Abstract
Filament-forming cytoskeletal proteins are key organizers of all cells. Bacterial homologs of the major eukaryotic cytoskeletal families have now been discovered, but studies suggest that yet more cytoskeletal proteins remain to be identified. Here we demonstrate that the metabolic enzyme CTP Synthase (CtpS) forms filaments in Caulobacter crescentus. These filaments are bifunctional and regulate Caulobacter curvature independently of CtpS catalytic activity. The morphogenic role of CtpS requires its functional interaction with the intermediate filament crescentin. Interestingly, the E. coli CtpS homolog also forms filaments both in vivo and in vitro, suggesting that CtpS polymerization may be widely conserved. E. coli CtpS can replace the enzymatic and morphogenic functions of Caulobacter CtpS, indicating that Caulobacter has adapted a conserved filament-forming protein for a secondary role. These results implicate CtpS as a novel bifunctional member of the bacterial cytoskeleton and suggest that localization and polymerization may be important properties of metabolic enzymes.
Cellular organization is fundamental to the viability of all cells. In eukaryotes, organization is largely achieved through a family of cytoskeletal proteins. These tubulin, actin, and intermediate filament cytoskeletal proteins assemble into linear polymers (filaments) that play important roles in a wide range of biological processes including cell shape, motility, transport, and division. While prokaryotes were classically believed to lack cytoskeletal proteins, bacterial homologs of all three canonical eukaryotic cytoskeletal families have now been identified. These bacterial cytoskeletons include the tubulin homolog FtsZ1,2, the actin homolog MreB3, and the intermediate filament-like protein crescentin4. The bacterial cytoskeleton, however, appears to be even more complex as electron cryotomographic studies of Caulobacter crescentus have shown that there are additional filament structures within cells whose molecular identities have yet to be determined5,6.
Caulobacter represents a powerful system for studying the bacterial cytoskeleton due to its unique curved morphology and asymmetric division cycle that produces one stalked cell and one swarmer cell from each predivisional progenitor. For example, crescentin was first identified in Caulobacter4. By light microscopy, crescentin appears to form filaments that extend along the inner curvature of the cell and are necessary for cell curvature4. In addition to their unique asymmetric morphology, Caulobacter cells are also thin enough to allow whole-cell electron cryotomography, enabling the direct visualization of cytoskeletal structures within intact cells5.
Here we demonstrate that the metabolic enzyme CTP synthase (CtpS) is a novel filament-forming protein in Caulobacter that is dynamically localized to the inner cell curvature during the cell cycle. Studies with the E. coli CtpS confirm that CtpS can polymerize independently of other factors and that filament formation is a conserved property of this enzyme. In Caulobacter, CtpS filaments interact with crescentin filaments and regulate cell curvature independently of enzymatic activity. These results address outstanding questions concerning the composition, function, and evolution of the bacterial cytoskeleton.
Results
mCherry-CtpS forms linear structures that co-localize with cellular filaments in Caulobacter
A recent electron cryotomography6 (ECT) study discovered a bundle of filamentous polymers along the inner curvature of Caulobacter5. These inner curvature filaments (hereafter referred to as ECT filaments) formed independently of MreB and crescentin and were also distinct from FtsZ5,7. In an effort to identify this elusive cytoskeletal protein, we examined a recently generated collection of Caulobacter proteins with non-diffuse subcellular localizations8, and found one protein with a similar distribution. Surprisingly, this protein encoded the sole Caulobacter homolog of the previously-characterized enzyme, CTP synthase (CtpS, CC1720), an enzyme that generates CTP from UTP, ATP, and glutamine9. To further examine CtpS localization in Caulobacter, we imaged an N-terminal mCherry-ctpS fusion (ZG153) and found that most cells in an asynchronous population exhibited linear fluorescent structures along the inner curvature of the cell (Figure 1a).
Figure 1. mCherry-CtpS is dynamic and co-localizes with linear filamentous structures along the inner curvature of Caulobacter cells.
(a) mCherry-CtpS localization in asynchronous Caulobacter cells. mCherry-ctpS expressing cells (ZG153) were induced for two hours with xylose and imaged. A merged phase and fluorescence image is shown. Scale bar represents 2 μm. (b) mCherry-CtpS localization in synchronized Caulobacter cells. mCherry-ctpS expressing cells (ZG153) were induced for two hours with xylose, synchronized, and imaged at 10-minute intervals in the presence of xylose. Merged phase and fluorescence (top), fluorescence (middle), and cartoon depictions (bottom) are shown for two fields of representative cells at 0, 50, 80, and 110 minutes from the beginning of the timelapse. Scale bar represents 2 μm. (c) Co-localization of mCherry-CtpS and filamentous structures in Caulobacter. mCherry-ctpS expressing cells (ZG153) were fixed on EM grids and imaged first by fluorescence light microscopy and then by ECT. Shown is an ECT slice, phase-fluorescence overlay (left inset), and cartoon depiction (right inset) of the same cells. These cells correspond to cells #15 from the field shown in Figure S2. Arrows point to the ends of the filaments in the ECT image, which correspond to the positions of the mCherry-CtpS structure shown in the inset. Scale bars represent 100 nm for EM and 1 μm for the inset LM images.
Time-lapse imaging of synchronized cultures expressing mCherry-CtpS revealed a dynamic, cell-cycle regulated localization pattern. Figure 1b shows representative images taken from a timelapse in which images were collected every 10 minutes throughout the cell cycle. Early in the Caulobacter cell cycle, in newly formed stalked cells, the mCherry-CtpS structures were generally short, sometimes even appearing as foci. As the stalked cell developed, the structures elongated to form a line roughly 500 nm long that initially did not associate with the cell periphery. At later times, the structure moved toward the inner cell curvature, where it remained throughout the rest of the cell cycle. The mCherry-CtpS structures were preferentially localized in the stalked compartment of late predivisional cells, such that little mCherry signal could generally be detected in the newly formed swarmer cell (Figure 1b). Despite these changes in localization, CtpS abundance did not dramatically change during the cell cycle (Figure S1).
In order to examine if the ECT filaments and mCherry-CtpS co-localize to the same region of individual cells, we used correlated fluorescence light microscopy-ECT10 (fLM-ECT). Inner curvature filaments were observed in 5 tomograms of cells that could be aligned with their corresponding fluorescence images. Strikingly, all 5 of these cells also displayed fluorescent mCherry-CtpS structures in the same subcellular position (Figure 1c, Figure S2).
