γ–Met-tRNAiMet complex model, generated as described in Figs. 4c–d. The RIT1 catalyzed 2′-O-ribosyl phosphate modification at residue 64 of tRNAiMet is shown in orange.
(b) Ribbons representation of EF-Tu–GDPNP–Phe-tRNAPhe complex (pdb code: 1TTT18). The site of the T394C mutation, which is analogous to the eIF2γ-K507C mutation, is shown as red spheres.
(c) Magnified view of proposed eIF2γ binding site for the 3′ end of Met-tRNAiMet (box in Fig. 5a). Locations of yeast eIF2γ mutations that impair eIF2 function in vivo are labeled and shown as blue sticks: Y142H (corresponds to S. sol. aIF2γ-Tyr51), E383K (Gly282), and G397S (Ala296).
(d,e) Purified WT, γY142H (D), or γK507C (E) mutant forms of eIF2 were assayed for eIF2 TC formation by filter binding assay. Fractions of [35S]Met-tRNAiMet bound to eIF2 were plotted as a function of eIF2 concentration; points and s.d. are averages of at least three independent experiments.
(f) Purified WT or T439C mutant forms of EF-Tu (T. therm) were assayed for TC formation by filter binding assay. Fractions of [14C]Phe-tRNAPhe bound to EF-Tu were plotted as a function of EF-Tu concentration; points and s.d. are averages of at least three independent experiments.
The results from the cleavage assays provide support for the model of the 40S–aIF2γ–Met-tRNAiMet complex. As shown in Figure 4c–d and Supplementary Figure 4b online, Lys507 (Ss Gly403) is located in the middle of its five cleavage sites and ~12 Å from the backbone of helix h44 and ~20Å from Met-tRNAiMet, consistent with the stronger cleavages observed in helix h44 than in Met-tRNAiMet (Figs. 3c and 4b). Likewise, Asp446 (Ss Met345) and Ala480 (Ss Lys375) are close to helix h44 and even farther from the Met-tRNAiMet, consistent with their very weak, or absence of, Met-tRNAiMet cleavage (data not shown). Finally, the model supports the Met-tRNAiMet cleavages generated by the γR118C (Ss Gln27) derivative (Figs. 4d–e).
eIF2γ binding site for the acceptor stem of Met-tRNAiMet
In our proposed model of the 40S–aIF2γ–Met-tRNAiMet complex, the 3′ CCA–Met acceptor end of Met-tRNAiMet binds in a pocket formed between the G domain and domain II of aIF2γ. The analogous pocket in EF-Tu binds the amino acid and aminoacyl end of elongator tRNAs in the EF-Tu–GTP–aminoacyl-tRNA complexes (Figs. 5a–b). As shown in Figure 5c, the eIF2γ residues Tyr142 (Ss Tyr51), Glu383 (Gly282) and Gly397 (Ala296) are positioned very close to the 3′ end of the tRNAiMet. Previously, mutations at these sites were reported to alter translational control of the GCN4 mRNA, a sensitive in vivo reporter of eIF2 function22. Production of GCN4, a transcriptional activator of amino acid biosynthetic enzyme genes, is constitutively repressed in gcn2Δ strains and cells cannot grow under amino acid starvation conditions, such as those imposed by the amino acid analogs 3-aminotriazole (3-AT), that impairs histidine biosynthesis, or sulfometuron methyl (SM), that impairs isoleucine and valine biosynthesis. Mutations that impair eIF2 function, such as Met-tRNAiMet or ribosome binding, enhance GCN4 expression, and enable gcn2Δ strains to grow on medium containing 3-AT or SM23. Accordingly, the eIF2γ mutations Y142H, E383K, and G397A derepressed GCN4 expression22,24, and the derepressed phenotype of the Y142H and G397A mutants was suppressed by overexpressing tRNAiMet (ref. 11,25), indicating that the mutations weakened Met-tRNAiMet binding to eIF2. In support of this latter notion, we confirmed that the Y142H mutation in yeast eIF2γ severely impaired TC formation by purified yeast eIF2 (Fig. 5d), consistent with the report by Erickson et al25. These genetic and biochemical results indicate that the aminoacyl-tRNA binding pocket is a major determinant for Met-tRNAiMet binding to eIF2.
Whereas the body of the tRNA contacts both domains II and III in the EF-Tu TC (Fig. 5b), in our model of the aIF2 TC the contact between the Met-tRNAiMet and aIF2γ is restricted to the acceptor stem-binding pocket described above (Fig. 5a). Previously, Uhlenbeck and colleagues reported that a critical contact for the stability of the EF-Tu TC occurs between the T-stem of tRNAs and domain III of EF-Tu26,27 (see Fig. 5b). Supporting this idea, we found that mutating Thr394 in domain III of T. thermophilus EF-Tu to Cys greatly impaired Phe-tRNAPhe binding (Figs. 5b and 5f, Kd value of 68 (±2) nM for WT EF-Tu and >1000 nM for EF-Tu-T394C). In contrast, the analogous K507C mutation in domain III of yeast eIF2γ (Gly403 in Ss aIF2γ) did not impair Met-tRNAiMet binding (Figs. 5a and e, Kd value of 14 (± 4) nM for WT eIF2 and 20 (± 2) nM for eIF2-γK507C). Importantly, the affinity of EF-Tu for GDP was not impaired by the T394C mutation (Kd value of 7 (± 2) nM for WT EF-Tu and 11 (± 5) nM for EF-Tu-T394C; data not shown), indicating that the structural integrity of EF-Tu was not altered by the T394C mutation. This significantly different impact of domain III mutations on EF-Tu versus eIF2 TC formation provides independent support for our structural model of the aIF2 TC bound to the 40S ribosome in which domain III of aIF2γ contacts the ribosome rather than Met-tRNAiMet.
