Abstract
A comparative analysis of 60 complete Burkholderia genomes was conducted to obtain insight in the evolutionary history behind the diversity and pathogenicity at species level. A concatenated multiprotein phyletic pattern and a dataset with Burkholderia clusters of orthologous genes (BuCOGs) were constructed. The extent of horizontal gene transfer (HGT) was assessed using a Markov based probabilistic method. A reconstruction of the gene gains and losses history shows that more than half of the Burkholderia genes families are inferred to have experienced HGT at least once during their evolution. Further analysis revealed that the number of gene gain and loss was correlated with the branch length. Genomic islands (GEIs) analysis based on evolutionary history reconstruction not only revealed that most genes in ancient GEIs were gained but also suggested that the fraction of the genome located in GEIs in the small chromosomes is higher than in the large chromosomes in Burkholderia. The mapping of coexpressed genes onto biological pathway schemes revealed that pathogenicity of Burkholderia strains is probably mainly determined by the gained genes in its ancestor. Taken together, our results strongly support that gene gain and loss especially in ancient evolutionary history play an important role in strain divergence, pathogenicity determinants of Burkholderia and GEIs formation.
Keywords: Burkholderia, gene gain/loss, Markov model, genomic island, coexpression
Introduction
The Burkholderia genus comprises more than 60 described species, which include zoonotic and plant pathogens as well as symbionts of fungi, insects and plants.1 Some rhizosphere-associated strains are beneficial to their plant hosts and several species display pollutant-degrading activities of interest for bioremediation.2 The ability of members of this bacterial genus to colonize such a wide range of ecological niches is attributed to their genomic plasticity and metabolic versatility.3–6
Burkholderia species can vary greatly in their gene contents and metabolic capabilities. Among 60 completely sequenced Burkholderia genomes, sizes range from 5.23 Mb to 9.73 Mb (Supplementary Table S1).7,8 The functional partitioning of genes between different chromosomes indicates distinct evolutionary origins.9 Previous reports have highlighted the key role played by horizontal gene transfer (HGT) in Burkholderia evolution. For example, Burkholderia xenovorans LB400, a polychlorinated biphenyl (PCB) degrader, apparently acquired several aromatic degradation capabilities by gene transfer, enabling its adaption to an environment with a recalcitrant carbon source.7 B. cenocepacia J2315, a human pathogen, contains 14 genomic islands (GEIs) which introduced functions promoting survival and pathogenesis in the cystic fibrosis (CF) lung.10
The availability of numerous complete genome sequences enables an assessment of the extent of HGT in bacterial genome evolution.11,12 Ussery and his colleagues investigated the species tree and analyzed the conserved genes by Blast matrix for 56 according to 56 Burkholderia genomes.13 However, reconstruction of ancestral states for gene repertoires along the evolution is still not carried out. As GEIs are important in bacterial pathogenicity, reconstruction of ancestral states for gene repertoires is important to identify highly variable GEIs that are not apparent through simple pairwise comparisons in previous Burkholderia analysis.
Using a multi-protein phyletic pattern and dedicated set of orthologs, the history of vertical transmission, gene acquisition, and gene loss was reconstructed by a Markov model. In addition, the impact of gene gain/loss on pathogenicity within Burkholderia was investigated by the GEIs analysis and the correspondent coexpression analysis.
Materials and Methods
Collection of genomic sequences
Complete genome sequences from 60 Burkholderia strains (representing 17 species) and five strains of other Burkholderiales species (Supplementary Table S1) were obtained from National Center for Biotechnology Information database (ftp://ftp.ncbi.nih.gov).
Clusters of orthologous genes in Burkholderia
A total of 416,696 open reading frames from the chromosomal regions of all the 65 strains were selected for homology searches. OrthoMCL was used to generate groups of orthologous proteins with default parameters.14 This resulted in a set of 52,031 Burkholdria clusters of orthologous genes (BuCOGs; Supplementary Table S2 and Supplementary Table S3), of which 416 were present as single copy genes within the 65 genomes. These single copy genes were selected to reconstruct the organism tree.
Each of the 416 orthologs was concatenated and aligned with MUSCLE using default parameters.15 Poorly aligned regions from this alignment were removed by trimAl V1.2.16 Phylogenetic trees were inferred by maximum likelihood with PHYML17 by 1,000 bootstrap replicates, using JTT substitution model18 as the model of amino acid evolution with a gamma distribution with 8 categories. Amino acid usage and the shape parameter were estimated from the data.
