Abstract
The capsid protein (CA) of the HIV-1 virus plays a significant role in the assembly of the immature virion, and is the critical building block of its mature capsid. Thus, there has been a significant interest in the CA protein as a target in the design of inhibitors of early and late stage events in the HIV-1 virus replication cycle. However, due to its inherent flexibility from the inter-domain linker and the monomer-dimer equilibrium in solution, HIV-1 wild-type CA monomer has defied structural determinations by X-ray crystallography and NMR spectroscopy. Here we report the detailed solution structure of the full length HIV-1 CA using a monomeric mutant that, though non-infective, preserves many of the critical properties of the wild-type protein. The structure shows independently folded N-terminal (NTD) and C-terminal domains (CTD) joined by a flexible linker. The CTD domain shows some differences from that of the dimeric wild-type CTD structures. This study provides insights into the molecular mechanism of the wild-type CA dimerization critical for capsid assembly. The monomeric mutant allows investigation of interactions of CA with human cellular proteins exploited by the HIV-1 virus, directly in solution without the complications associated with the monomer-dimer equilibrium of the wild-type protein. This structure also permits the design of inhibitors directed at a novel target, viz., interdomain flexibility, as well as inhibitors that target multiple interdomain interactions critical for assembly and interactions of CA with host cellular proteins that play significant roles within the replication cycle of the HIV-1 virus.
Retroviruses typically consist of a central capsid core particle encapsidating two copies of RNA and the viral enzymes. The capsids are composed of about 1500 copies of a capsid protein (CA) that is initially part of a Gag polyprotein synthesized in the infected host cell (1,2). The retroviral capsid proteins are typically ~24 to ~27 kDa in size, and are highly α-helical. The Gag proteins capture the viral RNA, assemble either in the cytosol (B and D-type retroviruses) or at the cell membrane (C-type, HTLV/BLV and lentiviruses) and bud into the enveloped immature virus particles (2). Gag is then proteolytically cleaved by the viral protease into the major structural proteins of the virus (2,3), followed by a maturation process in which the capsid proteins condense to form the mature capsid of the virus with a distinct shape characteristic of the genus (3). The HIV-1 capsid is a conical shaped fullerene structure (4).
The capsid protein (CA) of the HIV-1 virus plays a significant role in the early stages of the viral life cycle, controlling the virion size, morphology and Gag assembly (5–7). Electron cryotomography images of the immature virions have shown that along with the spacer SP1, CA domains also play an important role in the formation of the hexameric Gag lattice (5). Most importantly, intermolcular CTD-CTD interactions appear to be important in the assembly of the hexameric Gag lattice (5,7). Electron microscopy studies show that the mature capsid of HIV-1 is as fullerene cone, with its surface composed primarily of hexameric CA rings, with twelve pentameric rings of CA that allow the cone to close at both ends (4). The surface of the mature capsid of HIV-1 is composed of a hexameric (and pentameric) rings of the N-terminal domains (NTD) stabilized by NTD-NTD interactions, with each ring linked to neighboring hexamers through the inter-hexamer dimerization of the C-terminal domains (CTD). Additional intermolecular NTD-CTD and CTD-CTD interactions further stabilize the mature capsid surface lattice (1,3,8,9). Thus, because of the critical role of CA in the assembly of the immature particles and mature capsids, recently there has been a rather significant interest in the CA protein as an antiviral therapeutic target to design inhibitors of early and late stage events in the HIV-1 virus replication cycle (1,4,10–15). Thus the availability of the structure of the full-length HIV-1 CA monomer would be of critical importance for efforts in the structure-based design of inhibitors. Such a monomeric structure will also facilitate a structural biological characterization of the interactions of the HIV-1 capsid protein with host cell proteins exploited by the HIV-1 virus in its replication cycle, such as cyclophilin A and lysysl-tRNA synthetase.
However, HIV-1 wild-type full-length CA monomer protein has defied structural determinations by X-ray crystallography and NMR spectroscopy because of the high degree of flexibility of the inter-domain linker which made it difficult to crystallize, and the monomer-dimer equilibrium in solution which resulted in exchange-broadening and disappearance of many peaks from the CTD domain due its reversible CTD-CTD dimerization. Thus, efforts have focused on the structural determinations by crystallography or NMR spectroscopy of isolated domains, viz., the NTD domain (16,17), the dimeric structures of the isolated wild-type CTD domain (8,18), a domain-swapped CTD dimer (19), as well as the structure of the isolated monomeric mutants of the CTD domain (20,21). For the full-length wild-type CA protein, three crystallographic studies of CA dimers have been reported - - a parallel dimer of CA with NTD stabilized by complexation with Fab but with a disordered CTD (22), and antiparallel head-to-tail dimers of CA stabilized by complexation with Fab (23) and with triiodide (24). These dimer structures of the wt CA exhibit intermolecular CA-CA interactions and associated structural perturbations.
In this work we report the detailed solution structure of the full-length HIV-1 CA protein in a monomer state. To achieve this goal, we have utilized the strategy of making two critical mutations in the CTD domain of the wild-type (wt) CA that are known to disrupt the infectivity as well as the weak face-to-face dimerization in solution (18), viz., W184A/M185A, resulting in a monomeric form of the CA protein. Most importantly, these two mutations preserve many of the critical properties of the wt CA - - (i) unmyristoylated Gag protein containing these two mutations in CA is assembly-active and forms spherical virus-like particles in vitro and in mammalian cells (25), though the particles tend to be somewhat irregular in size and shape. (ii) Similarly, CA proteins with W184A and/or M185A mutations are assembly active though much less efficiently than the wild type. For example, the M185A-CA mutant has been shown to be assembly active but it assembles about 10 times slower than the wild type (26). The quadruple mutant protein A14C/E45C/W184A/M185A-CA polymerizes under reducing conditions (i.e., in the presence of β-mercaptoethanol to prevent spurious cross-linking of cysteines) and forms long tubes in vitro, though somewhat less efficiently than the wild-type (1) (it is only after polymerization that the cysteines were allowed in this study to oxidize so that stable CA-hexamers with correct cross-linking could be isolated for crystallization). The CTD-CTD dimer interface is preserved between these mutant CA hexamers despite the W184A/M185A mutations (1). Following a suggestion that our double mutant might in fact show such assembly activity at high concentrations (private communication, Peter Prevelige, Jr, 2011), we had undertaken in vitro assembly reactions. The data are shown in Figure 1, demonstrating again, and in agreement with the findings of Pornillos et al., (1) on the quadruple mutant, that our double mutant W184A/M185A-CA is indeed assembly active (but at higher concentrations than the wild type) and its assembly rate is much slower than that of the wild type (estimated to be about 28 times slower based on the data at 60 μM). (iii) The secondary structures of the double mutant Gag and the wt Gag (25) as well as of the double mutant CA and the wild-type CA (20) are also identical, as established by CD on very dilute solutions. (iv) Further, mutations at 184 and 185 which inhibit CTD-CTD dimerization in both CA and in Gag do not significantly alter the affinity of CA to host cell binding partners - - e.g., the binding properties of M185A-CA with prolyl isomerase cyclophilin A (27), and that of Gag polyprotein containing the W184A/M185A– CA domain with human lysyl t-RNA synthetase (28) are similar to that of the respective wild-type molecules. Taken together, the results of the above Gag and capsid assembly reactions, CD data, and binding assays with host cell binding partners demonstrate that the monomeric mutant CA, though not infective, retains many of the critical properties of the wild-type CA even though it exists as a monomer in solution. Residues W184 and M185 in HIV-1 CA appear to be important for infectivity (18), dimerization in solution, for improving the efficiency of assembly, and for providing uniformity in the size and shapes of the assembled capsids. However, as demonstrated by others previously (1) and by us in this work (Figure 1), the capsid protein is assembly active though much less efficiently than the wild type protein when these two residues are replaced by alanine. We note that these two residues are not conserved in other retroviruses (19).
