Abstract
Tyrosine kinases have a crucial role as key regulators of signaling pathways that influence cell differentiation and growth. Dysregulation of tyrosine kinase-mediated signaling is understood to be an important oncogenic driver. Genetic rearrangements involving the tyrosine kinase anaplastic lymphoma kinase (ALK) gene occur in non-small cell lung cancer (NSCLC), anaplastic large cell lymphomoas, inflammatory myofibroblastic tumors, and other cancers. Cells with abnormal ALK signaling are sensitive to ALK inhibitors such as crizotinib. This review will highlight the discovery of the fusion between echinoderm microtubule-associated protein-like 4 (EML4) and ALK as an oncogenic driver, recognition of other ALK gene rearrangements in NSCLC, and the confirmation that crizotinib is an effective treatment for patients with ALK-positive NSCLC. Work is underway to further define the role for crizotinib in the treatment of ALK-positive lung cancer and other cancers and to investigate the molecular mechanisms for resistance to ALK inhibition with crizotinib.
Keywords: anaplastic lymphoma kinase, carcinoma, crizotinib, non-small cell lung cancer, tyrosine kinase inhibitor
Introduction
Lung cancer has long been the most common cancer and cause of cancer-related deaths worldwide, with 1.61 million new cases and 1.38 million deaths in 2008 alone, representing 12.7% of new cancers and 18.2% of cancer mortality [Ferlay et al. 2010]. Approximately 85% of lung cancers are non-small cell lung cancer (NSCLC) and the majority of patients are diagnosed at an advanced stage [Subramanian and Govindan, 2008; Govindan et al. 2006; Yang et al. 2005]; 5-year survival for patients in the USA with NSCLC is approximately 16% [American Cancer Society, 2011]. Current treatments for NSCLC may extend survival but are rarely curative [Subramanian and Govindan, 2008; Greenlee et al. 2000].
NSCLC includes the adenocarcinoma, squamous-cell, and large-cell histological subtypes and, historically, these factors plus performance status have largely determined the choice of cytotoxic chemotherapy regimens for patients. NSCLC cytotoxic chemotherapy is currently focused on the use of platinum doublet chemotherapy, with or without bevacizumab [Langer et al. 2010].
Where NSCLC was previously considered to be a single disease treated with standard cytotoxic chemotherapy, it is now becoming more appropriate to consider NSCLC as a collection of disease subtypes according to the driving oncogenic aberration, and to select treatment accordingly. Crucially, the Iressa Pan Asia Study (IPASS), which compared the efficacy of the epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor (TKI) gefitinib with carboplatin plus paclitaxel in pulmonary adenocarcinoma, demonstrated not only that gefitinib was significantly more effective than chemotherapy first line in patients with EGFR mutations [hazard ratio (HR) for progression or death 0.75; p < 0.001] but also that, in patients without EGFR mutations, chemotherapy was superior to gefitinib [HR 2.85; 95% confidence interval (CI) 2.05 to 3.98; p < 0.001] [Mok et al. 2009]. Two Japanese studies have shown gefitinib to be more effective than platinum/taxane doublet chemotherapy as first-line treatment for patients with NSCLC harboring EGFR mutations; significant improvements in progression-free survival (PFS) were observed for gefitinib compared with both carboplatin/paclitaxel (PFS 10.8 versus 5.4 months; HR: 0.3; 95% CI 0.22 to 0.41; p < 0.001) [Maemondo et al. 2010] and cisplatin/docetaxel (PFS 9.2 versus 6.3 months; HR: 0.49; 95% CI 0.34 to 0.71; p < 0.001) [Mitsudomi et al. 2010]. The CALGB 30406 study is investigating the efficacy of the EGFR TKI erlotinib administered alone and in combination with carboplatin/paclitaxel in never/former light smokers with chemotherapy-naïve, advanced NSCLC, and has reported a significantly increased PFS for patients with EGFR mutations compared with wild-type EGFR [Janne et al. 2010]. Interestingly, in this study erlotinib monotherapy had similar efficacy to combination therapy in EGFR-mutated tumors. The OPTIMAL study has compared erlotinib with gemcitabine/carboplatin in Chinese patients with untreated EGFR-mutated NSCLC, finding a considerable PFS advantage for erlotinib compared with chemotherapy [median PFS 13.1 months (erlotinib) versus 4.6 months (gemcitabine/carboplatin); HR: 0.16; 95% CI 0.10 to 0.26; p < 0.0001; N = 154] [Zhou et al. 2010]. Thus, the selection of chemotherapy regimens according to tumor histology does not necessarily represent the best choice for patients, and the selection of a better treatment option requires screening to accurately identify the disease driver.
Despite the growth in understanding of tyrosine kinases, their role in tumor development, and the increasing success of tyrosine kinase-based therapeutic agents, the precise kinases that drive most solid tumors remain unclear. This limits the identification of drug targets and prediction of response. Research carried out over the last 15 years, however, has shed much light on the expression of these proteins in cancer. The nucleophosmin (NPM)–anaplastic lymphoma kinase (ALK) fusion protein was first identified as a neoplastic agent in patients with anaplastic large cell lymphoma (ALCL) [Shiota and Mori, 1996; Shiota et al. 1995; Morris et al. 1994], with the NPM–ALK protein resulting from the translocation t(2,5)(p23:q35), between the ALK gene on chromosome 2 and the NPM gene on chromosome 5 [Lamant et al. 1996; Shiota and Mori, 1996]. ALK has since been linked with many different fusion partners in different tumor types, including TRK-fused gene (TFG), tropomyosin (TPM)3, TPM4, 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase (ATIC), and moesin (MSN) in ALCL; TPM3, TPM4, cysteinyl tRNA synthetase (CARS), Ran-binding protein 2 (RANBP2), and clathrin heavy chain gene (CLTC) in inflammatory myofibroblastic tumor; a variety of ALK-amplifying point mutations in neuroblastoma; TPM4 in esophageal squamous tumor; SQSTM1–ALK fusion, CLTC, and NPM in diffuse large B-cell lymphoma; and echinoderm microtubule-associated protein-like 4 (EML4) and kinesin family member 5B (KIF5B) in NSCLC [Takeuchi et al. 2011, 2009; Palmer et al. 2009; Webb et al. 2009; Mossé et al. 2008; Coffin et al. 2007; Rikova et al. 2007; Soda et al. 2007; Jazii et al. 2006; Li et al. 2004; Ma et al. 2003; Onciu et al. 2003; Gascoyne et al. 2003; Drexler et al. 2000]. A further solid tumor ALK fusion (VCL–ALK in renal cell carcinoma) has also recently been reported [Debelenko et al. 2011]. In NSCLC, specifically, a large-scale survey of tyrosine kinase signaling across 41 cell lines and over 150 tumors identified known oncogenic kinases such as EGFR and c-MET, together with novel ALK and ROS fusion proteins [Rikova et al. 2007].