Probabilistic model for gene gain and loss
A binary character (0,1) table of gene absence and presence across the studied strains was used to estimate the phyletic patterns. The patterns were analyzed by probabilistic models, using the likelihood framework described by Csürös et al19,20 to study evolution of introns and gene presence/absence. Compared to the maximum parsimony method used in Prochlorococcus,21 the corresponding probabilistic model uses branch-specific gene gain and loss rate, with different species showing dramatic HGT frequency in the real evolution history.22
We used a rates-across-genes Markov model to trait gene evolution, with branch-specific gain and loss rates. Briefly, the procedure was as follows: it was assumed that gene sites evolved independently under a Markov model.23 The gene state (encoded by 0 and 1 for absence and presence) changed on each branch b of the phylogeny according to our modified probabilities formulas,
where λ denotes branch-specific gene gain rate, μ denotes branch-specific loss rate. The gene density at an ancestral node was computed as an expected value conditioned on the observed data, by summing posterior probabilities.19 The values of estimated gene gain and loss rate can be affected by several reasons, such as different standard of gene identification in each genome, and different reconstructed methods. In order to overcome the biases of gene numbers of different genome and different gene families, we bootstrapped the gene table, and generated 1,000 random gene tables by selecting genes of the original gene table independently. For each bootstrap table, the likelihood and evolutionary history are calculated. This procedure yields confidence levels for the estimates inferred in this study of about 1%. The mean values of estimate results were considered as final value. We also used the maximum parsimony method to estimate the gene numbers in each branch, and got the same tendency though the gained gene number in recent branches is slightly higher than with our methods.
The individual gain and loss history of gene groups was also analyzed. The posterior probability of gene presence and absence in each node was calculated according to the branch-specific gene gain/loss rate. Only when the gene presence probability was greater than 0.5, it would be considered as being present in the evolutionary node.
Each gene that was predicted to be transferred was compared with the NCBI nr database using BlastP with the expect value cutoff set at 10−10 to identify homologs in other organisms. For each protein, the first 100 hits were chosen for further potential HGTs analysis. The complete protein sequences of the 100 hits were extracted from the GenBank database, followed by multiple sequences alignment and phylogenetic tree generation by the same criterion described above. Based on tree topology, we identified the donor lineages, which naturally grouped with the Burkholderia genes. The candidate donor genus should have more than three genes and have strong bootstrap support values (>80).
COG classification and gene ontology analysis
The functional annotations of the Burkholderia genes were retrieved from the Burkholderia database24 except for B. phytofirmans PsJN and Burkholderia glumae BGR1, which were annotated by the NCBI COG database.25 Gene Ontology annotations are described at Pathema.26 In order to determine the overrepresented GO terms, we used the GOstats,27 taking P-values <1e−5 for significant.
Genomic islands (GEIs) in each evolutionary node
In this study, we first used a Markov model to predict the gained genes in 60 Burkholderia strains, which is one of the bacterial genera whose genomes have been most sequenced. Then, the transferred genes from different evolutionary nodes were mapped into the chromosome to investigate the existed GEIs. As we have the information about which genes are transferred, the criterion about GEIs was compared to the supplementary file in B. xenovorans LB400 genome analysis,7 for convenience, we defined minimum continuous four gained genes as a GEI.
Coexpression analysis
A Pearson correlation coefficient was calculated based on 47 arrays of B. pseudomallei K96243 downloaded from NCBI GEO GSE5495 dataset28 to investigate the function of transferred genes in different evolutionary time (different evolutionary node in phylogenetic tree). Background correction and data normalization was done by the RMA algorithm in Bioconductor29 and the poor annotated probe-sets were removed. Measurements for unique genes was calculated from means of the probes belonging to the same gene. The gene coexpressed with at least one of either gene in GEIs with the absolute correlation |r| > 0.6, which we expect to be co-regulated in our case, was further selected to analyze the function.
Mapping coexpressed genes onto existing biological pathways
For each gene in GEIs gained in the same node, the top 100 genes in each gained GEIs were selected. These genes were sorted by absolute correlation value |r| referred to as coexpressed genes in descending order. These coexpressed genes with GEIs genes in each node were then mapped onto the pathways from Biocyc.30 A score of each pathway was calculated by using following formula, which represent the extent of coexpression in each node:
, where G is the number of genes in a pathway present in the top 100 of all same node GEIs gene lists, and N is the number of lists in which gene i is present in the top 100 genes. Rij is the rank of the ith gene of the specified pathway in a list, rij is the absolute correlation value |r|.
