Abstract
Ionic flux in defined cell populations mediates essential physiological and behavioral functions. Cell type-specific activators of diverse ionic conductances are needed for probing these relationships. We combined chemistry and protein engineering to enable systematic creation of a toolbox of ligand-gated ion channels (LGICs) with orthogonal pharmacologic selectivity and divergent functional properties. The LGICs and their small molecule effectors can activate a range of ionic conductances in genetically-specified cell types. LGICs constructed for neuronal perturbation can be used to selectively manipulate neuron activity in mammalian brains in vivo. The diversity of ion channel tools accessible from this approach will be useful for examining the relationship between neuronal activity and animal behavior, as well as for cell biological and physiological applications requiring chemical control of ion conductance.
Ion channels are complex molecular machines with critical cell biological functions. Ligand-gated ion channels (LGICs) provide rapid, remote control over conductances for different ions. In neurons, LGICs can be exploited for stimulation or silencing to examine causal relationships between electrical activity and animal behavior.
Several neuron manipulation tools have been derived from LGICs and G-protein coupled receptors (1-4) that can be genetically targeted and are reported to be orthogonal to endogenous systems. These tools are useful (5-7) but also face limitations such as ligand instability and lack of brain access (2), slow pharmacokinetics (6), the need to knockout endogenous alleles (3), or reliance on complex intracellular signaling pathways (4). Optogenetic tools (8-10) activate conductances with millisecond precision, but optimization of ion conductance properties has been limited and light targeting is invasive.
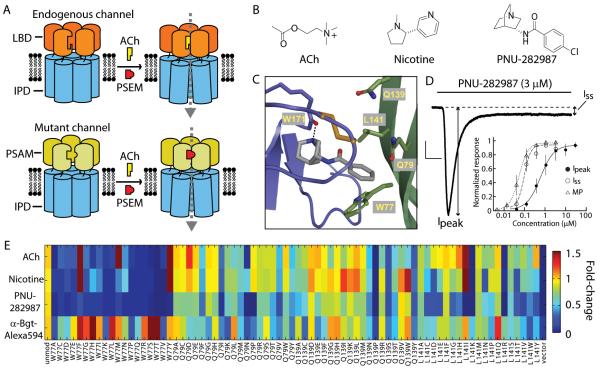
To overcome these limitations, we have developed a strategy to create chimeric LGICs with distinct conductance properties derived from modular combinations of pharmacologically-selective ligand binding domains (LBDs) and functionally diverse ion pore domains (IPDs). Within the Cys-loop receptor superfamily, the LBD of the α7 nicotinic acetylcholine receptor (nAChR) behaves as an independent actuator module that can be transplanted onto the IPDs of other Cys-loop receptors (11, 12). These include at least 43 ion channel subunits in vertebrates (13), and many additional invertebrate (14) and prokaryotic (15) subunits. Distinct IPDs confer selectivity for chloride or calcium as well as nonspecific cations. For example, splicing the α7 nAChR LBD to the IPDs of the serotonin receptor 3a or the glycine receptor produces chimeric channels (α7-5HT3 or α7-GlyR) with α7 nAChR pharmacology and cation or chloride conductance properties, respectively (11, 12). This modular property is a strong foundation for tailoring functional characteristics. However, the major challenge for using these ion channel LBDs and their ligands as genetically-targeted, cell type-selective perturbation tools is that they are already found in the brain; thus small molecule agonists will perturb electrical activity in untargeted cell populations. We addressed this challenge by developing an approach to modify the ligand recognition properties of the α7 nAChR LBD using a “bump-hole” strategy (Fig. 1A) (16-18).
Fig. 1.