Interaction of domain III of eIF2γ with 18S rRNA helix h44
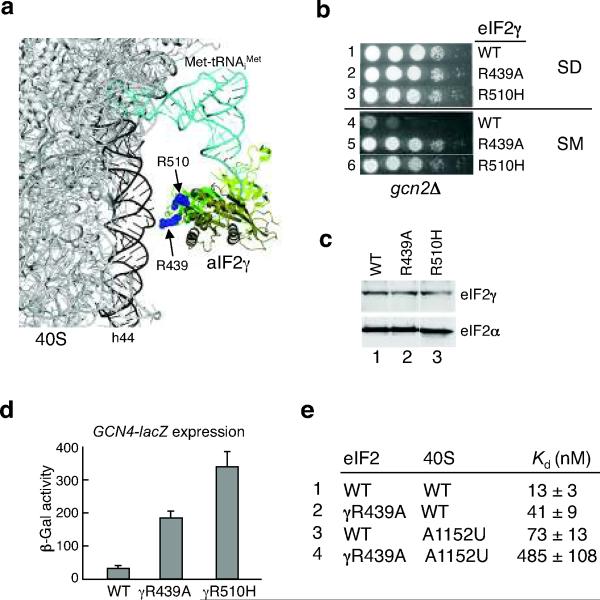
Residues Asp446, Ala480 and Lys507, the three sites in eIF2γ that yielded helix h44 cleavages, lie on the same face of domain III (Fig. 4d and Supplementary Fig. 4b online). To test the hypothesis that this face of domain III is a 40S ribosome interface, the conserved surface residues Arg439 and Arg510 in S. cerevisiae eIF2γ (Fig. 6a), that are not conserved in EF-Tu (Supplementary Fig. 1 online), were mutated to Ala and His, respectively. Whereas eIF2γ-R439A and eIF2γ-R510H mutations did not affect yeast cell growth on minimal SD medium (Fig. 6b, rows 1–3), these substitutions suppressed the SM-sensitive phenotype of a gcn2Δ strain (Fig. 6b, rows 4–6) and increased GCN4-lacZ expression 6- to 11-fold (Fig. 6d). This SM-resistant phenotype of the eIF2γ-R510H mutant is consistent with its previous isolation as a spontaneous 3-AT-resistant suppressor of a gcn2 gcn3 double mutant24. As eIF2γ-R439A and eIF2γ-R510H were expressed at levels equivalent to WT eIF2γ (Fig. 6c), the SM-resistant phenotype of these mutants indicates that they impair eIF2 function in vivo.
Figure 6.
eIF2γ domain III is involved in 40S binding.
(a) Locations of conserved Arg residues (blue spheres, labeled as in yeast eIF2γ) in the proposed 40S binding surface of eIF2γ domain III are shown in the 40S–aIF2γ–Met-tRNAiMet complex model.
(b) Altered GCN4 translational control in yeast cells expressing eIF2γ domain III mutants. Derivatives of the gcn2Δ yeast strain J551 expressing wild type or the indicated mutant forms of eIF2γ were spotted on minimal medium with essential nutrients (SD, rows 1–3) or SD medium containing 0.3 μg per ml sulfometuron methyl (SM, rows 4–6).
(c) Western blot analysis of eIF2γ expression. Whole-cell extracts of strains described in (b) were subjected to immunoblot analysis using anti-yeast eIF2γ (upper panel) or anti-yeast eIF2α (lower panel) antiserum. Immune complexes were visualized using enhanced chemiluminescence.
(d) Analysis of GCN4-lacZ expression. The GCN4-lacZ plasmid p18045 was introduced into derivatives of strain J551 expressing WT eIF2, eIF2γ-R439A, or eIF2γ-R510H. Cells were grown and β-galactosidase activities were determined as described previously45. The β-galactosidase activities are the averages of three independent transformants and the errors are s.d.
(e) Kd values and standard deviations for WT or γR439A mutant forms of eIF2 TC binding to WT or 18S-A1152U mutant forms of yeast 40S–eIF1–eIF1A complexes were measured by 43S gel shift assays. Fitting curves are shown in Supplementary Figure 5 online.
