Abstract
The modulation of DNA repair pathways for therapeutic benefit in cancer has now become a reality with the development of poly (ADP-ribose) polymerase inhibitors (PARPi). PARP is involved in single-strand DNA breaks, which in the presence of defective homologous recombination repair lead to double-strand DNA breaks, the most lethal form of DNA damage. These agents therefore may be the drugs of choice for BRCA mutant breast and ovarian cancers. PARPi result in synergistic antitumor effects when combined with cisplatin, temozolomide, topoisomerase inhibitors and ionizing radiation. The indications for PARPi lie beyond BRCA mutations and may include genomic and functional defects in DNA repair and damage response pathways. Several PARPi are in the clinical development phase at this time and, given the recent failure of a phase III clinical trial of iniparib in triple-negative breast cancer, the identification of structural and functional differences between these inhibitors becomes critical. Acquired resistance to PARPi is being noted and represents an important limitation in this field. A concise review of the literature in this field is presented.
Keywords: BRCA2 gene, DNA repair-deficiency disorders, poly (ADP-ribose) polymerases
Genomic instability: an ‘enabling’ characterisation of cancer
The human genome is continually exposed to potentially deleterious genotoxic events. These events could be endogenous, including oxidative stress from normal metabolism and DNA replication and recombination aberrations or exogenous, resulting from exposure to genotoxic agents such as chemical mutagens and ultraviolet light. Several detection and signaling pathways are activated by DNA damage, resulting in the recruitment and activation of groups of proteins to repair the incurred damage by the appropriate DNA repair pathway. This results either in cell cycle arrest, thereby allowing sufficient time for repair to take place, or in the case of irreparable damage, programmed cell death.
It follows therefore that the genes responsible for DNA damage detection and repair essentially behave as tumor suppressors and their defects would enable tumorigenesis in the presence of ongoing genotoxic stress. The consequent genetic or epigenetic alterations such as mutations, methylation or histone modifications result in genomic instability, which can lead to the evolution of a subclone of cells with a selective growth advantage. These variant subclones may outgrow normal, unaffected tissues and successive clonal expansions of mutated cells may be responsible for the multistep process of tumorigenesis. While hematologic malignancies have only a limited number of genetic alterations, in the case of solid tumors, myriad genetic alterations lead to genetic heterogeneity. Genomic instability has been described as an enabling characteristic of cancer and is broadly classified into microsatellite instability associated with the mutator phenotype, and chromosome instability recognized by gross chromosomal abnormalities [Hanahan and Weinberg, 2011].
Several DNA repair pathways are responsible for the maintenance of genomic integrity, each of which repairs a specific type of DNA damage. These include nonhomologous end joining (NHEJ), homologous recombination (HR), base excision repair (BER, also called single-strand break repair [SSBR]), nucleotide excision repair (NER), mismatch repair (MMR) and translesion synthesis (TLS). Each of these pathways repairs a specific DNA defect. For instance, MMR is involved in the detection and repair of base mismatches, insertions and deletions, BER/SSBR removes incorrect bases and repairs resultant nicks while NHEJ and HR are involved in the repair of DNA double-strand breaks (DSBs) and repair of DNA crosslinks is complex involving NER, HR and possibly other pathways. Interestingly, although DNA repair defects lead to increased tumorigenesis, a paradoxical situation arises from the fact that in cancer cells, sustained replication in the presence of genotoxic stress also requires some intact DNA repair pathways. Thus, cancers with aberrant DNA repair pathways become ‘addicted’ to one or more retained intact repair pathways to preserve their growth. This can represent a resistance mechanism to certain types of DNA damaging chemotherapy and radiotherapy. Targeting the upregulated DNA repair pathway can enhance the DNA damage and antitumor activity induced by radiotherapy and chemotherapy. These upregulated DNA damage signaling and repair pathways that cancer cells are addicted to may also represent the cancer’s ‘Achilles’ heel’. A specific inhibitor to the pathway could potentially lead to a selective antitumor effect by preventing the repair of intrinsic DNA damage by exploiting the principle of synthetic lethality.
Synthetic lethality
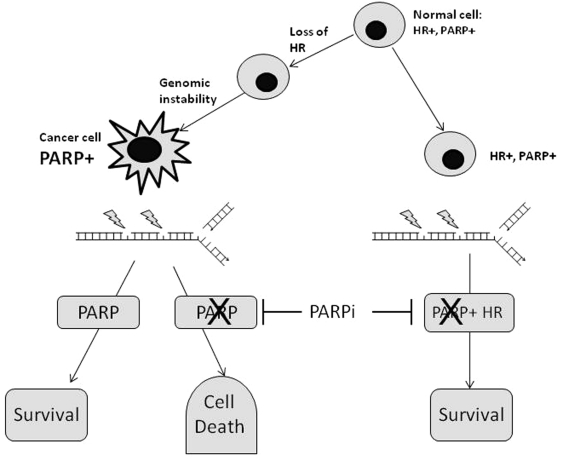
Originally described by the geneticist Dobzhansky in the 1940s, synthetic lethality refers to an interaction in which the individual deletion of either of two genes has no effect but the combined deletion of both genes is cytotoxic (Figure 1). Synthetic lethality can also be exploited in the treatment of cancer as in the case with cancer susceptibility syndromes, such as BRCA1 or BRCA2 mutations. The latter genes play a key role in maintaining genomic integrity due to their involvement in HR, an important repair pathway for DNA DSBs. Cancer cells with aberrant HR secondary to BRCA mutations are critically dependent on BER/SSBR for viability. The enzyme, poly (ADP-ribose) polymerase 1 (PARP-1) is critical for BER/SSBR (described below). Inhibition of PARP-1 leads to an accumulation of unrepaired SSBs and therefore is synthetically lethal in the case of BRCA1 or BRCA2 mutations due to the accumulation of fatal replication fork collapse and DSBs as was demonstrated by two independent groups [Bryant et al. 2005; Farmer et al. 2005]. Recent evidence suggests that activation of NHEJ is necessary for synthetic lethality, suggesting that the error-prone repair of replication-associated DSBs is associated with the cytotoxicity of PARP inhibitors (PARPi) in HR-defective cells [Patel et al. 2011]. While PARPi are effective in the case of BRCA1 or BRCA2 mutations, the paradigm of synthetic lethality can also be extended to other cancers, including sporadic cases. HR is a complex process involving many components including ATM, ATR, CHK1, RAD51 and its homologues, the FANC proteins, MRE11/RAD50/NBS1 (MRN) and loss of function in any of these components may confer sensitivity to PARPi [Mccabe et al. 2006]. PARPi may also be synthetically lethal in cases where epigenetic silencing of BRCA occurs [Drew et al. 2010]. This effect in sporadic breast and ovarian cancers was referred to as ‘BRCA-ness’ [Ashworth, 2008] but it is now apparent that this BRCA-centric view is misleading as defects in other HR components are associated with a variety of cancers, e.g. ATM defects in mantle-cell lymphoma, which may also benefit from PARPi therapy [Williamson et al. 2010]. EMSY and PTEN have also been implicated as they regulate the activity of other components of HR [Cousineau and Belmaaza, 2011; Mcellin et al. 2010].