The EML4–ALK fusion gene was identified as tumorigenic in NSCLC in 2007 [Soda et al. 2007; Rikova et al. 2007] and the development of a clinic-ready inhibitor targeting it, crizotinib (PF-02341066), has been rapid, with the first clinical trial data for crizotinib in patients who are ALK fusion gene-positive published in 2010 [Kwak et al. 2010]. Crizotinib is also a highly specific inhibitor of the receptor tyrosine kinase c-MET (hepatocyte growth factor receptor), and was previously studied as a c-MET inhibitor [Rodig and Shapiro, 2010]. Here I discuss ALK in NSCLC and look at the potential which crizotinib has to make a difference in the treatment of NSCLC, and the issues surrounding the emergence of this ALK inhibitor for the treatment of NSCLC in the future.
EML4–ALK in NSCLC and mode of action of crizotinib
EML4–ALK is an inversion, where the EML4 gene is disrupted at intron 13 and is linked to intron 19 of the ALK gene [Ensembl Genome Browser, 2011] (previously identified as ‘upstream of exon 20’), producing a gene of 3926 base pairs coding for a protein of 1059 amino acids [Soda et al. 2007].
Incorporation of EML4–ALK into mouse fibroblasts resulted in tumor formation when these cells were injected into mice. Subsequently, a number of variants of the fusion gene were identified, differing in the point at which the EML4 gene was disrupted [Soda et al. 2007]. Transgenic mice expressing alveolar epithelial EML4–ALK developed hundreds of adenocarcinoma nodules in both lungs within weeks of birth, and subsequent treatment with an ALK inhibitor rapidly reduced tumor size [Soda et al. 2008]. Similarly, injection of fibroblasts expressing EML4–ALK into nude mice led to tumor development and fatal respiratory failure, whereas treatment with the ALK inhibitor resulted in a reduction in tumor burden and prolonged survival [Soda et al. 2008].
The precise intracellular signaling pathways by which the EML4–ALK fusion protein induces tumor growth and development have not been fully characterized. Likewise, the physiological function of ALK is currently unknown but there is evidence that the ALK gene plays a role in the development of the nervous system [Pulford et al. 2004] and, in humans, ALK protein expression is confined to scattered cells within the central nervous system [Pulford et al. 1997]. Studies of NPM–ALK in ALCL have implicated RAS–mitogen-activated protein kinase, phospholipase C gamma–Src homology 2, phosphoinositide 3-kinase–AKT, signal transducer and activator of transcription 3, and nuclear interacting partner of ALK [Pulford et al. 2004; Ouyang et al. 2003]. It has recently been reported that interactions between NPM–ALK and growth factor receptor-bound protein 2 play a key role in the regulation of ALCL cell signaling and growth [Riera et al. 2010]. There is therefore abundant potential for ALK fusion proteins to generate tumor growth and proliferation.
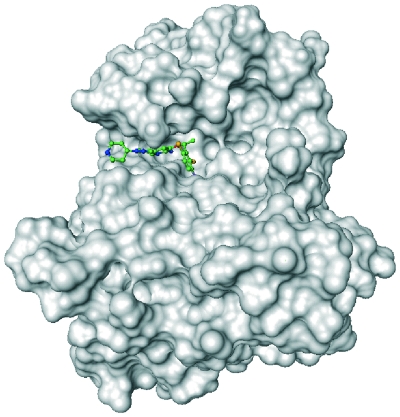
Crizotinib is an orally active small-molecule inhibitor of ALK and the c-Met receptor tyrosine kinase [Christensen et al. 2007; Zou et al. 2007], and belongs to the 3-benzyloxy-2-aminopyridine series of kinase inhibitors [Timofeevski et al. 2009]. Crizotinib is highly selective for ALK and c-Met kinases, with 50% inhibitory concentration values for ALK and c-Met of 5–20 nM, compared with values at least 20-fold higher for other kinases [Christensen et al. 2007; Zou et al. 2007]. Crizotinib acts by binding to the adenosine triphosphate (ATP) binding site (the ‘ATP binding pocket’) of the ALK enzyme (Figure 1), thereby preventing binding of ATP and subsequent autophosphorylation, which is required for activation of the enzyme. Crizotinib and the ALK inhibitor, TAE684, have been shown to inhibit tumor growth in the H2228 and H3122 NSCLC tumor models [Li et al. 2011; McDermott et al. 2008].
Figure 1.
Crizotinib in the anaplastic lymphoma kinase ATP binding pocket [Camidge et al. 2010a].
Detection methods for ALK fusion genes
The optimum detection method for ALK has yet to be established. The most common methods currently in use include break-apart fluorescence in situ hybridization (FISH), immunohistochemistry (IHC), and reverse transcription polymerase chain reaction (RT-PCR). Break-apart FISH has to date been the usual standard method for confirmation of ALK status in clinical trials [Yi et al. 2011; Kwak et al. 2010].
Break-apart FISH employs a green centromeric probe and a red telomeric probe which bind to regions of the chromosome flanking the ALK gene. Thus, if the gene is split, as in the EML4–ALK fusion gene, the red and green signals will be separated; if not, a yellow signal will be seen. The assay is considered positive if fusion is detected in 15% or more of cells in the sample [Shaw et al. 2009]. However, break-apart FISH may require an experienced operator and this technique may not be widely available [Horn and Pao, 2009]. Fusion FISH can also be performed, using specific probes for the EML4 and ALK genes [Sakairi et al. 2010].
IHC is a ubiquitous technique, in which antibodies labeled with markers are used to localize specific antigens in tissue samples. Detection with IHC can be difficult when the target antigen is expressed at low levels (as with ALK), and intermediate levels of staining may be open to interpretation. Furthermore, results can also be influenced by the way in which the sample is prepared and the detection system used [Koh et al. 2011; Ikeda et al. 2010; Takeuchi et al. 2009].
RT-PCR is a variant of PCR, in which reverse transcriptase is used to convert an RNA sequence into cDNA, which is then amplified using PCR [Bustin, 2000]. This technique provides an accurate indication of the presence of ALK abnormalities, and can be readily automated, making it suitable for high-throughput screening [Hirsch et al. 2010]. However, it requires predefined primers raised against known fusion genes, and hence previously unrecognized fusion partners may not be detected [Hirsch et al. 2010]. RT-PCR is technically difficult using formalin-fixed paraffin-embedded samples, although it may be feasible following recent advances in methodology [Danenberg et al. 2010; Hirsch et al. 2010; Mano, 2008].
Notably in this context, the chromosomal inversion leading to the formation of the EML4–ALK fusion gene does not always take place in the same location, and multiple EML4–ALK variants have been identified. All involve the tyrosine kinase domain of ALK, but there is variation in the truncation of EML4. At least 11 variants have been identified (see above), most of which are oncogenic [Sasaki et al. 2010b]. For example, the NSCLC cell lines H3122 and DFCI032 contain variants with fusions at EML4 exon 13, whereas H2228 has a variant fused at EML4 exon 6a/b [Koivunen et al. 2008]. In addition, as mentioned briefly earlier, EML4 is not the only fusion partner for ALK in ALK-translocated NSCLC: TFG and KIF5B have also been identified in NSCLC tumor samples [Takeuchi et al. 2009; Rikova et al. 2007]. These variations in EML4 fusion and the presence of non-EML4 fusion partners for ALK carry implications for clinical testing and characterization of ALK-translocated NSCLC.