“Bump-hole” approach to engineer selective ion channel-ligand interactions. (A) LGICs composed of ligand binding domain (LBD) and ion pore domain (IPD) modules. LBD mutations yield a Pharmacologically Selective Actuator Module (PSAM) that selectively binds Pharmacologically Selective Effector Molecules (PSEMs, red) but not the endogenous ligand (acetylcholine, ACh, yellow). PSEMs do not bind the unmodified LBD. (B) Chemical structures of ACh, nicotine, and PNU-282987. (C) Homology model of the α7 nAChR LBD with a docked agonist at the interface between two protomers (purple and green). Residues W77, Q79, Q139, and L141 were targeted for mutagenesis. (D) Current evoked by PNU-282987 application to an HEK 293 cell expressing α7-5HT3. A large peak current (Ipeak) rapidly decays to a persistent, steady state current (Iss). Black line indicates time course of ligand application. Scale bars: 200 pA, 0.5 s. Inset, Dose response curves (normalized to maximum response) from MP assay correspond to Iss (n = 3). (E) Color map showing activity and cell surface expression of mutated α7-5HT3 channels. Responses to ACh (100 μM), nicotine (100 μM), PNU-282987 (10 μM), and binding of α-Bungarotoxin (Bgt)-Alexa594 were normalized to the response of the unmodified channel to ACh (100 μM) or α-Bgt-Alexa594 binding. Error bars are s.e.m.
α7 nAChRs are homopentameric channels that form ligand binding sites at the protein-protein interfaces (19), typically a difficult environment for designing small molecule interactions. We required ion channel activation following ligand binding, necessitating that molecular alterations be compatible with the operation of these complex molecular machines. Thus, our strategy differed from previous efforts to devise “pharmacological alleles”, which used structure-based design to accommodate rigid purine-based antagonists or cofactors into well-defined binding sites in enzymes (17, 18) or small molecule-binding proteins (16). To address the additional challenges of pharmacologically engineering ion channels, we used structural models to predict molecular interactions and compensated for uncertainty in these models with functional assays to screen libraries of molecules and mutant ion channels for agonist activity.
First, we modeled the small molecule-protein interaction by homology to the x-ray crystal structure of the snail acetylcholine binding protein (19) complexed to nicotine (Fig. 1B and C). We selected the quinuclidinyl benzamide, PNU-282987 (Fig. 1B), as the starting point for ligand design because it was reported to cross the blood-brain barrier (20) and to be highly selective for α7 nAChR (21).
(R)-3-aminoquinuclidine benzamide, a PNU-282987 analog, was docked with a binding mode analogous to nicotine, where the protonated tertiary amine interacts with W171 and the benzamide interacts primarily with residues on the complementary face of the adjacent protomer (Fig. 1C). Because bulky benzamide substituents were detrimental to activity at the endogenous receptor (21), we mutated amino acids proximal to this group: W77, Q79, Q139, L141 (Fig. 1C and fig. S1) to 19 alternative amino acids for a total of 76 mutant channels. Mutant α7 nAChR LBDs were spliced to the IPD of the 5HT3a receptor (fig. S1) and pre-screened for cell surface expression and channel activation. Whole cell voltage clamp recordings of α7-5HT3 responses to PNU-282987 in transfected HEK 293 cells showed an initial peak current (Ipeak), which desensitized (t1/2 = 0.26 ± 0.07 s, mean ± s.d.) to a steady state current (Iss), where Iss/Ipeak = 0.09 ± 0.07 (Fig. 1D). Because we aimed to activate conductances for minutes, we used Iss to assess ligand potency. Dose responses from a fluorescence plate reader-compatible, membrane potential (MP) assay reflected Iss resulting from sustained ligand activation of the channel (Fig. 1D and Table S1). Mutations at Q79, Q139, and L141 were tolerated functionally, while for W77 only Tyr, Phe, and Met mutations produced acetylcholine (ACh)-responsive channels (Fig. 1E). In total, 43 mutated ion channels were selected for additional dose response assays.
We synthesized a focused chemical library of 71 enantiomerically pure 3-aminoquinuclidine benzamide candidate ligands to test against mutated α7-5HT3 channels (fig. S2). Because benzamide substitutuents at C2 and C4 can adversely affect α7-5HT3 channel activation (21), our library emphasized these substitution patterns. Compounds that exhibited low activity at α7-5HT3 (Fig. 2A) were selected for further analysis.
Fig. 2.