CtpS abundance and activity affect the cellular filaments
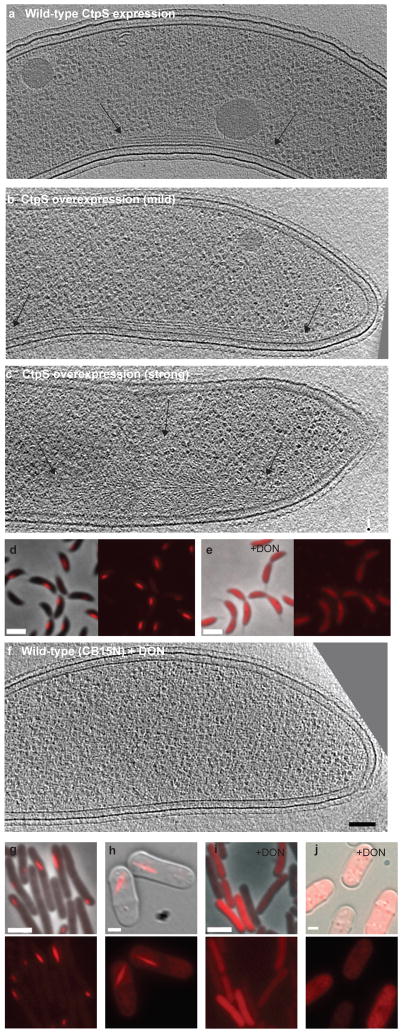
The mCherry-CtpS fusion co-localized with the ECT filaments, but could not fully substitute for the essential enzymatic activity of native CtpS (data not shown). Similarly, most other fusions to cytoskeletal proteins are not functional but do represent accurate reporters of protein localization4,11,12. We thus sought further support for the conclusion that the ECT filaments were composed of the native CtpS protein by altering the levels of untagged CtpS to assess the corresponding effect on the ECT filaments. At endogenous expression levels ECT filaments are found along the inner cell curvature in 23 ± 6% of wild-type Caulobacter cells5 (CB15N, 6% = standard error of the proportion, n=50) and averaged 390 nm in length (Figure 2a). In contrast, mild ctpS overexpression from a single-copy chromosomal locus (ZG215, ~1.5 fold overexpression, Figure S3) resulted in ECT filaments that were longer (790 nm on average) and more prevalent (found in 63 ± 17% of cells, n=8, Figure 2b). Similar levels of mCherry-CtpS overexpression resulted in similarly elongated mCherry-CtpS structures (Figure S4). Strong ctpS overexpression from a multi-copy plasmid (ZG208, ~4 fold overexpression, Figure S3) also increased the prevalence of ECT filaments to 70 ± 10%, but furthermore dramatically altered filament morphology, leading to structures that in most cases (76 ± 11%, n=20) resembled large collections of splayed filaments radiating from the inner curvature of the cell (Figure 2c).
Figure 2. CtpS forms filaments in Caulobacter.
(a–c) Tomographic slices through wild-type Caulobacter with endogenous ctpS levels (a, CB15N) and strains with mild (b, ZG215) and strong (c, ZG208) ctpS overexpression. Arrows point to the filament ends. (d–e) Merged phase and fluorescence (left) and fluorescence (right) images of cells expressing mCherry-ctpS (ZG153) in the absence (d) or presence (e) of 1 μM DON. (f) Tomographic slice of CB15N grown in 1 μM DON. (g–h) Heterologous expression of Caulobacter mCherry-ctpS in E. coli (g, ZG219) and S. pombe (h, ZG222). The mCherry-CtpS structures are delocalized by DON in both E.coli (i) and S. pombe (j). Phase-fluorescent merge (top) and fluorescent images (bottom) are shown in each case. Black scale bars (ECT slices) represent 100 nm and white scale bars (LM images) represent 2 μm.
We were unable to delete the Caulobacter ctpS gene, indicating that CtpS is essential. We were able to generate a depletion strain (further characterized below), but this strain grew very poorly, making it difficult to image by ECT. We thus sought a rapid method to acutely perturb CtpS, focusing on an irreversible small molecule CtpS inhibitor, 6-diazo-5-oxo-L-norleucine (DON)9,13. Treating mCherry-CtpS cells (ZG153) with 1 μM or higher concentrations of DON led to a rapid and complete disruption of the mCherry-CtpS fluorescent structures (n > 100) (Figure 2d, e). Similarly, after treatment with DON, the ECT filaments were only observed in 1 of 40 wild-type cells (CB15N, 2.5 ± 2% n=40, Figure 2f, as compared to 23 ± 6% of untreated cells examined), confirming that DON treatment indeed disrupts both mCherry-CtpS structures and the ECT filaments (Figure 2e–f). Thus, the inner curvature ECT filaments both co-localize with and depend upon CtpS. These data suggest that in Caulobacter crescentus, CtpS forms the inner curvature bundle, polymerizing into stacks of long filaments along the inner membrane.
Caulobacter CtpS can self-associate and form filaments upon heterologous expression
To determine if CtpS can itself form polymers, we first attempted to purify Caulobacter CtpS to study its polymerization in vitro, but the protein proved insoluble, preventing further analysis. We were able to demonstrate that CtpS monomers can self-associate in an Escherichia coli BACTH bacterial two-hybrid assay14 (Figure S5a). Though the ability to self-associate is an expected property of polymers, the BACTH assay does not distinguish dimerization from filament formation. We thus examined whether CtpS can form filaments in the absence of other Caulobacter proteins by expressing it in heterologous systems, as described previously for other bacterial cytoskeletons15,16. We expressed Caulobacter mCherry-ctpS in E. coli and in the eukaryote, Schizosaccharomyces pombe (Figure 2g–h). 48 ± 2% of the E. coli cells (n=328, ZG219) and 29 ± % of the S. pombe cells (n=135, ZG222) expressing mCherry-ctpS contained linear fluorescent structures. These mCherry-CtpS filamentous structures did not resemble any known E. coli or S. pombe structures or localization patterns, indicating that mCherry-CtpS is not assembling on a known scaffold. Furthermore, the mCherry-CtpS structures appeared to retain at least some of their native properties since they were still delocalized by DON treatment in all cases (Figures 2e, i–j). Thus, CtpS appears to be capable of forming filamentous structures in the absence of any bacterial-specific cofactors.