Figure 1.

In vitro assembly of the HIV-1 capsid protein : (A) wild-type CA (top) and (B) the double mutant W184A/M185A-CA (bottom) at a final concentration of 2.25 M NaCl. The concentrations of the proteins are indicated on the right side of each panel.
Materials and Methods
To construct the vector pET20bHIV-1CA for expression HIV-1CA with C-terminal His Tag, plasmid WISP98-85 (from Peter Prevelige, Jr) encoding 184A/185A double mutated HIV-1 CA was used as a template. The coding sequence of double mutant HIV-1CA was amplified with PCR to introduce NdeI/SalI cutting sites at 5′ and 3′ ends respectively, and ligated with a NdeI and XhoI cut pET20B (EMD). This new construct, pET20bHIV-1CA (Figure S5 in Supplementary Information) was transformed into E. coli Rosetta2 (DE3) (EMD) for expression. Auto-induction medium P5052N25 was used to produce 15N-HIV-1CA (Supplementary Information). For 15N/13C labeling of HIV-1 CA bacteria were grown in medium P040 (Supplementary Information), with 25mM 15N NH4Cl and 0.4% 13C glucose. Overexpression was induced with 0.8 mM IPTG at OD600nm 1.2–1.8 for 8 hours at 37°C.
For triple labeling, E. coli adapted in 100% D2O were transferred to P040 containing 75% D2O for 2H/15N/13C labeling of HIV-1CA. Protein was induced as above, for 12 hours. Ammonium sulfate precipitation and Q-column absorption were used to minimize DNA contamination in our preparations. Moreover, Co2+-bound HIV-1 CA was intensively washed with IEX buffer (1.0 M NaCl in binding buffer) to remove nonspecific binding via ion-exchange. Binding buffer containing 10% glycerol was used to remove nonspecifically bound molecules via the interaction with HIV-1 CA. The ratio of OD UV280nM/UV260 was determined to be close to 2.0, indicative of minimal nucleic acid contamination. Recombinant proteins were eluted with eluate buffer (50 mM NaCl, 250 mM imidazole, 50 mM Tris pH7.0) to obtain high purity recombinant protein (Figure S6 in Supplementary Information). The resulting protein used for NMR studies consisted of the start codon Met (which is not numbered) at the N-terminus followed by the CA sequence with W184A/M185A mutations, and a C-terminal His-tag.
The NMR sample conditions were as follows - - 1.25 mM protein concentration, pH 5.5, 1.25mM proteins, 25mM NaOAc (99.93%, D4), 25mM NaCl, 1 mM EDTA, 1.5mM DTT (98%, D10), 0.02% NaN3, and 100 nM AEBSF (4-(2-Aminoethyl) benzenesulfonyl fluoride hydrochloride). The H2O samples contained 10% D2O for field lock. For 13C-NOESYand HCCH-TOCSY/COSY, the samples were in 100% D2O. 15N-HSQC spectra at 0.5 mM and 2 mM concentrations (Fig. S7) are virtually identical indicating that there are no dimerization-induced shifts in the CTD domain resonances and that the protein is a monomer.
The following 3D-NMR measurements were performed at 303 K on a Bruker AVANCE 600 MHz NMR system equipped with a TCI-CryoProbe : HNCA (2048 × 40 × 100), HNCO (2048 ×, 40 × 100), HNCACB (2048 × 40 × 100), CBCA(CO)NH (2048 × 40 × 128), HCCH-TOCSY (2048 × 64 × 128), HCCH-COSY (2048 × 64 × 128), 15N-NOESY-HSQC (2048 × 40 × 128, 125 ms), 13C-NOESY-HSQC (2048 × 64 × 128, 175 ms, 250 ms). Data were processed with NMRPipe (29), and analyzed with XEASY (30). Structural calculations were performed using CYANA 2.1 (31). Dihedral restraints were obtained from the HNCACB, HNCA, and HNCO experiments, using TALOS+ (32). Distance restraints were obtained from NOESY data. Hydrogen bond restraints were obtained from the 15N-SOFAST-HMQC identification of slowly exchanging amide protons in H/D exchange experiment.
To identify flexible regions (including the interdomain linker) we have also measured the 15N longitudinal (R1 (s−1)) and transverse relaxation rates (R2(s−1)), and NOEs on a 0.5 mM 15N-labeled monomeric mutant CA sample using standard Bruker TROSY-based pulse programs for relaxation measurements. The delay times for R1 experiment were 10 ms, 50 ms, 150 ms, 250 ms, 500 ms, 1000 ms, 1500 ms and 2500 ms. The delay times for R2 experiment were 12 ms, 36 ms, 48 ms, 60 ms, 72 ms, 84 ms, and 96 ms. The delays were entered in an interleaved manner. The relaxation delays for R1 and R2 measurements were 5 sec each. For 15N NOE measurements, a relaxation delay of 8 sec was used. The relaxation data were fitted to single exponentials using NMRView to compute the R1 and R2 values for each residue.
In vitro CA assembly reactions
Assembly reactions were performed using the turbidity assay as previously described by other laboratories (26). Purified wild type (WT) and the W184A/M185A double mutant (DM) capsid proteins were dialyzed in 10 mM β-mercaptoethanol and 50 mM Tris-HCl at pH 8.0 (buffer D). Proteins were concentrated with Amicon Ultra 10K (Millipore, Billerica, MA) to 1 mM for WT and 2 mM for DM. Various concentrations of proteins were diluted and the volumes of solutions were adjusted with buffer D to 100 μl. Within five minutes of solution preparation, 900 μl of 2.5 M NaCl in buffer D was added to trigger the polymerization (final salt concentration of 2.25 M). Capsid assembly was monitored every minute for one hour at OD 350 nm on a Varian Cary 50 UV-Visible Spectrophotometer controlled by Kinetics software in WinUV package (Agilent, Santa Clara, CA). The delay between adding the salt and the first OD measurement was between 9 and 15 seconds. The in vitro assembly reaction data are shown in Figure 1.
Results and Discussion
The monomeric mutant protein with the critical mutations W184A/M185A was cloned and expressed using the procedures described in the Methods section. The detailed sequence-specific assignments for the backbone nuclei (NH, 15N, 13Cα, αH, 13Cβ, βH, 13CO) were completed by following one-bond and two-bond correlations in 3D-NMR spectra recorded using the methods described above. Figure 2 shows the 15N-HSQC spectrum of monomeric mutant with all the peaks identified. Side chain assignments were completed by a combination of HCCH-TOCSY, HCCH-COSY, 13C-NOESY-HSQC and 15N-NOESY-HSQC measurements on appropriate samples. The distance, torsion angle and hydrogen bond constraints were used to generate the final set of 500 structures, and the 20 lowest energy structures were retained and minimized using CYANA 2.1, until no further reductions in the target function were evident. The Structural Statistics for the 20 best structures are given in Table 1. The sequential and short range NOE connectivities identified in the monomeric mutant CA are shown in Fig. S1 in the Supplementary Information. Representative panels from 3D-NMR data are shown in Figs. S2 and S3.
Figure 2.
15N-HSQC spectrum of the HIV-1 monomeric mutant CA showing the assignments for the non-proline residues. The inset shows the assignments in the crowded region. The NMR assignments for the monomeric mutant HIV-1 CA protein are consistent with that of the isolated NTD (16) and CTD domains (20) and with the solid state NMR studies of capsid assemblies (47,48).
Table 1.