Selective interactions between ligands and mutated α7-5HT3 chimeric ion channels. (A) Color map showing EC50s for mutated α7-5HT3 with ACh, nicotine, and aminoquinuclidine benzamides. In order to highlight molecules that are highly selective for mutated α7-5HT3 chimeric receptors, only the activity of molecules with EC50 > 30 μM against unmodified α7-5HT3 (top row) are shown for the mutated receptors. (B) Dose response curves for compounds against mutated channels and showing negligible activation of unmodified α7-5HT3. EC50MP in parentheses. Normalization is to the maximum response to each compound or to ACh (for unmodified α7-5HT3). (C) Dose response curves for mutated ion channel-ligand interactions that show orthogonal pharmacology. Normalization is to maximum response for each compound across mutated channels. (D) Chemical structures of ligands in B and C. Error bars are s.e.m.
Using the MP assay, we measured 1118 dose response curves for 43 mutated α7-5HT3 channels against twenty-three 3-aminoquinuclidine benzamides as well as ACh, nicotine, and PNU-282987 (Fig. 2A). Three classes of mutated ion channels (W77F, Q79G, and L141F) showed selective activity for their cognate ligands over unmodified α7-5HT3 receptor and EC50s < 5 μM (Fig. 2B and Table S1). Channels with the W77F mutation were activated by quinuclidinyl ligands with 4-aromatic benzamide substitutions. The Q79G mutation, which likely expands the ligand binding pocket, accommodated larger ligands such as 9S, 22S, and 38R. A third set of mutations, L141F and L141P, both showed selectivity for the 2-methoxy substituted benzamide, 19S.
To improve the selective interaction of 19S with α7-5HT3 L141F, we synthesized and tested additional alkoxybenzamide analogs (Table S2). The dimethoxy derivatives, 88S and 89S, exhibited 4- to 10-fold improvement in potency against this mutant receptor, and did not activate the unmodified α7-5HT3 receptor (Tables S1 and S2).
Several ligand and ion channel combinations were also selective relative to each other (Fig. 2C). Compound 132S, a fluorinated analog of 28S (Fig. 2D), selectively activated α7-5HT3 W77F over receptors with the Q79G and L141F mutations. Similarly, 22S and 89S were orthogonally selective for Q79G and L141F, respectively. Such selectivity is critical for developing tools that allow separate manipulation of multiple ionic conductances in the same organism. These selective LBDs were used as Pharmacologically Selective Actuator Modules (PSAMs) in conjunction with their cognate Pharmacologically Selective Effector Molecule (PSEM) agonists.
To develop PSAMs for use in the brain, we also reduced responsiveness to the endogenous ligand, ACh. We focused on shifting EC50Ipeak because steady state concentrations of ACh are reported not to rise above 1 μM (22), which is below the activation threshold for these channels. Inspection of data in Fig. 2A revealed mutations on the complementary face of the binding pocket that diminish ACh responsiveness, which were combined with selectivity-conferring mutations to generate ligand binding domains that retained ligand selectivity but were unresponsive to physiological levels of ACh. PSAMQ79G,Q139G and PSAMQ79G,L141S were responsive to 22S and 9S, respectively, but ACh potency was reduced 8- to 10-fold relative to PSAMQ79G (fig. S3 and Table S1). For PSAML141F, Tyr→Phe mutation (23) at the ligand binding site (PSAML141F,Y115F) reduced ACh responsiveness while leaving the potency of PSEM89S largely unaffected (Table S1).
We combined PSAM, IPD, and PSEM modules to build tools for neuronal perturbations such as activation, Ca2+ flux, and silencing. For a robust neuron activating cation channel, we further developed the α7-5HT3 channel by incorporating three mutations (fig. S1) reported to increase single channel conductance (24). Addition of these high conductance (HC) mutations increased membrane current noise amplitude upon PSEM application (fig. S4), consistent with higher conductance properties (25).
The chimeric channels were tested in brain slices as neuronal activation tools for layer 2/3 cortical neurons. These channels distributed to somato-dendritic compartments (Fig. 3A). The membrane properties of neurons expressing these channels were not significantly different from control neurons lacking them (fig. S5). Under conditions that block neuron and network activity (tetrodotoxin and CNQX), these channels depolarized neurons in the presence of the cognate PSEMs (Fig. 3B). PSEMs showed negligible effects on membrane potential in neurons lacking the channels (Fig. 3B).
Fig. 3.