CtpS regulates Caulobacter cell shape independently of its enzymatic activity
Since CtpS localizes to the inner cell curvature, we examined whether CtpS might also regulate Caulobacter shape. Whereas wild-type Caulobacter cells have a characteristic curved morphology, cells overexpressing CtpS (ZG208) were significantly straighter. In this population of cells overexpressing CtpS, some cells were completely straight while other cells appeared hooked with reduced curvature at one end (Figure 3a). Though CtpS is essential, we were able to reduce CtpS levels by replacing the native CtpS promoter with a leaky xylose-inducible promoter and growing the resulting strain (ZG215) in the absence of xylose. These cells mildly depleted of CtpS grew very slowly but displayed a sharp kink near the middle of the cell consistent with this region of the cell “over-curving” (Figure 3a). These results implicate CtpS as a negative regulator of curvature.
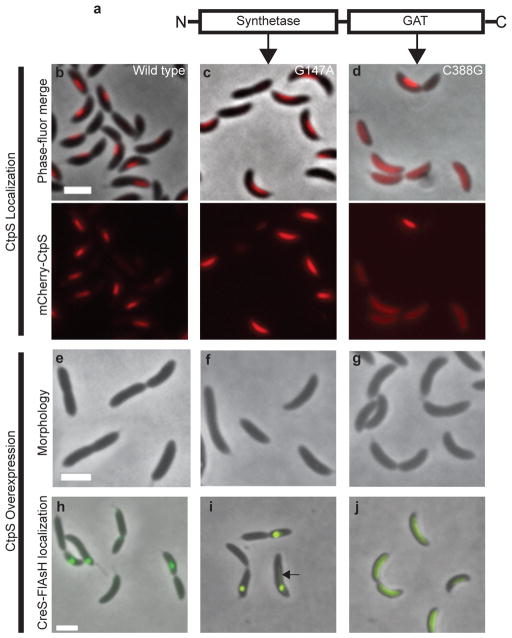
Figure 3. CtpS regulates Caulobacter cell shape independently of enzymatic activity.
(a) Morphological effect of overexpression and depletion of CtpS. Shown are phase images of wild type Caulobacter (left), and Caulobacter overexpressing (middle) or depleted (right) for CtpS. Above panels b–j is a cartoon representation of the synthetase and GAT domains of Caulobacter CtpS (not to scale). (b–d) Localization of wild-type and mCherry-CtpS point mutants. Merged phase-fluorescence (top) and fluorescence (bottom) images of cells expressing wild-type mCherry-CtpS (b, ZG153), synthetase domain mutant (c, ZG154), and GAT domain mutant (d, ZG155). (e–j) Effects of overexpression of wild-type CtpS (e, ZG208) and (h, ZG212), the synthetase mutant (f, ZG209) and (i, ZG213), and the GAT mutant (g, ZG210) and (j, ZG214) on cell shape (e–g) and CreS-tc localization (h–j). The arrow in (i) points to faint residual CreS-tc localization along the side of the cell. Scale bars are 2 μm.
We next examined whether CtpS filament formation and cell shape regulation are coupled to its enzymatic activity. The CtpS enzyme is comprised of two domains, a synthetase domain and a glutamine amidotransferase (GAT) domain17,18 (Figure 3). Based on homologous mutations that had been previously characterized, we constructed catalytic residue point mutations that are predicted to inactivate the catalytic sites of the synthetase (G147A) and GAT (C388G) domains19,20. Compared to wild-type mCherry-ctpS, the G147A synthetase domain mutation produced no detectable change in the frequency or morphology of filamentous structures (ZG154, Figure 3b–c). In contrast, the C388G GAT domain mutation dramatically perturbed the ability of mCherry-CtpS to form linear structures (in ZG155, only 13 ± 0.3% of cells retained filamentous localization in contrast to 79 ± 3% of wild-type cells, Figure 3d). None of the point mutations in CtpS affected protein stability, as mCherry-CtpS protein levels were similar in all strains (Figure S6). The GAT mutant’s impaired assembly is consistent with the fact that the GAT domain is also the binding target for the CtpS delocalizing agent, DON.
Whereas the C388G mutation implicates the GAT domain in CtpS assembly, the G147A synthetase domain mutant that still forms filaments provided us with an opportunity to functionally separate the filament-forming properties of CtpS from its enzymatic activity. We thus tested if these mutants retained the ability to regulate cell shape upon overexpression. Overexpression of the filament-forming G147A synthetase mutant resulted in straightened cells that were largely indistinguishable from those overexpressing wild-type CtpS (ZG208 and ZG209, Figure 3e–f). By contrast, overexpression of the C388G GAT mutant that can neither function as a CTP synthase nor form filaments had no detectable effect on cell shape (ZG210, Figure 3g). Consequently, the effects of CtpS on Caulobacter cell shape appear to require GAT-domain-dependent filamentation, but not enzymatic activity.
CtpS functionally interacts with crescentin to regulate Caulobacter curvature
Since CtpS regulates Caulobacter curvature, we examined whether it might function through the other known curvature regulator, the intermediate filament-like protein crescentin4,21. We found that ctpS overexpression dramatically disrupted the localization of a fully-functional CreS fusion to a tetra-cysteine FlAsH-binding epitope (CreS-tc). Rather than localizing CreS to an inner curvature structure that stretched from pole to pole, these cells primarily localized CreS to a single focus (ZG212, Figure 3h). The straightest cells retained no detectable CreS along the inner curvature, while the less affected cells retained faint inner curvature CreS, providing a potential explanation for this strain’s heterogeneous morphology (Figure 3e, h). Consistent with the effects on morphology, the mislocalization of crescentin depended upon the CtpS GAT domain but not the synthetase domain (Figure 3i–j). The cell straightening and CreS mislocalization observed upon CtpS overexpression closely resemble the phenotypes induced by a dominant negative creS mutant that inhibits CreS assembly22. Neither CtpS overexpression nor depletion was associated with changes in crescentin abundance (Figure S3a). We also determined whether the presence of crescentin is required for the hyper-curved cell shape of the CtpS depletion strain. Indeed, ΔcreS strains depleted for ctpS remained straight and did not kink (ZG216, Figure 4b, d). These results support the hypothesis that CtpS acts through crescentin to regulate cell curvature.