NMR and refinement statistics for the W184A/M185A-CA structures
|
| |
| NMR distance and dihedral constraints | |
| Distance constraints | |
| Total NOE | 3052 |
| Intra-residue | 1005 |
| Inter-residue | |
| Sequential (|i – j| = 1) | 907 |
| Medium-range (|i – j| < 4) | 720 |
| Long-range (|i – j| > 5) | 420 |
| Hydrogen bonds | 190 |
| Total dihedral angle restraints | |
| θ | 192 |
| ψ | 193 |
| ω | 1 |
| Structure statistics | |
| Violations (mean and s.d.) | |
| Distance constraints (>0.1Å) | 4 (+/− 3) |
| Dihedral angle constraints (>5.0°) | 0 |
| Max. dihedral angle violation (°) | 2.70 (+/− 0.10) |
| Max. distance constraint violation (Å) | 0.16 (+/− 0.05) |
| Average Target function | 1.93 |
| Average pairwise r.m.s. deviation** (Å) | |
| NTD/CTD domains1 | |
| Heavy | 1.84 (+/−0.21), 1.53 (+/− 0.24) |
| Backbone | 1.57 (+/− 0.24), 1.01 +/− 0.22) |
| NTD/CTD cores2 | |
| Heavy | 1.17 (+/−0.19), 1.06 (+/− 0.16) |
| Backbone | 0.71 (+/− 0.20), 0.53 (+/− 0.16) |
| Comparison of HIV1 W184A/M185A-CA with other similar structures (Å) | ||||
|---|---|---|---|---|
| NTD3 | NTD (all helices) | CTD4 | CTD (all helices)5 | |
| HIV1-CA NTD (NMR (16)) | 1.58 | 1.75 | - | |
| HIV1-CA CTD (NMR (20)) | - | - | 1.17 | 1.27 |
| HIV1-CA (Crystal (24)) | 1.81 | 1.77 | 1.69 | 1.83 |
| HIV1-CA NTD (Crystal(17)) | 1.85 | 1.82 | ||
| HIV1-CA CTD (2KOD(8)) | 2.19 | 2.21 | ||
| HIV1-CA CTD (2JO0 (21)) | 6.53 | 7.01 | ||
500 structures were calculated within CYANA 2.1: Pairwise r.m.s. deviation was calculated among 20 refined structures.
Domains are defined as residues 1–144 for the NTD, and 150–221 for the CTD.
Cores are defined as the major helices for the NTD (helices 1, 2, 3, 4, 7) and CTD (helices 9, 10, 11, 12).
Superposition of helices 1, 2, 3, 4, and 7
Superposition of helices 9, 10, 11,12
Superposition of all helices, including the 310 helix (helix 8)
Ramachandran analysis:
| Residues in favored regions (%) | 84.5 |
| Residues in additional allowed regions (%) | 14.5 |
| Residues in generously allowed regions (%) | 0.8 |
| Residues in disallowed regions (%) | 0.1 |
Solution Structure of the Full-length Monomeric Mutant CA
The individual NTD and CTD domains in the family of 20 NMR structures are shown in Fig. 3 while Fig. 4 shows a representative structure for the full-length protein from this family. Figures S4a and S4b in the Supplementary Information show a superposition of the twenty full-length protein structures, with the NTD aligned and CTD aligned, respectively, in separate panels. The NMR structure exhibits the distinct independently folded NTD and CTD domains joined by a highly flexible five-residue linker. No interdomain NOE contacts could be unequivocally established in the NOESY data. The highly flexible nature of the interdomain linker can be judged from the spatial distribution of domains that are compatible with the NMR data in Figures S4a and S4b, as well as from the 15N relaxation data in Figure 5. This figure also identifies other flexible regions such as the loops between helices, and the C-terminal tail following residue 220. The NTD domain consists of seven distinct helices spanning residues 17–29, 36–43, 51–57, 63–83, 101–104, 111–118, and 126–144. A short β-hairpin structure (16) involving residues 2 to 12 at the N-terminal has also been identified (Fig. 4). Proline 122 has been identified to be a cis-proline (16) by cisprocheck based on its unique 13Cβ and 13Cγ chemical shifts. The CTD domain in the full-length monomeric mutant CA consists of a short 310 helix (helix-8) formed by residues 150–152 followed by four helices (9 to 12) spanning residues 161–175, 185–193, 196–205, and 211–220 (note: some of the references in literature did not label the 310 helix as helix-8, and so they report only helices 8 to 11 in the CTD domain which correspond to 9 to 12 in our numbering scheme).
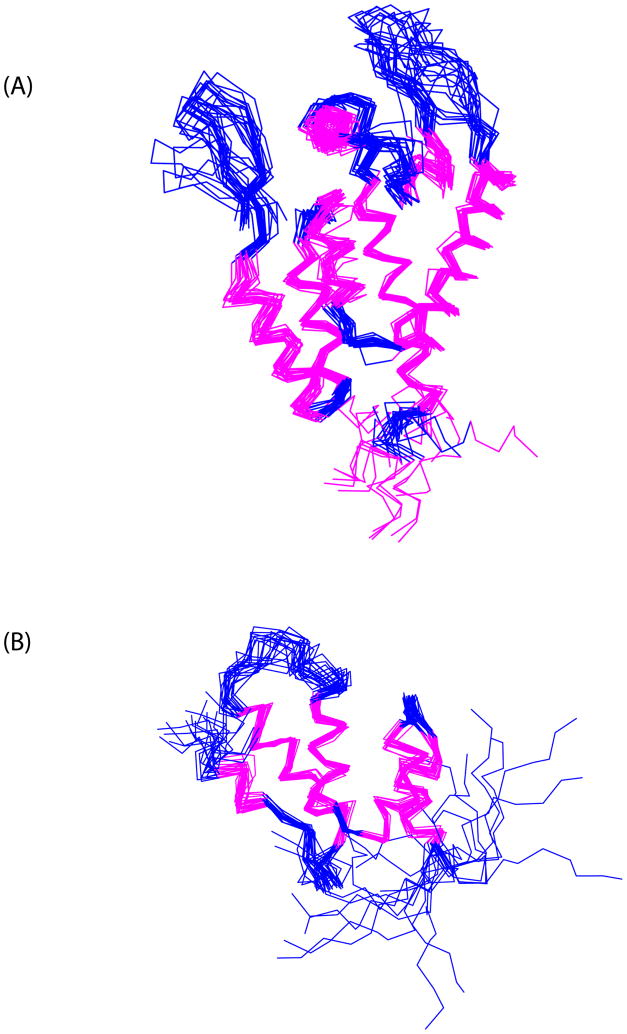
Figure 3.
Superposition of the NTD and CTD domains of HIV-1 CA monomeric mutant from the 20 best structures. (a) Superposition of the NTD domains. The helices are in magenta color. The structures also include the flexible linker (residues 144–147) at its C-terminus, as well as residues 148 to 151 constituting the 310 helix of the CTD domain, to show the highly disordered nature of the flexible linker. Figure S4a in the Supplementary Information shows the distribution of the CTD domains when the NTD domains are aligned. A comparison of the monomeric mutant CA’s NTD domain with that of the isolated NTD domain NMR structure (16) shows an RMSD of 1.58 Ǻ for the alignment of the five major helices (H1, H2, H3, H4 and H7). (b) Superposition of the CTD domains from the 20 best NMR structures. Figure S4b in the Supplementary Information shows the distribution of the NTD domains when the CTD domains are aligned. A comparison of the monomeric mutant CA’s CTD domain with that of the isolated CTD domain NMR structure (20) shows an RMSD of 1.17 Ǻ for the alignment of the four helices (H9, H10, H11 and H12). The structural statistics are contained in Table 1.