As purified WT eIF2 (Kd = 20 ± 2 nM) and eIF2 complexes containing the eIF2γ-R439A mutant subunit (Kd = 23 ± 4 nM) bound Met-tRNAiMet with similar affinities (data not shown), the γR439A mutation does not affect eIF2 TC formation. In contrast, a gel shift assay7 revealed that the γR439A mutation impaired eIF2 TC binding to 40S subunits in the absence of mRNA. As shown in Fig. 6e (rows 1 and 2), the γR439A mutation impaired TC binding to the 40S subunit and increased the Kd value for ribosome binding by 3-fold (see Supplementary Fig. 7 online). A previous study identified mutations in 18S rRNA helix 28 near the P-site of the yeast 40S subunit that impaired eIF2 TC binding28. Consistent with our model (Fig. 6a), these mutations indicate that the interaction between the Met-tRNAiMet and the P-site also contributes to the binding of the eIF2 TC to the 43S preinitiation complex. As observed previously28, we found that the A1152U mutation in 18S rRNA helix 28 increased the Kd value for eIF2 TC binding to 40S subunits by 5-fold (Fig. 6e, rows 1 and 3). Interestingly, the R439A mutation in domain III of eIF2γ further increased the Kd value by 6-fold (Fig. 6e, rows 3 and 4). Taken together, the in vivo GCN4 expression defects and the in vitro ribosome binding defects associated with the R439A mutation support the results of the hydroxyl radical probing experiments and indicate that domain III of eIF2γ makes important contacts with the 40S subunit that facilitate 43S complex formation.
43S [40S–aIF2αβγ–Met-tRNAiMet–eIF1–eIF1A] complex modeling
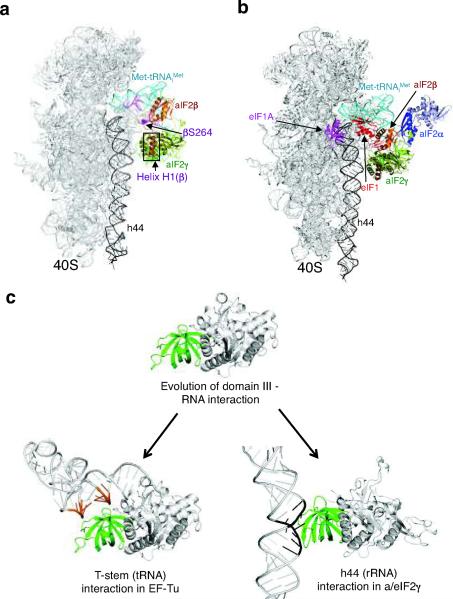
To dock aIF2β onto the 43S complex model, the structure of the aIF2βγ heterodimer (pdb code: 2QMU16) was aligned to the aIF2γ subunit in the model (Fig. 7a). Only the N-terminal γ-helix H1 of aIF2β forms a rigid body interaction with aIF2γ, so only this portion of aIF2β can be definitively positioned in the model. The remainder of aIF2β including the central and zinc-binding domains (ZBD) is known to be flexible14, and thus could adopt many conformations. The position of aIF2β in the model, close to the Met-tRNAiMet but distant from the ribosome, is consistent with the cleavage on one side of the Met-tRNAiMet when Fe(II)-BABE was tethered to eIF2ΔC-βS264C (Figs. 3c,e and 7a) and with the finding that Fe(II)-BABE tethered to multiple positions in eIF2β yielded no cleavages in the 18S rRNA.
Figure 7.
43S complex model.
(a) Docking aIF2β on the 40S–aIF2γ–Met-tRNAiMet complex model. aIF2β from the aIF2βγ heterodimer structure (pdb code: 2QMU16) was docked on the 40S–aIF2γ–Met-tRNAiMet complex in Fig. 4d. Helix H1 of aIF2β, which forms the only rigid body interaction with aIF2γ, is boxed. The aIF2β location corresponding to eIF2β-S264 is shown as purple spheres, and the Met-tRNAiMet residues cleaved by Fe(II)-BABE linked to eIF2ΔC-βS264C are colored purple.
(b) Docking of aIF2α, eIF1 and eIF1A on the 40S–aIF2βγ–Met-tRNAiMet complex. aIF2α is from the aIF2αγ heterodimer structure (pdb code: 2AHO15); eIF1 is from the 40S–eIF1 co-crystal structure (pdb code: 2XZM17); and eIF1A (pdb code: 1D7Q) was positioned based on the bacterial 30S–IF1 structure (pdb code: 1HR09) and hydroxyl radical mapping data8. Only the eIF1A core structure is shown.
(c) Schematic depicting the alternate tRNA T-stem (orange) and rRNA helix h44 (black) interactions of domain III (green) from EF-Tu (left) and aIF2γ(eIF2γ) (right).
The position of aIF2α in the 43S complex can be predicted by aligning the structure of the aIF2αγ heterodimer (pdb code: 2AHO15) with aIF2γ in the model (Fig. 7b). The aIF2α–aIF2γ interactions are restricted to the C-terminal domain III of aIF2α, and domains I and II of aIF2α appear to move as a rigid body independent of domain III14. As we failed to detect any cleavages of either 18S rRNA or Met-tRNAiMet by Fe(II)-BABE tethered at various residues in eIF2α, the N-terminal portion of aIF2α is depicted as in the aIF2αγ heterodimer structure. However, it is noteworthy that Pestova and colleagues cross-linked eIF2α in 48S complexes to the mRNA at the −3 position relative to the AUG start codon29. It is unclear whether yeast eIF2α undergoes rearrangements that could allow this contact to occur.