Figure 1.
Poly (ADP-ribose) polymerase (PARP) is upregulated in conditions causing genotoxic stress, leading to increased single-strand break repair. In cases of homologous recombination (HR) deficiency, this becomes the main pathway for DNA repair and therefore its inhibition leads to synthetic lethality. PARPi, PARP inhibitor.
PARP structure–function relationship
At the current time, a total of 16 PARP family members have been identified of which PARP1, PARP2, PARP4 (Vault-PARP), Tankyrase-1 and 2 have confirmed poly (ADP)-ribosylating activity and only PARP-1 and PARP-2 are involved in DNA repair [Schreiber et al. 2006]. Recently, PARP-3 was identified as cooperating with PARP-1 in DNA DSB repair, but deletion of PARP-3 alone does not compromise survival after DNA damage and the mechanisms remain to be fully elucidated [Boehler et al. 2011]. PARP-1 was the first member of this family to be discovered and its function in the maintenance of genomic integrity has been well documented. In response to DNA single-strand breaks resulting from genotoxic stimuli, the PARP reaction uses nicotinamide adenine dinucleotide (NAD) as a substrate to generate poly (ADP-ribose) (PAR) [De Murcia, G. and Menissier De Murcia, J. 1994; De Murcia, G et al. 1994].
PARP-1 and PARP-2 form homodimers and heterodimers at DNA breaks catalyzing the formation of long PAR chains covalently attached to PARP-1 itself (automodification) or other nuclear proteins, e.g. histone H1 heteromodification adjacent to the DNA breaks. These negatively charged polymers form a scaffold and recruit other proteins that are critical in BER [De Murcia et al. 1994] and chromatin remodeling [Ahel et al. 2009; Timinszky et al. 2009].
PARP activity also promotes the activation of mitotic recombination 11 (MRE11) and Nijmegen breakage syndrome (NBS), members of the DNA damage-sensing MRN complex which activates ATM to sites of double-stand DNA damage [Haince et al. 2007]. Thus, the role of PARP-1 in DNA repair extends beyond the repair of DNA single-strand breaks. PARP-1 not only plays a critical role in genomic maintenance but is also involved in transcriptional regulation, energy metabolism and cell death and these roles are discussed below.
PARP-1 has three distinct domains: an amino terminal DNA-binding domain, a nuclear localization signal, an automodification domain and a carboxy-terminal catalytic ‘PARP-signature’ domain that is responsible for PAR formation [De Murcia et al. 1994]. The DNA-binding domain also contains two zinc fingers that are required for the detection of DNA strand breaks resulting eventually in PARP-1 activation while a third zinc finger motif coordinates DNA-dependent enzyme activation.
The baseline activity of PARP-1 is low, but is stimulated by DNA strand breaks. PARP is upregulated in several cancers, implying its possible role in cancer growth and survival [Virag and Szabo, 2002]. In colorectal cancer, for instance, PARP-1 mRNA overexpression was detected in over 70% of colorectal cancers and correlated with the expression of beta-catenin, c-myc, cyclin D1 and MMP-7 [Nosho et al. 2006]. Inhibition of PARP is detrimental to cancer cells. However, PARP inhibition may not result in critical injury to normal cells. PARP-1 knockout mice have been reported to grow normally [Shall and De Murcia, 2000], however, the inactivation of both PARP-1 and PARP-2, confers embryonic lethality [Schreiber et al. 2002]. Owing to the very close structural homology of the catalytic domains of PARP-1 and PARP-2, it is thought that most PARPi inhibit both enzymes. Therefore, PARP inhibition in the clinical setting could potentially cause serious adverse effects but the experience to date suggests that profound PARP inhibition is associated with very mild toxicity. The clinical application of PARPi is therefore an active area of research and development.