Treatment paradigm and screening
Two groups have investigated the correlation between IHC and FISH detection results as a way to facilitate ALK screening [Paik et al. 2011; Yi et al. 2011]. Both groups found 100% correlation between IHC score 0 and FISH negativity, and between IHC score 3+ and FISH positivity. Both studies concluded that samples with IHC scores of 1+ and 2+ would require confirmation by FISH for a reliable assessment of ALK status. Any practically useful screening process would need to demonstrate a low rate of false negatives, low variability in sensitivity and specificity of the antibody used, and robust reliability across multiple users and sites. Furthermore, such a system would need to retain flexibility to identify ‘atypical’ FISH-negative cases which may be positive using other screening techniques, and allow for FISH testing based on clinical suspicion [Camidge et al. 2011]. The use of IHC for ALK screening is desirable due to its practicality and wide availability, and further research into the use of this diagnostic technique is ongoing.
The challenge of finding an effective and widely implementable screening method for the ALK fusion gene is representative of the challenges facing the widespread implementation of personalized medicine in NSCLC. In order to ensure that patients receive the most appropriate treatment for their tumors, a robust and locally available screening system is required. Such a system must not only provide detection of EGFR, KRAS, ALK, and any other aberration that influences treatment choice, but also do so quickly enough to give patients the best chance of a successful outcome from treatment; the collection of sufficient tissue for the various required tests should therefore be standard procedure [Hirsch et al. 2010]. The implementation of a successful screening system will therefore necessitate the close coordination of clinical, surgical, and pathology services.
Prevalence of ALK fusion genes in NSCLC
The ALK fusion gene has been reported to be present in up to 11.6% of patients with NSCLC [Zhang et al. 2010], depending on the population studied and screening methods used. The average prevalence appears to be approximately 3% in unselected populations and 4.5% in populations that have been ‘enriched’ by selection of patients with adenocarcinoma (Table 1).
Table 1.
Prevalence of anaplastic lymphoma kinase (ALK) fusion gene in unselected and adenocarcinoma-enriched patient populations with non-small cell lung cancer.
| Study | Country | Unselected populations |
Adenocarcinoma-enriched populations |
||
|---|---|---|---|---|---|
| Total, n | ALK-positive, n (%) | Total, n | ALK-positive, n (%) | ||
| [Soda et al. 2007] | Japan | 75 | 5 (6.7) | ||
| [Rikova et al. 2007] | China | 103 | 4 (3.9) | ||
| [Koivunen et al. 2008] | USA/Korea | 305 | 8 (2.6) | 200 | 8 (4.0) |
| [Inamura et al. 2008] | Japan | 221 | 5 (2.3) | 149 | 5 (3.4) |
| [Takeuchi et al. 2008] | Japan | 343 | 11 (3.2) | 253 | 11 (4.3) |
| [Perner et al. 2008] | USA/Switzerland | 603 | 16 (2.7) | ||
| [Shinmura et al. 2008] | Japan | 77 | 2 (2.6) | ||
| [Boland et al. 2009] | USA | 335 | 6 (1.8) | 185 | 5 (2.7) |
| [Wong et al. 2009] | China | 266 | 13 (4.9) | 209 | 11 (5.3) |
| [Martelli et al. 2009] | EU | 120 | 9 (7.5) | 63 | 3 (4.8) |
| [Rodig et al. 2009] | USA | 358 | 20 (5.6) | ||
| [Takeuchi et al. 2009] | Japan | 130 | 4 (3.1) | ||
| [Takahashi et al. 2010] | Japan | 313 | 5 (1.6) | 211 | 5 (2.4) |
| [Zhang et al. 2010] | China | 103 | 12 (11.7) | 62 | 10 (16.1) |
| Total | 2864 | 96 (3.4) | 1820 | 82 (4.5) | |
Clinicopathological studies have identified patients with ALK-positive NSCLC as clinically distinct from those with EGFR-positive (based on age) and EGFR wild-type (based on age, smoking status, and tumor differentiation/grade) disease [Koh et al. 2011; Takahashi et al. 2010; Rodig et al. 2009; Shaw et al. 2009; Wong et al. 2009]. The ALK fusion gene appears to be more common in patients who have never smoked, or those who are light smokers, than in people who smoke [Zhang et al. 2010; Rodig et al. 2009; Wong et al. 2009]. For example, in a study of 266 NSCLC tumors taken from Chinese patients, the ALK fusion gene was present in 8.5% of tumors from patients who had never smoked compared with only 0.8% of tumors from patients who had smoked at some time during their lives [Wong et al. 2009]: a statistically significant difference (p = 0.009). Similarly, in a US study, 70% of patients with ALK-positive NSCLC had never smoked [Rodig et al. 2009]. Again, this association was highly statistically significant (p < 0.0001; N = 358). Patients with ALK-positive disease also tend to be younger than those with ALK-negative disease. In a study by Koh and colleagues, patients with ALK-positive disease were significantly younger than those with ALK-negative disease, with a median age of 49 years compared with 61 years (p < 0.001; N = 221) [Koh et al. 2011]. Similarly, the median age of patients with ALK-positive NSCLC in the US study above was 51 years, versus 66 years in those with ALK-negative disease (p = 0.0002 for the difference; N = 358) [Rodig et al. 2009]. In addition to age, Koh and colleagues found that signet ring cell components were frequently observed (p = 0.056, N = 221) in ALK-positive tumors and TTF-1 expression was observed in all ALK-positive tumors for which IHC data were available [Koh et al. 2011]. In addition, ALK status of patients with NSCLC does not seem to influence response to platinum-based doublet chemotherapy [Koh et al. 2011; Shaw et al. 2009].
To date, data suggest that the ALK fusion gene appears to be more common in younger patients, patients who have never smoked, and patients with adenocarcinoma. However, the various published studies to date are subject to their own limitations with the consequent possibility of selection bias and, in lieu of a comprehensive study, we must consider that any patient with NSCLC can carry the ALK fusion gene. For example, the literature includes a case of ALK-positive NSCLC in an elderly patient (aged 76 years) with a history of smoking [Rodig et al. 2009]. Clinical characteristics alone are therefore not sufficient to identify patients with ALK-positive disease [Rodig et al. 2009]. Further studies are needed to better understand the clinical characteristics of patients with ALK-positive disease.