To complete the model of the 43S complex, the structures of the core domains of eIF1 and eIF1A were docked on the ribosome. Aligning the Tetrahymena eIF1–40S co-crystal structure (pdb code: 2XZM17) with the structure of the 40S–aIF2αβγ–Met-tRNAiMet model placed Tetrahymena eIF1 at the top of 18S rRNA helix 44 above aIF2γ and immediately below the Met-tRNAiMet (Fig. 7b). This binding site is compatible with the hydroxyl radical probing data for the complexes of human and yeast eIF1 with the 40S subunit5 (Supplementary Fig. 1d–e online). In the bacterial IF1–30S co-crystal structure (1HR09) IF1 binds near the base of the A-site, and hydroxyl radical mapping experiments placed the folded core of the orthologous human eIF1A in a similar position on mammalian ribosomes8. Accordingly, human eIF1A (1D7Q30) was docked in the A-site of the 43S complex model (Fig. 7b). In support of the final 40S–aIF2αβγ–Met-tRNAiMet–eIF1–eIF1A (43S complex) model, it is noteworthy that, aside from a small clash between the positions of Met-tRNAiMet and eIF117, there is little conflict between the positions of the aIF2 TC, eIF1, and eIF1A (Fig. 7b).
Discussion
Position of eIF2 on the 40S ribosome
In addition to the conserved G domain, the translational GTPases share a conserved β-barrel domain II. Whereas EF-Tu(eEF1), EF-G(eEF2), RF-3(eRF3), and IF2(eIF5B) bind near the A-site of the intact ribosome, eIF2γ binds Met-tRNAiMet to the P-site of the 40S ribosomal subunit. Consistent with the notion that eIF2 makes different contacts with the ribosome than the other translational GTPases, the face of domain II of EF-Tu1, EF-G31–33 and IF2(eIF5B)34–36 that contacts the small ribosomal subunit forms the docking site for eIF2α on eIF2γ11,15,37. The directed hydroxyl radical probing experiments reported here reveal that domain III of eIF2γ docks near helix h44 of the 40S ribosome. As mutation of conserved, surface exposed Arg residues in eIF2γ domain III impaired binding of the eIF2 TC to the 40S ribosome (Fig. 6e and Supplementary Fig. 5 online), we conclude that eIF2 binds to helix h44 on the intersubunit face of the 40S ribosome.
In the 43S complex model (Fig. 7b) the only contacts between the TC and the 40S subunit are through Met-tRNAiMet and domain III of aIF2γ. However, it is possible that eIF3 and eIF5, factors that were not included in the experiments in this paper, as well as other elements in eIF2 contribute to binding the TC to the 40S subunit. In particular, the N-terminus of eIF2β contains three lysine-rich elements (K-boxes) that have been shown to support both protein-protein and protein-nucleic acid interactions38,39, and eIF2α was reported to contact mRNA near the AUG start codon29. Additional studies are needed to determine whether eIF2α, the eIF2β K-boxes, eIF3, and eIF5 contribute to 48S complex formation and the positioning of eIF2 on the 40S subunit.
Structure of the eIF2 TC
It has generally been assumed that the orientation of the aminoacyl-tRNA in the eIF2 TC would resemble the structure of the EF-Tu TC, in which the T-stem of the tRNA contacts domain III of aIF2γ (Fig. 1). Docking aIF2γ in the EF-Tu TC configuration on Met-tRNAiMet in the P-site, placed aIF2γ remote from the ribosome (Supplementary Fig. 4a online). This model is at odds with the results from hydroxyl radical probing experiments (Fig. 4) that place domain III of eIF2γ in the vicinity of helix h44. Consistent with the structure of the EF-Tu TC, the T394C mutation in domain III of EF-Tu significantly impaired aminoacyl-tRNA binding (Fig. 5b,f). In contrast, mutation of the corresponding Lys507 residue in eIF2γ (Supplementary Fig. 1 online, and Fig. 5a) did not impair Met-tRNAiMet binding (Fig. 5e). Rather, directed hydroxyl radical probing experiments indicated that Lys507 binds the 40S subunit near helix h44 (Fig. 4). Taken together, we propose that as the unique translation initiation pathway evolved in eukaryotes, the tRNA-binding interface on domain III of the bacterial GTPase EF-Tu was adapted in the GTPase eIF2γ to bind rRNA (Fig. 7c).