Development of PARP Inhibitors
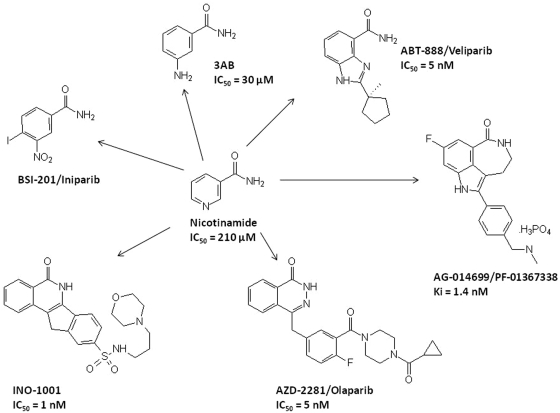
The first-generation PARPi included nicotinamide, benzamides and substituted benzamides such as 3-aminobenzamide (3-AB). These agents had relatively low potency and specificity and, therefore, second-generation benzamides and more recently, third-generation inhibitors, most of which are competitive NAD+ inhibitors and based on 3-AB structure, such as the nicotinamide pharmacophore have been developed (Figure 2). The preclinical/clinical development of PARPi has been as: (a) single agents in cases of deficient DNA repair mechanisms such as BRCA1 or BRCA2 mutations; (b) combined with cytotoxics (PARPi sensitize tumor cells to chemotherapeutic anticancer agents that induce DNA damage through BER; these agents include temozolomide, platinum analogues and topoisomerase-1 inhibitors); or (c) as radiation sensitizers [Calabrese et al. 2004]. Radiosensitization by PARP inhibition is enhanced in the presence of defective DNA repair and is more pronounced in rapidly dividing cancer tissues in the S phase as compared with normal, noncycling cells and may thus result in an enhanced safety ratio [Dungey et al. 2008]. PARP knockout models have been utilized to confirm the chemo- and radio-potentiation of the PARPi. Both PARP-1 knockout and PARP-2 knockout mice are hypersensitive to ionizing radiation (IR) and DNA alkylating agents (as reviewed by Rouleau et al. [2010]). In preclinical cancer models, Tentori and colleagues noted that melanoma models with stably silenced PARP-1 expression were highly sensitive to temozolomide. In the same study, decreased tumorigenicity and angiogenesis were noted in the PARP-1−/− melanoma models [Tentori et al. 2008]. On the other hand, Chalmers and colleagues, noted that while chemical inhibition of PARP-1 markedly enhanced the efficacy of low-dose radiation, such an effect was lacking in PARP-1 knockout models. The latter may be explainable on the basis of PARP-2 upregulation, which may compensate for the absence of PARP-1 [Chalmers et al. 2004]. Thus PARPi, by inhibiting both PARP-1 and PARP-2, are likely to have more profound effects than indicated by genetic knockout of only one of the two enzymes. Select PARP-1 inhibitors in clinical trials are discussed below.
Figure 2.
Structure of 3-aminobenzamide (3-AB) and the recent poly (ADP-ribose) polymerase inhibitors (PARPi) developed from the nicotinamide pharmacophore. The Ki of AG014699/PF-01367388 and the IC50 values for most other inhibitors are presented to show potency, but it should be noted that these values are not directly comparable due to different experimental conditions, e.g. substrate concentrations.
AG014699 (PF-01367338)
AG014699 is a phosphate salt of AG14447 with aqueous solubility and was selected as suitable for clinical trial from a panel of 42 potential PARPi based on its chemo- and radio-potentiating effect [Thomas et al. 2007]. This PARPi, and its forerunner AG14361, showed spectacular activity in xenograft models in combination with temozolomide, resulting in complete and durable tumor regression [Thomas et al. 2007; Calabrese et al. 2004]. AG14361 also caused a twofold to threefold enhancement of irinotecan-induced and radiation-induced tumor growth delay. AG014699 was the first PARPi to enter clinical trial for cancer therapy and has been studied in phase I and phase II clinical trials in combination with temozolomide for the treatment of metastatic melanoma. In the phase I study, dose escalation was driven by pharmacodynamic measurement of PARP inhibition and the PARP inhibitory dose (PID) was estimated at 12 mg/m2 based on 74–97% inhibition of PARP activity in peripheral lymphocytes and a >50% PARP inhibition in tumor biopsies posttreatment. AG014699 showed linear pharmacokinetics with no interaction with temozolomide [Plummer et al. 2008]. The recommended phase II dose was 200 mg/m2 of temozolomide with 12 mg/m2 of AG014699. In the phase II study, a doubling of response rate and time to tumor progression were noted as compared with temozolomide alone, but at the cost of significantly higher myelosuppression in the combination arm. At the present time, single-agent studies in ovarian or breast cancers in BRCA mutation carriers and combination studies with cisplatin, pemetrexed and epirubicin are being conducted. The combination of AG014699 with these latter drugs not classically associated with PARP may be based on the observation that AG014699 is vasoactive, potentially increasing drug delivery to the tumor [Ali et al. 2009].
Veliparib (ABT-888)
Veliparib has been developed as a PARP-1 and PARP-2 inhibitor with K(i)s of 5.2 and 2.9 nmol/l, respectively [Penning et al. 2009]. It is orally bioavailable and crosses the blood–brain barrier. ABT-888 potentiated the cytotoxic effect of temozolomide in several human tumor models and IR in HCT116 human colon cancers [Clarke et al. 2009; Palma et al. 2009; Donawho et al. 2007]. The activity of platinum analogues and cyclophosphamide were also enhanced by ABT-888 in the BRCA1 and 2 defective MX-1 xenografts but ABT-888 had no single-agent activity in this model in the schedule used [Donawho et al. 2007]. Velaparib was investigated in an innovative phase 0 trial, the first such study in oncology [Kummar et al. 2009]. The primary study endpoint was target modulation by the PARPi. PARP activity, when measured after a single dose of veliparib was significantly inhibited at the 25 and 50 mg doses. There is an extensive clinical trial program associated with this agent with 32 ongoing clinical trials of velaparib in combination with cytotoxics in ovarian, breast, colorectal, prostate, liver cancers, neurologic malignancies and leukemias.