PSAM/IPD chimeric ion channels for neuron activation and silencing. (A) Confocal projection image of α-Bgt-Alexa594 labeling in a cortical brain slice expressing PSAMQ79G,Q139G-5HT3 HC receptors. Left, Laminar boundaries. (B) Depolarization of layer 2/3 cortical neurons (in tetrodotoxin and CNQX) via PSAM-5HT3 HC channels by PSEMs applied at 10 μM. PSEM application (30 μM) to control neurons electroporated with GFP-expressing vector (green) did not depolarize cells. Sample sizes in parentheses. (C) Cell attached recordings of PSAM-5HT3 HC channel activation of layer 2/3 cortical neurons. Scale bar: 50 pA. Ligand application, 120 s. (D) Ligand selectivity: layer 2/3 cortical neurons expressing PSAML141F,Y115F-5HT3 HC are activated by PSEM89S but not PSEM22S. (E) Top, Confocal projection image of α-Bgt labeling in a cortical brain slice expressing PSAML141F,Y115F-GlyR receptors. Bottom, dendritic segment. (F,G) PSAML141F,Y115F-GlyR and PSEM89S (10 μM, black, n = 16), but not PSEM89S alone (30 μM, white, n = 10), reversibly reduce cellular input resistance, Rin. Shown in (G) is Rin normalized to pre-PSEM89S application (PRE); values are displayed for PSEM89S application and after 3 min wash-out (WASH). (H) PSAML141F,Y115F-GlyR reversibly reduces excitability of cortical neurons to depolarizing current injection in the presence of PSEM89S. (I) Rheobase, normalized to pre-PSEM89S application. Error bars are s.e.m.
We used cell-attached recordings, which do not perturb neuronal composition, to show that neurons expressing different PSAM-5HT3 HC channels sustainably increased their firing rate during PSEM application (Fig. 3C). Thus, several PSAM-5HT3 HC channels and their ligands are effective neuronal activators. The selectivity of PSEM/PSAM interactions was evident from activation of PSAML141F,Y115F-5HT3 HC by PSEM89S but not PSEM22S (Fig. 3D and fig. S6). Moreover, these PSEMs did not displace radioligand binding in a panel of 23 mammalian ion channels, G-protein coupled receptors, and transporters (Table S3). PSEM89S and PSEM22S also showed good brain penetrance in mice after minimally invasive intraperitoneal administration (fig. S7).
PSAMs can also be applied to generate pharmacologically-selective ligand-gated Ca2+ channels. We combined PSAMQ79G,L141S with the α7-nAChR IPD, a high conductance, primarily Ca2+-selective channel (26). Because α7 nAChR desensitizes rapidly, we tested channel pore mutations reported to reduce desensitization properties of these channels. We found that α7 nAChR Q79G L141S modified with the channel pore mutation V13’T (27) (here V274T, fig. S8) was non-desensitizing in response to PSEM9S, showing sustained activation and Ca2+influx (fig. S9, Movie 1). Such channels are potentially useful for inducing Ca2+-dependent processes in specific cell populations.
To construct neuron silencing tools, we generated chimeric channels using PSAMQ79G or PSAML141F fused to the glycine receptor (GlyR) chloride-selective IPD (12) (fig. S10). A similar channel was constructed using PSAML141F fused to the GABA C receptor IPD (fig. S11), illustrating the relative ease of generating new chimeric channels by combinations of PSAMs with IPDs. These channels exhibit a slow time course for activation (fig. S12) and low ACh potency (Table S1); both properties have been described previously for α7-GlyR (12). ACh-responsiveness was further diminished by modifying PSAML141F-GlyR with the Y115F mutation (Table S1).
Expression of PSAML141F,Y115F-GlyR in layer 2/3 cortical neurons showed somato-dendritic distribution (Fig. 3E) and was well tolerated (fig. S5). PSEM89S (10 μM) reversibly silenced transfected neurons by reducing cellular input resistance (Fig. 3, F and G). This shunt contributed to a nearly 7-fold reversible increase in the magnitude of injected current required to fire an action potential (rheobase) (Fig. 3, H and I), and some cells did not fire even with 500 pA injected current. In contrast, vector-transfected control neurons (n = 11) were unaffected by a higher concentration of PSEM89S (30 μM, Fig. 3, G and I). Thus, PSAML141F,Y115F-GlyR and PSEM89S constitute a powerful neuronal silencing system. Furthermore, the pharmacology of this neuron silencing system is orthogonal to the PSAMQ79G,Q139G-5HT3 HC/PSEM22S neuronal activator system (fig. S6 and S13), indicating the feasibility of using these tools in concert.
