Abstract
Current flu vaccines provide only limited coverage against seasonal strains of influenza viruses. The identification of VH1-69 antibodies that broadly neutralize almost all influenza A group 1 viruses constituted a breakthrough in the influenza field. Here we report the isolation and characterization of a human monoclonal antibody CR8020 with broad neutralizing activity against most group 2 viruses, including H3N2 and H7N7, which cause severe human infection. The crystal structure of Fab CR8020 with the 1968 pandemic H3 hemagglutinin (HA) reveals a highly conserved epitope in the HA stalk distinct from the epitope recognized by the VH1-69 group 1 antibodies. Thus, a cocktail of two antibodies may be sufficient to neutralize most influenza A subtypes and, hence, enable development of a universal flu vaccine and broad spectrum antibody therapies.
Influenza viruses cause millions of cases of severe illness each year, thousands of deaths, and considerable economic losses. Currently, two main countermeasures are used against flu. First, small molecule inhibitors of the neuraminidase surface glycoprotein and the viral ion channel M2 have been widely used and proven to be quite effective against susceptible strains (1). However, resistance to these antivirals has reduced their 9effectiveness and mutations associated with oseltamivir and amantadine are widespread (2–4). The second main countermeasure is vaccination. Current vaccines based upon inactivated virus elicit a potent immune response against viruses which are closely matched to the vaccine strain (5). However, predicting which strain(s) will dominate annually is difficult, and mismatches between the vaccine and circulating viruses lead to little or no protective effect (6, 7). A vaccine that stimulates production of a robust, broadly neutralizing antibody response would be a considerable advance.
Hemagglutinin (HA) is the major envelope glycoprotein of influenza A viruses and the target of almost all neutralizing antibodies. HA is synthesized as an immature polypeptide chain called HA0, which is activated upon cleavage by host proteases to yield two subunits, HA1 and HA2. The HA1 “head” subunit of HA mediates attachment of the virus to target cells through interactions with sialic acid receptors. After endocytosis of the virus, the low pH triggers conformational changes in HA2, leading to fusion of the viral and endosomal membranes and release of the viral genome into the cytoplasm. Most neutralizing antibodies bind to the exposed loops that surround the receptor binding site and interfere with attachment (8–12). Since these loops are highly variable, antibodies targeting these regions are strain-specific, explaining why immunity by natural exposure or vaccination is typically restricted to the current circulating strains.
Recently, we described the isolation and characterization of CR6261, a broadly neutralizing antibody with activity against group 1 influenza viruses (13–15). Similar antibodies using the same VH1-69 germline heavy chain have also been reported (16, 17). The discovery of such antibodies has raised hopes for the development of mAb-based immunotherapy and a universal vaccine (18–26). Crystal structures of CR6261 in complex with H1 and H5 HAs revealed a highly conserved epitope in the HA stalk (13). CR6261 neutralizes most group 1 HAs including H1, H5, H9, and some H2s, but has no activity against group 2 viruses (14). Group 2 includes the currently circulating human H3N2 viruses and H7N7 viruses, which sporadically cross from birds into humans and have the potential to develop into a future pandemic. Consequently, antibodies complementary to CR6261 and related VH1-69 antibodies, but with broad activity against group 2 viruses, are critical for the formulation of antibody-based therapies.
Isolation and characterization of CR8020 activity in vitro
Using a recently described method to generate stable monoclonal antibody–producing, B-cell receptor–positive, human memory B cells, we isolated H3 HA reactive clones from the blood of donors recently vaccinated against influenza (27). Subsequent screening of the resulting immunoglobulins (Ig’s) for reactivity with other HA subtypes led to identification of mAb CR8020, which recognizes H3 and H7 HAs, as well as representative HAs from other group 2 subtypes, but not group 1 HAs (Fig. 1, A and C). In this sense, CR8020 is complementary to previously described mAbs, such as CR6261, which neutralizes most group 1 HAs (14), but not group 2 (Fig. 1A and C). CR8020 binds most group 2 HAs with high affinity, including H3 isolates spanning 50 years of virus evolution (KD~1–35nM for H3, H7, and H10 HAs) (Fig. 1C). Consistent with binding to an apparently highly conserved epitope, CR8020 potently neutralizes a wide spectrum of H3N2 influenza strains as well as H7 and H10 viruses (Fig. 1B) (28). In contrast, a control mAb against the HA1 head neutralizes only a narrow spectrum of H3 viruses (Table S1).
