Abstract
Estrogenic compounds are an important class of hormonal substances that can be found as environmental contaminants, with sources including pharmaceuticals, human and animal waste, the chemical industry, and microbial metabolism. Here we report the creation of a biosensor useful for monitoring such compounds, based on complementation of fluorescent protein fragments. A series of sensors were made consisting of fragments of a split mVenus fluorescent protein fused at several different N-terminal and C-terminal positions flanking the ligand binding domain of the estrogen receptor alpha. When expressed in HeLa cells, sensor 6 (ERα 312-595) showed a nine-fold increase in fluorescence in the presence of estrogen receptor agonists or antagonists. Sensor 2 (ERα 281-549) discriminated between agonists and antagonists by showing a decrease in fluorescence in the presence of agonists while being induced by antagonists. The fluorescent signal of sensor 6 increased over a period of 24 hours, with a twofold induction visible at 4 hours and four-fold at 8 hours of ligand incubation. Ligand titration showed a good correlation with the known relative binding affinities of the compound. The sensor could detect a number of compounds of interest that can act as environmental endocrine disruptors. The lack of a substrate requirement, the speed of signal development, the potential for high throughput assays, and the ability to distinguish agonists from antagonists make this an attractive sensor for widespread use.
Keywords: molecular biosensor, estrogen receptor, estrogenic compounds, fluorescence complementation, split proteins
Introduction
As a member of the nuclear hormone receptor superfamily, the estrogen receptor alpha (ERα) acts as a ligand-regulated transcription factor that is involved in the control of a wide range of physiological processes, such as mammary gland development, fertility, bone growth and maintenance, and metabolism (Nilsson and Gustafsson 2010). Its canonical ligand is 17β-estradiol (E2), which binds to the ligand binding domain (LBD) and induces a conformational change in the position of helix 12 that stimulates dimerization, the recruitment of coactivators or corepressors, and alters the rate of gene transcription (Huang et al. 2010). ERα is also bound and influenced by a multitude of other steroid hormones and pharmaceuticals (Fig. 1), as well as natural products and industrial chemicals (Fig. 2) (Katzenellenbogen and Muthyala 2003; Kuiper et al. 1997; Kuiper et al. 1998). As a result, there has been much interest in the development of methods for identifying compounds that interact with ERα, which can be used to measure their concentrations, and also elucidate the conformational changes they induce.
Fig. 1.
Endogenous estrogens, pharmaceuticals, and steroid hormones.
Fig. 2.
Natural products and industrial chemicals discussed, some of which are estrogenic.
The ligands capable of binding to the ERα ligand binding pocket and the structural mechanisms underlying binding have been extensively investigated. An agonistic ligand stabilizes a conformation of the receptor where helix H12 lies across the binding pocket and completes the region that binds coactivators, while an antagonistic ligand obstructs this H12 position and causes it to bind in a manner that prevents coactivators binding, and selective estrogen receptor modulators act as antagonists in some tissues but allow activation in others (Anstead et al. 1997; Pike 2006). The primary endogenous compounds are 17β-estradiol, estriol, and estrone (along with its sulfated form), as well as 2- and 4-hydroxyestradiol (Gruber et al. 2002). Pharmaceuticals that interact with the estrogen receptor include diethylstilbestrol, 4-hydroxytamoxifen, raloxifene, dienestrol, and ICI 182780. Some natural products and industrial chemicals also exhibit estrogenic activity. Phytoestrogens such as genistein and daidzein from soy, and resveratrol from grapes, have been proposed to have beneficial health effects via an interaction with the estrogen receptor (Moutsatsou 2007). Alternatively, there is a growing concern over the effect of organic compounds produced by the chemical industry. While the presence of a single phenol group may not be sufficient to engender estrogenic activity, larger organic compounds such as bisphenol A and the nonionic detergent byproducts tert-octylphenol and nonylphenol do show such effects (Katzenellenbogen and Muthyala 2003).
Many of these estrogenic compounds can be found in the environment, where microbial organisms can act as either a source or a sink. A range of pharmaceuticals, hormones, and organic compounds can be detected in streams (Kolpin et al. 2002), with sources such as agricultural runoff, wastewater treatment, and industrial discharges. Estrogens also find their way into soil through the use of sewage sludge as fertilizers and can be degraded by some microorganisms (Combalbert and Hernandez-Raquet 2010). Microbes can synthesize estrogenic compounds such as zearalenone produced by Fusarium fungal contamination of corn and grain, or modify them as in the production of equol from soy isoflavones by the action of intestinal bacteria (Katzenellenbogen and Muthyala 2003). It is of interest to develop methods to monitor the levels of the many estrogenic compounds present in the environment.
Molecular biosensors are detection systems composed of engineered proteins that can be used to monitor for particular ligands or metabolites (East et al. 2008). Ligand controlled transcription factors like the ERα can be converted into biosensors for the activating ligand. This can be beneficial for investigating the range of ligands capable of interacting with hormonal signaling systems, for engineering specificity to new ligands, or for metabolic engineering. Here we develop a biosensor for estrogenic ligands by taking advantage of the bimolecular fluorescence complementation technique, a recently developed method for analyzing protein interactions based on the reconstitution of a functional fluorescent protein from non-fluorescent fragments (Hu et al. 2002). By fusing the two halves of a split Venus fluorescent protein to either end of the ERα ligand binding domain, the conformational change induced by ligand binding can be assayed by the complementation of the fragments leading to the formation of a fluorescent signal.
Materials and Methods
Reagents
All DNA polymerases, restriction enzymes, and T4 DNA ligase were from New England Biolabs (Ipswich, MA). Cell media, fetal bovine serum, and charcoal dextran treated calf serum were obtained from the University of Illinois cell media facility (Urbana, IL). Opti-MEM media, lipofectamine 2000, and trypsin / EDTA were from Invitrogen (Carlsbad, CA). Carbamyl and carbofuran were from Chem Service (West Chester, PA), and raloxifene was from Tocris Bioscience (St Louis, MO). Other chemicals were from Sigma (St Louis, MO).