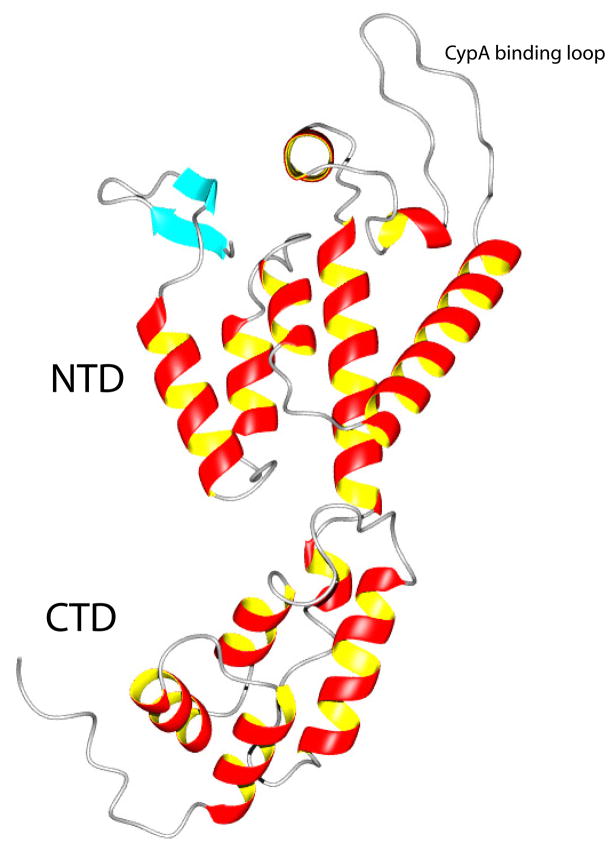
Figure 4.
A representative member of the NMR structures of the full-length monomeric mutant CA as a ribbon diagram (MolMol). The secondary structure consists of a short β-hairpin (aqua blue color), with helices 1 to 7 in the NTD domain at the top and helices 8 to 12 (including the short 310 - helix as helix 8) in the CTD domain at the bottom. Even though our construct has the start codon residue Met (unassigned) prior to Pro1, the general arrangement of the β-hairpin is very similar to that seen in the isolated NTD structure (16). The linker between the NTD and CTD domains is five residues long (residues 145 to 149 corresponding to YSPTS) and highly flexible in solution (see Figure 5). In the crystal structure of the antiparallel dimer of wt CA, it is two residues long (SP) (24). The cyclophilin A (CypA) binding loop (16) is located between helices 4 and 5 at upper right.
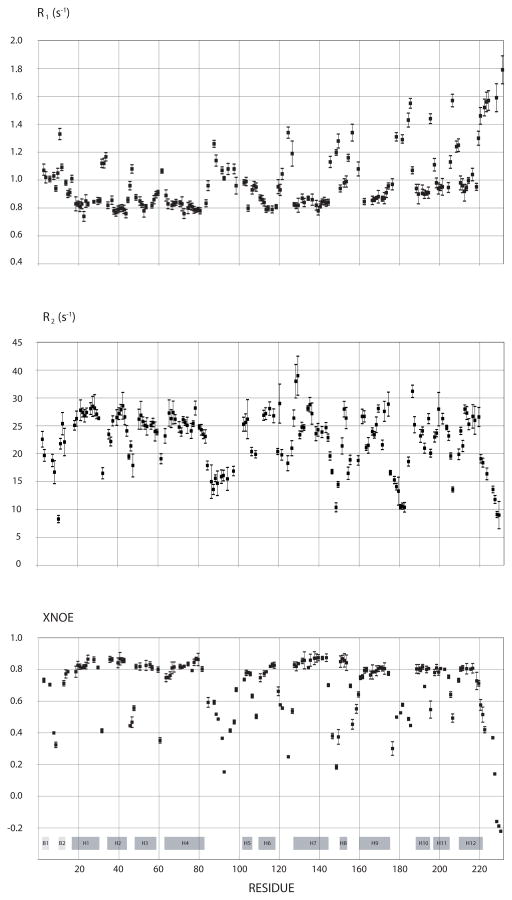
Figure 5.
15N- longitudinal (R1) and transverse (R2) relaxation rates and NOEs for the W184A/M185A-CA measured at 600 MHz. A total of 158 peaks without overlap were selected from the 2D-NMR spectra, and their relaxation data were fitted to single exponentials. The locations of the secondary structures are identified at the bottom, with B standing for beta strand and H for helices. Helix-8 refers to the short 310 helix. The flexible interdomain linker is located between H7 and H8. Residues 220 and 221 in helix-12 exhibited biexponential behavior and as a result show larger error bars when fitted to a single exponential. The error bars in R1 and R2 refer to the deviations of data points with the single exponential fitted curve. For the NOEs, the error bars refer to the noise in the spectrum.
Table 1 shows the RMSD values for the best-fit alignment of the individual NTD and CTD domains with previously published crystal and NMR structures of isolated domains, as well as with the domains in the recent crystal structure of the triiodide-stabilized antiparallel dimer of the full-length wild type CA. The NTD domain in the full length monomeric mutant CA shows excellent agreement with the NMR structure of the isolated NTD domain (16) with an RMSD of 1.58 Å. Similarly, the CTD domain of the monomeric mutant CA shows excellent agreement with the NMR structure of the isolated W184A/M185A mutant of the CTD domain (20) with an RMSD of 1.17 Å. Figure 6 shows a comparison of the CTD domains of the monomeric mutant CA with those from the wt CA in the antiparallel dimer crystal structure (24) and the wt CTD-dimer NMR structure (8). Most noteworthy is the difference in helix-10 between these structures. In the crystal structure and the CTD-dimer NMR structure (8), helix-10 is longer and shows a bend, whereas in the monomeric mutant it is shorter and straight.
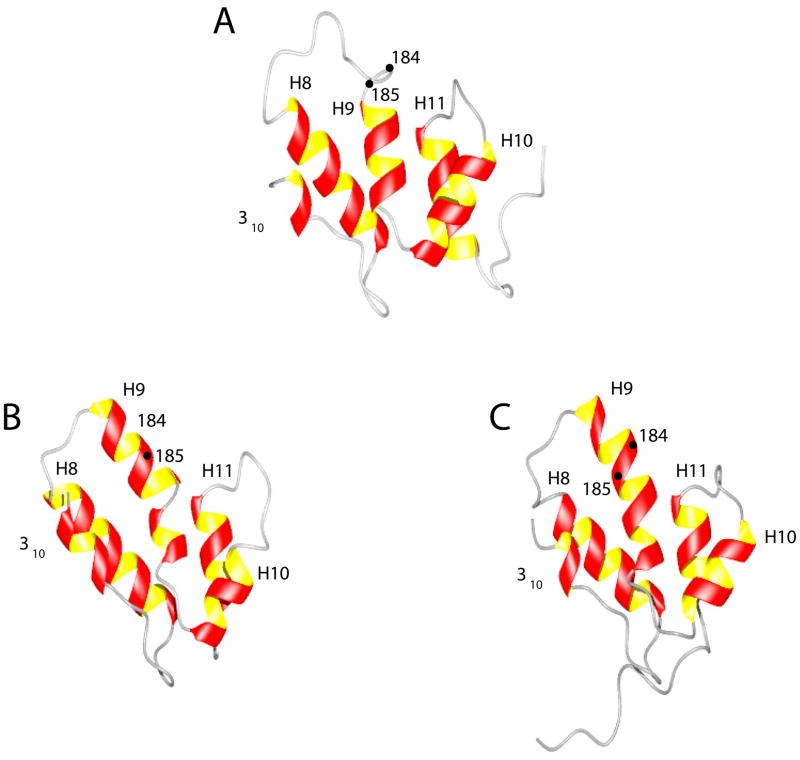
Figure 6.