Interestingly, a consistent finding has been that the ALK fusion gene is largely exclusive of EGFR and KRAS mutations: in other words, patients with this fusion gene seldom have concomitant mutations in EGFR or KRAS [Sun et al. 2010; Yoshida et al. 2010; Zhang et al. 2010; Camidge et al. 2010b; Rodig et al. 2009; Shaw et al. 2009], and patients with EGFR or KRAS mutations do not generally have concurrent EML4–ALK rearrangement, although comutation with EGFR has been reported [Tiseo et al. 2011]. Also, ALK fusion is not limited to adenocarcinoma, having been reported in patients with squamous cell lung carcinoma histology [Boland et al. 2009; Wong et al. 2009; Soda et al. 2007].
Clinical data for crizotinib in NSCLC
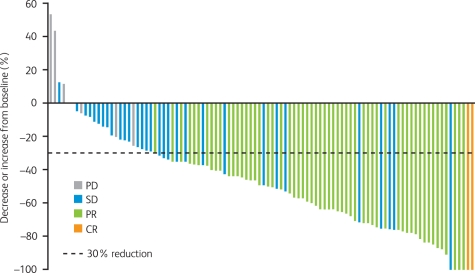
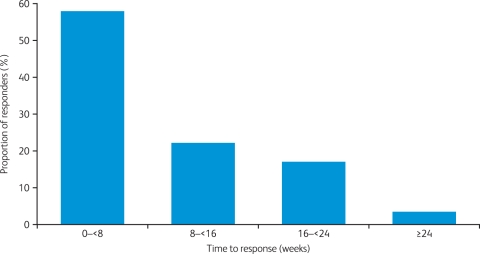
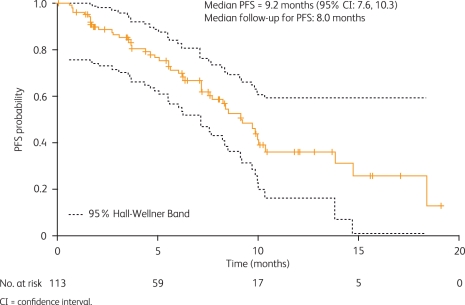
Current clinical data supporting the efficacy of crizotinib in patients with ALK-positive NSCLC come from an expanded cohort study (A8081001 [ClinicalTrials.gov identifier: NCT00585195]), which enrolled patients with ALK-positive disease regardless of prior therapy [Kwak et al. 2010]. ALK positivity was confirmed by break-apart FISH in all cases. Patients received crizotinib 250 mg twice daily continuously. Data from the first 82 patients with NSCLC were presented at the 2010 American Society of Clinical Oncology Annual Meeting [Bang et al. 2010] and subsequently reported in the New England Journal of Medicine [Kwak et al. 2010]. An update to these data with 113 enrolled patients and a median follow up of 8 months was presented at the 2010 European Society of Medical Oncology Congress [Camidge et al. 2010a]. Patients were younger than is typical for patients with NSCLC, tended to have never smoked, and usually had adenocarcinoma tumors (Table 2). As of August 2010, there were 105 patients evaluable for response, including two complete responses and 57 partial responses (objective response rate 56%), plus a further 33 patients (31%) with stable disease [Solomon et al. 2010]. The best reduction in tumor size is shown in Figure 2, from which it can be seen that most patients showed a decrease from baseline in target lesion tumor size of 30% or more. Responses to crizotinib occurred quickly, with 56% of responses occurring by week 8 of treatment (Figure 3) [Solomon et al. 2010]. Median PFS was 9.2 months (95% CI 7.6 to 10.3; Figure 4), with a probability of being progression free at 6 months of 71.3% (95% CI 60.3 to 79.7) [Camidge et al. 2010a].
Table 2.
Baseline characteristics of patients with non-small cell lung cancer participating in a study of crizotinib (N = 113) [Camidge et al. 2010a].
| Characteristic | |
|---|---|
| Number of patients | 113 |
| Mean age (range), years | 52 (21–79) |
| Gender (male/female), n (%) | 57/56 (50.4/49.6) |
| ECOG performance status, n (%) | |
| 0 | 38 (33.6) |
| 1 | 60 (53.1) |
| 2 | 15 (13.3) |
| Ethnicity, n (%) | |
| White | 69 (61.1) |
| Asian | 34 (30.1) |
| Other | 10 (8.8) |
| Smoking history, n (%) | |
| Never | 82 (72.6) |
| Former | 30 (26.5) |
| Current | 1 (<1.0) |
| Histology, n (%) | |
| Adenocarcinoma | 109 (96.5) |
| Squamous cell carcinoma | 1 (<1.0) |
| Large cell | 1 (<1.0) |
| Other | 2 (1.8) |
| Previous treatment regimens, n (%) | |
| 0 | 6 (5.3) |
| 1 | 32 (28.3) |
| 2 | 21 (18.6) |
| 3 | 17 (15.0) |
| >3 | 35 (31.0) |
| Not reported | 2 (1.8) |
ECOG, Eastern Cooperative Oncology Group.
Figure 2.
Waterfall plot of best percentage change from baseline in target lesions by patient in patients with anaplastic lymphoma kinase (ALK)-positive non-small cell lung cancer who received crizotinib (excluding those with early death and indeterminate or unavailable response from the 105 evaluable patients; August 2010) [Camidge et al. 2010a]. CR, complete response; PD, progressive disease; PR, partial response; SD, stable disease.
Figure 3.
Time to response with crizotinib for responding patients (n = 59 of 105 evaluable patients; August 2010) [Solomon et al. 2010].
Figure 4.
Progression-free survival (PFS) following treatment with crizotinib (August 2010; N = 113) [Camidge et al. 2010a]. CI, confidence interval.
The A8081001 protocol made a provision for patients to continue to receive crizotinib after Response Evaluation Criteria in Solid Tumors (RECIST) version 1.0-defined disease progression if the investigator felt that the patient would derive clinical benefit. Interestingly, of 36 patients who experienced RECIST-defined progression, 15 received postprogression crizotinib for more than 2 weeks and five received crizotinib for longer than 6 months [Camidge et al. 2010a].
The most common treatment-related adverse events (AEs) in this analysis (August 2010; N = 113) were grade 1/2 gastrointestinal AEs (nausea, 52%; diarrhea, 50%; vomiting, 42%; all were manageable with standard antiemetic or antidiarrheal treatments) and grade 1 visual disturbance in 45% of patients (Table 3) [Camidge et al. 2010a]. Visual disturbances with crizotinib have been described as ‘trails of light’ when accommodating from dark to light [Clinical Care Options 2010; Kwak et al. 2010]. Grade 3/4 treatment-related AEs included increases in alanine aminotransferase (4% grade 3; 1% grade 4) and aspartate aminotransferase (4% grade 3). Neutropenia and lymphopenia were also reported at grade 3 severity (3% and 2% respectively). One patient experienced grade 3 pneumonitis, but this was not considered to be related to treatment [Camidge et al. 2010a]. A total of 36 patients (31.9%) had discontinued study treatment by August 2010 (median follow up 8 months; progressive disease, n = 23; death, n = 7 unrelated to treatment; AEs, n = 3 with one considered treatment related; withdrawal of consent, n = 1; and other, n = 2) [Camidge et al. 2010a].