Cloning of the sensor constructs
A monomeric version of Venus fluorescent protein was constructed by mutagenesis of the yellow fluorescent protein EYFP. Primers mVenus-Asmbl F1, R2, F3, and R4 were assembled using overlap extension PCR, and the product was used as a megaprimer on EYFP plasmid to extend to the C-terminus with mVenus-Rev-BamHI. This C-terminal fragment was used as a megaprimer with mVenus F64L-For on EYFP. The N-terminal fragment was created by PCR with primers mVenus-For-KpnI and mVenus F46L-Rev, and was then used with the C-terminal fragment to fill in the complete mVenus with EYFP as a template. The gene was digested with KpnI and BamHI and ligated into the vector pCMV5. The mVenus gene was split into fragments Vn (1-154) and Vc (155-239) by PCR of Vn with mVenus-For-KpnI and mVn154-Stop-Rev-BamHI, and PCR of Vc with mVc155-For-KpnI and mVenus-Rev-BamHI, both of which were cloned into pCMV5. For cloning of the sensors, the Vn-SalI-Vc-pCMV5 vector was constructed to insert the SalI site into mVenus-pCMV5 with mVenus-For-KpnI + mVenus154noStopRevSalI; and mVenus155-SalI-For + mVenus-Rev-BamHI. The estrogen receptor α ligand binding domain regions were amplified by PCR with combinations of the forward primer ER281-For-SalI only or ER312-For-SalI, and reverse primer ER532-noStop-Rev-SalI, ER549-noStop-Rev-SalI, or ER595-noStop-Rev-SalI, and were cloned into the SalI site of Vn-SalI-Vc-pCMV5. See Table S1 for primer sequences.
Cell culture and transfection
HeLa cells were grown in minimal essential medium (MEM) / 1 mM sodium pyruvate / 10% fetal bovine serum in a humidified incubator at 37 °C with 5% carbon dioxide. When cells were 80–90% confluent, they were trypsinized and split into 12-well plates with 1 mL per well of MEM / 1 mM sodium pyruvate / 5% charcoal dextran treated calf serum (CDCS). Cells were grown for 24 hours until they were over 90% confluent and transfected using lipofectamine 2000. Typically, 1.5 μL lipofectamine 2000 was resuspended in 48.5 μL OptiMEM and incubated at room temperature for 10 min, then 50 ng of the sensor plasmid with 750 ng of the plasmid encoding β-galactosidase resuspended in 50 μL Opti-MEM was added, and the mixture was incubated for 30 minutes and added to a well of HeLa cells. Cells were grown for 18 hours after which the media was changed to MEM media / 1 mM sodium pyruvate / 5% CDCS plus the desired ligand. After a further 30 hours of growth, cells were analyzed by flow cytometry.
Flow cytometry
Adherent cells were collected by trypsinization, resuspended in MEM + 5% CDCS, pelleted by centrifugation for 5 min at 800 ×g, and resuspended in 300–400 μL PBS / 5 mM EDTA. Cells were measured on a BD LSRII flow cytometer (Becton Dickinson, Franklin Lakes, NJ) using standard GFP/FITC filter sets. Analysis was performed using FCS Express 3 software (De Novo Software, Los Angeles, CA). Events were gated on the region corresponding to single whole cells and the mean fluorescence for 10000 cells was recorded.
Agonist and antagonist discrimination
Sensors 1–6 were transiently transfected by lipofection into HeLa cells. Samples were treated with no ligand, agonists (17β estradiol, diethylstilbestrol, estriol, or β-zearalanol), or antagonists / selective estrogen receptor modulators (ICI 182780, 4-hydroxytamoxifen, or raloxifene). Flow cytometry was performed and the average population fluorescence determined. Data was normalized by dividing by the average of the six no ligand conditions.
Time course of fluorescence signal generation
Sensor 6 was transiently transfected by lipofection into HeLa cells. After 18 hours, the media was replaced with fresh media containing 10−7 M 17β-estradiol or 0.1% ethanol control. After 0, 1, 2, 4, 8, or 24 hours of ligand exposure, cells were trypsinized, pelleted, resuspended in 200 μl PBS, and 1 mL of 3.6 % formaldehyde in PBS was added. Cells were fixed for 10 minutes, after which they were pelleted, resuspended in 300–400 μL PBS/5 mM EDTA, and stored at 4 °C until analyzed by flow cytometry.
Ligand titration
Sensor 6 was transiently transfected by lipofection into HeLa cells. After 18 hours, the media was changed and varying concentrations of the following ligands were added: 17β-estradiol, diethylstilbestrol, genistein, dienestrol, ICI 182780, progesterone, testosterone, 4-hydroxyestradiol, 2-hydroxyestradiol, estriol, norethindrone, estrone sulfate, 4-hydroxytamoxifen, and 17α-estradiol. After 30 hours, cells were analyzed by flow cytometry to measure the average population fluorescence, which was normalized such that the 10−7 M 17β-estradiol value was equal to 1. The data was plotted using OriginPro 8 (OriginLab Corporation, Northampton, MA), and curves were fitted to the sigmoidal dose response function to determine the concentrations of half-maximal induction (EC50). Relative binding affinities for the compounds were taken from published data (Kuiper et al. 1997) or determined in a similar manner by Kathryn Carlson using radioligand competition experiments with recombinant ERα (Carlson and Katzenellenbogen, unpublished data).