Figure 4. CtpS regulates cell shape through an interaction with crescentin.
(a–d) Cell morphology of Caulobacter cells with wild-type (a and b) and depleted (c and d) CtpS levels in a background either with creS (a and c, creS+, ZG215) or without creS (b and d, ΔcreS, ZG216). (e–f) Localization of Caulobacter mCherry-CtpS (red) in creS+ (e, ZG287) and ΔcreS (f, ZG285) backgrounds. Merged fluorescent images are shown, and the cell periphery is marked with a gfp fusion (green) to the periplasmic protein encoded by CC2395. (g) ECT filaments in ΔcreS cells (ZG17). Arrows point to filaments ends. (h–i) Co-localization of Caulobacter mCherry-CtpS (red) and CreS-tc (green) in both Caulobacter (h, ZG218) and upon heterologous co-expression in E.coli (i, ZG221). White scale bars represent 2 μm, black scale bars represent 100 nm.
Since CtpS can affect crescentin localization, we also examined whether crescentin can affect CtpS localization. In ΔcreS cells (ZG285), we found that mCherry-CtpS still formed linear structures, but these structures no longer associated with the cell periphery in most cells. Only 19 ± 2% of cells retained membrane-associated localization (Figure 4e, f). Similarly, in most of the ΔcreS cells examined by ECT (5 of 6 cases), filament bundles resembling the inner curvature bundle were seen extending through the cytoplasm away from the membrane (Figure 4g). In the previous ECT analysis of Caulobacter cytoskeletal filaments, an inner curvature bundle was found in one ΔcreS cell, proving that this particular filament bundle type is not crescentin5. While in the absence of specific ECT labels, we cannot confirm that these ECT filaments are CtpS, taken together, the ECT and mCherry-CtpS data show that CtpS forms filaments in the absence of crescentin but requires crescentin for proper localization along the inner curvature. This membrane tethering of CtpS filaments by crescentin could explain the CtpS cell-cycle localization dynamics wherein CtpS first localizes in the cytoplasm and then moves to the inner curvature (Figure 1b).
Consistent with a potential interaction between the two proteins, mCherry-CtpS and CreS-tc co-localize to the inner curvature of Caulobacter cells (ZG218), though the crescentin structures extend beyond those of CtpS (Figure 4h). To determine if the colocalization of CtpS and CreS is mediated by other Caulobacter-specific proteins, we expressed both mCherry-CtpS and CreS-tc in E.coli, where each of these proteins can form filaments on its own22 (ZG221, Figure 2g). When co-expressed in E. coli, mCherry-CtpS and CreS-tc always co-localized (Figure 4i), and as in Caulobacter, the crescentin structures extended beyond those of CtpS (Figure 4h, i). While not all cells co-expressed both proteins, they colocalized in each case in which both proteins could be detected. The fact that crescentin localization extends beyond that of CtpS in both Caulobacter and E. coli suggests that this difference may reflect intrinsic properties of these filaments. We also found that CtpS and crescentin can interact with one another in an E. coli BACTH assay (supplemental information, Figure S5b–c). These results support the conclusion that crescentin and CtpS functionally interact, though it is unclear whether the interaction is direct.
E. coli CtpS forms filaments both in vivo and in vitro
CtpS forms filaments and regulates cell shape in Caulobacter, but since CtpS is a universally conserved enzyme, CtpS could also form filaments in other organisms. We thus examined the localization of an mCherry fusion to the E. coli homolog of CtpS (also known as PyrG, but hereafter referred to as EcCtpS). In E. coli, the mCherry-EcCtpS fusion (ZG283) formed filamentous structures that closely resembled those of Caulobacter mCherry-CtpS (Figure 5a). To assay the localization of the endogenous EcCtpS protein, we purified and raised an antibody to EcCtpS and performed immunofluorescence microscopy (IF). We found that in wild-type E. coli (NCM3722), CtpS filaments could be detected by IF in 72 ± 5% of cells (n=365), indicating that the native CtpS protein can form linear structures in vivo (Figure 5b). Though the E. coli CtpS antibody could weakly detect Caulobacter CtpS on Western blots, IF with Caulobacter cells proved unsuccessful.
Figure 5. The E. coli CtpS homolog forms filaments both in vivo and in vitro.
(a) Localization of an mCherry fusion to the E. coli CtpS homolog (referred to here as EcCtpS but also known as pyrG) in E. coli. mCherry-EcCtpS expressing cells (ZG283) were induced for two hours with IPTG and imaged. Merged phase fluorescent (left) and fluorescent (right) images are shown. (b) Localization of the endogenous, untagged E.coli CtpS. Immunofluorescence microscopy was performed on wild type E. coli cells (NCM3722) probed with an α CtpS antibody. Merged phase and fluorescent (left) and fluorescent (right) images are shown. (c) Electron microscopy image of purified CtpS filaments observed in vitro after 10 minutes of incubation in CtpS activity buffer. (d) Localization of E.coli mCherry-EcCtpS in Caulobacter. A Caulobacter strain where Caulobacter CtpS was replaced with EcCtpS at the endogenous ctpS locus and mCherry-EcCtpS was expressed under the xylose promoter (ZG286) were induced with xylose and imaged. Shown are the phase fluorescent merged (left) and fluorescent (right) images. Black scale bars represent 2 μm; white scale bars represent 100 nm.
Having purified CtpS protein from E. coli, we were able to directly test whether CtpS, like other cytoskeletal proteins, can polymerize on its own in vitro. Indeed, examining the purified CtpS protein by electron microscopy (EM) revealed clear linear filaments (Figure 5c). The filaments were approximately 200–400 nm in length and appeared in stacks of roughly 3–5 filaments with a gap of approximately 8–9 nm between adjacent filaments (Figure 5c). These characteristics of EcCtpS observed in vitro were comparable to those of Caulobacter CtpS observed by ECT (~390 nm long in stacks of ~3, spaced ~10 nm apart)5. As a second assay for polymerization, we found that much of the CtpS protein sediments upon ultracentrifugation (Figure S7). The activity buffer used for the CtpS in vitro assays includes 10 mM MgCl2. Since high levels of Mg2+ can affect protein interactions, we confirmed that CtpS still polymerizes in the presence of 2 mM MgCl2, as assayed by both EM and sedimentation (Figure S7).