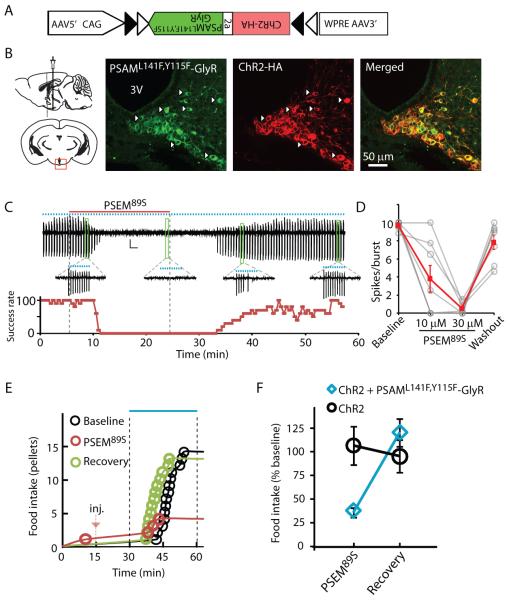
To test the effectiveness of a PSEM/PSAM-IPD in vivo, we examined the capacity of a neuronal silencer to influence behavior in mice. We used PSAML141F,Y115F-GlyR and PSEM89S in hypothalamic Agouti-related protein-expressing (AGRP) neurons to suppress voracious eating evoked by photostimulation of these neurons co-expressing the light-activated cation channel, channelrhodopsin-2 (ChR2) (8, 9). The stringency of this behavioral assay as a test for a neuronal silencer tool is due to the strong synchronous input from ChR2 activation. Moreover, because the magnitude of AGRP neuron-evoked food consumption varies continuously with the level of AGRP neuron activation, nearly full suppression of activity is required to block the evoked feeding response (28).
AGRP neurons in Agrp-cre mice (29) were transduced using a bicistronic Cre recombinase (Cre)-dependent viral vector (30) (Fig. 4A). AGRP neurons co-expressing ChR2 and PSAML141F,Y115F-GlyR (Fig. 4B) could be activated with light and were reversibly silenced by PSEM89S during photostimulation in brain slices (Fig. 4, C and D). Mice co-expressing ChR2 and PSAML141F,Y115F-GlyR or expressing ChR2 alone in AGRP neurons ate voraciously in response to photostimulation following intraperitoneal (i.p.) saline injection, and, for each mouse, this consumption was used as the baseline for subsequent treatments. After i.p. administration of PSEM89S, photostimulation-evoked feeding was strongly suppressed in mice expressing PSAML141F,Y115F-GlyR but not in mice expressing only ChR2 (Fig. 4, E and F). Twenty-four hours later, photostimulation-evoked food intake recovered to baseline levels (2-way ANOVA, 1-factor repeated measure, ±PSEM89S: F1,9 = 12.9, P = 0.006; ±PSAML141F,Y115F-GlyR: F1,9 = 1.3, P = 0.30; interaction: F1,9 = 22.4, P < 0.001; Fig. 4, E and F). Moreover after photostimulation, Fos, a marker of neuron activation (31), was almost completely suppressed in ChR2-expressing neurons from mice administered PSEM89S (fig. S14). Thus, PSAML141F,Y115F-GlyR and PSEM89S are an effective neuronal silencer system in vivo, even for strong, synchronous depolarizing currents that result from ChR2 photoactivation.
Fig. 4.