Environmental compound detection
Sensor 6 was transiently transfected by lipofection into HeLa cells. After 18 hours, the media was changed and varying concentrations of the following ligands were added: daidzein, equol, resveratrol, β-zearalenol, p-tert-octylphenol, nonylphenol, bisphenol A, o,p’-DDT, 2,2’,5-trichloro-4-hydroxy-PCB, phenol, 1-naphthol, 2-naphthol, carbaryl, or carbofuran. After 30 hours, cells were analyzed by flow cytometry.
Results
Cloning of mVenus and sensors
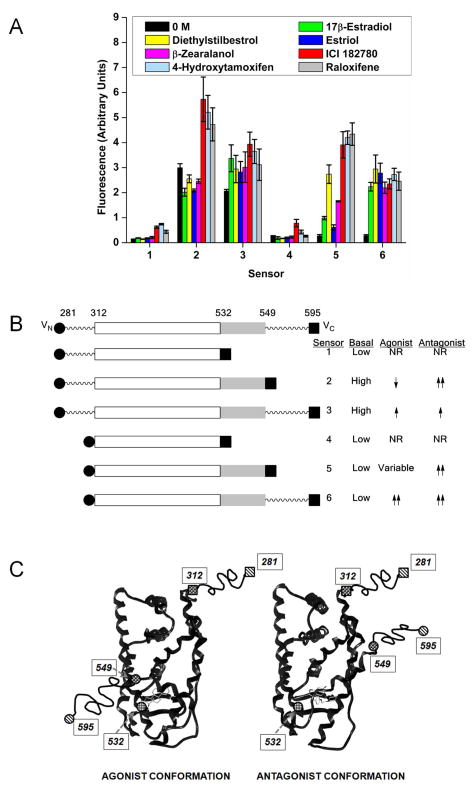
To create the estrogenic compound biosensor, the fluorescent protein Venus was used because it is bright, shows fast maturation at 37 °C (Nagai et al. 2002), and has been shown to perform well in fluorescence complementation applications (Shyu et al. 2006). The mutations F46L, F64L, M153T, V163A, and S175G were introduced into the EYFP plasmid to convert EYFP to Venus, as well as the A206K mutation which inhibits the weak dimerization activity of GFP variants (Zacharias et al. 2002), thus producing the monomeric mVenus construct. This was then split at position 155 to produce the two non-fluorescent fragments Vn (amino acids 1-154) and Vc (amino acids 155-239). Transfection studies confirmed that mVenus was expressed and fluorescent in HeLa cells. The two fragments, Vn and Vc, were not fluorescent when expressed independently but when co-expressed at high levels, they exhibited spontaneous association to give a fluorescent signal (Fig. S1). Sensors 1–6 were made by inserting regions of the ERα ligand binding domain between Vn and Vc (Table I). The N-terminal points for the LBD were amino acid positions 281 (including most of the upstream hinge region) and 312 (a few residues downstream of the LBD starting position). The C-terminal points of the LBD were 532 (removing helix 12 of the ligand binding domain), 549 (removing the C-terminal F domain), or 595 (extending to the end of ERα).
Table I.
Description of the sensors.
| Sensor | Description |
|---|---|
| 1 | Vn(1-154)-SalI-ER(281-532)-SalI-Vc(155-239) |
| 2 | Vn(1-154)-SalI-ER(281-549)-SalI-Vc(155-239) |
| 3 | Vn(1-154)-SalI-ER(281-595)-SalI-Vc(155-239) |
| 4 | Vn(1-154)-SalI-ER(312-532)-SalI-Vc(155-239) |
| 5 | Vn(1-154)-SalI-ER(312-549)-SalI-Vc(155-239) |
| 6 | Vn(1-154)-SalI-ER(312-595)-SalI-Vc(155-239) |
Agonist and antagonist compound evaluation
Transfection of the six sensors into HeLa cells followed by exposure to 100 nM of known agonists (17β-estradiol, diethylstilbestrol, estriol, or β-zearalanol), the antagonist ICI 182780, or the selective estrogen receptor modulators 4-hydroxytamoxifen and raloxifene revealed that each sensor had different properties (Fig. 3). Sensors 1 and 4 had low basal fluorescence with minimal induction in the presence of ligand. Sensor 3 also performed poorly, having a high basal fluorescence and only a slight increase upon the addition of ligand. Sensor 6 had a low basal fluorescence and showed 9-fold induction with both agonists and antagonists. Sensor 2 had high basal expression and a 15–30% decrease in the presence of agonists, but an increase of 50–90% in the presence of antagonists. Sensor 5 had a low basal signal with strong induction of 16-fold in the presence of the three antagonists, with a variable but lower induction to the agonists.
Fig. 3.
(A) Sensor performance with agonists and antagonists. Sensors 1–6 were transiently transfected into HeLa cells and exposed to 0.1% ethanol control or 100 nM of the agonists 17β-estradiol, diethylstilbestrol, estriol, or β-zearalanol, or the antagonist/selective estrogen receptor modulators ICI 182780, 4-hydroxytamoxifen, or raloxifene. Data shown is the mean and standard error of the mean for two or more independent experiments. (B) Schematic representation of the sensors. The portion of the ERα LBD found highly structured in crystal structures is shown as a boxed area (roughly from 305 to 549), with the portion representing helix-12 shown in light gray. The sites of truncation at the N-terminus (281 and 312) and at the C-terminus (532, 549, and 595) are indicated. (C) Cartoon representing the proposed localization of the split sensor fragments in response to agonist or antagonist bound ER ligand binding domain, and helix-12 (roughly position 537-549) conformation change. Because the sequence positions at the far N- and C-termini (281 and 595) are beyond that of known crystal structures, these are connected to the structured core by wavy lines. Representative structures of the estrogen receptor with agonist or antagonist bound can be found in the protein data bank under accession numbers 1ERE and 1ERR.