Studies on the RIT1 enzyme in yeast provide additional support for the idea that the conformation of Met-tRNAiMet in the eIF2 TC is different than the conformation of aminoacyl-tRNAs in the EF-Tu TC. The RIT1-catalyzed addition of a 2′-O-ribosyl phosphate modification at position 64 in the T stem prevents Met-tRNAiMet from functioning in translation elongation40. Yeast have separate genes for initiator and elongator tRNAMet and these tRNAs are used exclusively in translation initiation and elongation, respectively41. Yeast cells lacking elongator tRNAMet are inviable; however, deletion of RIT1 restores viability and enables tRNAiMet to function in both translation initiation and elongation40. As shown in Figures 3f and 5a, the 2′-O-ribosyl phosphate modification occurs in the T stem of Met-tRNAiMet and would be predicted to interfere with Met-tRNAiMet binding to eEF1A (the eukaryotic ortholog of EF-Tu) as well as with Met-tRNAiMet binding to eIF2 in the EF-Tu mode. On the other hand, the 2′-O-ribosyl phosphate modification should have no impact on Met-tRNAiMet binding to eIF2 in the altered mode shown in the TC (Fig. 5a) and 43S complex (Fig. 7b) models.
Whereas EF-Tu interacts with both the amino acid and the body of aminoacyltRNAs26,27,42,43, in our TC model (Fig. 5a) eIF2 interacts exclusively with the Met residue and acceptor stem of Met-tRNAiMet. Elegant biochemical experiments by Uhlenbeck and colleagues have shown that by balancing independent interactions with the tRNA body and the esterified amino acid, EF-Tu binds different aminoacyl-tRNAs with comparable affinities42,43. Moreover, they showed that residues in domain III of EF-Tu that interact with the T stem of aminoacyl-tRNA play critical roles in tRNA binding26. So how does eIF2 achieve comparably strong binding of Met-tRNAiMet despite its limited contact with the Met residue and 3′ end of the tRNA? One possibility is that eIF2α and eIF2β provide additional Met-tRNAiMet binding contacts in the eIF2 TC. Consistently, linkage of Fe(II)-BABE to eIF2β-S264C yielded cleavages in both the T and D stems of Met-tRNAiMet (Fig. 3c,e). Alternatively, and in contrast to EF-Tu, which must bind a variety of aminoacyl-tRNAs, the Met-tRNAiMet binding pocket in eIF2γ might be optimized to tightly bind Met and the 3′ end of tRNAiMet. Consistent with this proposal, it has been shown that methionine contributes 13-fold to the binding of Met-tRNAiMet to eIF244. Taken together, we propose that very tight and highly specific interactions between eIF2γ and Met linked to the 3′end of tRNAiMet promote eIF2 TC formation.
Methods
Plasmids and yeast strains
Plasmids used in this study are listed in Supplementary Table 1. Please see Supplementary Methods for details on plasmid and yeast strain construction.
Protein purification and Fe(II)-BABE derivatization
Please see Supplementary Methods.
Hydroxyl radical cleavage of 18S rRNA
For hydroxyl radical cleavage of 18S rRNA, 48S complexes were assembled by incubating 0.5 μM Fe(II)-BABE-eIF2, 1 mM GDPNP-Mg2+, 1 μM Met-tRNAiMet, 1 μM eIF1, 1 μM eIF1A, 1 μM model unstructured mRNA (5−-GGAA(UC)7UAUG(CU)10C-3−; Dharmacon), and 0.5 μM 40S ribosomal subunits in cleavage buffer (30 mM HEPES [pH 7.5], 100 mM potassium acetate, 3 mM magnesium acetate). Following incubation at 26 °C for 15 min, reactions were chilled on ice. To examine tRNAiMet cleavages, 48S complexes were assembled using 0.5 μM Fe(II)-BABE-eIF2, 1 mM GDPNP-Mg2+, 0.1 μM Met-[5−-32P]tRNAiMet, 1 μM eIF1, 1 μM eIF1A, 1 μM mRNA, and 0.5 μM 40S ribosomal subunits. To generate hydroxyl radicals, a Fenton reaction was initiated by adding 1 μl of 100 mM ascorbic acid and 1 μl of 0.5% (v/v) H2O2> to 20 μl mixtures containing 48S complexes46. The reactions were incubated on ice for 10 min, and then quenched by addition of 2 μl of 100 mM thiourea. Following addition of 0.3 ml rRNA extraction buffer (30 mM sodium acetate [pH 5.3], 12.5 mM EDTA [pH 8.0], 0.5% (w/v) SDS), 18S rRNA was extracted once with phenol, once with phenol-chloroform, and then precipitated by adding 2.5 vol ethanol. For further information on preparation and hydroxyl radical cleavage of tRNAiMet, please see Supplementary Methods.
Primer Extension Analysis
Primer extension analysis to identify rRNA cleavage sites was performed as described previously47. Seven primers were used to detect the cleavage sites in 18S rRNA: P1, 5−-GGC CAT GCG ATT CGA AAA GTT-3−; P2, 5−-GTAT TTA CAT TGT ACT CAT TCC-3−; P3, 5−-GCTA ATA TAT TCG AGC AAT ACG-3−; P4, 5−-ATAG TTT ATG GTT AAG ACT ACG-3−; P5, 5−-TCA CTC CAC CAA CTA AGA ACG G-3−; P6, 5−-CAAT AAT TAC AAT GCT CTA TCC-3−; P7, 5−-TAA TGA TCC TTC CGC AGG TTC ACC-3−. Primer extension reactions were performed using either AMV Reverse Transcriptase (Roche) or Superscript II Reverse Transcriptase (Invitrogen). Sequencing ladders for 18S rRNA were obtained using intact 18S rRNA extracted from 40S subunits as a template for reverse transcription in the presence of small amounts of dideoxynucleotides as described by McPheeters et al.48. All cleavage assays were repeated three or more times to verify reproducibility of the results.