Table 3.
Most common treatment-related adverse events (≥10%) with crizotinib (N = 113) [Camidge et al. 2010a].
| Adverse event | Grade 1, n (%) | Grade 2, n (%) | Grade 3, n (%) | Grade 4, n (%) | Total, n (%) |
|---|---|---|---|---|---|
| Nausea | 58 (51.3) | 1 (0.9) | 0 | 0 | 59 (52.2) |
| Diarrhea | 55 (48.7) | 2 (1.8) | 0 | 0 | 57 (50.4) |
| Visual impairment | 51 (45.1) | 0 | 0 | 0 | 51 (45.1) |
| Vomiting | 46 (40.7) | 1 (0.9) | 0 | 0 | 47 (41.6) |
| Constipation | 22 (19.5) | 6 (5.3) | 0 | 0 | 28 (24.8) |
| Peripheral edema | 19 (16.8) | 3 (2.7) | 0 | 0 | 22 (19.5) |
| Decreased appetite | 21 (18.6) | 0 | 0 | 0 | 21 (18.6) |
| Dizziness | 21 (18.6) | 0 | 0 | 0 | 21 (18.6) |
| Fatigue | 14 (12.4) | 3 (2.7) | 1 (0.9) | 0 | 18 (15.9) |
| Alanine aminotransferase increased | 3 (2.7) | 6 (5.3) | 4 (3.5) | 1 (0.9) | 14 (12.4) |
Data indicate differences in the pharmacokinetics of crizotinib between Asian and non-Asian patients, with body weight and surface area accounting partially but not completely for the observed pharmacokinetic differences [Ou et al. 2010]. Steady-state mean area under the curve of crizotinib, adjusted for body weight and adjusted for body surface area, was found to be 26% and 46% higher respectively in Asian patients compared with non-Asian patients. A higher overall response rate to crizotinib was also seen in Asian patients, although clinically significant responses to crizotinib were also seen across all ethnicities. As such, current efficacy and safety results support clinical investigation of crizotinib with the same dose regimen in Asian and non-Asian patients.
Future research and clinical development for crizotinib and NSCLC
At this early stage in the development of crizotinib, there are several issues which have still to be fully explored. As with all targeted cancer therapies, resistance to crizotinib is likely to be a significant issue for therapy, and there is already a recorded case of crizotinib resistance in a young patient with EML4–ALK-positive NSCLC [Choi et al. 2010]. Two independent mutations were identified; a substitution of adenine for guanine at position 4374 of EML4–ALK, resulting in replacement of cysteine with tyrosine at position 1156 of ALK (C1156Y), and a substitution of adenine for cytosine at ALK position 4493, resulting in replacement of leucine with methionine at position 1196 of ALK (L1196M) [Choi et al. 2010]. A third mutation (F1174L) has been identified in a patient with RANBP2–ALK-positive inflammatory myofibroblastic tumor, and was associated with decreased sensitivity of Ba/F3 cells to crizotinib, although this mutation was unlikely to directly prevent binding of crizotinib to ALK [Sasaki et al. 2010a]. Further investigation of mechanisms of resistance to crizotinib and how to overcome it will be crucial [Butrynski et al. 2010]. The potential of combination therapy with different intracellular signaling inhibitors to target proliferation and resistance pathways simultaneously is also an option, as is the development of other agents to overcome crizotinib resistance.
As research into oncogenic drivers in NSCLC continues, it is inevitable that new molecular targets will be discovered for distinct patient subgroups. Experience with crizotinib shows that it is feasible to develop agents for new targets in NSCLC and subsequently progress to clinical trials within a relatively short timeframe. It should also be noted that crizotinib may show promise in other cancers harboring ALK gene rearrangements such as anaplastic large-cell lymphomas [Gambacorti-Passerini et al. 2011]. In summary, the integration of crizotinib and of future personalized therapies into standard treatment practice in NSCLC will rest on the widespread implementation of an effective screening system for newly diagnosed patients with NSCLC which is flexible enough to incorporate new targets as treatments are developed for them.
Acknowledgments
Medical writing support was provided by Martin Quinn at ACUMED® (Tytherington, UK) and was funded by Pfizer Inc.
Funding
Dr Bang has received honoraria and research funding from Pfizer Inc.
Conflict of interest statement
Dr Bang has performed a consultation/advisory role for Pfizer Inc.
References
- American Cancer Society (2011) Global Cancer: Facts and Figures, 2nd edn. American Cancer Society website: www.cancer.org/acs/groups/content/@epidemiologysurveilance/documents/document/acspc-027766.pdf [last accessed 27 July 2011]
- Bang Y., Kway E.L., Shaw A.T., Camidge D.R., Iafrate A.J., Maki R.G., et al. (2010) Clinical activity of the oral ALK inhibitor PF-02341066 in ALK-positive patients with non-small cell lung cancer (NSCLC). Presented at the 46th American Society of Clinical Oncology Annual Meeting. J Clin Oncol 28(Suppl 18s) (Abstract 3) [Google Scholar]
- Boland J.M., Erdogan S., Vasmatzis G., Yang P., Tillmans L.S., Johnson M.R., et al. (2009) Anaplastic lymphoma kinase immunoreactivity correlates with ALK gene rearrangement and transcriptional up-regulation in non-small cell lung carcinomas. Hum Pathol 40: 1152–1158 [DOI] [PubMed] [Google Scholar]
- Bustin S.A. (2000) Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol 25: 169–193 [DOI] [PubMed] [Google Scholar]
- Butrynski J.E., D’Adamo D.R., Hornick J.L., Dal C.P., Antonescu C.R., Jhanwar S.C., et al. (2010) Crizotinib in ALK-rearranged inflammatory myofibroblastic tumor. N Engl J Med 363: 1727–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camidge D.R., Bang Y.-J., Iafrate A.J., Kwak E.L., Maki R.G., Solomon B., et al. (2010) Clinical activity of crizotinib (PF-02341066) in ALK-positive patients with advanced non-small cell lung cancer. Presented at the 35th European Society of Medical Oncology Congress. Ann Oncol 21(Suppl 8): viii123–viii123, Milan, Italy, 8–12 October 2010 (Abstract 366PD) [Google Scholar]
- Camidge D.R., Hirsch F.R., Varella-Garcia M., Franklin W.A. (2011) Finding ALK-positive lung cancer: What are we really looking for?. J Thorac Oncol 6: 411–413 [DOI] [PubMed] [Google Scholar]