Since Caulobacter and E. coli CtpS appear to form similar filaments, we tested whether E. coli CtpS can substitute for Caulobacter CtpS. We found that Caulobacter cells in which the Caulobacter ctpS gene was replaced with the E. coli ctpS homolog (ZG286), were not only viable but had wild-type morphology. Furthermore, E. coli mCherry-EcCtpS localized to the inner curvature of these Caulobacter (Figure 5d). Thus, the abilities of Caulobacter CtpS to both form filaments and regulate cell curvature are conserved with E. coli CtpS.
Discussion
Here we demonstrate that CtpS forms filaments both in vivo and in vitro, and that filamentation is conserved in Caulobacter and E. coli. Furthermore, Caulobacter CtpS both forms linear filaments and regulates cell shape, thereby defining CtpS as a novel member of the bacterial cytoskeleton. CtpS forms filaments and regulates cell shape independently of enzymatic activity, indicating that Caulobacter CtpS is bifunctional with separable metabolic and cytoskeletal roles. While CtpS is the first reported example of a polymer-forming enzyme that regulates cell shape, other enzymes have been shown to either polymerize or structurally resemble known cytoskeletons23,24. Thus, CtpS may reflect a widespread cytoskeletal parallel to other bifunctional proteins like lens crystallins25, suggesting that protein polymerization potentially serves multiple functions in different cellular contexts.
Caulobacter CtpS filaments regulate cell curvature, but our work indicates that CtpS also forms filaments in E. coli, which are not curved. So what function might CtpS polymerization serve in E. coli? The previously characterized enzymatic activity of CtpS is to generate CTP from UTP, ATP, and glutamine. Polymerization could regulate enzymatic activity, perhaps coordinating the regulation of many CtpS subunits to cooperatively transition between active and inactive states. Such regulation of CtpS polymerization could explain why CtpS polymers are more prevalent in replicatively active stalked cells than in quiescent swarmer cells, despite the similar abundance of CtpS in these two cell types. Dynamic regulation of CtpS assembly could also explain why CtpS structures are more prevalent when observed by fluorescence microscopy than by ECT. Additionally, CtpS localization could restrict its enzymatic activity to specific subcellular regions, thereby enabling localized regulation of its substrate or product levels. CTP is required for DNA, RNA, and phospholipid synthesis so CtpS localization could preferentially enrich these products in the replicatively active stalked cell. Thus, protein localization may be a general and largely underappreciated mode of metabolic regulation.
CtpS limits the extent of Caulobacter cell curvature by interacting with another cytoskeletal element, crescentin. Specifically, crescentin tethers CtpS to the inner curvature membrane where CtpS in turn mitigates crescentin-mediated curvature. The details of how CtpS regulates crescentin assembly or localization remain unknown, but this activity is independent of CtpS enzymatic activity. In one model for how crescentin regulates curvature22, crescentin forms an elastic filament that acts like a spring. This spring locally compresses peptidoglycan, such that the cell grows with a circumferential gradient of peptidoglycan insertion. CtpS could decrease the stiffness of this crescentin spring, perhaps by loosening connections or destabilizing certain conformations. In this scenario, depleting CtpS hypercurves cells because the spring is unabated, while CtpS over-expression relaxes or disengages the spring. The fact that curvature formation depends on cell growth may also explain why the transient absence of CtpS filaments in many swarmer cells does not significant affect the morphology of these slow-growing cells.
The ability of E. coli CtpS to replace both the enzymatic and morphogenic functions of Caulobacter CtpS suggest that Caulobacter CtpS has not been specifically adapted for its role in regulating curvature. Thus, Caulobacter appears to have co-opted a conserved filament for a secondary structural role and adapted its other cell shape regulators to interact with CtpS. It is possible that many proteins polymerize, often for non-structural, perhaps regulatory functions. Once these filaments are present, they can be co-opted for additional structural purposes. Gene duplication and diversification would then allow some proteins to retain only their cytoskeletal characteristics. This hypothesis could explain the similarity shared by eukaryotic actin and bacterial MreB with several enzymes such as hexokinase and Hsp70 family chaperones26. Thus, our findings suggest a general pathway for the evolution of structural cytoskeletal functions from polymer-forming enzymes.
Methods
Bacterial strains and growth conditions
All Caulobacter crescentus, Escherichia coli, and Schizosaccharomyces pombe strains were derived from CB15N27, DH5α, NCM3722, and ySP228 respectively, and were grown at 30°C (Caulobacter and S. pombe) or 37°C (E. coli) in PYE, LB or EMM media supplemented with the appropriate antibiotics or amino acids. Full genotypes of all strains are detailed in Supplemental table 1. Plasmids (supplemental table 2) were introduced into E.coli and Caulobacter by electroporation as described previously29,30.
For imaging, all S. pombe strains were grown from single colonies in EMM media with the appropriate amino acids and thiamine when necessary. Caulobacter strains depleted for or overexpressing CtpS were grown overnight in PYE media containing the appropriate antibiotics and in the presence of glucose, xylose, or neither sugar. All other Caulobacter and E. coli strains were grown to stationary phase in PYE or LB, subcultured in media and grown for two hours, and induced with 0.03% xylose, 1mM vanillate, 0.01mM IPTG, or 0.2% arabinose for two hours31. Cells expressing CreS-tc fusions were incubated with 2 μM FlAsH reagent one hour after induction and were washed twice in media containing 1x BAL WASH buffer (Invitrogen) before being resuspended in media without WASH buffer prior to imaging.
Light microscopy
All cells were observed in exponential growth phase. For still images, cells were immobilized on pads of 1% agarose dissolved in water. Pads were supplemented with 1–2 μM DON when appropriate. For time-lapse images, cells were synchronized27 and immobilized on pads of 1% agarose dissolved in PYE and supplemented by 0.03% xylose. Pads were sealed by a 1:1:1 mixture of paraffin, Vaseline, and lanolin after coverslip placement. Images were captured with a Nikon90i epifluorescent microscope equipped with a 100 × 1.4 NA objective (Nikon), Rolera XR cooled CCD camera (QImaging), and NIS Elements software (Nikon).