Stringent neuronal silencing test of PSAML141F,Y115F-GlyR for suppressing AGRP neuron-evoked feeding behavior. (A) Construct for a Cre-dependent recombinant adeno-associated viral vector with an inverted bicistronic open reading frame for PSAML141F,Y115F-GlyR and ChR2 under control of a FLEX-switch [two antiparallel pairs of heterotypic loxP sites (triangles)]. (B) Diagram illustrating injection site (left) and confocal images (right) taken from brain slices of virally transduced Agrp-cre mice showing co-expression of PSAML141F,Y115F-GlyR (green, α-Bgt-Alexa488) and ChR2 (red, anti-HA). (C) Cell-attached recording showing suppression of light-evoked bursts of action potential currents (10 Hz light pulses for 1 s, represented by blue dots) following application and then wash-out of PSEM89S (above). Bursts are expanded for selected time points. Below is spike success rate (percent) before, during and after PSEM89S application. (D) Successful light-evoked spikes per burst (n = 8 cells). (E) Food intake on successive days resulting from photostimulation in an Agrp-cre mouse co-expressing ChR2 and PSAML141F,Y115F-GlyR after saline (Baseline and Recovery) or PSEM89S [intraperitoneal injection (inj.), marked with red arrow]. (F) Evoked food intake normalized to initial photostimulation session. Injection of PSEM89S reduced photostimulation-evoked eating in Agrp-cre mice co-expressing ChR2 and PSAML141F,Y115F-GlyR (30 mg/kg, blue diamond, n = 5 mice) but not in Agrp-cre mice expressing only ChR2 (50 mg/kg, black circle, n = 6 mice). Error bars are s.e.m.
Here we show concerted chemical and genetic engineering of a complex ligand binding interface to develop pharmacologically-selective actuators and small molecule effectors for construction of a LGIC toolbox. PSEMs, the agonists for the resulting ion channels, act rapidly in the brain after peripheral delivery. Together, these components enable combinatorial construction (fig. S15) of cell type-selective tools to control a range of conductances, which can be used to activate or silence neurons. These ion channels could be further elaborated by applying extensive structure-function relationships in Cys-loop receptors, including mutations that modify ion selectivity (27),(32-34), intracellular interactions (35-37), and desensitization (27, 38, 39). The pharmacologically orthogonal ion channels described here can also be used with each other or with existing tools such as channelrhodopsin, facilitating multiple perturbations in the same organism to investigate functions of ion flux in cell biology, physiology, and behavior.
Supplementary Material
Acknowledgements
This work was funded by the Howard Hughes Medical Institute. C.J.M. performed the electrophysiology and the imaging; P.H.L. synthesized the molecules and performed the MP screen; D.A. performed the behavioral experiments; H.H.S. made the mutated channels and other constructs; L.L.L. made the homology model; S.M.S. developed the mutant ion channel screen, planned the experiments, analyzed data, and wrote the paper with comments from all authors. We thank S. Winfrey and H. White for cell culture support. S.M.S., P.H.L., and L.L.L. are inventors on a patent application regarding combined use of these ligand gated ion channels and small molecule agonists.
References
- 1.Slimko EM, McKinney S, Anderson DJ, Davidson N, Lester HA. Selective electrical silencing of mammalian neurons in vitro by the use of invertebrate ligand-gated chloride channels. J. Neurosci. 2002;22:7373. doi: 10.1523/JNEUROSCI.22-17-07373.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tan EM, et al. Selective and quickly reversible inactivation of mammalian neurons in vivo using the Drosophila allatostatin receptor. Neuron. 2006;51:157. doi: 10.1016/j.neuron.2006.06.018. [DOI] [PubMed] [Google Scholar]