Results and Discussion
Reconstruction of gene gain and loss in the evolution of Burkholderia
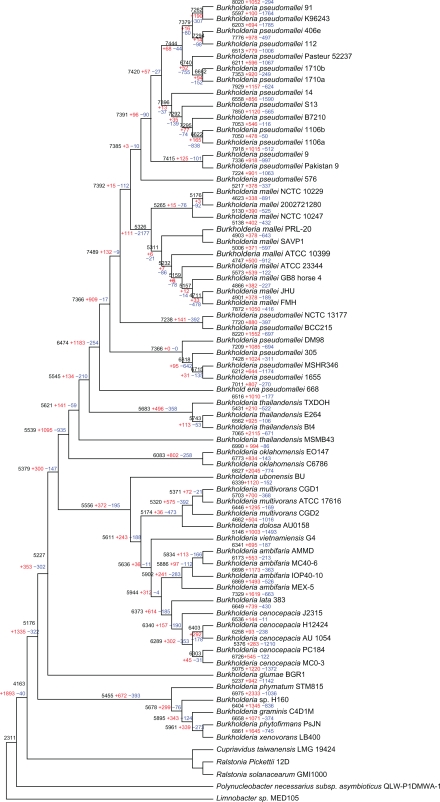
Gene gain and loss are ongoing processes in microbial genomes, resulting in diversity in genome sizes, even among closely related strains. We used the probabilistic model (details in Methods) to reconstruct the events that occurred during the evolution of this genus after its divergence from the common ancestor (Fig. 1). By mapping the gene distributions in a phylogenetic context, we could infer the ancestry and dynamics of genes among strains. The reconstruction shows that the gene gain and loss seems to be a prevailing trend in the evolution of Burkholderia. In the Burkholderia ancestor and the ancestral branches of the major clades (the clade definition was presented in Supplementary Table S1) a huge number of gene acquisitions have occurred, whereas gene loss seems to be occurring more frequently in evolution of closely related strains, generating large differences in a short divergence time.
Figure 1.
Phylogenetic tree of Burkholderiaceae based on concatenated amino acid alignments of one homolog gene and rooted by using proteins from other Burkholderiaceae species as the outgroup. All the evolutionary nodes in this tree were supported by bootstrap values higher than 90%. Reconstruction of gene content evolution based on BuCOGs in Burkholderia is shown. For each species and each internal node of a tree, the inferred number of BuCOGs present (numbers in black), and the numbers of BuCOGs lost (numbers in blue) or gained (numbers in red) along the branch leading to a given node (species) are indicated.
The common ancestor of Burkholderia had an estimated 5,176 genes, losing 322 genes and acquiring 1,335 genes compared with the outgroup of other Burkholderiaceae. Thus, gain of ancestral genes seems to be the prevailing trend in the evolution of Burkholderia (Fig. 1). This result is different from former similar analysis in other bacteria.31 Most of the changes mapped to this ancestral stage of evolution seem to be linked to metabolism. Abundant genes for membrane transport and metabolism were gained via HGT (see Table 1 for the GO over-represented categories). The subsequent evolution of the Burkholderia reveals a considerable number of strain-specific duplications and acquisitions of unique genes as well as ancestral node gene loss (Fig. 1). In order to confirm the ancestral gene aquisitions, we randomly selected 100 ancestral transfer gene families and manually checked the phylogenetic trees. 85 gene families were confirmed nested within other groups, while 8 gene families without enough homologs in other species and 17 other cannot be determined from the trees. For instance, the whole fabG (BuCOG8606) gene family in Burkholderia is clustered with other unrelated bacteria, and the copC (BuCOG142) gene family might have transferred from Enterobacteriaceae (Supplementary Figs. S1 and S2).
Table 1.
Over-represented GO categories among BuCOGs gained in the ancestor of the Burkholderia.