Electron cryotomography (ECT)
For ECT, Caulobacter cells were grown to exponential phase in PYE medium. 2 mL of cell culture were centrifuged for 5 minutes at 1500 × g and resuspended in ~50 μl supernatant. A solution of 10 nm colloidal gold was treated with BSA to prevent aggregation of the gold particles. The treated colloidal gold was added to the concentrated cells immediately before plunge freezing. 4 μl drops of gold-and-cell solutions were manually applied to glow-discharged R2/2 copper-Rhodium Quantifoil™ grids (Quantifoil Micro Tools, Jena, Germany), and then the grids were automatically blotted and plunged into liquid ethane or a liquid ethane-propane mixture using a Vitrobot (FEI Company, Hillsboro, OR)32,33. Frozen grids were stored under liquid nitrogen until use and kept below −165°C during loading and data collection. EM images were collected using an FEI Polara™ (FEI Company, Hillsboro, OR, USA), 300 kV FEG transmission electron microscope equipped with a Gatan energy filter (slit width 20 eV) on a 2 × 2 k Gatan Ultrascan CCD camera or later a lens-coupled 4 × 4 k Ultracam (Gatan, Pleasanton, CA). Pixels on the CCD represented between 0.67 and 1.2 nm on the specimen. Tilt-series from −60° to 60° with an increment between 0.5° and 1° were recorded semi-automatically around one or two axes34 at 10 and 12 μm underfocus using the predictive UCSF-Tomo package or Leginon 35,36. A cumulative dose of 200 e−/Å2 or less was used for the directly plunge frozen samples and 100 e−/Å2 or less for the correlated fLM-ECT tomograms. Three dimensional reconstructions were calculated using the IMOD package37 or RAPTOR38.
Correlated fluorescence light microscopy electron crytomography (fLM-ECT)
Caulobacter cells expressing mCherry-CtpS were induced with 0.03% xylose for two hours of growth in exponential phase. 800 μl of the induced cell culture were added to 200 μl 5x concentrated fixing solution (12.5% paraformaldehyde in 150mM Na-phosphate buffer, pH 7.5) and incubated for 15 min at room temperature. Cells were washed twice in Na-phosphate buffer and resuspended in ~40 μl fresh buffer. H2 gold finder TEM grids covered with R2/2 Quantifoil were glow discharged before 5 μl of 0.5 mg/ml sterile-filtered poly-L-lysine (Sigma P1524) was added to each grid. The grids were then dried in a 60°C oven before use. A 4 μl droplet of the fixed cell solution was added to a poly-L-lysine treated grid, blotted with filter paper and gently rinsed with buffer. The grid was immediately transferred onto a droplet of Na-phosphate buffer on a glass slide and covered with a coverslip. After imaging the grid using a Nikon 90i fluorescence microscope, the grid was carefully taken from the slide. A 4 μl droplet of BSA-treated 10 nm gold solution was added and the grid was plunge-frozen in a liquid ethane-propane mixture as described above. Tilt series were recorded of the same cells imaged by fluorescence light microscopy.
CtpS purification and antibody production
E.coli CtpS was purified from BL21 cells expressing an N terminal His-tagged EcCtpS in pET22HT, generously provided by the Williamson lab (Scripps). 2 liters of cells were grown to an OD600 of 0.5 and induced with 1 mM IPTG for 4 hours. Cells were harvested by centrifugation at 10,000 × g for 30 minutes and lysed in 200 mL lysis buffer (50 mM K-Hepes (pH 7.6), 1 M NaCl, 20 mM imidazole, 10% glycerol) by a cell cracker (Microfluidics). Lysates were centrifuged at 31,000 × g and loaded onto an Ni-NTA (Qiagen) column (25 mL resin) equilibrated with lysis buffer minus glycerol. The column was washed with six volumes of equilibration buffer and the protein was eluted with equilibration buffer with 250 mM imidazole. Purified protein was treated with AcTEV protease (Invitrogen) as per manufacturer’s instructions to remove the N terminal His tag. Cleaved protein was then dialyzed against 4 L of 50 mM K-Hepes (pH7.6), 20 mM β-mercaptoethanol, 5 mM glutamine with 50% glycerol using a 3,500 MWCO Slide-A-Lyzer dialysis cassette (Thermo Scientific). Purified protein was used to create polyclonal rabbit antibodies (Pocono Rabbit Farm and Laboratory).
Immunofluorescence
Immunofluorescence microscopy was performed as described previously39 with modifications. Cells were treated with a 1:1000 dilution of rabbit anti-CtpS antibody. A Goat anti rabbit Alexa Fluor 488 antibody (Invitrogen) was used at a 1:200 dilution to detect the primary antibody.
Electron microscopy of purified EcCtpS protein
5 μL of purified EcCtpS protein (≈2mg/mL) was incubated 10 minutes in 15 μL of a buffer described to enable CtpS enzymatic activity (50 mM Tris HCl pH 7.8, 1 mM UTP, 1 mM ATP, 0.2 mM GTP, 10 mM MgCl2, 10 mM glutamine)40 at room temperature and applied to 200 mesh thin-film carbon with nitrocellulose, glow-discharged grids and stained with 1% uranyl acetate. Grids visualized at 80kV on a Zeiss912AB Transmission Electron Microscope equipped with an Omega Energy Filter (Confocal and Electron Microscopy Core Facility Laboratory, Princeton University). Micrographs were captured using a digital camera from Advanced Microscopy Techniques.
Sedimentation of purified EcCtpS protein
5 μL of purified EcCtpS protein (≈2mg/mL) was incubated at room temperature for 10 minutes in 45 μL of a buffer described to enable CtpS enzymatic activity. Samples were spun at 55K RPM for 30 minutes at 4 C in a Beckman Optima TLA-100.2 rotor. After centrifugation the supernatant was removed and the pellet was resuspended in the same volume. Pellet and supernatant samples were analyzed for CtpS protein by Western blot.