Olaparib (AZD2281)
Olaparib (AZD2281) also inhibits PARP-1 and PARP-2 at nanomolar concentrations [Menear et al. 2008]. Preclinical studies have largely concentrated on investigations of synthetic lethality in BRCA1 or BRCA2 defective models or combinations with platinum in these models [Rottenberg et al. 2008]. Radiosensitization in a glioma model has also been demonstrated [Dungey et al. 2008]. Studies with human ovarian cancer xenografts demonstrated that olaparib had single-agent activity and increased the efficacy of carboplatin in xenografts defective in BRCA2 but not those with normal BRCA function [Kortmann et al. 2011]. Olaparib was observed to increase the toxicity of topotecan in animal models [Zander et al. 2010]. The first clinical study of PARP inhibition in BRCA mutant cancers was with this agent. In this phase I study which enrolled 60 patients, olaparib doses were escalated from 10 mg daily for 2 of every 3 weeks to 600 mg twice daily [Fong et al. 2009a]. The dose of 200 mg twice daily was selected for further study in a select cohort of 23 patients with BRCA mutations. In this group, nine had partial responses according to the NCI response evaluation criteria (RECIST). A total of 19 of the 23 patients had BRCA-associated tumors, including breast, ovarian, and prostate cancers.
Given these interesting preliminary data, two multicenter, international phase II studies of olaparib in patients with breast or ovarian cancers having BRCA1 or BRCA2 mutations were conducted [Fong et al. 2009b; Tutt et al. 2010]. Patients enrolled were refractory to standard chemotherapeutic regimens. A total of 27 patients in the first cohort received 400 mg of olaparib twice daily for 28 days, and 27 patients in the second cohort received 100 mg of olaparib twice daily. The overall response rate was 41% with 400 mg, and 22% with 100 mg olaparib. The median time to progression was 5.7 and 3.8 months, respectively. The common adverse effects were mild, including fatigue, nausea and vomiting. A parallel study using the two dosage regimens in 55 BRCA-mutated carriers with ovarian cancer confirmed an overall response rate of 33% in the 400 mg group, and 12.5% in the 100 mg group. These proof-of-concept studies confirmed that BRCA1 or BRCA2 mutational status serves as a predictive marker for PARPi.
Iniparib (BSI-201)
Unlike other PARPi that compete with NAD+ for the PARP catalytic site, iniparib (4-iodo, 3-nitrobenzamide) is unique in that it targets the zinc finger domain and prevents PARP-1 activation by DNA breaks [Mendeleyev et al. 1995]. Therefore, it may have differential effects as compared with other synthetic catalytic PARPi. Moreover, as this inhibitor has also been shown to inhibit other enzymes such as GAPDH [Bauer et al. 2002], it would be dangerous to conclude that its anticancer effects are solely attributable to PARP inhibition. This agent has been extensively investigated in triple-negative (TN) breast cancers.
TN breast cancers are believed to share the molecular characteristics of BRCA1-associated cancers [Vona-Davis et al. 2008]. Both, BRCA1-associated cancers and sporadic TN tumors share a high degree of genomic instability, implying an impaired ability to repair DNA damage. HR defects seen in TN breast cancer include BRCA1 methylation, overexpression of deregulators including ID4 and HMG as well as aberrations of MRE11, ATM and PALB2 [Alli et al. 2009; Alexander et al. 2010]. Iniparib (BSI-201), when combined with gemcitabine and carboplatin for the treatment of TN breast cancer, has been studied in a randomized phase II trial compared with the same chemotherapy alone. The addition of iniparib increased disease control rate (56% to 34%), response rate (52% to 32%), progression-free survival (5.9 to 3.6 months) and overall survival (12.3 to 7.7 months) without increasing toxicity [O'shaughnessy et al. 2011]. A follow-up phase III study, however, was negative as it did not meet the prespecified criteria for significance for coprimary endpoints of overall survival and progression-free survival. Given the structural and mechanistic differences between iniparib and other PARPi, these negative results do not necessarily imply a ‘class effect’ and further study of TN breast cancer with other PARPi should be encouraged.
INO-1001
This agent is an isoindolinone derivative and is being developed for both oncological and cardiovascular indications. Preclinical studies demonstrate its protective effect in models of cardiac dysfunction [Pacher et al. 2006] and reversal of temozolomide resistance in MMR-defective xenografts [Cheng et al. 2005]. This was the first PARP-1 inhibitor to be investigated for cardiovascular disease and has been granted orphan drug status by the US Food and Drug Administration for the prevention of postoperative aortic aneurysm repair complications. In this phase II study, INO-1001 may have reduced the plasma levels of C-reactive protein and the inflammatory marker interleukin-6, without reducing plasma markers of myocardial injury. No serious toxic events ensued in this trial [Morrow et al. 2009]. This agent is being developed in oncology in melanoma and glioma and as a single agent in cancer for BRCA1- and BRCA2-deficient tumors. Phase I studies of INO-001 at 100, 200 and 400 mg/m2 in combination with temozolomide indicated a short terminal half life and the dose-limiting toxicities observed at the highest dose were myelosuppression and elevated liver enzymes [Bedikian et al. 2009].
Other PARPi in preclinical and phase I trial stages include GPI21016 (MGI/Eisai), MK-4827 (Merck), BMN-673 (Biomarin) and CEP-9722 (Cephalon). Additional information on these inhibitors can be found in a review by Ferraris [Ferraris, 2010].
Resistance mechanisms for PARPi
Acquired resistance to targeted agents is common and PARPi are no exception in this regard. As the clinical development of PARPi is still at an early stage, the underlying resistance mechanisms have not yet been elucidated. However, preclinical studies offer interesting possibilities. CAPAN-1 pancreatic cancer cells lines are BRCA2 deficient secondary to a 6174delT frameshift mutation, which makes them exquisitely sensitive to PARPi. CAPAN-1 cells cannot form damage-induced RAD51 foci, as they are HR-defective. PARPi-resistant clones were highly resistant to the drug (over 1000-fold), and were also cross-resistant to the DNA crosslinking agent, cisplatin. Interestingly, these resistant clones acquired the ability to form RAD51 foci after PARPi treatment or exposure to irradiation suggesting that re-acquisition of HR capability may be the mechanism of acquired resistance. In support of this, DNA sequencing of PARP inhibitor-resistant clones revealed new BRCA2 isoforms as a result of intragenic deletion of the c.6174delT mutation and restoration of the open reading frame [Edwards et al. 2008; Sakai et al. 2008; Swisher et al. 2008]. Recently, 53BP1 has been shown to promote error-prone NHEJ in BRCA1 mutant cells and that loss of 53BP1 partially restores HR function and can rescue from DNA damaging agent and PARPi sensitivity [Bouwman et al. 2010; Bunting et al. 2010]. Loss of 53BP1 appears to be relatively common in TN and BRCA1-mutant breast cancer samples [Bouwman et al. 2010].