| GO ID | GO term | Count | Total | P-value | GO level |
|---|---|---|---|---|---|
| GO:0051234 | Establishment of localization | 124 | 726 | 1.64e-31 | 3 |
| GO:0006810 | Transport | 106 | 618 | 3.30e-27 | 3 |
| GO:0016020 | Membrane | 84 | 571 | 1.38e-15 | 3 |
| GO:0022891 | Substrate-specific transmembrane transporter activity | 37 | 179 | 2.83e-12 | 4 |
| GO:0022892 | Substrate-specific transporter activity | 42 | 237 | 8.03e-11 | 3 |
| GO:0044425 | Membrane part | 66 | 497 | 1.13e-9 | 3 |
| GO:0022804 | Active transmembrane transporter activity | 36 | 200 | 1.75e-9 | 4 |
| GO:0044238 | Primary metabolic process | 145 | 1521 | 1.35e-8 | 3 |
| GO:0005975 | Carbohydrate metabolic process | 44 | 290 | 1.69e-8 | 4 |
| GO:0044237 | Cellular metabolic process | 151 | 1661 | 1.62e-7 | 3 |
| GO:0044459 | Plasma membrane part | 45 | 327 | 4.22e-7 | 4 |
| GO:0005886 | Plasma membrane | 45 | 329 | 4.75e-7 | 4 |
| GO:0031224 | Intrinsic to membrane | 47 | 364 | 1.92e-6 | 4 |
| GO:0016746 | Transferase activity, transferring acyl groups | 23 | 130 | 8.15e-6 | 4 |
Moore et al32 have reported that B. thailandensis, a species closely related to of B. pseudomallei, is unable to cause disease. In comparison with B. pseudomallei, the attenuation of B. thailandensis is caused by a functional arabinose biosynthesis operon which is largely deleted in B. pseudomallei.32 In Burkholderia, the virulence divergence between different strains can be caused by mutation or loss of few genes.33 The concerted evolution trends in Burkholderia could be explained by the large-scale ancestral gene acquisition that might facilitate the acquisition of a generally pathogenic lifestyle of Burkholderia, and the strain specific pathogenic lifestyles can be modified by fine-tuning gene loss.
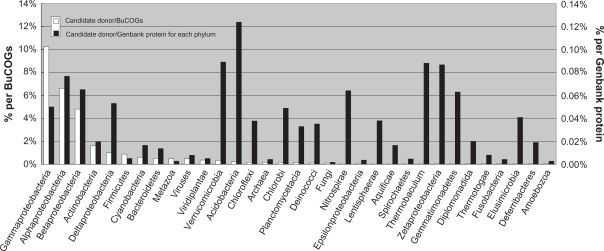
Overall, our results suggested that about 60% of the genes families in Burkholderia genus are inferred to have experienced HGT at least once during their evolution. As a large proportion of genes in Burkholderia are Burkholderia-specific, they could represent new birth of genes or caused by in silico prediction errors. Hence, the HGT percentage might be overestimated and the real HGT genes number in each Burkholderia strains could be less than what we predicted. The candidates for these genes outside the Burkholderia were divided according to taxonomic classification (Fig. 2). With more than 7,000 candidate donors in Proteobacteria, this represents the second largest gene reservoir for Burkholderia. Along with other Betaproteobacteria, there is a clear overrepresentation of Gammaproteobacteria and Alphaproteobacteria (Fig. 2). Most of the Burkholderia species are soil dwellers, and it has shown that the Burkholderia species are ancient symbionts of legumes,34 which created the opportunity to gain new genes from eukaryotes.35 Furthermore, more than two third of the gained genes were Burkholderia-only, which means these genes have no homolog outside this genus. This phenomenon may be attributed to the extinction of the donors followed by ancient gene transfer36 or gene transfer within Burkholderia lineage followed by new gene birth.
Figure 2.
Taxonomic distribution of candidate donors for putatively transferred Burkholderia genes. The taxonomy is sorted by the percentages representing numbers of hits relative to the BuCOGs total number (white bars). The percentages relative to the number of phylum-specific proteins in Genbank are shown as black bars. No matches were detected for Dictyoglomi, Elusimicrobia, Synergistetes and Tenericutes (data from 2, 2, 1, and 26 genomes, respectively). Data for Caldiserica, Chrysiogenetes were not available at the time of analysis.
To further understand the evolutionary history of Burkholderia, we selected the representative genome of B. pseudomallei K26943 to elucidate impact of gene gain and loss evolution on different evolutionary nodes. The GEIs of this strain in each evolutionary node is represented in Supplementary Fig. S3. This figure indicates that most of the GEIs in this strain were gained in the ancestor node while only a small fraction of GEIs were gained most recently (Supplementary Fig. S3). We also found that the percentage about transferred chromosome fragments were different between two chromosomes in B. pseudomallei K26943 with 15.58% in the large and 26.88% in the small chromosome, respectively. The result strongly suggests that the small chromosome was much more prone to HGT than the large chromosome in the evolutionary history. It has been reported that the small chromosomes in Burkholderia carry more accessory functions associated with survival and adaptation in different niches, whereas the large chromosomes encode most of the core genes (shared by all strains) involved in central metabolism and cell growth.9 As GEIs play key roles in Burkholderia pathogenicity, our results indicate that the ancient gene transfer played key roles in pathogenicity of Burkholderia.