Time course of fluorescence signal generation
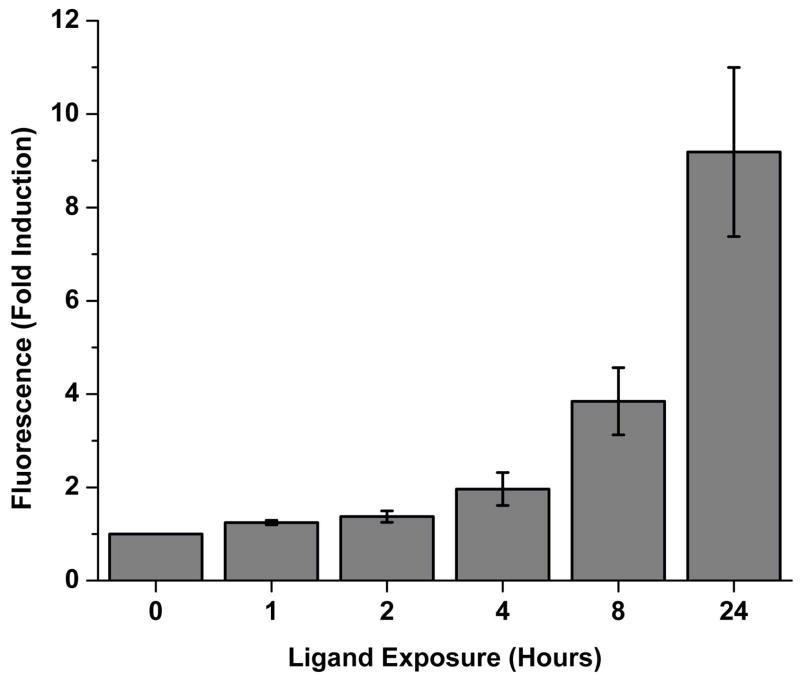
The time course of signal development for the fluorescence complementation biosensor was determined using sensor 6, as it showed the best induction properties. The sensor was transfected into HeLa cells and exposed to 100 nM 17β-estradiol for up to 24 hours before the cells were fixed in formaldehyde for analysis by flow cytometry. The results show a 25% ligand dependent induction within 1 hour, 2-fold induction by 4 hours, and 9-fold induction with a 24 hour incubation in the presence of ligand (Fig. 4).
Fig. 4.
Time course of fluorescence signal generation of sensor 6. HeLa cells were transiently transfected with sensor 6 and exposed to 100 nM 17β-estradiol or 0.1% ethanol control for up to 24 hours, followed by formaldehyde fixation and flow cytometry. Data is the mean fold induction between ligand and no ligand conditions at each time point, with the standard error of the mean for three independent experiments.
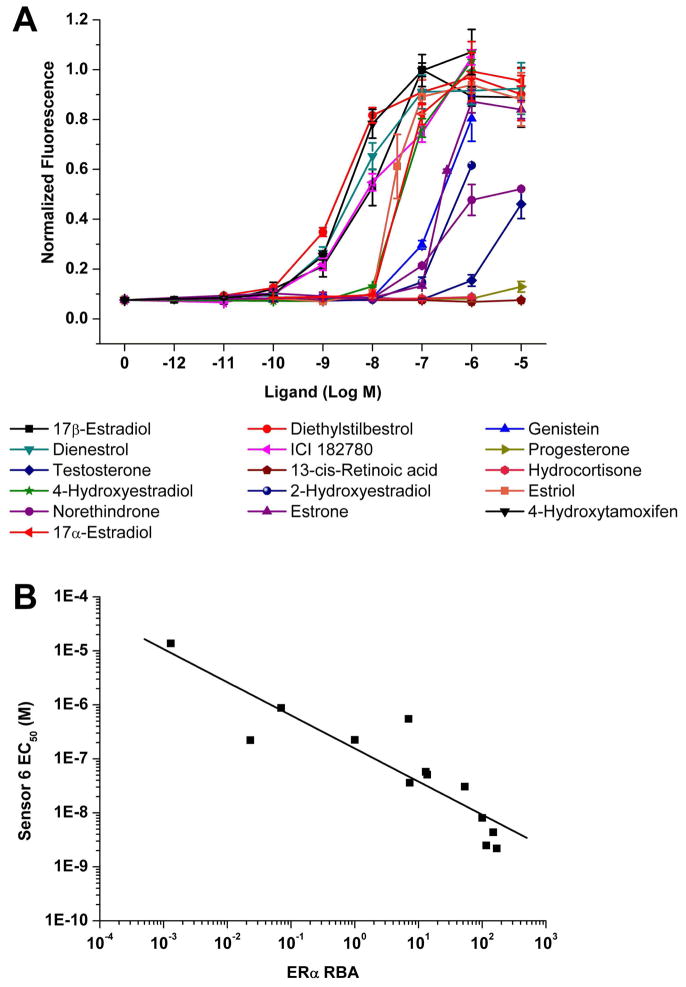
Ligand titration
As sensor 6 was seen to perform well, its sensitivity was examined to a number of different ligands whose relative binding affinities for the estrogen receptor was known. The ligand binding curves are shown in Fig. 5A, and for those compounds that showed induction the calculated concentration at half maximal induction (EC50) is shown in Table II. The EC50 values were plotted against known relative binding affinities (RBA) for the compounds (Carlson and Katzenellenbogen, unpublished data; and (Kuiper et al. 1997)) (Fig. 5B). A power function regression model fitted to the data using Microsoft Excel gave an r2 value of 0.81.
Fig. 5.
(A) Ligand titration of compounds with sensor 6. (B) Log-Log scatter plot of relative binding affinity (RBA) against EC50 determined by sensor 6. The RBA of 17β-estradiol is set to be 100, and a regression curve was fitted with an r2 value of 0.81.
Table II.