Biochemical assays
Filter binding assays to monitor TC formation (GTP-dependent aminoacylated-tRNA binding affinity) by eIF2 were performed as described previously49. TC formation by EF-Tu was monitored using [14C]Phe-tRNAPhe. To make [14C]Phe-tRNAPhe, yeast tRNAPhe (Chemblock) was aminoacylated with [14C]Phe (Perkin Elmer) using a crude yeast cell S100 extract. The S100 extract was prepared by growing yeast strain F353 to OD600 = 1.0, and the cells were transferred to 10 mM KHPO4 [pH 7.5] lysis buffer and lysed using liquid nitrogen and a Waring blender as described previously49. Following centrifugation at 20,000 g for 30 min, the supernatant was subjected to a second centrifugation at 100,000 g for 3 hrs. The resulting supernatant was incubated with DE52 resin (pre-swollen, Whatman) for 1 hr at 4 °C, and the mixture was then poured into column, washed with 5 column volumes of 10 mM KHPO4, and then eluted with 250 mM KHPO4 [pH 6.5]. Aliquots of the eluate were frozen and stored at −80 °C. To measure EF-Tu TC formation, two-fold serial dilutions (0 to 200 nM) of EF-Tu were mixed with 2 mM GDPNP–Mg2+ in TC Buffer (25 mM HEPES [pH 7.5], 2.5 mM magnesium acetate, 80 mM potassium acetate, 2 mM DTT) and incubated at 37 °C for 1 h to generate EF-Tu–GDPNP binary complexes. Next, 10 nM [14C]Phe-tRNAPhe was added to the EF-Tu–GDPNP mixture and incubated at 37 °C for 5 min. Finally, [14C]Phe-tRNAPhe bound to EF-Tu was measured by filter binding assay49. To measure GDP binding affinity to EF-Tu, two-fold serial dilution of EF-Tu (0 to 100 nM) in TC Buffer were incubated with 0.4 mM [3H]GDP at 37 °C for 1 h. The fraction of [3H]GDP bound to EF-Tu was then analyzed by nitrocellulose filter binding assay49.
Both 43S and 48S complex formation assays to monitor binding of eIF2 TC to 40S ribosomal subunits were performed as described previously49. Briefly, 40S subunits were mixed with eIF1, eIF1A, and eIF2–GDPNP–[35S]Met-tRNAiMet TCs in the presence (48S) or absence (43S) of the model unstructured mRNA described above. Final concentrations for each component are: 1 μM eIF1, 1 μM eIF1A, 0.8 μM eIF2, 1 mM GDPNP–Mg2+, 0.5 nM [35S]Met-tRNAiMet, 1 μM mRNA and 0.4 μM 40S subunits. Twofold serial dilutions of 40S subunits (0 to 400 nM) were used to measure 40S binding affinity. Reaction mixtures were separated on 4% (w/v) native polyacrylamide gels using 34 mM Tris, 57 mM HEPES, 0.1 mM EDTA, and 2.5 mM magnesium chloride as a gel and running buffer. Following electrophoresis, the gel was transferred to Whatman paper, and the fraction of free and ribosome-bound [35S]Met-tRNAiMet was determined by phosphorimage analysis. To calculate dissociation constants for TC formation, 43S or 48S complex formation, or GDP binding, binding curves for Met-tRNAiMet and GDP were fit using the program KaleidaGraph (Synergy) to the equation: Fraction bound = Bmax[S] / (Kd + [S]), where Bmax is the maximum fraction bound at excess [S].
Supplementary Material
Acknowledgments
We thank Alan Hinnebusch, Rachel Green, and our colleagues in the Dever, Lorsch and Hinnebusch laboratories for advice and helpful discussions. We thank Jeanne Fringer (National Institutes of Health), Dan Eyler, Shan He, Hani Zaher (all Johns Hopkins University), and Olke Uhlenbeck (Northwestern University) for protocols and reagents. This work was supported in part by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health (T.E.D.), and by the grant GM62128 from the NIH (J.R.L.).
Footnotes
Author Contributions B.-S.S. and J.-R.K. did the mutagenesis and protein purification, B.-S.S. performed the hydroxyl radical mapping experiments, biochemical analyses, and model building, S.E.W. did the toe-printing assay, and J.D. provided reagents. The manuscript was prepared by B.-S.S., S.E.W., J.R.L., and T.E.D.