Fig. 1.
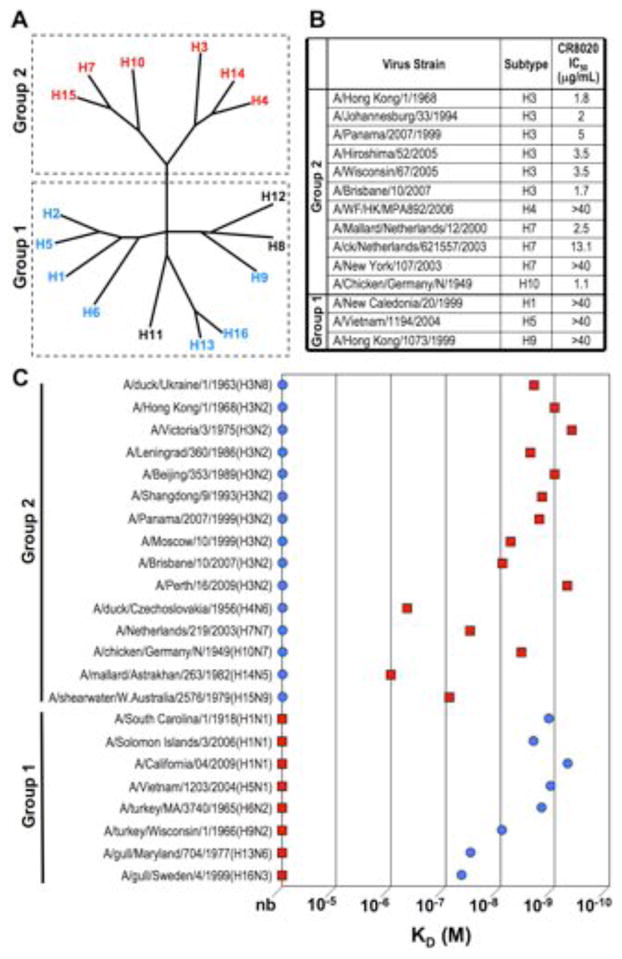
In vitro binding and neutralization of CR8020 and complementarity with CR6261. (A) Phylogenetic tree showing the relationships between the 16 HA subtypes and a summary of CR8020 and CR6261 activity. Red indicates positive binding by CR8020 while blue indicates positive binding by CR6261. Subtypes that have not been tested are indicated in black. (B) In vitro neutralization (IC50 in μg/ml) of CR8020 against a panel of influenza A viruses as determined by microneutralization assay. (C) Affinity measurements (KD) for binding of CR8020 and CR6261 to various H3 HAs and representative members of most of the other HA subtypes. nb indicates no detectable binding. Lowest affinity detectable under the experimental conditions was ~10−5 M.
Prophylactic and therapeutic efficacy of CR8020 in vivo
Prophylaxis using 3 mg/kg CR8020 protected mice against challenge with a high lethal dose of either mouse-adapted A/Hong Kong/1/68 (H3N2) or A/Ck/Netherlands/621557/03 (H7N7) virus (Fig. 2, A and B). Consistent with the absence of any signs of respiratory distress, groups of H3N2-challenged mice that received 30, 10 or 3 mg/kg CR8020 and H7N7-challenged mice receiving 10 or 3 mg/kg CR8020 all showed increases in bodyweight at the study end (Fig. 2, A and B) (29). In contrast, animals that received an irrelevant control antibody rapidly lost weight, showed signs of respiratory distress, and succumbed to infection or were euthanized within two weeks after challenge. A 1 mg/kg dose was only partially protective against mortality to either virus (Fig. 2, A and B), and the weight loss was not significantly different from the controls (p=0.666 and p=0.633 for groups challenged with H3N2 and H7N7 virus, respectively). Therapeutic treatment with 15 mg/kg of CR8020 up to 2 days after infection with H3N2 completely prevented mortality, whereas 50% of the mice treated 3 days after infection were protected (Fig. 2C). Treatment with CR8020 could be started even later after H7N7 challenge, as 100% of the mice treated 3 days post-infection, and 50% treated 4 days post-infection survived, although one animal treated 2 days after challenge succumbed to infection (Fig. 2D). Therapeutic treatment did not prevent morbidity as illustrated by initial weight loss, but all surviving animals appeared healthy and were regaining weight at the study end.