- 3.Wulff P, et al. From synapse to behavior: rapid modulation of defined neuronal types with engineered GABAA receptors. Nat. Neurosci. 2007;10:923. doi: 10.1038/nn1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armbruster BN, Li X, Pausch MH, Herlitze S, Roth BL. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc. Natl. Acad. Sci. U. S. A. 2007;104:5163. doi: 10.1073/pnas.0700293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gosgnach S, et al. V1 spinal neurons regulate the speed of vertebrate locomotor outputs. Nature. 2006;440:215. doi: 10.1038/nature04545. [DOI] [PubMed] [Google Scholar]
- 6.Lerchner W, et al. Reversible silencing of neuronal excitability in behaving mice by a genetically targeted, ivermectin-gated Cl-channel. Neuron. 2007;54:35. doi: 10.1016/j.neuron.2007.02.030. [DOI] [PubMed] [Google Scholar]
- 7.Ferguson SM, et al. Transient neuronal inhibition reveals opposing roles of indirect and direct pathways in sensitization. Nat. Neurosci. 2011;14:22. doi: 10.1038/nn.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 2005;8:1263. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 9.Li X, et al. Fast noninvasive activation and inhibition of neural and network activity by vertebrate rhodopsin and green algae channelrhodopsin. Proc. Natl. Acad. Sci. U. S. A. 2005;102:17816. doi: 10.1073/pnas.0509030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang F, et al. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446:633. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- 11.Eisele JL, et al. Chimaeric nicotinic-serotonergic receptor combines distinct ligand binding and channel specificities. Nature. 1993;366:479. doi: 10.1038/366479a0. [DOI] [PubMed] [Google Scholar]
- 12.Grutter T, et al. Molecular tuning of fast gating in pentameric ligand-gated ion channels. Proc. Natl. Acad. Sci. U. S. A. 2005;102:18207. doi: 10.1073/pnas.0509024102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lester HA, Dibas MI, Dahan DS, Leite JF, Dougherty DA. Cys-loop receptors: new twists and turns. Trends Neurosci. 2004;27:329. doi: 10.1016/j.tins.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 14.Dent JA. Evidence for a diverse Cys-loop ligand-gated ion channel superfamily in early bilateria. J. Mol. Evol. 2006;62:523. doi: 10.1007/s00239-005-0018-2. [DOI] [PubMed] [Google Scholar]
- 15.Tasneem A, Iyer LM, Jakobsson E, Aravind L. Identification of the prokaryotic ligand-gated ion channels and their implications for the mechanisms and origins of animal Cys-loop ion channels. Genome Biol. 2005;6:R4. doi: 10.1186/gb-2004-6-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hwang YW, Miller DL. A mutation that alters the nucleotide specificity of elongation factor Tu, a GTP regulatory protein. J. Biol. Chem. 1987;262:13081. [PubMed] [Google Scholar]
- 17.Bishop AC, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- 18.Lin Q, Jiang F, Schultz PG, Gray NS. Design of allele-specific protein methyltransferase inhibitors. J. Am. Chem. Soc. 2001;123:11608. doi: 10.1021/ja011423j. [DOI] [PubMed] [Google Scholar]
- 19.Celie PH, et al. Nicotine and carbamylcholine binding to nicotinic acetylcholine receptors as studied in AChBP crystal structures. Neuron. 2004;41:907. doi: 10.1016/s0896-6273(04)00115-1. [DOI] [PubMed] [Google Scholar]
- 20.Walker DP, et al. Design, synthesis, structure-activity relationship, and in vivo activity of azabicyclic aryl amides as alpha7 nicotinic acetylcholine receptor agonists. Bioorg. Med. Chem. 2006;14:8219. doi: 10.1016/j.bmc.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 21.Bodnar AL, et al. Discovery and structure-activity relationship of quinuclidine benzamides as agonists of alpha7 nicotinic acetylcholine receptors. J. Med. Chem. 2005;48:905. doi: 10.1021/jm049363q. [DOI] [PubMed] [Google Scholar]
- 22.Vinson PN, Justice JB., Jr.M Effect of neostigmine on concentration and extraction fraction of acetylcholine using quantitative microdialysis. J. Neurosci. Methods. 1997;73:61. doi: 10.1016/s0165-0270(96)02213-3. [DOI] [PubMed] [Google Scholar]
- 23.Galzi JL, et al. Functional significance of aromatic amino acids from three peptide loops of the alpha 7 neuronal nicotinic receptor site investigated by site-directed mutagenesis. FEBS Lett. 1991;294:198. doi: 10.1016/0014-5793(91)80668-s. [DOI] [PubMed] [Google Scholar]