Comparison of the CTD domains: (A) The CTD domain of the W184A/M185A-CA in this work. (B) The CTD domain from the wild-type antiparallel dimer CA crystal structure (24). (C) The CTD domain from the wild type CTD dimer NMR structure (8). The location of residues 184 and 185 is also shown. In the crystal structure and in the wt CTD-dimer NMR structure, helix-10 is longer and shows a bend. The hydrophobic groove formed by helices 9 and 10 has been a target for inhibitors such as CAI (14) and NYAD-13 (15). The CTD-CTD dimerization interface involving helix-10 has been targeted by inhibitors of dimerization (12,38). Further, helix-12 in the CTD domain has been suggested to be the binding site for human lysyl t-RNA synthetase (28) which is thought to play a critical role in targeting tRNALys for viral packaging.
The NTD and CTD domains are connected by a five-residue linker formed by residues 145 to 149 corresponding to the sequence YSPTS. Of these, Y145 and S149 are the partially unwound terminal residues of helices 7 and 8 (the 310-helix), respectively, on either side of the linker. The linker in the NMR structure is highly flexible, as evidenced by the random coil chemical shifts for the the central three residues, a lack of sequential NH(i)-NH(i+1) connectivities, and the 15N relaxation data (Figure 5). In contrast, in the triiodide-stabilized antiparallel dimer crystal structure of the wt CA, the linker is only two residues long (146 and 147 corresponding to SP). The flexibility of the linker is thought to have important consequences on the capsid shape and morphology of the mature capsids (vide infra).
Insight into the Detailed Molecular Mechanism of CA-CTD Dimerization During Capsid Assembly and the Critical Role of W184 and M185 Residues
Our structure of the monomeric mutant CA displays a helix-10 that is straight and shorter (residues 185–193) than that seen in crystallographic and NMR structures of wt CTD-dimers (8,18). Based on this observation, we can speculate the following detailed molecular mechanism for the dimerization of the wt CA during the assembly of the capsid - - we propose that just like the monomeric mutant CA, the wt CA in a monomeric state also exhibits a straight and short helix-10 with residues 179 to 184 forming part of the loop between helices 9 and 10. This is reasonable since the circular dichroism data are virtually identical for the wt CA and the monomeric mutant CA (20). During the assembly process, two wt CA molecules that diffuse toward each other may form an initial encounter complex wherein the helix-10 still is short as in the monomer but the complex involves a loose association of hydrophobic surfaces on each CTD. This initial encounter complex then transitions to a bound complex where the hydrophobic-residue-rich surfaces involving helix-10 and the 310-helix form a hydrophobic pocket with helix-10 experiencing a conformational switch involving an elongation at its N-terminus with residues 179 to 185 switching to a helical conformation. This seems reasonable since residues in this region such as Q, E, V, K, W and M gain in their helical propensity over A in a hydrophobic environment (33). We further suggest that this helix-10 elongation is accompanied by a kink in the helix to optimally accommodate the packing of residues across the hydrophobic dimer interface in the bound complex. The hydrophobic interactions identified in the wt CTD-dimer NMR structure (8) include residues Y145, T148, L151, V181, W184, M185 AND V191. Among these, W184 and M185 from each CTD participate in a pair of critical intermolecular W/M aromatic ring/methyl group hydrophobic interactions. With these additional intermolecular interactions in place to stabilize the dimeric interface, the wt CTD-dimer has much less variability in the helix-10 crossing angle across the CTD-CTD dimeric interface, thus resulting in more uniform formation of mature capsids. The self-association affinity of the wt CA with a Kd 18 μM (18) also facilitates the formation of encounter complexes thus leading higher efficiency in assembly. Mutations such as W184A and M185A (where A has a shorter side chain compared to W and M) results in a loss of some of these critical intermolecular hydrophobic interactions needed to stabilize the dimer interface in the bound complex under assembly conditions, thus leading to a wider variability in the helix-10 crossing angles for each CTD-CTD dimeric interface which in turn might contribute to the assembly of abnormal or defective capsids. Further, these mutations also significantly decrease the self-association affinity (18) there by leading to fewer encounter complexes. This in turn can result in inefficient assembly. Thus the loss of infectivity of HIV-1 CA due to W184A and M185A mutations (34) might be the combined result of both the above factors, viz., a decreased CTD dimerization and formation of defective capsids. Molecular dynamics simulations (35) show that W184A mutation is lot more detrimental to the stability of the dimeric interface than M185A mutation. We also note that W184 and M185 are unique for HIV-1 and are not conserved in other retroviruses (e.g., RSV and HTLV); as such, the fidelity of CTD-CTD dimerization during assembly of these other retroviruses is likely to be governed by other residues in the dimeric interface.
Because of the helical propensity (33) of residues 179–185 near its N-terminus (indeed, in the monomeric mutant we see a few weak sequential NH(i)-NH (i+1) contacts among these residues (Fig. S1 in Supporting Information)), helix-10 is dynamic and exhibits a high degree of plasticity (1,8,20,21) with these residues easily switching to helical conformations in response to hydrophobic associations. This plasticity can be easily appreciated in the crystal structures (PDB code 3H4E) of the A14C/E45C/W184A/M185A-CA hexamers (1) in the hexamer sheet where helix-10 exhibits considerable variations among the different CAs in the hexameric rings, ranging from an elongated 3 ½-turn bent helix (residues 179 to 189) to a 1- turn short helix (residues 189 to 192), presumably reflecting variations in the contacts with the neighboring hexamers in the lattice. Interestingly, the Ccmk4-templated CA with W184A/M185A mutations (PDB Code 3GV2) exhibits longer helix with a kink (despite the W184A/M185A mutations) in all the CAs in the hexamer sheet - - we speculate that this might be due to the Ccmk4 protein (to which the CA (1–226) is attached through the C-terminus of CTD via a 2-residue linker) which forms rigid hexamers itself (1), thus stabilizing the CTD domain orientations in the interhexamer interactions, resulting in a greater hydrphobic contact surface between the CTD-CTD pairs. This suggestion is supported by the observation that the elongation and bend in helix-10 is also seen in a monomeric-CTD/peptide complex (15) where the peptide is found to associate with helices 9 and 10 together.
Figure 6 shows a comparison of the CTD domains from the monomeric mutant CA NMR structure with that in the triiodide-stabilized antiparallel dimer crystal structure (24) and the wt CTD-dimer NMR structure (8). It is easily seen that helix-10 is longer and shows a bend (kink) in the antiparallel dimer crystal structure (24) presumably due to its hydyphobic intermolecular contact with helices-1and 2 in the NTD domain from the second CA molecule (Figure 6b). Similarly, this helix is also longer and bent in the wt CTD-dimer NMR structure (Figure 6C). Our above proposed molecular mechanism for CTD dimerization, viz., that the elongation accompanied by a kink for helix-10 in the wt CTD might be the result of a hydrophobic environment from the association of this helix with an interacting partner molecule, is supported by a number of recent studies in which helix-10 generally exhibits elongation and bending (or kink) in the wt CTD dimer in solution (8), in wt CTD-peptide complexes (14), as well as in crystal structures of the full-length wt CA head-to-tail antiparallel dimers where the helix-10 in the CA is interacting with a helix from the second CA (23,24), and in the crystal structures of hexameric assemblies where the wt CTD-CTD interface has been observed with sufficient resolution (1,3,8).