Fig. 2.
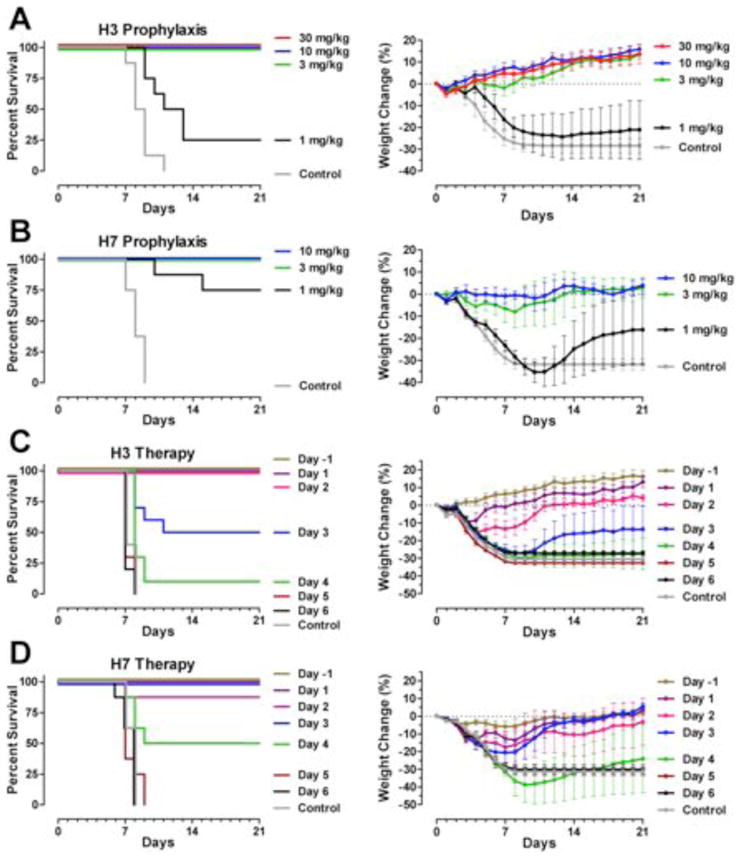
Prophylactic and therapeutic efficacy of CR8020 in mice. Prophylactic efficacy of CR8020 against lethal challenge with (A) mouse-adapted A/Hong Kong/1/1968 (H3N2) or (B) A/Ck/Netherlands/621557/2003 (H7N7) viruses. Survival curves (left) and weight loss (right) of mice treated with 30, 10, 3, or 1 mg/kg of CR8020 or 30 mg/kg control mAb 24 hours before lethal challenge by intranasal inoculation (at day 0). Therapeutic efficacy of CR8020 against lethal challenge with (C) mouse-adapted A/Hong Kong/1/1968 (H3N2) or (D) A/Ck/Netherlands/621557/2003 (H7N7) viruses. Survival curves (left) and weight loss (right) of mice treated with 15 mg/kg CR8020 at various time-points after inoculation (at day 0).
Determination of CR8020-H3 HA crystal structure
To elucidate how mAb CR8020 can neutralize multiple group 2 influenza virus subtypes, we determined the crystal structure of CR8020 Fab in complex with the HA from the 1968 H3N2 pandemic (A/Hong Kong/1/1968 (H3N2), “HK68”) at 2.85 Å resolution (Table S2). The overall structure of the HK68 HA in the complex (Fig. 3A) is similar to other unliganded H3’s (30), although some subtle pH-induced changes are discussed below (31).
Fig. 3.