Bacterial two-hybrid assays
N- and C-terminal fusions of CtpS and CreS to the T18 and T25 subunits of adenylate cyclase were constructed in vectors pUT18, pUT18C, pKT25, and pKNT25 (Euromedex). Two-hybrid strains were constructed by simultaneously electroporating pairs of vectors into BTH101. Single transformants were grown in selective LB media to stationary phase and were pinned in triplicate onto LB media with antibiotics, 40 μg/mL X-gal, and 0.25 mM IPTG. Plates were incubated at 30°C for 36 hours to allow blue color to develop on indicator media. The resulting colonies were imaged using a Sony Cyber-shot DSC-P8 digital camera (Sony).
Site directed mutagenesis
Site directed mutants of Caulobacter CtpS were constructed using the Stratagene quik-change site directed muatgenesis kit (Stratagene). Mutagenic primers were constructed using Stratagene’s quik change site directed mutagenesis program (http://www.stratagene.com/sdmdesigner/default.aspx). Specific primer sequences are available upon request. Site directed mutagenesis protocols provided by Stratagene were used with the exception of replacement of the DNA polymerase with KOD DNA polymerase (Novagen).
Protein abundance in CtpS, CreS, and site directed mutant strains
Appropriate strains (noted in Supplementary Figure legends) were grown overnight in the presence or absence of 0.3% xylose. Samples of each culture normalized by OD660 measurements were run on a PAGE gel and transferred to a nitrocellulose membrane (Whatman). For the cell cycle experiment, the same volume of cells was used for each lane. All quantitation was done by normalizing protein levels to MreB. CtpS levels were detected with an αCtpS antibody (1:2,500) raised against purified E.coli CtpS. CreS levels were detected with an αCreS antibody (1:15,000). MreB levels were detected with an αMreB antibody (1:15,000) and were used as a loading control.
Supplementary Material
Acknowledgments
We are grateful to Bonnie Bassler, Coleen Murphy, Eric Klein, and Kimberly Cowles for critical reading and suggestions on the manuscript. We thank Matthew Cabeen, Christine Jacobs-Wagner, Jamie Williamson, and the members of the Gitai lab for helpful reagents and thoughtful discussions. JNW is supported by a postdoctoral fellowship, Grant 1F32AI073043–01A1, from the National Institute of Allergy and Infectious Diseases. GJJ was supported in part by NIH grants R01 AI067548 and P50 GM082545, as well as the Howard Hughes Medical Institute, the Beckman Institute at Caltech, and gifts to Caltech from the Gordon and Betty Moore Foundation and Agouron Institute. ZG is supported by funding from Grant DE-FG02-05ER64136 from the U.S. Department of Energy Office of Science (Biological and Environmental Research), NIH grant 1DP2OD004389-01, the Human Frontiers Science Program, and the Beckman Foundation.
Footnotes
Supplemental information. Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions M.J.I. performed all experiments except for the ECT and LM-ECT experiments, which were performed by A.B. J.N.W. performed the initial screen that identified CtpS as a linearly localized protein. Experiments were conceived by M.J.I, A.B., G.J.J., and Z.G., and the manuscript was written by M.J.I. and Z.G. with significant input from A.B. and G.J.J.
References
- 1.Bi EF, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 2.Lowe J, Amos LA. Crystal structure of the bacterial cell-division protein FtsZ. Nature. 1998;391:203–206. doi: 10.1038/34472. [DOI] [PubMed] [Google Scholar]
- 3.Jones LJ, Carballido-Lopez R, Errington J. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell. 2001;104:913–922. doi: 10.1016/s0092-8674(01)00287-2. S0092-8674(01)00287-2 [pii] [DOI] [PubMed] [Google Scholar]
- 4.Ausmees N, Kuhn JR, Jacobs-Wagner C. The bacterial cytoskeleton: an intermediate filament-like function in cell shape. Cell. 2003;115:705–713. doi: 10.1016/s0092-8674(03)00935-8. S0092867403009358 [pii] [DOI] [PubMed] [Google Scholar]
- 5.Briegel A, et al. Multiple large filament bundles observed in Caulobacter crescentus by electron cryotomography. Mol Microbiol. 2006;62:5–14. doi: 10.1111/j.1365-2958.2006.05355.x. MMI5355 [pii] [DOI] [PubMed] [Google Scholar]
- 6.Li Z, Jensen GJ. Electron cryotomography: a new view into microbial ultrastructure. Curr Opin Microbiol. 2009;12:333–340. doi: 10.1016/j.mib.2009.03.007. S1369-5274(09)00042-3 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Z, Trimble MJ, Brun YV, Jensen GJ. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J. 2007;26:4694–4708. doi: 10.1038/sj.emboj.7601895. 7601895 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Werner JN, et al. Quantitative genome-scale analysis of protein localization in an asymmetric bacterium. Proc Natl Acad Sci U S A. 2009;106:7858–7863. doi: 10.1073/pnas.0901781106. 0901781106 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Long CW, Levitzki A, Koshland DE., Jr The subunit structure and subunit interactions of cytidine triphosphate synthetase. J Biol Chem. 1970;245:80–87. [PubMed] [Google Scholar]
- 10.Briegel A, et al. Location and architecture of the Caulobacter crescentus chemoreceptor array. Mol Microbiol. 2008;69:30–41. doi: 10.1111/j.1365-2958.2008.06219.x. MMI6219 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma X, Ehrhardt DW, Margolin W. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc Natl Acad Sci U S A. 1996;93:12998–13003. doi: 10.1073/pnas.93.23.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gitai Z, Dye N, Shapiro L. An actin-like gene can determine cell polarity in bacteria. Proc Natl Acad Sci U S A. 2004;101:8643–8648. doi: 10.1073/pnas.0402638101. 0402638101 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartman SC. The Interaction of 6-Diazo-5-Oxo-L-Norleucine with Phosphoribosyl Pyrophosphate Amidotransferase. J Biol Chem. 1963;238:3036–3047. [PubMed] [Google Scholar]