Implications for New Directions in the Design of Inhibitors
Previous attempts at structure-based design of CA inhibitors were based primarily on the structures of the isolated NTD and CTD domains because the full-length HIV-1 CA monomer structure was not available. Accordingly these efforts focused on developing inhibitors of interdomain interactions. Our structural determination of the full-length monomer as well as the availability of chemical shifts will facilitate inhibitor design for a totally new target, viz., interdomain flexibility, since such interdomain flexibility is a critical requirement for the productive assembly of fullerene cone shaped mature capsids (vide infra). For example, (i) it may be possible to identify or design small molecules that tightly bind directly to the flexible linker area and minimize the degree of interdomain flexibility. (ii) Alternately, by fragment-based design of high-affinity inhibitors by SAR-by-NMR (36) one can design inhibitors that simultaneously bind to both NTD and CTD domains (i.e., with one functional group binding to NTD and the second functional group binding to CTD), thereby tethering the CTD domain to the NTD, and significantly diminish interdomain flexibility. This approach might indeed be feasible because the free monomeric mutant exhibits considerable flexibility across the interdomain linker (Fig. 5 and Figs. S4a and S4b). Both the above approaches will benefit from a structural study of the complexes of the inhibitors (and initial leads) with the full-length monomeric mutant CA. Such a reduction in interdomain flexibility is likely to have profound detrimental effects on the assembly of mature capsids and immature virions. Thus our structural study in this work provides a firm foundation for undertaking these new directions that target the interdomain flexibility.
Additionally, using fragment based design of high affinity inhibitors by SAR-by-NMR, it may be feasible to simultaneously inhibit two or more separate interactions such as the intermolecular interdomain interactions (i.e., NTD-NTD, NTD-CTD, CTD-CTD) that play a role during the assembly of the mature capsid as well as the interactions of CA with host cell proteins such as cyclophilin A and lysyl tRNA synthetase. With NMR assignments and structure available from this study, they may facilitate a combined STD-NMR/CORCEMA-based (37) development of new leads for inhibitors.
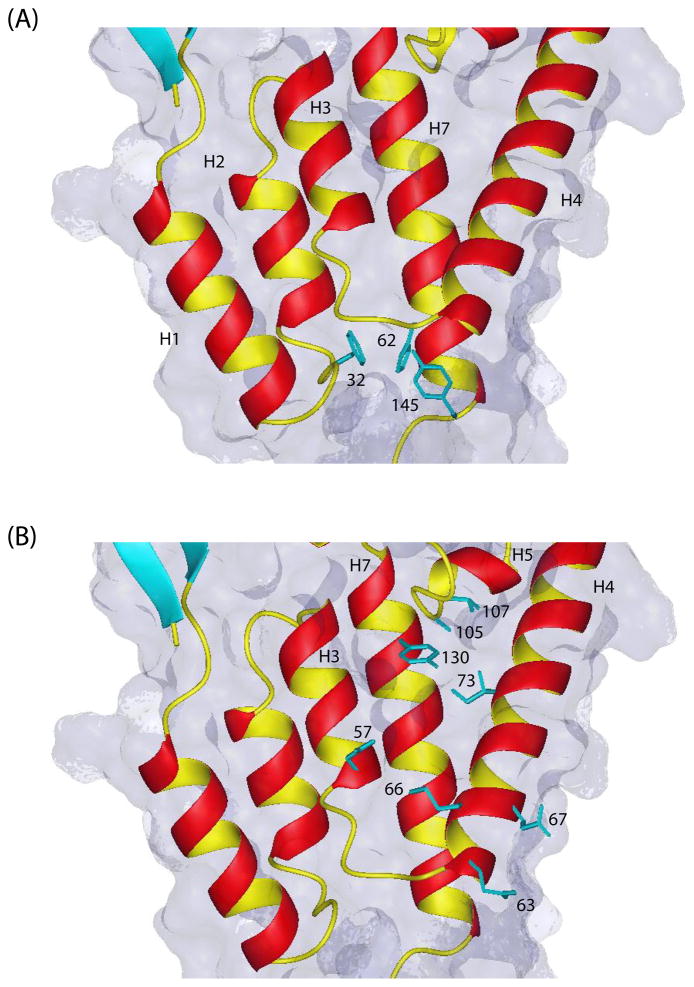
Previous work from other laboratories have identified the assembly inhibitor CAP-1 that binds to the NTD domain (13). It is thought CAP-1 functions by inhibiting the intermolecular NTD-CTD interactions during the assembly of the mature capsid. The binding site for CAP-1 is at the bottom end of the hydrophobic cavity formed by helices 1, 2, 4 and 7 in the NTD domain (Fig. 7a). In our NMR structures, though there is some distribution among their side chain orientations, the aromatic rings of Phe32, His62 and Tyr145 consistently tend to point inward toward the bottom of the hydrophobic cavity formed by these four helices (Figure 7a). A second inhibitor PF-3450074 (10) also has been identified that binds to the NTD domain at a different site, viz., to a hydrophobic cavity formed by helices 3, 4, 5 and 7 (Fig. 7b), and affects both early and late stage events in the HIV-1 replication cycle binds. It thus appears to act from a mechanism different from that of CAP-1.
Figure 7.
(a) Close-up view of the hydrophobic binding pocket on the NTD domain between helices 1, 2, 4 and 7 for the assembly inhibitor CAP-1 (13). Though there is some distribution in sidechain orientations among the 20 structures, the aromatic rings of Phe32, His62, and Tyr145 tend to point inwards toward the bottom of the cavity. The binding of the inhibitor is accompanied by a displacement of these aromatic sidechains. (b) The binding pocket for the antiviral compound PF-3450074 formed by helices 3, 4, 5 and 7 on the NTD domain (10). Some of the binding pocket residues involved in interactions with the inhibitor are also shown (from the crystal structure of the inhibitor with a mutant NTD (10)). This inhibitor targets both early and late events in the replication cycle. The cyclophilin-dependent mechanism of antiviral activity of PF-3450074 is different from that of CAP-1.
On the CTD domain, the hydrophobic pocket constituted by helix-10 and its neighboring helix-9 (Fig. 6) have been the focus of assembly inhibitors. These inhibitors include CAI (14), and the cell-penetrating hydrocarbon-stapled peptide NYAD-13 and its analogs (15). Interestingly, even though both these peptides bind essentially in the same site in the nonpolar groove involving helix-10 (that contributes to CTD-CTD dimerization), they do not appear to seriously alter the monomer-dimer equilibrium of the CTD domain in solution. Thus their ability to inhibit the formation of both immature and mature particles is thought to be due to the formati on of non-functional dimers upon peptide binding (14,15). The CTD-CTD dimerization interface involving helix-10 has also been targeted by designing peptide-based and dendrimer-based inhibitors that directly interfere with dimerization (12,38).
Implications for Studying Interactions with Host Cell Binding Partners
The HIV-1 virus exploits a large number of host cell proteins in its replication cycle (39, 40). For example, the capsid protein binds to human cyclophilin A (CypA) (41) and to lysyl-tRNA synthetase (28). The precise role of human CypA within HIV-1 life cycle remains poorly understood though it has been known for some time that CypA is required for the optimal infectivity of the virus and for viral replication (41,42). The binding site for cyclophilin A (41) is located in the loop between helices 4 and 5 on the NTD domain (Fig. 4). Even though the crystal structure for the NTD/CypA complex has been published (41), the structure of CypA complex with full-length CA is not yet available. Such a structural study with full-length CA is important to address the CypA effects on the CTD domain (43). Thus the monomeric mutant CA study described here will serve as a basis for the study of the CA/CypA complex in solution to probe the effects of CypA binding on the CTD domain under different solvent conditions. Human lysyl-tRNA synthetase (LysRS) is thought to play a critical role in targeting tRNALys for viral packaging (28). For lysyl-tRNA synthetase, helix-12 on the CTD domain has been suggested (28) as the binding site (Figure 6). Thus, these capsid-protein/human cellular protein interactions are also attractive targets for inhibitor design. The restriction factor rhTrim5α from rhesus macaque monkeys restricts HIV-1 infection by a mechanism that was thought to involve binding to the conical capsid and cause premature uncoating (44). Thus the design of rhTrim5α-mimetics that bind to HIV-1 CA is an attractive strategy in drug design (44). With the availability of the NMR structure of a full-length monomeric mutant CA, it is now possible to study the interaction of CA with these host cellular proteins directly in solution by NMR without the complications associated with monomer-dimer equilibrium (it is anticipated that crystallization of these complexes for X-ray crystallographic studies could still likely pose a problem due to the flexibility of the interdomain linker). The ability, provided by our NMR data and structure, to develop high affinity inhibitors by SAR-by-NMR (36) that simultaneously inhibit two or more of the above events within the HIV-1 replication cycle including various inter-domain interactions during the assembly of both immature virions and mature capsids, as well as interdomain flexibility, and interactions with host cell proteins exploited by the virus, adds yet another exciting dimension to future efforts in the development of antivirals for HIV-1.