- 24.Kelley SP, Dunlop JI, Kirkness EF, Lambert JJ, Peters JA. A cytoplasmic region determines single-channel conductance in 5-HT3 receptors. Nature. 2003;424:321. doi: 10.1038/nature01788. [DOI] [PubMed] [Google Scholar]
- 25.Rayes D, Spitzmaul G, Sine SM, Bouzat C. Single-channel kinetic analysis of chimeric alpha7-5HT3A receptors. Mol. Pharmacol. 2005;68:1475. doi: 10.1124/mol.105.015438. [DOI] [PubMed] [Google Scholar]
- 26.Seguela P, Wadiche J, Dineley-Miller K, Dani JA, Patrick JW. Molecular cloning, functional properties, and distribution of rat brain alpha 7: a nicotinic cation channel highly permeable to calcium. J. Neurosci. 1993;13:596. doi: 10.1523/JNEUROSCI.13-02-00596.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galzi JL, et al. Mutations in the channel domain of a neuronal nicotinic receptor convert ion selectivity from cationic to anionic. Nature. 1992;359:500. doi: 10.1038/359500a0. [DOI] [PubMed] [Google Scholar]
- 28.Aponte Y, Atasoy D, Sternson SM. AGRP neurons are sufficient to orchestrate feeding behavior rapidly and without training. Nat. Neurosci. 2011;14:351. doi: 10.1038/nn.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaelin CB, Xu AW, Lu XY, Barsh GS. Transcriptional regulation of agouti-related protein (Agrp) in transgenic mice. Endocrinology. 2004;145:5798. doi: 10.1210/en.2004-0956. [DOI] [PubMed] [Google Scholar]
- 30.Atasoy D, Aponte Y, Su HH, Sternson SM. A FLEX switch targets Channelrhodopsin-2 to multiple cell types for imaging and long-range circuit mapping. J. Neurosci. 2008;28:7025. doi: 10.1523/JNEUROSCI.1954-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morgan JI, Curran T. Stimulus-transcription coupling in the nervous system: involvement of the inducible proto-oncogenes fos and jun. Annu. Rev. Neurosci. 1991;14:421. doi: 10.1146/annurev.ne.14.030191.002225. [DOI] [PubMed] [Google Scholar]
- 32.Bertrand D, Galzi JL, Devillers-Thiery A, Bertrand S, Changeux JP. Mutations at two distinct sites within the channel domain M2 alter calcium permeability of neuronal alpha 7 nicotinic receptor. Proc. Natl. Acad. Sci. U. S. A. 1993;90:6971. doi: 10.1073/pnas.90.15.6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keramidas A, Moorhouse AJ, French CR, Schofield PR, Barry PH. M2 pore mutations convert the glycine receptor channel from being anion- to cation-selective. Biophys. J. 2000;79:247. doi: 10.1016/S0006-3495(00)76287-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gunthorpe MJ, Lummis SC. Conversion of the ion selectivity of the 5-HT(3a) receptor from cationic to anionic reveals a conserved feature of the ligand-gated ion channel superfamily. J. Biol. Chem. 2001;276:10977. [PubMed] [Google Scholar]
- 35.Temburni MK, Blitzblau RC, Jacob MH. Receptor targeting and heterogeneity at interneuronal nicotinic cholinergic synapses in vivo. J Physiol. 2000;525(Pt 1):21. doi: 10.1111/j.1469-7793.2000.00021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu J, Zhu Y, Heinemann SF. Identification of sequence motifs that target neuronal nicotinic receptors to dendrites and axons. J. Neurosci. 2006;26:9780. doi: 10.1523/JNEUROSCI.0840-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jansen M, Bali M, Akabas MH. Modular design of Cys-loop ligand-gated ion channels: functional 5-HT3 and GABA rho1 receptors lacking the large cytoplasmic M3M4 loop. J. Gen. Physiol. 2008;131:137. doi: 10.1085/jgp.200709896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Revah F, et al. Mutations in the channel domain alter desensitization of a neuronal nicotinic receptor. Nature. 1991;353:846. doi: 10.1038/353846a0. [DOI] [PubMed] [Google Scholar]
- 39.Breitinger HG, Villmann C, Becker K, Becker CM. Opposing effects of molecular volume and charge at the hyperekplexia site alpha 1(P250) govern glycine receptor activation and desensitization. J. Biol. Chem. 2001;276:29657. doi: 10.1074/jbc.M100446200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.