An alternative mechanism was described with olaparib [Rottenberg et al. 2008]. In this case, resistance may be related to the upregulation of the ABCB1a/b genes, which encode P-glycoprotein multidrug resistance drug efflux pumps; this effect could be reversed with the P-glycoprotein inhibitor, tariquidar. A recent study investigated the role of 6-thioguanine in reversing this resistance mechanism. Issaeva and colleagues first noted that BRCA1, BRCA2 or XRCC3 tumors are very sensitive to 6-thioguanine as HR is involved in the repair of 6-thioguanine-induced DSBs [Issaeva et al. 2010]. 6-thioguanine is not a substrate for p glycoprotein and is a potent cytotoxic in PARP-resistant tumors. Furthermore, these investigators noted that genetically reverted BRCA2 defective tumors also retain sensitivity to 6-thioguanine. Altogether, these findings suggest that 6-thioguanine may be efficient in also killing advanced and drug-resistant BRCA1 or BRCA2 defective tumors.
Patient selection for PARPi trials
A major challenge is to identify predictive markers for PARPi therapy for those sporadic cancers that may benefit due to functional defects in the HR pathways. Several surrogate markers have been described, none of which are widely available in the clinical setting at the current time. Gene-expression arrays have been investigated for their predictive value [Jazaeri et al. 2002]. Turner and colleagues hypothesized that a gene and protein expression signature may have the ability to identify the BRCA-ness profile in patients [Turner et al. 2004]. Konstantinopoulos and colleagues defined a BRCA-like gene expression profile that correlated with PARPi and platinum sensitivity [Konstantinopoulos et al. 2010]. Phosphorylation of the Ser-139 residue of the histone variant H2AX, forming γH2AX, is an early cellular response to the induction of DNA DSBs. Detection of this phosphorylation event has emerged as a highly specific and sensitive molecular marker for monitoring DNA damage initiation and resolution. This accumulation is detectable by immunofluorescence using an antibody to γH2AX. Rad51 is a crucial downstream protein involved in HR repair, which is relocalized within the nucleus in response to DNA damage. Rad51 foci can also be visualized by immunofluorescent microscopy and are thought to represent assemblies of proteins at these sites of HR repair. Combination γH2AX/RAD51 immunofluorescence has been investigated in primary ovarian cancer cell cultures [Mukhopadhyay et al. 2010] and primary acute myeloid leukemia (AML) cultures [Gaymes et al. 2009]. Both studies demonstrated that raised γH2AX and decreased RAD51 foci expression predicts PARPi sensitivity. Graeser and colleagues investigated RAD51 immunofluorescence in replicating (geminin positive) cells in breast cancer biopsy specimens from women receiving neoadjuvant anthracycline therapy. The RAD51 score was predictive of complete response in women receiving neoadjuvant therapy for breast cancer [Graeser et al. 2010]. Although cumbersome, these assays currently represent the most reliable way to identify HR defects, particularly in light of the recent studies showing that even in BRCA1-mutant tumors, coincident loss of 53BP1 can restore HR function and PARPi resistance [Bouwman et al. 2010; Bunting et al. 2010].
Conclusions
Genomic instability in cancer may result from an imbalance of DNA damage signaling and repair pathways; upregulated pathways may confer resistance to certain types of genotoxic agent but may also be necessary for the survival of the cancer cell. Targeting these pathways may result in single-agent activity specifically in the cancer cell where repair of endogenously generated DNA damage is inhibited as well as tumor-selective chemo and radio-sensitization. PARPi increase the anticancer activity of temozolomide, topoisomerase I poisons and IR in a wide range of tumor models, an approach that has been validated by genetic knockdown of PARP-1 and PARP-2. However, the most exciting aspect is the discovery that PARPi alone selectively kill cancer cells that lack HR without affecting repair competent cells. This observation has rapidly translated into clinical trials where PARPi have shown good anticancer activity in BRCA1 and BRCA2 patients with breast, ovarian and prostate cancer with only mild toxicities. HR is a complex and multicomponent pathway and preclinical data indicates that PARPi will be useful in tumors lacking any one of a number of these key proteins. Identification of these potentially PARPi-responsive tumors is the next challenge. Gene expression signatures and assays of HR function can fulfill this function but they are currently too expensive and cumbersome to become routine clinical practice.
Funding
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Conflict of interest statement
Milind Javle has no conflicts of interest to declare. Nicola Curtin receives current research support from Pfizer and BioMarin and consultancy fees from BioMarin and Eisai and has previously received research support and consultancy fees from BiPAR sciences.