Capsid Shapes and Polymorphism
Retroviral capsids such as HIV-1 and RSV exhibit considerable polymorphism in shape, and it has been suggested that this polymorphism might be a result of the flexible linker (1,45) which permits some degree of play in the relative orientations of two domains across the NTD-CTD and CTD-CTD interdomain interfaces during the assembly. The curvature of the fullerene cone-shaped HIV-1 capsid has been proposed (1) to result from CA hexamers consisting of a ring of NTD domains surrounded by a mobile belt of CTD domains (with mobility arising from the flexible linker) that interact through CTD-CTD dimerization with neighboring hexamers. Our NMR structural and relaxation data lend strong support to this notion, viz., a highly flexible linker can lead to variations in the various CA-CA interfaces that contribute to the variable curvature of the conical shaped mature HIV-1 capsid as well as the polymorphism in mature capsid shapes (1,45).
Supplementary Material
Acknowledgments
The authors thank Prof. Peter Prevelige, Jr., for the clone of the non-His-tag double mutant CA, and for discussions about the assembly properties of the wild type and mutant CA proteins of the HIV-1 virus, and Dr. Clemens Anklin of Bruker Biospin Inc for the SOFAST-HMQC sequence.
Abbreviations
- HIV-1
Human immunodeficiency virus-1
- CA
capsid protein
- NTD
N-terminal domain
- CTD
C-terminal domain
Footnotes
Support of this work by the NIAID Grants 1R21AI081591, 3R21AI081591-02S1, the NCI Grant 1P30 CA13148 that supported the NMR Facility, and the NCRR grant 1S10RR021064-01A1 for the 600 MHz CryoProbe is gratefully acknowledged.
Data Deposition: The coordinates for the family of 20 lowest energy structures (Figure 3) including the representative NMR structure (Figure 4) of the of the HIV-1 monomeric mutant CA have been deposited with the RCSB Protein Data Bank (PDB Code 2LF4). The detailed chemical shifts for the W184A/M185A-CA have been deposited with the BMRB bank at the University of Wisconsin-Madison (Accession Code 17738), and have been reported elsewhere (46).
Supporting Information: A description of the media used in protein expression, and Figures S1 to S7 are contained in the Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Pornillos O, Ganser-Pornillos BK, Kelly BN, Hua Y, Whitby FG, Stout CD, Sundquist WI, Hill CP, Yeager M. X-ray structures of the hexameric building block of the HIV capsid. Cell. 2009;137:1282–1292. doi: 10.1016/j.cell.2009.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campos-Olivas R, Newman JL, Summers MF. Solution structure and dynamics of the Rous sarcoma virus capsid protein and comparison with capsid proteins of other retroviruses. J Mol Biol. 2000;296:633–649. doi: 10.1006/jmbi.1999.3475. [DOI] [PubMed] [Google Scholar]
- 3.Ganser-Pornillos BK, Cheng A, Yeager M. Structure of full-length HIV-1CA: a model for the mature capsid lattice. Cell. 2007;131:70–79. doi: 10.1016/j.cell.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 4.Sundquist WI, Hill CP. How to assemble a capsid. Cell. 2007;131:17–19. doi: 10.1016/j.cell.2007.09.028. [DOI] [PubMed] [Google Scholar]
- 5.Wright ER, Schooler JB, Ding HJ, Kieffer C, Fillmore C, Sundquist WI, Jensen GJ. Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J. 2007;26:2218–2226. doi: 10.1038/sj.emboj.7601664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ako-Adjei D, Johnson MC, Vogt VM. The retroviral capsid domain dictates virion size, morphology, and coassembly of gag into virus-like particles. J Virol. 2005;79:13463–13472. doi: 10.1128/JVI.79.21.13463-13472.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Briggs JA, Riche JD, Glass B, Bartonov V, Zanetti G, Kräusslich HG. Structure and assembly of immature HIV. Proc Nat’l Acad Sci U S A. 2009;106:11090–11095. doi: 10.1073/pnas.0903535106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Byeon IJ, Meng X, Jung J, Zhao G, Yang R, Ahn J, Shi J, Concel J, Aiken C, Zhang P, Gronenborn AM. Structural convergence between Cryo-EM and NMR reveals intersubunit interactions critical for HIV-1 capsid function. Cell. 2009;139:780–790. doi: 10.1016/j.cell.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lanman J, Lam TT, Barnes S, Sakalian M, Emmett MR, Marshall AG, Prevelige PE., Jr Identification of novel interactions in HIV-1 capsid protein assembly by, high-resolution mass spectrometry. J Mol Biol. 2003;325:759–772. doi: 10.1016/s0022-2836(02)01245-7. [DOI] [PubMed] [Google Scholar]
- 10.Blair WS, Pickford C, Irving SL, Brown DG, Anderson M, Bazin R, Cao J, Ciaramella G, Isaacson J, Jackson L, Hunt R, Kjerrstrom A, Nieman JA, Patick AK, Perros M, Scott AD, Whitby K, Wu H, Butler SL. HIV capsid is a tractable target for small molecule therapeutic intervention. PLoS Pathog. 2010;6:1–10. doi: 10.1371/journal.ppat.1001220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adamson CS, Freed EO. Novel approaches to inhibiting HIV-1 replication. Antiviral Res. 2010;85:119–141. doi: 10.1016/j.antiviral.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neira JL. The capsid protein of human immunodeficiency virus: designing inhibitors of capsid assembly. FEBS J. 2009;276:6110–6117. doi: 10.1111/j.1742-4658.2009.07314.x. [DOI] [PubMed] [Google Scholar]
- 13.Kelly BN, Kyere S, Kinde I, Tang C, Howard BR, Robinson H, Sundquist WI, Summers MF, Hill CP. Structure of the antiviral assembly inhibitor CAP-1 complex with the HIV-1 CA protein. J Mol Biol. 2007;373:355–366. doi: 10.1016/j.jmb.2007.07.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ternois F, Sticht J, Duquerroy S, Kräusslich HG, Rey FA. The HIV-1 capsid protein C-terminal domain in complex with a virus assembly inhibitor. Nat Struct Mol Biol. 2005;12:678–682. doi: 10.1038/nsmb967. [DOI] [PubMed] [Google Scholar]
- 15.Bhattacharya S, Zhang H, Debnath AK, Cowburn D. Solution structure of ahydrocarbon stapled peptide inhibitor in complex with monomeric C-terminal domainof HIV-1 capsid. J Biol Chem. 2008;283:16274–16278. doi: 10.1074/jbc.C800048200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gitti RK, Lee BM, Walker J, Summers MF, Yoo S, Sundquist WI. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science. 1996;273:231–235. doi: 10.1126/science.273.5272.231. [DOI] [PubMed] [Google Scholar]
- 17.Kelly BN, Howard BR, Wang H, Robinson H, Sundquist WI, Hill CP. Implications for viral capsid assembly from crystal structures of HIV-1 Gag(1–278) and CA(N)(133–278) Biochemistry. 2006;45:11257–11266. doi: 10.1021/bi060927x. [DOI] [PubMed] [Google Scholar]