GEIs are discrete DNA segments which contain a number of genes acquired by HGT that might be beneficial to hosts under certain conditions.37 Two major methods which can be broadly grouped into sequence-based methods and comparative genomics/phylogeny based methods, have been developed to predict the GEIs computationally.38–40 The first method can only predict recently acquired GEIs while the prediction accuracy of the latter method is based on the number of related species. In this study, 109 GEIs were found in B. pseudomallei K26943 (Supplementary Table S4). The results indicate that evolutionary history reconstruction is a powerful method to investigate the GEIs, especially if based on a large enough number of closely related species.
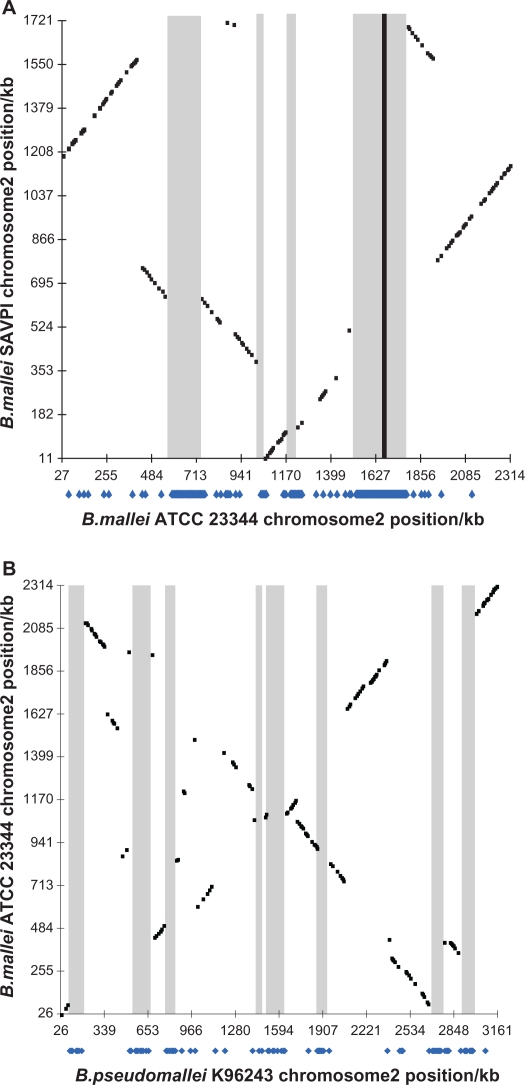
Comparison of the number of genes gained or lost on a particular evolutionary scale and the length of the corresponding branch revealed a pattern different from that described for Prochlorococcus (cyanobacteria) and lactic acid bacteria.21 Similar to the Prochlorococcus, the number of gene loss is significantly correlated with the branch length (r = 0.576, P < 8 × 10−5).21 However, the number of gene gains also has such a correlation (r = 0.500, P < 0.0009). The clock-like behaviour of gene loss and gain could be explained by a large number of small-scale events, which might be randomly distributed along the evolutionary path. However, the gene acquisition and removal model points to a mixed pattern, combining disruption events and complete acquisition/removal. Apparently, multiple type III secretion systems (T3SS) in Burkholderia were acquired from HGT and most of these genes were located as clusters.8–10,41 The acquisition stage analysis revealed that these genes were gained in the same period, indicating that these T3SS might be integrated through the entire fragment being transferred. Such GEIs probably played a crucial role and could facilitate the evolution by ‘quantum leaps’ as their gain or loss could rapidly and dramatically alter the genome and lifestyle of a bacterium.37 The transfers of large fragments were frequently associated with particular physiologic adaptation such as virulence, catabolism, or resistance to a toxic compound.37 Mutated pseudogenes likely accounted for B. mallei being nonmotile or nonflagellated.34 The presence of numerous insertion sequence elements mediated extensive deletions and rearrangements of the genome. These deletions likely accounted for the large gene variation among closely related strains. For example, comparing the B. mallei SAVP1 and B. mallei ATCC 23344 genomes revealed a large T3SS-encoding fragment lost in B. mallei SAVP1 (Fig. 3). This loss likely accounted for the difference in virulence between B. mallei SAVP1 and B. mallei ATCC 23344.8 By comparing B. mallei ATCC 23344 with B. pseudomallei K96243, the same event of large T3SS-encoding fragment loss was also observed (Fig. 3).