References
- Ahel D., Horejsi Z., Wiechens N., Polo S.E., Garcia-Wilson E., Ahel I., et al. (2009) Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science 325: 1240–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander B.M., Sprott K., Farrow D.A., Wang X., D'andrea A.D., Schnitt S.J., et al. (2010) DNA repair protein biomarkers associated with time to recurrence in triple-negative breast cancer. Clin Cancer Res 16: 5796–5804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali M., Telfer B.A., Mccrudden C., O'rourke M., Thomas H.D., Kamjoo M., et al. (2009) Vasoactivity of AG014699, a clinically active small molecule inhibitor of poly(ADP-ribose) polymerase: a contributory factor to chemopotentiation in vivo?. Clin Cancer Res 15: 6106–6112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alli E., Sharma V.B., Sunderesakumar P., Ford J.M. (2009) Defective repair of oxidative DNA damage in triple-negative breast cancer confers sensitivity to inhibition of poly(ADP-ribose) polymerase. Cancer Res 69: 3589–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashworth A. (2008) A synthetic lethal therapeutic approach: poly(ADP) ribose polymerase inhibitors for the treatment of cancers deficient in DNA double-strand break repair. J Clin Oncol 26: 3785–3790 [DOI] [PubMed] [Google Scholar]
- Bauer P.I., Mendeleyeva J., Kirsten E., Comstock J.A., Hakam A., Buki K.G., et al. (2002) Anti-cancer action of 4-iodo-3-nitrobenzamide in combination with buthionine sulfoximine: inactivation of poly(ADP-ribose) polymerase and tumor glycolysis and the appearance of a poly(ADP-ribose) polymerase protease. Biochem Pharmacol 63: 455–462 [DOI] [PubMed] [Google Scholar]
- Bedikian A.Y., Papadopoulos N.E., Kim K.B., Hwu W.J., Homsi J., Glass M.R., et al. (2009) A phase Ib trial of intravenous INO-1001 plus oral temozolomide in subjects with unresectable stage-III or IV melanoma. Cancer Invest 27: 756–763 [DOI] [PubMed] [Google Scholar]
- Boehler C., Gauthier L.R., Mortusewicz O., Biard D.S., Saliou J.M., Bresson A., et al. (2011) Poly(ADP-ribose) polymerase 3 (PARP3), a newcomer in cellular response to DNA damage and mitotic progression. Proc Natl Acad Sci U S A , in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman P., Aly A., Escandell J.M., Pieterse M., Bartkova J., Van Der Gulden H., et al. (2010) 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol 17: 688–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant H.E., Schultz N., Thomas H.D., Parker K.M., Flower D., Lopez E., et al. (2005) Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434: 913–917 [DOI] [PubMed] [Google Scholar]
- Bunting S.F., Callen E., Wong N., Chen H.T., Polato F., Gunn A., et al. (2010) 53BP1 inhibits homologous recombination in BRCA1-deficient cells by blocking resection of DNA breaks. Cell 141: 243–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese C.R., Almassy R., Barton S., Batey M.A., Calvert A.H., Canan-Koch S., et al. (2004) Anticancer chemosensitization and radiosensitization by the novel poly(ADP-ribose) polymerase-1 Inhibitor AG14361. J Natl Cancer Inst 96: 56–67 [DOI] [PubMed] [Google Scholar]
- Chalmers A., Johnston P., Woodcock M., Joiner M., Marples B. (2004) PARP-1, PARP-2, and the cellular response to low doses of ionizing radiation. Int J Radiat Oncol Biol Phys 58: 410–419 [DOI] [PubMed] [Google Scholar]
- Cheng C.L., Johnson S.P., Keir S.T., Quinn J.A., Ali-Osman F., Szabo C., et al. (2005) Poly(ADP-Ribose) polymerase-1 inhibition reverses temozolomide resistance in a DNA mismatch repair-deficient malignant glioma xenograft. Mol Cancer Ther 4: 1364–1368 [DOI] [PubMed] [Google Scholar]
- Clarke M.J., Mulligan E.A., Grogan P.T., Mladek A.C., Carlson B.L., Schroeder M.A., et al. (2009) Effective sensitization of temozolomide by ABT-888 is lost with development of temozolomide resistance in glioblastoma xenograft lines. Mol Cancer Ther 8: 407–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousineau I., Belmaaza A. (2011) EMSY overexpression disrupts the BRCA2/RAD51 Pathway in the DNA-damage response: implications for chromosomal instability/recombination syndromes as checkpoint diseases. Mol Genet Genomics , in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Murcia G., Menissier De Murcia J. (1994) Poly(ADP-ribose) polymerase: a molecular nick-sensor. Trends Biochem Sci 19: 172–176 [DOI] [PubMed] [Google Scholar]
- De Murcia G., Schreiber V., Molinete M., Saulier B., Poch O., Masson M., et al. (1994) Structure and function of poly(ADP-ribose) polymerase. Mol Cell Biochem 138: 15–24 [DOI] [PubMed] [Google Scholar]
- Donawho C.K., Luo Y., Penning T.D., Bauch J.L., Bouska J.J., Bontcheva-Diaz V.D., et al. (2007) ABT-888, an orally active poly(ADP-ribose) polymerase inhibitor that potentiates DNA-damaging agents in preclinical tumor models. Clin Cancer Res 13: 2728–2737 [DOI] [PubMed] [Google Scholar]
- Drew Y., Mulligan E.A., Vong W.T., Thomas H.D., Kahn S., Kyle S., et al. (2010) Therapeutic potential of poly(ADP-ribose) polymerase inhibitor AG014699 in human cancers with mutated or methylated BRCA1 or BRCA2. J Natl Cancer Inst 103: 334–346 [DOI] [PubMed] [Google Scholar]
- Dungey F.A., Loser D.A., Chalmers A.J. (2008) Replication-dependent radiosensitization of human glioma cells by inhibition of poly(ADP-ribose) polymerase: mechanisms and therapeutic potential. Int J Radiat Oncol Biol Phys 72: 1188–1197 [DOI] [PubMed] [Google Scholar]