- 18.Gamble TR, Yoo S, Vajdos FF, von Schwedler UK, Worthylake DK, Wang H, McCutcheon JP, Sundquist WI, Hill CP. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science. 1997;278:849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- 19.Ivanov D, Tsodikov OV, Kasanov J, Ellenberger T, Wagner G, Collins T. Domain-swapped dimerization of the HIV-1 capsid C-terminal Domain. Proc Nat’l Acad Sci USA. 2007;104:4353–4358. doi: 10.1073/pnas.0609477104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong HC, Shin R, Krishna NR. Solution structure of a double mutant of the carboxy-terminal dimerization domain of the HIV-1 capsid protein. Biochemistry. 2008;47:2289–2297. doi: 10.1021/bi7022128. [DOI] [PubMed] [Google Scholar]
- 21.Alcaraz LA, del Alamo M, Barrera FN, Mateu MG, Neira JL. Flexibility in HIV-1 assembly subunits: solution structure of the monomeric C- terminal domain of the capsid protein. Biophys J. 2007;93:1264–1276. doi: 10.1529/biophysj.106.101089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Momany C, Kovari LC, Prongay AJ, Keller W, Gitti RK, Lee BM, Gorbalenya AE, Tong L, McClure J, Ehrlich LS, Summers MF, Carter C, Rossmann MG. Crystal structure of dimeric HIV-1 capsid protein. Nat Struct Biol. 1996;3:763–770. doi: 10.1038/nsb0996-763. [DOI] [PubMed] [Google Scholar]
- 23.Berthet-Colominas C, Monaco S, Novelli A, Sibaï G, Mallet F, Cusack S. Head-to-tail dimers and interdomain flexibility revealed by the crystal structure of HIV-1 capsid protein (p24) complexed with a monoclonal antibody Fab. The EMBO J. 1999;18:1124–1136. doi: 10.1093/emboj/18.5.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du S, Betts L, Yang R, Shi H, Concel J, Ahn J, Aiken C, Zhang P, Yeh JI. Structure of the HIV-1 full-length capsid protein in a conformationally trapped unassembled state induced by small-molecule binding. J Mol Biol. 2011;406:371–386. doi: 10.1016/j.jmb.2010.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Datta SA, Curtis JE, Ratcliff W, Clark PK, Crist RM, Lebowitz J, Krueger S, Rein A. Conformation of HIV-1 Gag Protein in Solution. J Mol Biol. 2007;365:812–824. doi: 10.1016/j.jmb.2006.10.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lanman J, Sexton J, Sakalian M, Prevelige PE., Jr Kinetic analysis of the role of intersubunit interactions in human immunodeficiency virus type 1 capsid protein assembly in vitro. J Virol. 2002;76:6900–6908. doi: 10.1128/JVI.76.14.6900-6908.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bosco DA, Kern D. Catalysis and binding of cyclophilin A with different HIV-1 capsid constructs. Biochemistry. 2004;43:6110–6119. doi: 10.1021/bi049841z. [DOI] [PubMed] [Google Scholar]
- 28.Kovaleski BJ, Kennedy R, Hong MK, Datta SA, Kleiman L, Rein A, Musier-Forsyth K. In vitro characterization of the interaction between HIV-1 Gag and human lysyl-tRNA synthetase. J Biol Chem. 2006;281:19449–19456. doi: 10.1074/jbc.M601189200. [DOI] [PubMed] [Google Scholar]
- 29.Delaglio F, Grzesiek S, Vuister GW, Bax A. NMRPipe – a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 30.Bartels C, Xia T, Billeter M, Guentert P, Wuethrich K. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J Biomol NMR. 1995;6:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 31.Güntert P. Automated NMR structure calculation with CYANA. Method Mol Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 32.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NM. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krittanai C, Johnson WC. Proteins, The relative order of helical propensity of amino acids changes with solvent environment. Proteins: Structure, Function and Genetics. 2000;39:132–141. doi: 10.1002/(sici)1097-0134(20000501)39:2<132::aid-prot3>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 34.Von Schwedler UK, Stray KM, Garrus JE, Sundquist WI. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J Virology. 2003;77:5439–5450. doi: 10.1128/JVI.77.9.5439-5450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu X, Wang Q, Yang JC, Buch I, Tsai CJ, Ma B, Cheng SZ, Nussinov R, Zheng J. Mutational analysis and allosteric effects in the HIV-1 capsid proteincarboxyl-terminal dimerization domain. Biomacromolecules. 2009;10:390–399. doi: 10.1021/bm801151r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shuker SB, Hajduk PJ, Meadows RP, Fesik SW. Discovering high-affinity ligands for proteins: SAR by NMR. Science. 1996;274:1531–1534. doi: 10.1126/science.274.5292.1531. [DOI] [PubMed] [Google Scholar]
- 37.Krishna NR, Jayalakshmi V. Complete relaxation and conformational exchange matrix analysis of STD-NMR spectra of ligand-receptor complexes. Prorg NMR Spectrosc. 2006;49:1–25. [Google Scholar]
- 38.Doménech R, Abian O, Bocanegra R, Correa J, Sousa-Herves A, Riguera R, Mateu MG, Fernandez-Megia E, Velázquez-Campoy A, Neira JL. Dendrimers as potential inhibitors of the dimerization of the capsid protein of HIV-1. Biomacromolecules. 2010;11:2069–2078. doi: 10.1021/bm100432x. [DOI] [PubMed] [Google Scholar]
- 39.Freed EO. HIV-1 and the host cell: an intimate association. Trends in Microbiology. 2004;12:170–177. doi: 10.1016/j.tim.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 40.Ott DE. Potential roles of cellular proteins in HIV-1. Rev Med Virol. 2002;12:359–74. doi: 10.1002/rmv.367. [DOI] [PubMed] [Google Scholar]
- 41.Gamble TR, Vajdos FF, Yoo S, Worthylake DK, Houseweart M, Sundquist WI, Hill CP. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell. 1996;87:1285–1294. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- 42.Thali M, Bukovsky A, Kondo E, Rosenwirth B, Walsh CT, Sodroski J, Göttlinger HG. Functional association of cyclophilin A with HIV-1 virions. Nature. 1994;372:363–365. doi: 10.1038/372363a0. [DOI] [PubMed] [Google Scholar]
- 43.Bon Homme M, Carter C, Scarlata S. The cysteine residues of HIV-1 capsid regulate oligomerization and cyclophilin A-induced changes. Biophys J. 2005;88:2078–2088. doi: 10.1529/biophysj.104.053298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mascarenhas AP, Musier-Forsyth K. The capsid protein of human immunodeficiency virus: interactions of HIV-1 capsid with host protein factors. FEBS Journal. 2009;276:6118–6127. doi: 10.1111/j.1742-4658.2009.07315.x. [DOI] [PubMed] [Google Scholar]
- 45.Cardone G, Purdy JG, Cheng N, Craven RC, Steven AC. Visualization of a missing link in retrovirus capsid assembly. Nature. 2009;457:694–698. doi: 10.1038/nature07724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shin R, Tzou Y-M, Wong HC, Krishna NR. 1H, 15N and 13C resonance assignments for a monomeric mutant of the HIV-1 capsid protein. Biomol NMR Assign. 2011 doi: 10.1007/s12104-011-9340-3. in press (on line September 20, 2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Han Y, Ahn J, Concel J, Byeon IJ, Gronenborn AM, Yang J, Polenova T. Solid-state NMR studies of HIV-1 capsid protein assemblies. J Am Chem Soc. 2010;132:1976–1987. doi: 10.1021/ja908687k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen B, Tycko R. Structural and dynamical characterization of tubular HIV-1 capsid protein assemblies by solid state nuclear magnetic resonance and electron microscopy. Protein Sci. 2010;19:716–730. doi: 10.1002/pro.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.