Figure 3.
Profiling of the B. mallei SAVP1 and ATCC 23344 lost genes and regions. (A) Graphs represent the best protein hits scatter plot (black dots) and B. mallei SAVP1 chromosome 2 recent lost genes (blue dots under x axis) in B. mallei ATCC 23344 chromosome 2. (B) Graphs represent the best protein hits scatter plot (black dots) and B. mallei ATCC 23344 chromosome 2 B. mallei lost genes (blue dots under x axis) in B. pseudomallei K96243 chromosome 2. Black scatter plot represents the best protein hits of two genomes chromosome according to the Pathema Scatter Plot Results. The lost gene homologues were plotted onto the another genome’s chromosome under the bar with blue dots. The shaded regions represent the lost regions. The black rectangle region represents the T3SS location.
Expression and coexpression analysis
We analyzed the proportion of putative horizontally transferred genes in growth-regulated B. pseudomallei genes by using expression data reported by Rodrigues et al.28 In three major growth phases (early, log and stationary phase), about 900 genes were differentially expressed, among which 229 were predicted to be horizontally transferred. Nearly 50% of the latter (115 out of 229) displayed differential expression during exponential growth. Rodrigues et al (2006) noted that genes differentially expressed in stationary phase had a close relationship with strain pathogenicity, which confirmed that HGT genes might contribute to their host adaption. A total of 90 acquired genes that were differentially expressed in stationary phase could be assigned to the Burkholderia origin. This result suggests that the ancestor gene acquisition shaped the major characteristics of different Burkholderia species.
As many transferred genes in this strain are functionally unknown, based on co-regulation and coexpression of a set of genes in the same pathway, gene function can be predicted by quantitatively transforming the coexpression correlations as the degree of function similarity. Significant difference was observed after quantitatively mapping the coexpressed genes in different evolutionary nodes onto existing biological pathways (Supplementary Table S5). The transferred genes in each node involved in several important regulation and metabolism pathways, such as sphingolipid metabolism, exhibited a high degree of coexpression based on the correlation coefficients (Supplementary Table S5). By contrast, plotting Spathway score values to recently transferred and anciently transferred genes, we found that the scores for bacterial secretion system and protein export pathway were several fold higher in case of anciently transferred genes than for recently transferred genes, which means the function of anciently acquired genes are more likely related with the secretion system and export protein pathway (Supplementary Table S5). The result is consistent with the GEIs analysis reported above that the pathogenicity of Burkholderia was mainly determined by ancient gene transfer.
The mapping of coexpressed genes onto biological pathway schemes provides a comprehensive way to identify previously unknown functional patterns in sets of genes with known functions. The results revealed a complex pattern across many biological pathways, indicating that in different evolutional stage, the genes in GEIs are linked to different biological processes. The results presented here indicate that the integration of gene gain history and expression patterns is valuable.
The diversity of the Burkholderia genus is reflected in the diversity of ecological niches occupied by the different species, ranging from soil to aqueous environments, associations with plants, fungi, amoeba, animals, and human, from saprophytes to endosymbionts and from biocontrol agents to pathogens. Our results suggest that HGT events occurred extensively in the adaptive evolution of Burkholderia. As a majority of these acquired genes encode hypothetical proteins or Burkholderia-specific proteins of unknown function, coexpression analysis of these gene products will be instrumental for a better understanding of their role in adaptation and strains divergence. This analysis also suggests that HGT played an important role in adaption and pathogenicity. Most of the transferred genes are located in the small chromosomes and were gained in ancient time, which has resulted in the major differences between different species. However, gene loss and large chromosome fragment rearrangements are also major causes for the diversity and adaption between these closely related strains.
Supplementary Materials
Acknowledgments
We gratefully acknowledge the support of the National Natural Science Foundation of China (30871655, 30671397), Agricultural Ministry of China (nyhyzx07-056) and 863 project of China (2006 AA10 A211).
Footnotes
Author Contributions
BZ conceived the study. All authors contributed to data collection. BZ, SZ, ML and GJ analyzed the data and prepared the report. All authors provided critical review of the draft and approved the final version.
Disclosures
Author(s) have provided signed confirmations to the publisher of their compliance with all applicable legal and ethical obligations in respect to declaration of conflicts of interest, funding, authorship and contributorship, and compliance with ethical requirements in respect to treatment of human and animal test subjects. If this article contains identifiable human subject(s) author(s) were required to supply signed patient consent prior to publication. Author(s) have confirmed that the published article is unique and not under consideration nor published by any other publication and that they have consent to reproduce any copyrighted material. The peer reviewers declared no conflicts of interest.