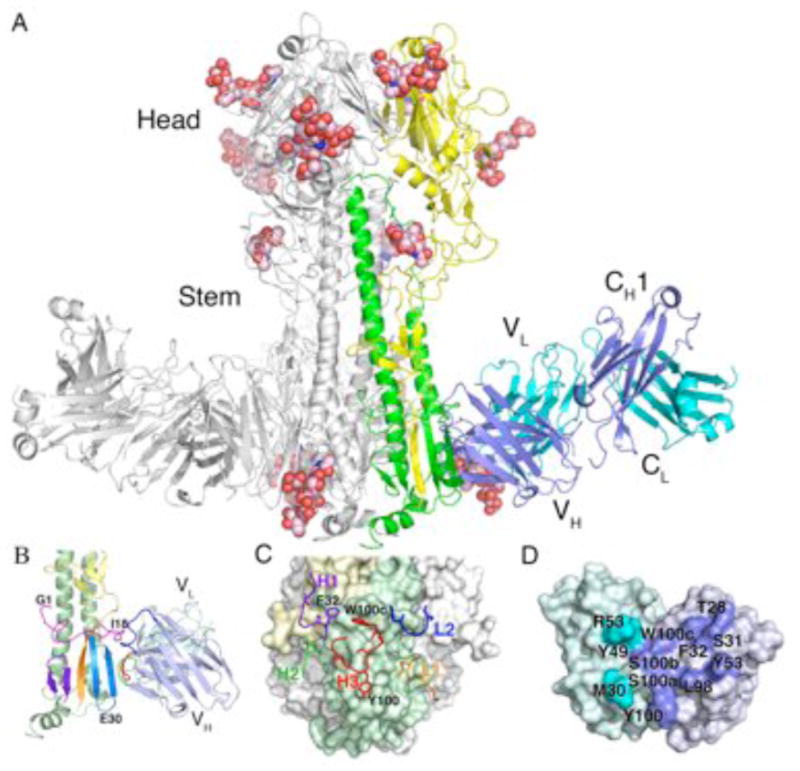
CR8020 binds an epitope in the HA stem close to the virus membrane. (A) Crystal structure of HK68/H3 HA in complex with broadly neutralizing antibody CR8020. One HA/Fab protomer of the trimeric complex is colored with HA1 in yellow, HA2 in green, the Fab heavy chain in blue, and the Fab light chain in cyan. The other two protomers are colored gray. Glycans are shown as spheres (carbon in light pink, oxygen in red and nitrogen in blue). (B) Closer view of the interaction between HK68 HA and CR8020. The coloring is essentially as in (A,) but with the fusion peptide, which forms part of the epitope, in magenta, and the three segments of the small -sheet in purple, orange, and light blue (derived from the C-terminus of HA2, the N-terminus of HA1, and the N-terminus of HA2, respectively). On the Fab, HCDR1 is blue, HCDR2 is green and HCDR3 is red. For clarity, light chain CDRs are not highlighted here (see part C). (C) Interaction of CR8020 CDRs with the membrane proximal region of HA. CDRs are shown as ribbons and sticks (H1 in purple, H2 in green, H3 in red, L1 in orange, and L2 in blue. L3 makes no contacts). CR8020 binds HA with both chains, using 5 of the 6 CDRs (LCDR3 makes no contacts). Key antibody side chains are shown. (D) Footprint of HA on CR8020 combining site, highlighting antibody residues contacting HA. Note the usage of both the heavy and light chains in recognition (blue and cyan, respectively).
Fab CR8020 binds HK68 HA at the base of the stem in close proximity (~15–20Å) to the viral membrane (Fig. 1, A and B) (32), lower down the stalk than any other flu antibody characterized to date (Fig. 4A). Despite its proximity to the viral membrane, this epitope is accessible on virions (Fig. S1), in accord with the in vitro and in vivo potency of this mAb. The epitope consists of two main components: 1) the outermost strand (HA2 residues 30–36) of the 5-stranded β-sheet near the base of the stalk and 2) the C-terminal portion (HA2 residues 15–19) of the fusion peptide (Fig. 3B), as well as a few peripheral contacts with other surrounding residues (33). Compared to CR6261, CR8020 recognizes its epitope in a more conventional manner, using both heavy and light chains (Fig. 3, C and D). A total surface area of 1280Å2 is buried, of which 81% arises from binding of the heavy chain and 19% from the light chain. The fusion peptide component accounts for ~50% of the van der Waals’ contacts between Fab and HA, and is specifically recognized by HCDRs 1 and 3, where Phe32 (HCDR1) and Trp100c (HCDR3) make many of the key hydrophobic interactions (Fig. 3C). HCDR3 also interacts extensively with the outermost strand of the -sheet, in a pseudo -strand-like interaction with HA2 residues 32–36 (fig. S2). Together, the CR8020 interactions with the fusion peptide and edge of the β-sheet, account for > 80% of the van der Waals’ contacts and 6 out of 7 hydrogen bonds.
Fig. 4.