- Edwards S.L., Brough R., Lord C.J., Natrajan R., Vatcheva R., Levine D.A., et al. (2008) Resistance to therapy caused by intragenic deletion in BRCA2. Nature 451: 1111–1115 [DOI] [PubMed] [Google Scholar]
- Farmer H., Mccabe N., Lord C.J., Tutt A.N., Johnson D.A., Richardson T.B., et al. (2005) Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434: 917–921 [DOI] [PubMed] [Google Scholar]
- Ferraris D.V. (2010) Evolution of poly(ADP-ribose) polymerase-1 (PARP-1) inhibitors. From concept to clinic. J Med Chem 53: 4561–4584 [DOI] [PubMed] [Google Scholar]
- Fong P.C., Boss D.S., Yap T.A., Tutt A., Wu P., Mergui-Roelvink M., et al. (2009a) Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med 361: 123–134 [DOI] [PubMed] [Google Scholar]
- Fong P.C., Yap T.A., Boss D.S., Carden C.P., Mergui-Roelvink M., Gourley C., et al. (2009b) Poly(ADP)-ribose polymerase inhibition: frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. J Clin Oncol 28: 2512–2519 [DOI] [PubMed] [Google Scholar]
- Gaymes T.J., Shall S., Macpherson L.J., Twine N.A., Lea N.C., Farzaneh F., et al. (2009) Inhibitors of poly ADP-ribose polymerase (PARP) induce apoptosis of myeloid leukemic cells: potential for therapy of myeloid leukemia and myelodysplastic syndromes. Haematologica 94: 638–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graeser M., Mccarthy A., Lord C.J., Savage K., Hills M., Salter J., et al. (2010) A marker of homologous recombination predicts pathologic complete response to neoadjuvant chemotherapy in primary breast cancer. Clin Cancer Res 16: 6159–6168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haince J.F., Kozlov S., Dawson V.L., Dawson T.M., Hendzel M.J., Lavin M.F., et al. (2007) Ataxia telangiectasia mutated (ATM) signaling network is modulated by a novel poly(ADP-ribose)-dependent pathway in the early response to DNA-damaging agents. J Biol Chem 282: 16441–16453 [DOI] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R.A. (2011) Hallmarks of cancer: the next generation. Cell 144: 646–674 [DOI] [PubMed] [Google Scholar]
- Issaeva N., Thomas H.D., Djureinovic T., Jaspers J.E., Stoimenov I., Kyle S., et al. (2010) 6-Thioguanine selectively kills BRCA2-defective tumors and overcomes PARP inhibitor resistance. Cancer Res 70: 6268–6276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazaeri A.A., Yee C.J., Sotiriou C., Brantley K.R., Boyd J., Liu E.T. (2002) Gene expression profiles of BRCA1-linked, BRCA2-linked, and sporadic ovarian cancers. J Natl Cancer Inst 94: 990–1000 [DOI] [PubMed] [Google Scholar]
- Konstantinopoulos P.A., Spentzos D., Karlan B.Y., Taniguchi T., Fountzilas E., Francoeur N., et al. (2010) gene expression profile of BRCAness that correlates with responsiveness to chemotherapy and with outcome in patients with epithelial ovarian cancer. J Clin Oncol 28: 3555–3561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortmann U., Mcalpine J.N., Xue H., Guan J., Ha G., Tully S., et al. (2011) Tumor growth inhibition by olaparib in BRCA2 germline-mutated patient-derived ovarian cancer tissue xenografts. Clin Cancer Res 17: 783–791 [DOI] [PubMed] [Google Scholar]
- Kummar S., Kinders R., Gutierrez M.E., Rubinstein L., Parchment R.E., Phillips L.R., et al. (2009) Phase 0 clinical trial of the poly (ADP-ribose) polymerase inhibitor ABT-888 in patients with advanced malignancies. J Clin Oncol 27: 2705–2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mccabe N., Turner N.C., Lord C.J., Kluzek K., Bialkowska A., Swift S., et al. (2006) Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res 66: 8109–8115 [DOI] [PubMed] [Google Scholar]
- Mcellin B., Camacho C.V., Mukherjee B., Hahm B., Tomimatsu N., Bachoo R.M., et al. (2010) PTEN loss compromises homologous recombination repair in astrocytes: implications for glioblastoma therapy with temozolomide or poly(ADP-ribose) polymerase inhibitors. Cancer Res 70: 5457–5464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendeleyev J., Kirsten E., Hakam A., Buki K.G., Kun E. (1995) Potential chemotherapeutic activity of 4-iodo-3-nitrobenzamide. Metabolic reduction to the 3-nitroso derivative and induction of cell death in tumor cells in culture. Biochem Pharmacol 50: 705–714 [DOI] [PubMed] [Google Scholar]
- Menear K.A., Adcock C., Boulter R., Cockcroft X.L., Copsey L., Cranston A., et al. (2008) 4-[3-(4-Cyclopropanecarbonylpiperazine-1-carbonyl)-4-fluorobenzyl]-2H-PHTH alazin-1-one: a novel bioavailable inhibitor of poly(ADP-ribose) polymerase-1. J Med Chem 51: 6581–6591 [DOI] [PubMed] [Google Scholar]
- Morrow D.A., Brickman C.M., Murphy S.A., Baran K., Krakover R., Dauerman H., et al. (2009) A randomized, placebo-controlled trial to evaluate the tolerability, safety, pharmacokinetics, and pharmacodynamics of a potent inhibitor of poly(ADP-ribose) polymerase (INO-1001) in patients with ST-elevation myocardial infarction undergoing primary percutaneous coronary intervention: results of the TIMI 37 Trial. J Thromb Thrombolysis 27: 359–364 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay A., Elattar A., Cerbinskaite A., Wilkinson S.J., Drew Y., Kyle S., et al. (2010) Development of a functional assay for homologous recombination status in primary cultures of epithelial ovarian tumor and correlation with sensitivity to poly(ADP-ribose) polymerase inhibitors. Clin Cancer Res 16: 2344–2351 [DOI] [PubMed] [Google Scholar]