Concentration of ligand required to reach half maximal induction of sensor 6 (nd = not determined, SEM = Standard Error of the Mean).
| Ligand | EC50 ± SEM (nM) |
|---|---|
| 17β-Estradiol | 8.1 ± 1.5 |
| Diethylstilbestrol | 2.2 ± 0.37 |
| Genistein | 220 ± 11 |
| Dienestrol | 4.4 ± 0.87 |
| ICI 182780 | 31 ± 13 |
| Progesterone | nd |
| 13-cis Retinoic acid | nd |
| Hydrocortisone | nd |
| Testosterone | 14000 ± 3700 |
| 4-Hydroxyestradiol | 58 ± 4.7 |
| 2-Hydroxyestradiol | 550 ± 100 |
| Estriol | 36 ± 11 |
| Norethindrone | 880 ± 260 |
| Estrone sulfate | 230 ± 27 |
| 4-Hydroxytamoxifen | 2.5 ± 0.73 |
| 17α-Estradiol | 51 ± 1.7 |
Detection of exogenous endocrine active substances
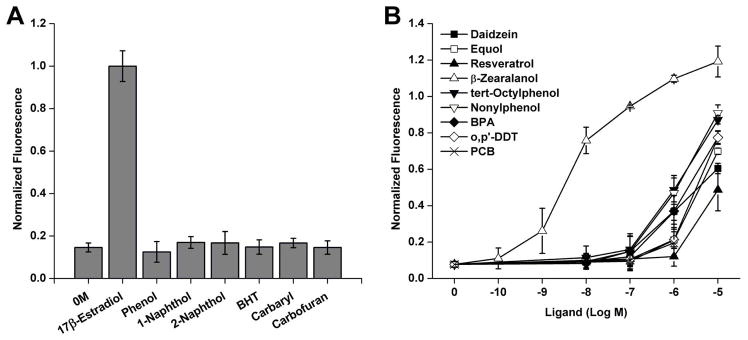
After showing that sensor 6 functioned well with a range of compounds, a number of environmental compounds were examined. Figure 6 shows the results for compounds reported to be negative for estrogenic activity (Fig. 6A) and also those expected to produce a response (Fig. 6B). Phenol, 1-naphthol, and 2-naphthol are commonly used in organic synthesis and the plastic industry; butylated hydroxytoluene is an antioxidant, while carbofuran and carbaryl are insecticides. All of these have aromatic ring structures but do not show estrogenic activity (Soto et al. 1995). When tested here using sensor 6, none showed an increase in fluorescence at 10 μM concentration. Phytoestrogens are compounds derived from plants and can be consumed in amounts sufficient to produce plasma concentrations up to 10 μM (Moutsatsou 2007). Daidzein is found in soy and is acted on by microorganisms in the gut to produce equol, both of which caused an increase in fluorescence at micromolar concentrations. Resveratrol is found in grapes and has been proposed to have many beneficial health effects but only weakly activated the sensor at 10 μM. The Fusarium fungus can grow on corn and other crops and produces zearalanone, from which the more active β-zearalanol, which is strongly estrogenic (Katzenellenbogen and Muthyala 2003), can be prepared, as seen in Figure 6B. The nonionic detergent byproducts tert-octylphenol and nonylphenol showed strong induction at micromolar levels, along with bisphenol A. The pesticide o,p’-DDT, and a PCB compound also displayed activation beginning at micromolar levels.
Fig. 6.
Detection of environmental chemicals using sensor 6. (A) Compounds reported to not show estrogenic activity were tested at 10 μM, along with 0.1% ethanol control (0 M) and 0.1 μM 17β-estradiol as controls (BHT = butylated hydroxytoluene). (B) Compounds reported to show estrogenic activity: daidzein, equol, resveratrol, β-zearalanol, tert-octylphenol, nonylphenol, bisphenol A (BPA), o,p’-DDT, and 2,2’,5-trichloro-4-hydroxy-PCB (PCB) were tested at concentrations up to 10 μM. Data was normalized to the 100 nM estradiol condition and is shown as the mean and standard error of the mean.
Discussion
The fluorescent complementation biosensor for estrogenic compounds described here proved to perform well. It was capable of detecting the binding of ligands whose affinity varied from nanomolar to tens of micromolar. Sensor 6 could detect both agonists and antagonists from a range of sources, while sensor 2 could distinguish antagonists from agonists. The six sensors we investigated differed in their overall activity, their basal activity, their responsiveness to ligands, and their ability to discriminate between agonist vs. antagonist ligands. Speculation on the cause of these differences begins by reference to the portion of the ERα sequence used in their construction, which is illustrated in a linear sense in Figure 3B and a structural sense in Figure 3C.
The two sensors that show essentially no complementation, regardless of presence or absence of ligand (sensors 1 and 4), both terminate at 532, at the end of helix-11. Being at the end of a highly structured region, this site may provide insufficient topological flexibility for the Vc fragment to access the Vn fragment, regardless of where it is attached (281 or 312). Of the remaining four sensors, the two that begin at 281 (sensors 2 and 3) have high basal activity. The 281 site is in the “hinge region” of ERα, thought to be an unstructured sequence linking the DNA binding domain to the ligand binding domain. The high levels of complementation observed suggest that it provides good access of the Vn fragment to the Vc fragment, regardless of whether the latter is positioned at 549 or 595. On the other hand, sensors 5 and 6, which begin at 312, have low basal activity, the Vn fragment now being constrained so as to be able to access the Vc region only under certain states of ligand occupancy of the LBD.