- Camidge D.R., Kono S.A., Flacco A., Tan A.C., Doebele R.C., Zhou Q., et al. (2010b) Optimizing the detection of lung cancer patients harboring anaplastic lymphoma kinase (ALK) gene rearrangements potentially suitable for ALK inhibitor treatment. Clin Cancer Res 16: 5581–5590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y.L., Soda M., Yamashita Y., Ueno T., Takashima J., Nakajima T., et al. (2010) EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N Engl J Med 363: 1734–1739 [DOI] [PubMed] [Google Scholar]
- Christensen J.G., Zou H.Y., Arango M.E., Li Q., Lee J.H., McDonnell S.R., et al. (2007) Cytoreductive antitumor activity of PF-2341066, a novel inhibitor of anaplastic lymphoma kinase and c-Met, in experimental models of anaplastic large-cell lymphoma. Mol Cancer Ther 6: 3314–3322 [DOI] [PubMed] [Google Scholar]
- Clinical Care Options (2010) ALK inhibitor crizotinib safe and highly active in ALK-positive NSCLC. http://www.clinicaloptions.com/Oncology/Conference%20Coverage/Clin%20Onc%20June%202010/Tracks/Lung%20Cancer/Capsules/3.aspx [last accessed 27 July 2011]
- Coffin C.M., Hornick J.L., Fletcher C.D. (2007) Inflammatory myofibroblastic tumor: Comparison of clinicopathologic, histologic, and immunohistochemical features including ALK expression in atypical and aggressive cases. Am J Surg Pathol 31: 509–520 [DOI] [PubMed] [Google Scholar]
- Danenberg P.V., Stephens C., Cooc J., Gandara D.R., Mack P.C., Grimminger P.P., et al. (2010) A novel RT-PCR approach to detecting EML4-ALK fusion genes in archival NSCLC tissue. J Clin Oncol 28(Suppl 15s) (Abstract 10535) [Google Scholar]
- Debelenko L.V., Raimondi S.C., Daw N., Shivakumar B.R., Huang D., Nelson M., Bridge J.A. (2011) Renal cell carcinoma with novel VCL-ALK fusion: New representative of ALK-associated tumor spectrum. Mod Pathol 24: 430–432 [DOI] [PubMed] [Google Scholar]
- Drexler H.G., Gignac S.M., von Wasielewski R., Werner M., Dirks W.G. (2000) Pathobiology of NPM-ALK and variant fusion genes in anaplastic large cell lymphoma and other lymphomas. Leukemia 14: 1533–1559 [DOI] [PubMed] [Google Scholar]
- Ensembl Genome Browser (2011) Transcript: ALK-001 (ENST00000389048). http://www.ensemblorg/Homo_sapiens/Transcript/Idhistory?db=core;g=ENSG00000171094;r=2:29415640-30144432;t=ENST00000389048 [last accessed 27 July 2011]
- Ferlay J., Shin H.R., Bray F., Forman D., Mathers C., Parkin D.M. (2010) GLOBOCAN 2008, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 10, International Agency for Research on Cancer [Google Scholar]
- Gambacorti-Passerini C., Messa C., Pogliani E.M. (2011) Crizotinib in anaplastic large-cell lymphoma. N Engl J Med 364: 775–776 [DOI] [PubMed] [Google Scholar]
- Gascoyne R.D., Lamant L., Martin-Subero J.I., Lestou V.S., Harris N.L., Muller-Hermelink H.K., et al. (2003) ALK-positive diffuse large B-cell lymphoma is associated with Clathrin-ALK rearrangements: report of 6 cases. Blood 102: 2568–2573 [DOI] [PubMed] [Google Scholar]
- Govindan R., Page N., Morgensztern D., Read W., Tierney R., Vlahiotis A., et al. (2006) Changing epidemiology of small-cell lung cancer in the United States over the last 30 years: Analysis of the surveillance, epidemiologic, and end results database. J Clin Oncol 24: 4539–4544 [DOI] [PubMed] [Google Scholar]
- Greenlee R.T., Murray T., Bolden S., Wingo P.A. (2000) Cancer statistics, 2000. CA Cancer J Clin 50: 7–33 [DOI] [PubMed] [Google Scholar]
- Hirsch F.R., Wynes M.W., Gandara D.R., Bunn P.A., Jr (2010) The tissue is the issue: Personalized medicine for non-small cell lung cancer. Clin Cancer Res 16: 4909–4911 [DOI] [PubMed] [Google Scholar]
- Horn L., Pao W. (2009) EML4-ALK: Honing in on a new target in non-small-cell lung cancer. J Clin Oncol 27: 4232–4235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda S., Takabe K., Inagaki M., Funakoshi N., Suzuki K. (2010) [Detection of ALK positive pulmonary adenocarcinoma using immunostaining.]. Rinsho Byori 58: 565–570 [PubMed] [Google Scholar]
- Inamura K., Takeuchi K., Togashi Y., Nomura K., Ninomiya H., Okui M., et al. (2008) EML4-ALK fusion is linked to histological characteristics in a subset of lung cancers. J Thorac Oncol 3: 13–17 [DOI] [PubMed] [Google Scholar]
- Janne P.A., Wang X.F., Socinski M.A., Crawford J., Capelletti M., Edelman M.J., et al. (2010) Randomized phase II trial of erlotinib (E) alone or in combination with carboplatin/paclitaxel (CP) in never or light former smokers with advanced lung adenocarcinoma: CALGB 30406. J Clin Oncol 28(Suppl 15s) (Abstract 7503) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazii F.R., Najafi Z., Malekzadeh R., Conrads T.P., Ziaee A.A., Abnet C., et al. (2006) Identification of squamous cell carcinoma associated proteins by proteomics and loss of beta tropomyosin expression in esophageal cancer. World J Gastroenterol 12: 7104–7112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh Y., Kim D.-W., Kim T.M., Lee S.-H., Jeon Y.K., Chung D.H., et al. (2011) Clinicopathologic characteristics and outcomes of patients with anaplastic lymphoma kinase-positive advanced pulmonary adenocarcinoma. J Thorac Oncol 6: 905–912 [DOI] [PubMed] [Google Scholar]
- Koivunen J.P., Mermel C., Zejnullahu K., Murphy C., Lifshits E., Holmes A.J., et al. (2008) EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin Cancer Res 14: 4275–4283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak E.L., Bang Y.J., Camidge D.R., Shaw A.T., Solomon B., Maki R.G., et al. (2010) Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 363: 1693–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamant L., Meggetto F., al Saati T., Brugieres L., de Paillerets B.B., Dastugue N., et al. (1996) High incidence of the t(2;5)(p23;q35) translocation in anaplastic large cell lymphoma and its lack of detection in Hodgkin’s disease. Comparison of cytogenetic analysis, reverse transcriptase-polymerase chain reaction, and P-80 immunostaining. Blood 87: 284–291 [PubMed] [Google Scholar]
- Langer C.J., Besse B., Gualberto A., Brambilla E., Soria J.C. (2010) The evolving role of histology in the management of advanced non-small-cell lung cancer. J Clin Oncol 28: 5311–5320 [DOI] [PubMed] [Google Scholar]