- 14.Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci U S A. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Srinivasan R, Mishra M, Murata-Hori M, Balasubramanian MK. Filament formation of the Escherichia coli actin-related protein, MreB, in fission yeast. Curr Biol. 2007;17:266–272. doi: 10.1016/j.cub.2006.11.069. S0960-9822(06)02590-5 [pii] [DOI] [PubMed] [Google Scholar]
- 16.Srinivasan R, Mishra M, Wu L, Yin Z, Balasubramanian MK. The bacterial cell division protein FtsZ assembles into cytoplasmic rings in fission yeast. Genes Dev. 2008;22:1741–1746. doi: 10.1101/gad.1660908. 22/13/1741 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levitzki A, Koshland DE., Jr Cytidine triphosphate synthetase. Covalent intermediates and mechanisms of action. Biochemistry. 1971;10:3365–3371. doi: 10.1021/bi00794a008. [DOI] [PubMed] [Google Scholar]
- 18.Endrizzi JA, Kim H, Anderson PM, Baldwin EP. Crystal structure of Escherichia coli cytidine triphosphate synthetase, a nucleotide-regulated glutamine amidotransferase/ATP-dependent amidoligase fusion protein and homologue of anticancer and antiparasitic drug targets. Biochemistry. 2004;43:6447–6463. doi: 10.1021/bi0496945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lunn FA, Macleod TJ, Bearne SL. Mutational analysis of conserved glycine residues 142, 143 and 146 reveals Gly(142) is critical for tetramerization of CTP synthase from Escherichia coli. Biochem J. 2008;412:113–121. doi: 10.1042/BJ20071163. BJ20071163 [pii] [DOI] [PubMed] [Google Scholar]
- 20.Paluh JL, Zalkin H, Betsch D, Weith HL. Study of anthranilate synthase function by replacement of cysteine 84 using site-directed mutagenesis. J Biol Chem. 1985;260:1889–1894. [PubMed] [Google Scholar]
- 21.Charbon G, Cabeen MT, Jacobs-Wagner C. Bacterial intermediate filaments: in vivo assembly, organization, and dynamics of crescentin. Genes Dev. 2009;23:1131–1144. doi: 10.1101/gad.1795509. 23/9/1131 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cabeen MT, et al. Bacterial cell curvature through mechanical control of cell growth. EMBO J. 2009;28:1208–1219. doi: 10.1038/emboj.2009.61. emboj200961 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kleinschmidt AK, Moss J, Lane DM. Acetyl coenzyme A carboxylase: filamentous nature of the animal enzymes. Science. 1969;166:1276–1278. doi: 10.1126/science.166.3910.1276. [DOI] [PubMed] [Google Scholar]
- 24.Carrey EA, et al. Detection and location of the enzymes of de novo pyrimidine biosynthesis in mammalian spermatozoa. Reproduction. 2002;123:757–768. [PubMed] [Google Scholar]
- 25.Zigler JS, Jr, Rao PV. Enzyme/crystallins and extremely high pyridine nucleotide levels in the eye lens. FASEB J. 1991;5:223–225. doi: 10.1096/fasebj.5.2.2004667. [DOI] [PubMed] [Google Scholar]
- 26.Bork P, Sander C, Valencia A. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc Natl Acad Sci U S A. 1992;89:7290–7294. doi: 10.1073/pnas.89.16.7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Evinger M, Agabian N. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J Bacteriol. 1977;132:294–301. doi: 10.1128/jb.132.1.294-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pinter SF, Aubert SD, Zakian VA. The Schizosaccharomyces pombe Pfh1p DNA helicase is essential for the maintenance of nuclear and mitochondrial DNA. Mol Cell Biol. 2008;28:6594–6608. doi: 10.1128/MCB.00191-08. MCB.00191-08 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ely B. Genetics of Caulobacter crescentus. Methods Enzymol. 1991;204:372–384. doi: 10.1016/0076-6879(91)04019-k. [DOI] [PubMed] [Google Scholar]
- 30.Ellgaard AK, DAM, Minnich SA. Abstr Annu Meeting Am Soc Microbiol. 1989;209 [Google Scholar]
- 31.Meisenzahl AC, Shapiro L, Jenal U. Isolation and characterization of a xylose-dependent promoter from Caulobacter crescentus. J Bacteriol. 1997;179:592–600. doi: 10.1128/jb.179.3.592-600.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iancu CV, et al. Electron cryotomography sample preparation using the Vitrobot. Nat Protoc. 2007;1:2813–2819. doi: 10.1038/nprot.2006.432. [DOI] [PubMed] [Google Scholar]
- 33.Tivol W, Briegel A, Jensen GJ. An Improved Cryogen for Plunge Freezing. Microsc Micoanal. 2008;14:375–379. doi: 10.1017/S1431927608080781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iancu CV, et al. A “flip-flop” rotation stage for routine dual-axis electron cryotomography. J Struct Biol. 2005;151:288–297. doi: 10.1016/j.jsb.2005.07.004. S1047-8477(05)00143-7 [pii] [DOI] [PubMed] [Google Scholar]
- 35.Zheng QS, Braunfeld MB, Sedat JW, Agard DA. An improved strategy for automated electron microscopic tomography. J Struct Biol. 2004;147:91–101. doi: 10.1016/j.jsb.2004.02.005. S1047847704000413 [pii] [DOI] [PubMed] [Google Scholar]
- 36.Suloway C, et al. Fully automated, sequential tilt-series acquisition with Leginon. J Struct Biol. 2009;167:11–18. doi: 10.1016/j.jsb.2009.03.019. S1047-8477(09)00097-5 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mastronarde DN. Dual-axis tomography: an approach with alignment methods that preserve resolution. J Struct Biol. 1997;120:343–352. doi: 10.1006/jsbi.1997.3919. S1047-8477(97)93919-8 [pii] [DOI] [PubMed] [Google Scholar]
- 38.Amat F, et al. Markov random field based automatic image alignment for electron tomography. J Struct Biol. 2008;161:260–275. doi: 10.1016/j.jsb.2007.07.007. S1047-8477(07)00155-4 [pii] [DOI] [PubMed] [Google Scholar]
- 39.Domian IJ, Quon KC, Shapiro L. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell. 1997;90:415–424. doi: 10.1016/s0092-8674(00)80502-4. S0092-8674(00)80502-4 [pii] [DOI] [PubMed] [Google Scholar]
- 40.Simard D, Hewitt KA, Lunn F, Iyengar A, Bearne SL. Limited proteolysis of Escherichia coli cytidine 5′-triphosphate synthase. Identification of residues required for CTP formation and GTP-dependent activation of glutamine hydrolysis. Eur J Biochem. 2003;270:2195–2206. doi: 10.1046/j.1432-1033.2003.03588.x. 3588 [pii] [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.