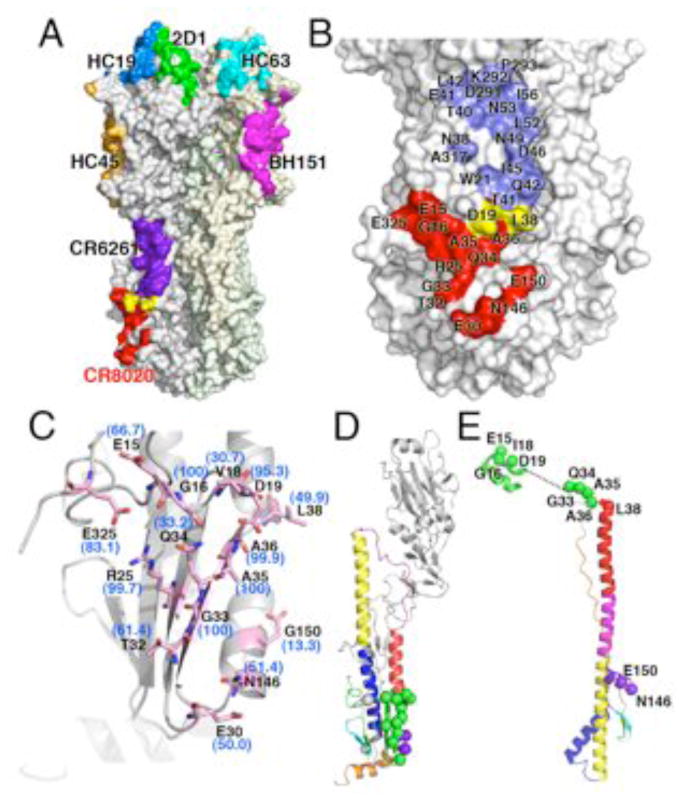
CR8020 binds a unique, highly conserved site in the stem. (A) Comparison of CR8020 binding site to the epitopes of all other structurally characterized antibodies. All antibody footprints are mapped onto the surface of HK68 HA, although they are derived from different structures (and subtypes): Red, CR8020 (PDB code: XXXX); Purple, CR6261 (PDB codes 3GBM and 3GBN); green, 2D1 (PDB code: 1LFZ); blue, HC19 (PDB code: 2VIR); orange, HC45 (PDB code: 1QFU); cyan, HC63 (PDB code: 1KEN); pink, BH151 (PDB code: 1EO8). Note that mAb F10 and CR6261 bind the same epitope in the stem, but only CR6261 is illustrated here for clarity. (B) Comparison of the broadly neutralizing CR8020 and CR6261 epitopes, which constitute discrete surfaces on on group 2 and group1 HAs, respectively. Coloring is similar to (A), with CR8020 epitope in red, CR6261 epitope in blue, and shared epitope residues in yellow. (C) Conservation of CR8020 epitope across group 2 HAs. Residues comprising the epitope are shown as sticks (carbon in light pink, oxygen in red and nitrogen in blue). Percent identity with the group 2 consensus sequence is indicated alongside each residue. Note that the residue label reflects the most common residue across group 2 HAs, which is not always identical to the residue at that position in the HK68 crystal structure. View is similar to that of Fig. 4B, looking from the Fab (not shown) towards the epitope. (D) Location of CR8020 epitope residues in the pre-fusion and (E) post-fusion state, where they reside on critical regions of the fusion peptide and helix-capping region of HA2. For D and E, CR8020 epitope residues are indicated as spheres, with green mapping to the fusion peptide and N-cap region. Additional contact residues are purple (HA2) and gray (HA1, not present in available post-fusion structures). Residue positions were mapped onto the structures from PDB codes 1QU1 and 2KXA.
CR8020 binds a novel, conserved epitope on the HA stem
The crystal structures of CR6261 and F10 revealed a neutralizing epitope on the HA stem that is conserved and accessible in most group 1 influenza viruses (13, 17). However, an N-linked glycosylation at HA1:Asn38 in most group 2 HAs may restrict antibody access to this epitope. Consequently, antibodies with broad neutralizing activity against group 2 viruses would be expected to recognize an epitope that would be spatially distinct from that recognized by CR6261. Indeed, of the 15 and 20 residues that constitute the epitopes targeted by CR8020 and CR6261, respectively, only two residues (Asp 19 and Leu 38) are in common (Fig. 4, A and B). Thus, CR8020 defines a second, neutralizing epitope in the HA stem that is present in all group 2 HAs tested thus far.