- Li X.Q., Hisaoka M., Shi D.R., Zhu X.Z., Hashimoto H. (2004) Expression of anaplastic lymphoma kinase in soft tissue tumors: An immunohistochemical and molecular study of 249 cases. Hum Pathol 35: 711–721 [DOI] [PubMed] [Google Scholar]
- Li Y., Ye X., Liu J., Zha J., Pei L. (2011) Evaluation of EML4-ALK fusion proteins in non-small cell lung cancer using small molecule inhibitors. Neoplasia 13: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z., Hill D.A., Collins M.H., Morris S.W., Sumegi J., Zhou M., et al. (2003) Fusion of ALK to the Ran-binding protein 2 (RANBP2) gene in inflammatory myofibroblastic tumor. Genes Chromosomes Cancer 37: 98–105 [DOI] [PubMed] [Google Scholar]
- Maemondo M., Inoue A., Kobayashi K., Sugawara S., Oizumi S., Isobe H., et al. (2010) Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 362: 2380–2388 [DOI] [PubMed] [Google Scholar]
- Mano H. (2008) Non-solid oncogenes in solid tumors: EML4-ALK fusion genes in lung cancer. Cancer Sci 99: 2349–2355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martelli M.P., Sozzi G., Hernandez L., Pettirossi V., Navarro A., Conte D., et al. (2009) EML4-ALK rearrangement in non-small cell lung cancer and non-tumor lung tissues. Am J Pathol 174: 661–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott U., Iafrate J., Gray N.S., Shioda T., Classon M., Maheswaran S., et al. (2008) Genomic alterations of anaplastic lympohoma kinase may sensitize tumors to anaplastic lymphoma kinase inhibitors. Cancer Res 68: 3389–3395 [DOI] [PubMed] [Google Scholar]
- Mitsudomi T., Morita S., Yatabe Y., Negoro S., Okamoto I., Tsurutani J., et al. (2010) Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): An open label, randomised phase 3 trial. Lancet Oncol 11: 121–128 [DOI] [PubMed] [Google Scholar]
- Mok T.S., Wu Y.L., Thongprasert S., Yang C.H., Chu D.T., Saijo N., et al. (2009) Gefitinib or carboplatin–paclitaxel in pulmonary adenocarcinoma. N Engl J Med 361: 947–957 [DOI] [PubMed] [Google Scholar]
- Morris S.W., Kirstein M.N., Valentine M.B., Dittmer K.G., Shapiro D.N., Saltman D.L., et al. (1994) Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin’s lymphoma. Science 263: 1281–1284 [DOI] [PubMed] [Google Scholar]
- Mossé Y.P., Laudenslager M., Longo L., Cole K.A., Wood A., Attiyeh E.F., et al. (2008) Identification of ALK as a major familial neuroblastoma predisposition gene. Nature 455: 930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onciu M., Behm F.G., Downing J.R., Shurtleff S.A., Raimondi S.C., Ma Z., et al. (2003) ALK-positive plasmablastic B-cell lymphoma with expression of the NPM-ALK fusion transcript: report of 2 cases. Blood 102: 2642–2644 [DOI] [PubMed] [Google Scholar]
- Ou S.I., Salgia R., Clark J., Kwak E., Camidge D.R., Maki R., et al. (2010) Comparison of crizotinib (PF-02341066) pharmacokinetics between Asian and non-Asian patients with advanced malignancies. Presented at the 4th Asia Pacific Lung Cancer Conference (APLCC) Seoul, South Korea, 2–4 December 2010 [Google Scholar]
- Ouyang T., Bai R.Y., Bassermann F., von Klitzing C., Klumpen S., Miething C., et al. (2003) Identification and characterization of a nuclear interacting partner of anaplastic lymphoma kinase (NIPA). J Biol Chem 278: 30028–30036 [DOI] [PubMed] [Google Scholar]
- Paik J.H., Choe G., Kim H., Choe J.Y., Lee H.J., Lee C.T., et al. (2011) Screening of anaplastic lymphoma kinase rearrangement by immunohistochemistry in non-small cell lung cancer: Correlation with fluorescence in situ hybridization. J Thorac Oncol 6: 466–472 [DOI] [PubMed] [Google Scholar]
- Palmer R.H., Vernersson E., Grabbe C., Hallberg B. (2009) Anaplastic lymphoma kinase: Signalling in development and disease. Biochem J 420: 345–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perner S., Wagner P.L., Demichelis F., Mehra R., Lafargue C.J., Moss B.J., et al. (2008) EML4-ALK fusion lung cancer: A rare acquired event. Neoplasia 10: 298–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulford K., Lamant L., Morris S.W., Butler L.H., Wood K.M., Stroud D., et al. (1997) Detection of anaplastic lymphoma kinase (ALK) and nucleolar protein nucleophosmin (NPM)-ALK proteins in normal and neoplastic cells with the monoclonal antibody ALK1. Blood 89: 1394–1404 [PubMed] [Google Scholar]
- Pulford K., Morris S.W., Turturro F. (2004) Anaplastic lymphoma kinase proteins in growth control and cancer. J Cell Physiol 199: 330–358 [DOI] [PubMed] [Google Scholar]
- Riera L., Lasorsa E., Ambrogio C., Surrenti N., Voena C., Chiarle R. (2010) Involvement of Grb2 adaptor protein in nucleophosmin-anaplastic lymphoma kinase (NPM-ALK)-mediated signaling and anaplastic large cell lymphoma growth. J Biol Chem 285: 26441–26450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikova K., Guo A., Zeng Q., Possemato A., Yu J., Haack H., et al. (2007) Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 131: 1190–1203 [DOI] [PubMed] [Google Scholar]
- Rodig S.J., Mino-Kenudson M., Dacic S., Yeap B.Y., Shaw A., Barletta J.A., et al. (2009) Unique clinicopathologic features characterize ALK-rearranged lung adenocarcinoma in the western population. Clin Cancer Res 15: 5216–5223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodig S.J., Shapiro G.I. (2010) Crizotinib, a small-molecule inhibitor of the c-MET and ALK receptor tyrosine kinases. Curr Opin Investig Drugs 11: 1477–1490 [PubMed] [Google Scholar]
- Sakairi Y., Nakajima T., Yasufuku K., Ikebe D., Kageyama H., Soda M., et al. (2010) EML4-ALK fusion gene assessment using metastatic lymph node samples obtained by endobronchial ultrasound-guided transbronchial needle aspiration. Clin Cancer Res 16: 4938–4945 [DOI] [PubMed] [Google Scholar]