- Nosho K., Yamamoto H., Mikami M., Taniguchi H., Takahashi T., Adachi Y., et al. (2006) Overexpression of poly(ADP-ribose) polymerase-1 (PARP-1) in the early stage of colorectal carcinogenesis. Eur J Cancer 42: 2374–2381 [DOI] [PubMed] [Google Scholar]
- O'shaughnessy J., Osborne C., Pippen J.E., Yoffe M., Patt D., Rocha C., et al. (2011) Iniparib plus chemotherapy in metastatic triple-negative breast cancer. N Engl J Med 364: 205–214 [DOI] [PubMed] [Google Scholar]
- Pacher P., Liaudet L., Mabley J.G., Cziraki A., Hasko G., Szabo C. (2006) Beneficial effects of a novel ultrapotent poly(ADP-ribose) polymerase inhibitor in murine models of heart failure. Int J Mol Med 17: 369–375 [PMC free article] [PubMed] [Google Scholar]
- Palma J.P., Wang Y.C., Rodriguez L.E., Montgomery D., Ellis P.A., Bukofzer G., et al. (2009) ABT-888 confers broad in vivo activity in combination with temozolomide in diverse tumors. Clin Cancer Res 15: 7277–7290 [DOI] [PubMed] [Google Scholar]
- Patel A.G., Sarkaria J.N., Kaufmann S.H. (2011) Nonhomologous end joining drives poly(ADP-ribose) polymerase (PARP) inhibitor lethality in homologous recombination-deficient cells. Proc Natl Acad Sci U S A 108: 3406–3411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penning T.D., Zhu G.D., Gandhi V.B., Gong J., Liu X., Shi Y., et al. (2009) Discovery of the poly(ADP-ribose) polymerase (PARP) inhibitor 2-[(R)-2-methylpyrrolidin-2-Yl]-1h-benzimidazole-4-carboxamide (ABT-888) for the treatment of cancer. J Med Chem 52: 514–523 [DOI] [PubMed] [Google Scholar]
- Plummer R., Jones C., Middleton M., Wilson R., Evans J., Olsen A., et al. (2008) Phase I study of the poly(ADP-ribose) polymerase inhibitor, AG014699, in combination with temozolomide in patients with advanced solid tumors. Clin Cancer Res 14: 7917–7923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottenberg S., Jaspers J.E., Kersbergen A., Van Der Burg E., Nygren A.O., Zander S.A., et al. (2008) High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc Natl Acad Sci U S A 105: 17079–17084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouleau M., Patel A., Hendzel M.J., Kaufmann S.H., Poirier G.G. (2010) PARP inhibition: PARP1 and beyond. Nat Rev Cancer 10: 293–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai W., Swisher E.M., Karlan B.Y., Agarwal M.K., Higgins J., Friedman C., et al. (2008) Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature 451: 1116–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber V., Ame J.C., Dolle P., Schultz I., Rinaldi B., Fraulob V., et al. (2002) Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J Biol Chem 277: 23028–23036 [DOI] [PubMed] [Google Scholar]
- Schreiber V., Dantzer F., Ame J.C., De Murcia G. (2006) Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol 7: 517–528 [DOI] [PubMed] [Google Scholar]
- Shall S., De Murcia G. (2000) Poly(ADP-ribose) polymerase-1: what have we learned from the deficient mouse model?. Mutat Res 460: 1–15 [DOI] [PubMed] [Google Scholar]
- Swisher E.M., Sakai W., Karlan B.Y., Wurz K., Urban N., Taniguchi T. (2008) Secondary BRCA1 mutations in BRCA1-mutated ovarian carcinomas with platinum resistance. Cancer Res 68: 2581–2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tentori L., Muzi A., Dorio A.S., Bultrini S., Mazzon E., Lacal P.M., et al. (2008) Stable depletion of poly (ADP-ribose) polymerase-1 reduces in vivo melanoma growth and increases chemosensitivity. Eur J Cancer 44: 1302–1314 [DOI] [PubMed] [Google Scholar]
- Thomas H.D., Calabrese C.R., Batey M.A., Canan S., Hostomsky Z., Kyle S., et al. (2007) Preclinical selection of a novel poly(ADP-ribose) polymerase inhibitor for clinical trial. Mol Cancer Ther 6: 945–956 [DOI] [PubMed] [Google Scholar]
- Timinszky G., Till S., Hassa P.O., Hothorn M., Kustatscher G., Nijmeijer B., et al. (2009) A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat Struct Mol Biol 16: 923–929 [DOI] [PubMed] [Google Scholar]
- Turner N., Tutt A., Ashworth A. (2004) Hallmarks of 'BRCAness' in sporadic cancers. Nat Rev Cancer 4: 814–819 [DOI] [PubMed] [Google Scholar]
- Tutt A., Robson M., Garber J.E., Domchek S.M., Audeh M.W., Weitzel J.N., et al. (2010) Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet 376: 235–244 [DOI] [PubMed] [Google Scholar]
- Virag L., Szabo C. (2002) The therapeutic potential of poly(ADP-ribose) polymerase inhibitors. Pharmacol Rev 54: 375–429 [DOI] [PubMed] [Google Scholar]
- Vona-Davis L., Rose D.P., Hazard H., Howard-Mcnatt M., Adkins F., Partin J., et al. (2008) Triple-negative breast cancer and obesity in a rural Appalachian population. Cancer Epidemiol Biomarkers Prev 17: 3319–3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson, C.T., Muzik, H., Turhan, A.G., Zamo, A., O'connor, M.J., Bebb, D.G. et al. (2010) Atm Deficiency Sensitizes Mantle Cell Lymphoma Cells to Poly(Adp-Ribose) Polymerase-1 Inhibitors. Mol Cancer Ther 9: 347–357. [DOI] [PMC free article] [PubMed]
- Zander S.A., Kersbergen A., Van Der Burg E., De Water N., Van Tellingen O., Gunnarsdottir S., et al. (2010) Sensitivity and acquired resistance of BRCA1;P53-deficient mouse mammary tumors to the topoisomerase I inhibitor topotecan. Cancer Res 70: 1700–1710 [DOI] [PubMed] [Google Scholar]