To investigate the breadth of CR8020’s cross-neutralizing activity, we examined the epitope conservation across all 16 influenza A virus subtypes by examining all full-length, non-redundant HA sequences in the NCBI FLU database (34, 35). Around half of the contact residues are either identical (>95%- HA2 residues 16, 33, 35, and 36), or conserved (>99%- HA2 residues 18, 30, and 146) across all 16 subtypes (Fig. 4C and Tables S3 and S4)(36), whereas other contact residues are conserved only across group 2 (>95% identity: HA2 25; >99% conserved: HA1 325, HA2 15 and 19). Thus, 11 of 15 residues contacting CR8020 are >99% conserved across all group 2 HAs, whereas the remaining residues are more variable (HA2 32, 34, 38, and 150 are conserved in only ~56–81% of isolates, Fig. 4C and Table S3). However, virus neutralization and in vitro binding data suggest that most natural variation is well tolerated by CR8020 (Table 1). Essentially all natural variation in the epitope commonly observed in group 2 viruses is represented in the extensive panel of wild-type H3, H4, H7, H10, H14, and H15 isolates and engineered variants of HK68 that were tested for binding and/or neutralization (Table 1, Fig 1, B and C). CR8020 binds nearly all of these naturally occurring HA variants with similar high affinity (KD ~1–10nM) to the HAs from group 2 viruses that are neutralized by CR8020 ( ~1–35nM). However, one variant (Asp19Asn) correlates with markedly reduced affinity and loss of neutralizing activity in vitro, and is discussed below. Overall, these results show that the core of the CR8020 epitope is highly conserved across all group 2 viruses, while natural variation in the surrounding residues is well tolerated and does not adversely impact binding and most likely neutralization.
Table 1. Most natural variation in CR8020 epitope in group 2 HAs has little effect on binding.
Dissociation constants (KD), presence of a glycosylation site at HA1 position 21 (predicted to block binding in group 1 HAs), and protein sequence of CR8020 contact residues are shown. Data presented here include targeted mutants of HK68 and wild-type isolates selected to probe the impact of natural variation in epitope on CR8020 activity. Wild-type HK68 is included at the top, as a reference. The bottom row summarizes the most common naturally occurring variants at each position (present at least in >1% of all group 2 sequences), with tested variants in bold/underlined. Residues that differ from the HK68 sequence are in black boxes.
| Subtype | Isolate | Mutationa | KD(nM) | HA1 | HA2 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NX(T/S)b | 325 | 15 | 16 | 18 | 19 | 25 | 30 | 32 | 33 | 34 | 35 | 36 | 38 | 146 | 150 | ||||
| H3N2 | A/Hong Kong/1/1968 | WT | 1.0 | - | E | E | G | I | D | R | E | T | G | Q | A | A | L | N | E |
| H3N8 | A/duck/Ukraine/1/1963 | WT | 2.4 | - | G | E | G | I | D | R | E | T | G | Q | A | A | L | N | E |
| H3N2 | A/Hong Kong/1/1968 | E15Q | 9.3 | - | E | Q | G | I | D | R | E | T | G | Q | A | A | L | N | E |
| H3N2 | A/Panama/2007/1999 | WT | 1.9 | - | E | E | G | V | D | R | E | T | G | Q | A | A | L | N | G |
| H3N2 | A/Hong Kong/1/1968 | I18M | 5.0 | - | E | E | G | M | D | R | E | T | G | Q | A | A | L | N | E |
| H3N2 | A/Hong Kong/1/1968 | D19N | 310 | - | E | E | G | I | N | R | E | T | G | Q | A | A | L | N | E |
| H3N2 | A/Hong Kong/1/1968 | E30Q | 3.0 | - | E | E | G | I | D | R | Q | T | G | Q | A | A | L | N | E |
| H3N2 | A/Hong Kong/1/1968 | T32Q | 3.0 | - | E | E | G | I | D | R | E | Q | G | Q | A | A | L | N | E |
| H3N2 | A/Hong Kong/1/1968 | T32E | 3.6 | - | E | E | G | I | D | R | E | E | G | Q | A | A | L | N | E |
| H3N2 | A/Hong Kong/1/1968 | T32I | 5.5 | - | E | E | G | I | D | R | E | I | G | Q | A | A | L | N | E |
| H3N2 | A/Hong Kong/1/1968 | T32R | 2.3 | - | E | E | G | I | D | R | E | R | G | Q | A | A | L | N | E |