The responses of sensors 2, 3, 5, and 6 to ligands are also different. Of the sensors having high basal activity (sensors 2 and 3), the one with the Vn and Vc fragments attached by flexible tethers (sensor 3) shows minimal response to either agonist or antagonist ligands beyond the high basal activity, the high flexibility of this construct probably masking the different positions of helix-12 in such complexes. The sensor that terminates at 549, however, has the Vc fragment attached precisely at the LBD site that undergoes maximal conformation change with agonist vs. antagonist ligands bound (Pike 2006). The agonist conformation, in which helix-12 is folded back over the ligand, toward helix-11, gives a signal reduced from basal, whereas the antagonist conformation, having helix-11 extended outward as it binds into the coactivator binding groove, places position 549 closer to the site of Vn attachment (281). Thus, this is the sensor that provides the clearest discrimination between agonist and antagonist ligands, although the high basal activity of the unliganded sensor suggests that it adopts, on the average, a conformation more like that of an agonist than an antagonist structure.
The final two sensors, 5 and 6, show good response to most ligands. Sensor 6 gives equivalent, enhanced signals to both agonists and antagonists. Presumably, the extra flexibility that results from extending the Vc fragment from 549 to 595 mutes the conformational differences of helix-12 in agonist vs. antagonist complexes, making sensor 6 a convenient universal sensor of ligand binding to ERα. Sensor 5, having the Vc fragment attached to 549, regains sensitivity to agonists vs. antagonists, though with lower basal activity than sensor 2, because the Vn fragment in sensor 5 is attached to the structured 312 site, not the flexible 281 site. However, although sensor 5 shows a large response to antagonists, akin to that of sensor 2, its response to agonists, though evident, is variable. Even though there are canonical conformations of the ERα LBD associated with antagonists (helix-12 extended) and agonists (helix-12 folded back), as in Figure 3C, it is known that there are more subtle differences even among structures of different ligands within the antagonist and agonist classes (Pike 2006). Sensor 5, being based on the shorter sequence of the LBD, appears to be the one most able to discriminate among different agonist conformations.
Multiple biosensors for nuclear hormone-like compounds have been developed that differ in the reporter used, and also in the host species (Gillies et al. 2008). Assays have been developed based on the natural activity of 17β-estradiol, such as measuring an increase in prepubertal mouse uterine weight or the growth of the estradiol responsive MCF-7 tumor cell line (Soto et al. 1995). While these have the benefit of being highly relevant physiologically, they take longer than other assays and are not useful from an engineering perspective. As ERα is a transcription factor, biosensors have been constructed simply by cloning a reporter gene such as β-galactosidase, luciferase, or EGFP under the control of estrogen receptor response elements and expressing these in yeast (Bovee et al. 2004; Coldham et al. 1997; Sanseverino et al. 2005). Other studies have replaced the natural DNA binding domain with that of the yeast GAL4 transcription factor (Wilkinson et al. 2008). Yeast one- or two-hybrid assays have been developed that rely on either a β-galactosidase reporter or a growth-based assay (Chen et al. 2004; Chen and Zhao 2003; Chockalingam et al. 2005; Lee et al. 2006; McLachlan et al. 2009). Fluorescently tagged estrogen receptors or ligand binding domains have been used to monitor ligand binding based on sensor stabilization or Förster resonance energy transfer (FRET) (De et al. 2005; Muddana and Peterson 2003; Umezawa 2005) or bioluminescence resonance energy transfer (BRET) between a luciferase donor and a fluorescent acceptor (Michelini et al. 2004). Other systems have been developed based on complementation of split luciferase fragments expressed in mammalian cells (Paulmurugan and Gambhir 2006), or ligand-stimulated splicing out of inteins from a thymidylate synthase gene in bacteria (Skretas and Wood 2005).
The properties of fluorescence complementation used in the current sensor system compare favorably to previously reported sensors. It has the benefit of requiring no exogenous substrates to produce a signal, as opposed to the luciferase complementation sensor (Paulmurugan and Gambhir 2006) or BRET sensor (Michelini et al. 2004). The signal generated is simple to measure by the increase in fluorescence although the current implementation with flow cytometry does limit throughput. The sensor is expressed in vivo within mammalian cells, and while this increases the complexity of culturing conditions compared to yeast or bacteria, the transport of ligands into the cell is more appropriate for monitoring effects on mammalian systems.
The time for signal generation is intermediate compared to other reported biosensors, partly due to fluorescence complementation requiring a period of time after ligand binding for the generation of the Venus fluorophore. The signal from a FRET sensor is very fast since the fluorophores are already formed, so the time taken to measure a change in emission ratio reflects only the time for ligand binding and the conformation change of helix 12 and can be detected within 4–20 minutes (Umezawa 2005). Split luciferase systems in general are also capable of signal changes on the timescale of minutes (Fan et al. 2008), however the estrogen biosensor by Paulmurugan et al. using a split luciferase (Paulmurugan and Gambhir 2006) took between 6 and 12 hours to give a measurable signal, with maximal induction after 24 hours, which is similar to the sensor described here. The 24 hours required for maximal induction of the fluorescence and luciferase complementation signal suggest that these assays are not simply monitoring a conformation change. It is possible that the sensors are stabilized by ligand binding, but that the length of time to reach full signal strength reflects the growing accumulation of newly synthesized sensor. The fluorescence complementation assay is faster than growth based assays for estrogenic compounds such as the E-SCREEN method which monitors growth of the human cell line MCF-7 after six days (Soto et al. 1995). Another advantage of the fluorescence complementation assay is that after cell exposure and sensor signal generation, cells can be fixed with formaldehyde, and flow cytometry can be performed at a convenient later time.