- Sasaki T., Okuda K., Zheng W., Butrynski J., Capelletti M., Wang L., et al. (2010a) The neuroblastoma-associated F1174L ALK mutation causes resistance to an ALK kinase inhibitor in ALK-translocated cancers. Cancer Res 70: 10038–10043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T., Rodig S.J., Chireac L.R., Jänne P.A. (2010b) The biology and treatment of EML4-ALK non-small cell lung cancer. Eur J Cancer 46: 1773–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw A.T., Yeap B.Y., Mino-Kenudson M., Digumarthy S.R., Costa D.B., Heist R.S., et al. (2009) Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol 27: 4247–4253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinmura K., Kageyama S., Tao H., Bunai T., Suzuki M., Kamo T., et al. (2008) EML4-ALK fusion transcripts, but no NPM-, TPM3-, CLTC-, ATIC-, or TFG-ALK fusion transcripts, in non-small cell lung carcinomas. Lung Cancer 61: 163–169 [DOI] [PubMed] [Google Scholar]
- Shiota M., Mori S. (1996) The clinicopathological features of anaplastic large cell lymphomas expressing p80NPM/ALK. Leuk Lymphoma 23: 25–32 [DOI] [PubMed] [Google Scholar]
- Shiota M., Nakamura S., Ichinohasama R., Abe M., Akagi T., Takeshita M., et al. (1995) Anaplastic large cell lymphomas expressing the novel chimeric protein p80NPM/ALK: A distinct clinicopathologic entity. Blood 86: 1954–1960 [PubMed] [Google Scholar]
- Soda M., Choi Y.L., Enomoto M., Takada S., Yamashita Y., Ishikawa S., et al. (2007) Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 448: 561–566 [DOI] [PubMed] [Google Scholar]
- Soda M., Takada S., Takeuchi K., Choi Y.L., Enomoto M., Ueno T., et al. (2008) A mouse model for EML4-ALK-positive lung cancer. Proc Natl Acad Sci U S A 105: 19893–19897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon B., Bang Y.-J., Camidge D.R., Iafrate A.J., Kwak E.L., Maki R.G., et al. (2010) Timing of responses to crizotinib (PF-02341066) in anaplastic lymphoma kinase-positive patients with advanced non-small cell lung cancer. Presented at the 22nd EORTC-NCI-AACR Symposium on Molecular Targets and Cancer Therapeutics; Berlin, Germany, 16–19 November 2010 [Google Scholar]
- Subramanian J., Govindan R. (2008) Lung cancer. Govindan R. The Washington Manual® of Oncology, 2nd, Lippincott Williams and Wilkins: Philadelphia, PA, pp. 134–148 [Google Scholar]
- Sun Y., Ren Y., Fang Z., Li C., Fang R., Gao B., et al. (2010) Lung adenocarcinoma from East Asian never-smokers is a disease largely defined by targetable oncogenic mutant kinases. J Clin Oncol 28: 4616–4620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T., Sonobe M., Kobayashi M., Yoshizawa A., Menju T., Nakayama E., et al. (2010) Clinicopathologic features of non-small-cell lung cancer with EML4-ALK fusion gene. Ann Surg Oncol 17: 889–897 [DOI] [PubMed] [Google Scholar]
- Takeuchi K., Choi Y.L., Soda M., Inamura K., Togashi Y., Hatano S., et al. (2008) Multiplex reverse transcription-PCR screening for EML4-ALK fusion transcripts. Clin Cancer Res 14: 6618–6624 [DOI] [PubMed] [Google Scholar]
- Takeuchi K., Choi Y.L., Togashi Y., Soda M., Hatano S., Inamura K., et al. (2009) KIF5B-ALK, a novel fusion oncokinase identified by an immunohistochemistry-based diagnostic system for ALK-positive lung cancer. Clin Cancer Res 15: 3143–3149 [DOI] [PubMed] [Google Scholar]
- Takeuchi K., Soda M., Togashi Y., Ota Y., Sekiguchi Y., Hatano S., et al. (2011) Identification of a novel fusion, SQSTM1-ALK, in ALK-positive large B-cell lymphoma. Haematologica 96: 464–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timofeevski S.L., McTigue M.A., Ryan K., Cui J., Zou H.Y., Zhu J.X., et al. (2009) Enzymatic characterization of c-Met receptor tyrosine kinase oncogenic mutants and kinetic studies with aminopyridine and triazolopyrazine inhibitors. Biochemistry 48: 5339–5349 [DOI] [PubMed] [Google Scholar]
- Tiseo M., Gelsomino F., Boggiani D., Bortesi B., Bartolotti M., Bozzetti C., et al. (2011) EGFR and EML4-ALK gene mutations in NSCLC: A case report of erlotinib-resistant patient with both concomitant mutations. Lung Cancer 71: 241–243 [DOI] [PubMed] [Google Scholar]
- Webb T.R., Slavish J., George R.E., Look A.T., Xue L., Jiang Q., et al. (2009) Anaplastic lymphoma kinase: Role in cancer pathogenesis and small-molecule inhibitor development for therapy. Expert Rev Anticancer Ther 9: 331–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong D.W., Leung E.L., So K.K., Tam I.Y., Sihoe A.D., Cheng L.C., et al. (2009) The EML4-ALK fusion gene is involved in various histologic types of lung cancers from nonsmokers with wild-type EGFR and KRAS. Cancer 115: 1723–1733 [DOI] [PubMed] [Google Scholar]
- Yang P., Allen M.S., Aubry M.C., Wampfler J.A., Marks R.S., Edell E.S., et al. (2005) Clinical features of 5,628 primary lung cancer patients: Experience at Mayo Clinic from 1997 to 2003. Chest 128: 452–462 [DOI] [PubMed] [Google Scholar]
- Yi E.S., Boland J.M., Maleszewski J.J., Roden A.C., Oliveira A.M., Aubry M.C., et al. (2011) Correlation of IHC and FISH for ALK gene rearrangement in non-small cell lung carcinoma: IHC score algorithm for FISH. J Thorac Oncol 6: 459–465 [DOI] [PubMed] [Google Scholar]
- Yoshida A., Tsuta K., Watanabe S.I., Sekine I., Fukayama M., Tsuda H., et al. (2010) Frequent ALK rearrangement and TTF-1/p63 co-expression in lung adenocarcinoma with signet-ring cell component. Lung Cancer 72: 309–315 [DOI] [PubMed] [Google Scholar]
- Zhang X., Zhang S., Yang X., Yang J., Zhou Q., Yin L., et al. (2010) Fusion of EML4 and ALK is associated with development of lung adenocarcinomas lacking EGFR and KRAS mutations and is correlated with ALK expression. Mol Cancer 9: 188–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Wu Y.L., Chen C., Feng J., Liu X., Wang C., et al. (2010) Efficacy results from the randomised phase III OPTIMAL (CTONG 0802) study comparing first-line erlotinib versus carboplatin (CBDCA) plus gemcitabine (GEM), in Chinese advanced non small-cell lung cancer (NSCLC) patients (PTS) with EGFR activating mutations. Ann Oncol 21(Suppl. 8): viii6–viii6 (abstract LBA13) [Google Scholar]
- Zou H.Y., Li Q., Lee J.H., Arango M.E., McDonnell S.R., Yamazaki S., et al. (2007) An orally available small-molecule inhibitor of c-Met, PF-2341066, exhibits cytoreductive antitumor efficacy through antiproliferative and antiangiogenic mechanisms. Cancer Res 67: 4408–4417 [DOI] [PubMed] [Google Scholar]