References
- 1.Compant S, Nowak J, Coenye T, Clement C, Barka EA. Diversity and occurrence of Burkholderia spp. in the natural environment. FEMS Microbiol Rev. 2008;32:607–26. doi: 10.1111/j.1574-6976.2008.00113.x. [DOI] [PubMed] [Google Scholar]
- 2.Chiarini L, Bevivino A, Dalmastri C, Tabacchioni S, Visca P. Burkholderia cepacia complex species: health hazards and biotechnological potential. Trends Microbiol. 2006;14:277–86. doi: 10.1016/j.tim.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 3.Coenye T, Vandamme P. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ Microbiol. 2003;5:719–29. doi: 10.1046/j.1462-2920.2003.00471.x. [DOI] [PubMed] [Google Scholar]
- 4.Mahenthiralingam E, Urban TA, Goldberg JB. The multifarious, multireplicon Burkholderia cepacia complex. Nat Rev Microbiol. 2005;3:144–56. doi: 10.1038/nrmicro1085. [DOI] [PubMed] [Google Scholar]
- 5.Zhang L, Xie G. Diversity and distribution of Burkholderia cepacia complex in the rhizosphere of rice and maize. FEMS Microbiol Lett. 2007;266:231–5. doi: 10.1111/j.1574-6968.2006.00530.x. [DOI] [PubMed] [Google Scholar]
- 6.Lou MM, Fang Y, Zhang GQ, Xie GL, Zhu B, et al. Diversity of Burkholderia cepacia Complex from the Moso Bamboo (Phyllostachys edulis) Rhizhosphere Soil. Curr Microbiol. 2011;62:650–8. doi: 10.1007/s00284-010-9758-3. [DOI] [PubMed] [Google Scholar]
- 7.Chain PSG, Denef VJ, Konstantinidis KT, Vergez LM, Agullo L, et al. Burkholderia xenovorans LB400 harbors a multi-replicon, 9.73-Mbp genome shaped for versatility. Proc Natl Acad Sci U S A. 2006;103:15280–7. doi: 10.1073/pnas.0606924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schutzer SE, Schlater LRK, Ronning CM, DeShazer D, Luft BJ. Characterization of Clinically-Attenuated Burkholderia mallei by Whole Genome Sequencing: Candidate Strain for Exclusion from Select Agent Lists. PLoS ONE. 2008;3:e2058. doi: 10.1371/journal.pone.0002058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holden MTG, Titball RW, Peacock SJ, Cerdeno-Tarraga AM, Atkins T, et al. Genornic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc Natl Acad Sci U S A. 2004;101:14240–5. doi: 10.1073/pnas.0403302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holden MTG, Seth-Smith H, Crossman LC, Sebaihia M, Bentley SD, et al. The genome of Burkholderia cenocepacia J2315, an epidemic pathogen of cystic fibrosis patients. J Bacteriol. 2009;191:261–77. doi: 10.1128/JB.01230-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doolittle WF. Lateral genomics. Trends Genet. 1999;15:5–8. [PubMed] [Google Scholar]
- 12.Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 13.Ussery DW, Kiil K, Lagesen K, Sicheritz-Pontén T, Bohlin J, et al. The Genus Burkholderia: Analysis of 56 Genomic Sequences. Genome Dyn. 2009;6:140–57. doi: 10.1159/000235768. [DOI] [PubMed] [Google Scholar]
- 14.Li L, Stoeckert CJ, Roos DS. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–89. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–3. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 18.Jones DT, Taylor WR, Thornton JM. A mutation data matrix for transmembrane proteins. FEBS Lett. 1994;339:269–75. doi: 10.1016/0014-5793(94)80429-x. [DOI] [PubMed] [Google Scholar]
- 19.Csürös M, Holey JA, Rogozin IB. In search of lost introns. Bioinformatics. 2007;23:i87–96. doi: 10.1093/bioinformatics/btm190. [DOI] [PubMed] [Google Scholar]
- 20.Csürös M, Rogozin IB, Koonin EV. Extremely intron-rich genes in the alveolate ancestors inferred with a flexible maximum-likelihood approach. Mol Biol Evol. 2008;25:903–11. doi: 10.1093/molbev/msn039. [DOI] [PubMed] [Google Scholar]
- 21.Kettler GC, Martiny AC, Huang K, Zucker J, Coleman ML, et al. Patterns and implications of gene gain and loss in the evolution of Prochlorococcus. PLoS Genet. 2007;3:e231. doi: 10.1371/journal.pgen.0030231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cohen O, Pupko T. Inference and characterization of horizontally transferred gene families using stochastic mapping. Mol Biol Evol. 2010;27:703–13. doi: 10.1093/molbev/msp240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steel M. Recovering a tree from the leaf colourations it generates under a Markov model. Appl Math Lett. 1994;7:19–24. [Google Scholar]