There is a reasonable correlation between the sensitivity of sensor 6 and previously determined binding data, as can be seen in Figure 5B. The sensitivity toward 17β-estradiol was lower than some measurements, although the value depends on the assay used, ranging from 6 pM when monitoring MCF-7 cell growth, up to nM values (Olsen et al. 2005). The 8 nM EC50 is the same (within experimental error) as that reported with FRET biosensors (De et al. 2005; Umezawa 2005). For diethylstilbestrol, 4-hydroxytamoxifen, ICI 182780, and estriol (the compounds tested by both methods), De et al. reported RBAs of 127.3, 62.7, 10.3, and 2.9 respectively, compared to the current complementation sensor’s more sensitive values of 368, 324, 26, and 23 (obtained by calculation from Table II). Umezawa et al. reported a lower RBA for diethylstilbestrol and a higher RBA for genistein (60 and 12) than the complementation sensor (368 and 3.7) for their FRET sensor and was more sensitive toward nonylphenol and bisphenol-A. Progesterone, 13-cis retinoic acid, and hydrocortisone were not expected to bind and indeed gave minimal response. Some low affinity ligands showed detectable binding, such as norethindrone and testosterone at micromolar and higher, and genistein at 100 nM and higher. These compounds are typically not detected by some sensors, such as genistein by the luciferase complementation system (which did not report relative binding affinity data) (Paulmurugan and Gambhir 2006). The dynamic range of the complementation assay was around four orders of magnitude, allowing the measurement of ligands whose binding affinity was from nanomolar to tens of micromolar. For any particular ligand, the complementation assay tended to be sensitive to a concentration range of two to three orders of magnitude around the EC50. These characteristics generally compare well to other bioassays although the FRET assay from Umezawa et al. had a wider range over which a given ligand responded (4–5 orders of magnitude) (Umezawa 2005).
Sensors 2 and 6 described here are useful novel biosensors for estrogenic compounds. They could be used for screening programs aiming to identify novel drugs targeting the ERα, or for monitoring environmental contamination. Although the compounds regarded as environmental contaminants that were examined here led to fluorescence at concentrations much higher than found in the environment, it is conceivable that multiple such compounds could lead to a cumulative estrogenic effect. The data presented here is from transiently transfected cells, a technique which has been used in other sensor studies such as (Paulmurugan and Gambhir 2006) and (Umezawa 2005), gives reproducible results, and allows for the easy choice of different target cells. Alternatively, stable integration into a human cell line such as HEK 293 would simplify screening by avoiding the need for transfection.
Supplementary Material
Acknowledgments
This work was supported by the National Science Foundation CAREER Award (BES-0348107) and the Centennial Chair Professorship at the Department of Chemical and Biomolecular Engineering at the University of Illinois at Urbana-Champaign (to H. Z.), and the National Institutes of Health (5R37DK015556 to J. A. K.). We thank Kathy Carlson for providing data on relative binding affinity of ligands to the estrogen receptor alpha and assistance with ligands, John Comninos for assistance with ligands and figures, and Zhanar Abil for assistance in DNA cloning.
References
- Anstead GM, Carlson KE, Katzenellenbogen JA. The estradiol pharmacophore: Ligand structure-estrogen receptor binding affinity relationships and a model for the receptor binding site. Steroids. 1997;62:268–303. doi: 10.1016/s0039-128x(96)00242-5. [DOI] [PubMed] [Google Scholar]
- Bovee TF, Helsdingen RJ, Koks PD, Kuiper HA, Hoogenboom RL, Keijer J. Development of a rapid yeast estrogen bioassay, based on the expression of green fluorescent protein. Gene. 2004;325:187–200. doi: 10.1016/j.gene.2003.10.015. [DOI] [PubMed] [Google Scholar]
- Chen Z, Katzenellenbogen BS, Katzenellenbogen JA, Zhao H. Directed evolution of human estrogen receptor variants with significantly enhanced androgen specificity and affinity. J Biol Chem. 2004;279(32):33855–64. doi: 10.1074/jbc.M402118200. [DOI] [PubMed] [Google Scholar]
- Chen Z, Zhao H. A highly efficient and sensitive screening method for trans-activation activity of estrogen receptors. Gene. 2003;306:127–134. doi: 10.1016/s0378-1119(03)00431-1. [DOI] [PubMed] [Google Scholar]
- Chockalingam K, Chen Z, Katzenellenbogen JA, Zhao H. Directed evolution of specific receptor-ligand pairs for use in the creation of gene switches. Proc Natl Acad Sci U S A. 2005;102:5691–6. doi: 10.1073/pnas.0409206102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coldham NG, Dave M, Sivapathasundaram S, McDonnell DP, Connor C, Sauer MJ. Evaluation of a recombinant yeast cell estrogen screening assay. Environ Health Perspect. 1997;105:734–742. doi: 10.1289/ehp.97105734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combalbert S, Hernandez-Raquet G. Occurrence, fate, and biodegradation of estrogens in sewage and manure. Appl Microbiol Biotechnol. 2010;86:1671–1692. doi: 10.1007/s00253-010-2547-x. [DOI] [PubMed] [Google Scholar]
- De S, Macara IG, Lannigan DA. Novel biosensors for the detection of estrogen receptor ligands. J Steroid Biochem Mol Biol. 2005;96:235–244. doi: 10.1016/j.jsbmb.2005.04.030. [DOI] [PubMed] [Google Scholar]
- East AK, Mauchline TH, Poole PS. Biosensors for ligand detection. Adv Appl Microbiol. 2008;64:137–166. doi: 10.1016/S0065-2164(08)00405-X. [DOI] [PubMed] [Google Scholar]