References
- 1.Schmeing TM, et al. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science. 2009;326:688–94. doi: 10.1126/science.1179700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V. The mechanism for activation of GTP hydrolysis on the ribosome. Science. 2010;330:835–8. doi: 10.1126/science.1194460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–45. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fekete CA, et al. N-and C-terminal residues of eIF1A have opposing effects on the fidelity of start codon selection. EMBO. 2007;26:1602–1614. doi: 10.1038/sj.emboj.7601613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lomakin IB, Kolupaeva VG, Marintchev A, Wagner G, Pestova TV. Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes Dev. 2003;17:2786–97. doi: 10.1101/gad.1141803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Passmore LA, et al. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol Cell. 2007;26:41–50. doi: 10.1016/j.molcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 7.Maag D, Fekete CA, Gryczynski Z, Lorsch JR. A conformational change in the eukaryotic translation preinitiation complex and release of eIF1 signal recognition of the start codon. Mol Cell. 2005;17:265–75. doi: 10.1016/j.molcel.2004.11.051. [DOI] [PubMed] [Google Scholar]
- 8.Yu Y, et al. Position of eukaryotic translation initiation factor eIF1A on the 40S ribosomal subunit mapped by directed hydroxyl radical probing. Nucleic Acids Res. 2009;37:5167–5182. doi: 10.1093/nar/gkp519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carter AP, et al. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science. 2001;291:498–501. doi: 10.1126/science.1057766. [DOI] [PubMed] [Google Scholar]
- 10.Saini AK, Nanda JS, Lorsch JR, Hinnebusch AG. Regulatory elements in eIF1A control the fidelity of start codon selection by modulating tRNAiMet binding to the ribosome. Genes Dev. 2010;24:97–110. doi: 10.1101/gad.1871910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roll-Mecak A, Alone P, Cao C, Dever TE, Burley SK. X-ray structure of translation initiation factor eIF2γ: implications for tRNA and eIF2α binding. J Biol Chem. 2004;279:10634–42. doi: 10.1074/jbc.M310418200. [DOI] [PubMed] [Google Scholar]
- 12.Schmitt E, Blanquet S, Mechulam Y. The large subunit of initiation factor aIF2 is a close structural homologue of elongation factors. EMBO J. 2002;21:1821–1832. doi: 10.1093/emboj/21.7.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sokabe M, Yao M, Sakai N, Toya S, Tanaka I. Structure of archaeal translational initiation factor 2βα-GDP reveals significant conformational change of the β-subunit and switch 1 region. Proc Natl Acad Sci U S A. 2006;103:13016–21. doi: 10.1073/pnas.0604165103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stolboushkina E, et al. Crystal structure of the intact archaeal translation initiation factor 2 demonstrates very high conformational flexibility in the α- and β-subunits. J Mol Biol. 2008;382:680–91. doi: 10.1016/j.jmb.2008.07.039. [DOI] [PubMed] [Google Scholar]
- 15.Yatime L, Mechulam Y, Blanquet S, Schmitt E. Structural switch of the γ subunit in an archaeal aIF2αγ heterodimer. Structure. 2006;14:119–28. doi: 10.1016/j.str.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 16.Yatime L, Mechulam Y, Blanquet S, Schmitt E. Structure of an archaeal heterotrimeric initiation factor 2 reveals a nucleotide state between the GTP and the GDP states. Proc Natl Acad Sci U S A. 2007;104:18445–50. doi: 10.1073/pnas.0706784104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science. 2011;331:730–6. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- 18.Nissen P, et al. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science. 1995;270:1464–1472. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- 19.Basavappa R, Sigler PB. The 3 Å crystal structure of yeast initiator tRNA: functional implications in initiator/elongator discrimination. EMBO J. 1991;10:3105–3111. doi: 10.1002/j.1460-2075.1991.tb07864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2010;330:1203–9. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- 21.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 22.Dorris DR, Erickson FL, Hannig EM. Mutations in GCD11, the structural gene for eIF-2γ in yeast, alter translational regulation of GCN4 and the selection of the start site for protein synthesis. EMBO J. 1995;14:2239–2249. doi: 10.1002/j.1460-2075.1995.tb07218.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 24.Harashima S, Hinnebusch AG. Multiple GCD genes required for repression of GCN4, a transcriptional activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:3990–3998. doi: 10.1128/mcb.6.11.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Erickson FL, Hannig EM. Ligand interactions with eukaryotic translation initiation factor 2: role of the γ-subunit. EMBO J. 1996;15:6311–6320. [PMC free article] [PubMed] [Google Scholar]