- 24.Winsor GL, Khaira B, Van Rossum T, Lo R, Whiteside MD, et al. The Burkholderia Genome Database: facilitating flexible queries and comparative analyses. Bioinformatics. 2008;24:2803–4. doi: 10.1093/bioinformatics/btn524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tatusov RL, Natale DA, Garkavtsev IV, Tatusova TA, Shankavaram UT, et al. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 2001;29:22–8. doi: 10.1093/nar/29.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brinkac LM, Davidsen T, Beck E, Ganapathy A, Caler E, et al. Pathema: a clade-specific bioinformatics resource center for pathogen research. Nucleic Acids Res. 2010;38:D408–14. doi: 10.1093/nar/gkp850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23:257–8. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- 28.Rodrigues F, Sarkar-Tyson M, Harding SV, Sim SH, Chua HH, et al. Global map of growth-regulated gene expression in Burkholderia pseudomallei, the causative agent of melioidosis. J Bacteriol. 2006;188:8178–88. doi: 10.1128/JB.01006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5 doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karp PD, Ouzounis CA, Moore-Kochlacs C, Goldovsky L, Kaipa P, et al. Expansion of the BioCyc collection of pathway/genome databases to 160 genomes. Nucleic Acids Res. 2005;33:6083–9. doi: 10.1093/nar/gki892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Makarova K, Slesarev A, Wolf Y, Sorokin A, Mirkin B, et al. Comparative genomics of the lactic acid bacteria. Proc Natl Acad Sci U S A. 2006;103:15611–6. doi: 10.1073/pnas.0607117103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore RA, Reckseidler-Zenteno S, Kim H, Nierman W, Yu Y, et al. Contribution of gene loss to the pathogenic evolution of Burkholderia pseudomallei and Burkholderia mallei. Infect Immun. 2004;72:4172–87. doi: 10.1128/IAI.72.7.4172-4187.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adler NRL, Govan B, Cullinane M, Harper M, Adler B, et al. The molecular and cellular basis of pathogenesis in melioidosis: how does Burkholderia pseudomallei cause disease? FEMS Microbiol Rev. 2009;33:1079–99. doi: 10.1111/j.1574-6976.2009.00189.x. [DOI] [PubMed] [Google Scholar]
- 34.Bontemps C, Elliott GN, Simon MF, Dos Reis FBD, Gross E, et al. Burkholderia species are ancient symbionts of legumes. Mol Ecol. 2010;19:44–52. doi: 10.1111/j.1365-294X.2009.04458.x. [DOI] [PubMed] [Google Scholar]
- 35.Brown JR. Ancient horizontal gene transfer. Nat Rev Genet. 2003;4:121–32. doi: 10.1038/nrg1000. [DOI] [PubMed] [Google Scholar]
- 36.Fournier GP, Huang J, Gogarten JP. Horizontal gene transfer from extinct and extant lineages: biological innovation and the coral of life. Philos Trans R Soc Biol Sci. 2009;364:2229–39. doi: 10.1098/rstb.2009.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Juhas M, van der Meer JR, Gaillard M, Harding RM, Hood DW, et al. Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol Rev. 2009;33:376–93. doi: 10.1111/j.1574-6976.2008.00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mantri Y, Williams KP. Islander: a database of integrative islands in prokaryotic genomes, the associated integrases and their DNA site specificities. Nucleic Acids Res. 2004;32:D55–8. doi: 10.1093/nar/gkh059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Merkl R. SIGI: score-based identification of genomic islands. BMC Bioinformatics. 2004;5 doi: 10.1186/1471-2105-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langille MGI, Hsiao WWL, Brinkman FSL. Evaluation of genomic island predictors using a comparative genomics approach. BMC Bioinformatics. 2008;9 doi: 10.1186/1471-2105-9-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nierman WC, DeShazer D, Kim HS, Tettelin H, Nelson KE, et al. Structural flexibility in the Burkholderia mallei genome. Proc Natl Acad Sci U S A. 2004;101:14246–51. doi: 10.1073/pnas.0403306101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.