- Fan F, Binkowski BF, Butler BL, Stecha PF, Lewis MK, Wood KV. Novel genetically encoded biosensors using firefly luciferase. ACS Chem Biol. 2008;3:346–351. doi: 10.1021/cb8000414. [DOI] [PubMed] [Google Scholar]
- Gillies AR, Skretas G, Wood DW. Engineered systems for detection and discovery of nuclear hormone-like compounds. Biotechnol Prog. 2008;24:8–16. doi: 10.1021/bp070144i. [DOI] [PubMed] [Google Scholar]
- Gruber CJ, Tschugguel W, Schneeberger C, Huber JC. Production and actions of estrogens. N Engl J Med. 2002;346:340–352. doi: 10.1056/NEJMra000471. [DOI] [PubMed] [Google Scholar]
- Hu CD, Chinenov Y, Kerppola TK. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell. 2002;9:789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- Huang P, Chandra V, Rastinejad F. Structural overview of the nuclear receptor superfamily: insights into physiology and therapeutics. Annu Rev Physiol. 2010;72:247–272. doi: 10.1146/annurev-physiol-021909-135917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzenellenbogen JA, Muthyala R. Interactions of exogenous endocrine active substances with nuclear receptors. Pure Appl Chem. 2003;75:1797–1817. [Google Scholar]
- Kolpin DW, Furlong ET, Meyer MT, Thurman EM, Zaugg SD, Barber LB, Buxton HT. Pharmaceuticals, hormones, and other organic wastewater contaminants in U.S. streams, 1999–2000: a national reconnaissance. Environ Sci Technol. 2002;36(6):1202–1211. doi: 10.1021/es011055j. [DOI] [PubMed] [Google Scholar]
- Kuiper GG, Carlsson B, Grandien K, Enmark E, Haggblad J, Nilsson S, Gustafsson JA. Comparison of the ligand binding specificity and transcript tissue distribution of estrogen receptors alpha and beta. Endocrinology. 1997;138:863–870. doi: 10.1210/endo.138.3.4979. [DOI] [PubMed] [Google Scholar]
- Kuiper GG, Lemmen JG, Carlsson B, Corton JC, Safe SH, van der Saag PT, van der Burg B, Gustafsson JA. Interaction of estrogenic chemicals and phytoestrogens with estrogen receptor beta. Endocrinology. 1998;139:4252–4263. doi: 10.1210/endo.139.10.6216. [DOI] [PubMed] [Google Scholar]
- Lee HS, Sasagawa S, Kato S, Fukuda R, Horiuchi H, Ohta A. Yeast two-hybrid detection systems that are highly sensitive to a certain kind of endocrine disruptors. Biosci Biotechnol Biochem. 2006;70:521–524. doi: 10.1271/bbb.70.521. [DOI] [PubMed] [Google Scholar]
- McLachlan MJ, Chockalingam K, Lai KC, Zhao HM. Directed evolution of orthogonal ligand specificity in a single scaffold. Angew Chem Int Edit. 2009;48:7783–7786. doi: 10.1002/anie.200903413. [DOI] [PubMed] [Google Scholar]
- Michelini E, Mirasoli M, Karp M, Virta M, Roda A. Development of a bioluminescence resonance energy-transfer assay for estrogen-like compound in vivo monitoring. Anal Chem. 2004;76:7069–7076. doi: 10.1021/ac048914h. [DOI] [PubMed] [Google Scholar]
- Moutsatsou P. The spectrum of phytoestrogens in nature: our knowledge is expanding. Hormones. 2007;6:173–193. [PubMed] [Google Scholar]
- Muddana SS, Peterson BR. Fluorescent cellular sensors of steroid receptor ligands. Chembiochem. 2003;4:848–855. doi: 10.1002/cbic.200300606. [DOI] [PubMed] [Google Scholar]
- Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- Nilsson S, Gustafsson JA. Estrogen receptors: their actions and functional roles in health and disease. In: Bunce CM, Campbell MJ, editors. Nuclear Receptors. Netherlands: Springer; 2010. pp. 91–141. [Google Scholar]
- Olsen CM, Meussen-Elholm ET, Hongslo JK, Stenersen J, Tollefsen KE. Estrogenic effects of environmental chemicals: an interspecies comparison. Comp Biochem Physiol C Toxicol Pharmacol. 2005;141:267–274. doi: 10.1016/j.cca.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Paulmurugan R, Gambhir SS. An intramolecular folding sensor for imaging estrogen receptor-ligand interactions. Proc Natl Acad Sci U S A. 2006;103:15883–15888. doi: 10.1073/pnas.0607385103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike AC. Lessons learnt from structural studies of the oestrogen receptor. Best Pract Res Clin Endocrinol Metab. 2006;20:1–14. doi: 10.1016/j.beem.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Sanseverino J, Gupta RK, Layton AC, Patterson SS, Ripp SA, Saidak L, Simpson ML, Schultz TW, Sayler GS. Use of Saccharomyces cerevisiae BLYES expressing bacterial bioluminescence for rapid, sensitive detection of estrogenic compounds. Appl Environ Microbiol. 2005;71:4455–4460. doi: 10.1128/AEM.71.8.4455-4460.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu YJ, Liu H, Deng X, Hu CD. Identification of new fluorescent protein fragments for bimolecular fluorescence complementation analysis under physiological conditions. BioTechniques. 2006;40:61–66. doi: 10.2144/000112036. [DOI] [PubMed] [Google Scholar]
- Skretas G, Wood DW. A bacterial biosensor of endocrine modulators. J Mol Biol. 2005;349:464–474. doi: 10.1016/j.jmb.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Soto AM, Sonnenschein C, Chung KL, Fernandez MF, Olea N, Serrano FO. The E-SCREEN assay as a tool to identify estrogens: an update on estrogenic environmental pollutants. Environ Health Perspect. 1995;103(Suppl 7):113–122. doi: 10.1289/ehp.95103s7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa Y. Genetically encoded optical probes for imaging cellular signaling pathways. Biosens Bioelectron. 2005;20:2504–2511. doi: 10.1016/j.bios.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Wilkinson JM, Hayes S, Thompson D, Whitney P, Bi K. Compound profiling using a panel of steroid hormone receptor cell-based assays. J Biomol Screen. 2008;13:755–765. doi: 10.1177/1087057108322155. [DOI] [PubMed] [Google Scholar]
- Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.