- 26.Sanderson LE, Uhlenbeck OC. Directed mutagenesis identifies amino acid residues involved in elongation factor Tu binding to yeast Phe-tRNAPhe. J Mol Biol. 2007;368:119–30. doi: 10.1016/j.jmb.2007.01.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanderson LE, Uhlenbeck OC. The 51–63 base pair of tRNA confers specificity for binding by EF-Tu. RNA. 2007;13:835–40. doi: 10.1261/rna.485307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dong J, et al. Genetic identification of yeast 18S rRNA residues required for efficient recruitment of initiator tRNAMet and AUG selection. Genes Dev. 2008;22:2242–55. doi: 10.1101/gad.1696608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pisarev AV, et al. Specific functional interactions of nucleotides at key −3 and +4 positions flanking the initiation codon with components of the mammalian 48S translation initiation complex. Genes Dev. 2006;20:624–36. doi: 10.1101/gad.1397906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Battiste JB, Pestova TV, Hellen CUT, Wagner G. The eIF1A solution structure reveals a large RNA-binding surface important for scanning function. Mol Cell. 2000;5:109–119. doi: 10.1016/s1097-2765(00)80407-4. [DOI] [PubMed] [Google Scholar]
- 31.Agrawal RK, Penczek P, Grassucci RA, Frank J. Visualization of elongation factor G on the Escherichia coli 70S ribosome: the mechanism of translocation. Proc Natl Acad Sci U S A. 1998;95:6134–6138. doi: 10.1073/pnas.95.11.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Connell SR, et al. Structural basis for interaction of the ribosome with the switch regions of GTP-bound elongation factors. Mol Cell. 2007;25:751–64. doi: 10.1016/j.molcel.2007.01.027. [DOI] [PubMed] [Google Scholar]
- 33.Gao YG, et al. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326:694–9. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shin BS, et al. rRNA suppressor of a eukaryotic translation initiation factor 5B/initiation factor 2 mutant reveals a binding site for translational GTPases on the small ribosomal subunit. Mol Cell Biol. 2009;29:808–21. doi: 10.1128/MCB.00896-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simonetti A, et al. Structure of the 30S translation initiation complex. Nature. 2008;455:416–20. doi: 10.1038/nature07192. [DOI] [PubMed] [Google Scholar]
- 36.Unbehaun A, et al. Position of eukaryotic initiation factor eIF5B on the 80S ribosome mapped by directed hydroxyl radical probing. Embo J. 2007;26:3109–23. doi: 10.1038/sj.emboj.7601751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yatime L, Schmitt E, Blanquet S, Mechulam Y. Functional molecular mapping of archaeal translation initiation factor 2. J Biol Chem. 2004;279:15984–15993. doi: 10.1074/jbc.M311561200. [DOI] [PubMed] [Google Scholar]
- 38.Asano K, et al. Multiple roles for the carboxyl terminal domain of eIF5 in translation initiation complex assembly and GTPase activation. EMBO J. 2001;20:2326–2337. doi: 10.1093/emboj/20.9.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laurino JP, Thompson GM, Pacheco E, Castilho BA. The β subunit of eukaryotic translation initiation factor 2 binds mRNA through the lysine repeats and a region comprising the C2-C2 motif. Mol Cell Biol. 1999;19:173–181. doi: 10.1128/mcb.19.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Åström S, Byström AS. Rit1, a tRNA backbone-modifying enzyme that mediates initiator and elongator tRNA discrimination. Cell. 1994;79:535–546. doi: 10.1016/0092-8674(94)90262-3. [DOI] [PubMed] [Google Scholar]
- 41.Astrom SU, von Pawel-Rammingen U, Bystrom AS. The yeast initiator tRNAMet can act as an elongator tRNAMet in vivo. J. Mol. Biol. 1993;233:43–58. doi: 10.1006/jmbi.1993.1483. [DOI] [PubMed] [Google Scholar]
- 42.Dale T, Sanderson LE, Uhlenbeck OC. The affinity of elongation factor Tu for an aminoacyl-tRNA is modulated by the esterified amino acid. Biochemistry. 2004;43:6159–66. doi: 10.1021/bi036290o. [DOI] [PubMed] [Google Scholar]
- 43.LaRiviere FJ, Wolfson AD, Uhlenbeck OC. Uniform binding of aminoacyl-tRNAs to elongation factor Tu by thermodynamic compensation. Science. 2001;294:165–8. doi: 10.1126/science.1064242. [DOI] [PubMed] [Google Scholar]
- 44.Kapp LD, Lorsch JR. GTP-dependent recognition of the methionine moiety on initiator tRNA by translation factor eIF2. J Mol Biol. 2004;335:923–36. doi: 10.1016/j.jmb.2003.11.025. [DOI] [PubMed] [Google Scholar]
- 45.Hinnebusch AG. A hierarchy of trans-acting factors modulate translation of an activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol Cell Biol. 1985;5:2349–2360. doi: 10.1128/mcb.5.9.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Culver GM, Noller HF. Directed hydroxyl radical probing of RNA from iron(II) tethered to proteins in ribonucleoprotein complexes. Methods Enzymol. 2000;318:461–75. doi: 10.1016/s0076-6879(00)18070-x. [DOI] [PubMed] [Google Scholar]
- 47.Lowe TM, Eddy SR. A computational screen for methylation guide snoRNAs in yeast. Science. 1999;283:1168–71. doi: 10.1126/science.283.5405.1168. [DOI] [PubMed] [Google Scholar]
- 48.McPheeters DS, Christensen A, Young ET, Stormo G, Gold L. Translational regulation of expression of the bacteriophage T4 lysozyme gene. Nucleic Acids Res. 1986;14:5813–26. doi: 10.1093/nar/14.14.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Acker MG, Kolitz SE, Mitchell SF, Nanda JS, Lorsch JR. Reconstitution of yeast translation initiation. Methods Enzymol. 2007;430:111–45. doi: 10.1016/S0076-6879(07)30006-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.