| H3N2 | A/Hong Kong/1/1968 | Q34T | 7.7 | - | E | E | G | I | D | R | E | T | G | T | A | A | L | N | E |
| H3N2 | A/Hong Kong/1/1968 | L38Y | 3.2 | - | E | E | G | I | D | R | E | T | G | Q | A | A | Y | N | E |
| H3N2 | A/Hong Kong/1/1968 | N146D | 3.5 | - | E | E | G | I | D | R | E | T | G | Q | A | A | L | D | E |
| H3N2 | A/Hong Kong/1/1968 | E150A | 2.0 | - | E | E | G | I | D | R | E | T | G | Q | A | A | L | N | A |
| H3N2 | A/Victoria/3/1975 | WT | 0.5 | - | E | E | G | I | D | R | E | T | G | Q | A | A | L | N | G |
| H4N6 | A/duck/Czechoslovakia/1956 | WT | ~500 | - | E | Q | G | I | D | R | E | T | G | T | A | A | L | N | E |
| H7N7 | A/Netherlands/219/2003 | WT | 36 | - | E | E | G | I | D | R | Q | E | G | T | A | A | Y | D | A |
| H10N7 | A/chicken/Germany/n/1949 | WT | 4.1 | - | E | E | G | V | D | R | Q | T | G | Q | A | A | Y | D | E |
| H14N5 | A/mallard/Astrakhan/263/1982 | WT | ~1000 | - | G | Q | G | I | D | R | E | T | G | T | A | A | L | N | E |
| H15N9 | A/shearw/W.Australia/2576/1979 | WT | 87 | - | E | E | G | I | D | R | Q | Q | G | T | A | A | Y | D | E |
| Most common natural variation (present in >1% of sequences analyzed; bold/underlined if tested for binding): | E |
E Q |
G |
V I M |
D N |
R |
E Q |
T E I R |
G |
Q T |
A | A |
L Y |
N D |
G E A |
||||
Indicates whether HA tested is wild-type (wt) or a mutant
Presence (+) or absence (−) of an NX(T/S) glycosylation motif on Asn21. Glycosylation of this site is predicted to block CR8020 binding in group 1 HAs. The glycosylation site is absent from all group 2 HAs analyzed, including those tested above.
The structural constraints imposed by the membrane fusion activity enforce strict conservation of many regions of HA2. To understand why the CR8020 contact residues are so well conserved in group 2 viruses, we examined their potential role in membrane fusion. The fusion peptide (HA2 1–25) is critical for membrane fusion and is nearly invariant across all influenza A viruses (Fig. S3) (37, 38). The fusion peptide adopts an unusually tight, helical-hairpin conformation (34), which provides an elegant, structural explanation for conservation of most residues, including CR8020 contact residues HA2 16 and 18 (Fig. 4, C–E and Fig. S4A). After exiting the membrane and traversing a short linker region, residues close to HA2:31–38 cap the ends of the long, three-helix bundle of the post-fusion structure (Fig. 4, D and E, and Fig. S4, A and B), thereby constraining the identity of the amino acids in the cap, which are thought to make a significant energetic contribution to the membrane fusion process (39). Therefore, elements of the CR8020 epitope likely have critical roles in driving the fusion process.
Group 2 restriction and antibody escape
Antibody escape variants selected by CR8020 in an H3N2 virus exhibited mutations in HA2 Asp19Asn or Gly33Glu close to the CR8020 epitope center. A recombinant H7N7 virus with the Asp19Asn mutation also escaped neutralization by CR8020 (Table S5 and S6). This mutation disrupts a possible salt bridge to VL Arg53, presumably leading to destabilization of the antibody-HA interaction (40). The Gly33Glu mutation inserts a large side chain into a highly confined space in the antibody-antigen interface. However, both variants are relatively rare (Fig. S5), particularly in human isolates. Glycine is strongly preferred at position 33 (8716/8720 sequences, group 1 and 2 HAs), whereas position 19 is an Asp in ~95% of all group 2 viruses and in 1534/1537 of human H3 viruses (41). Whether these mutations negatively impact viral fitness is not known, but other antibodies may recognize the CR8020 epitope in subtly different ways that would render them less sensitive to these substitutions. Notwithstanding, our results suggest that CR8020 will neutralize most viruses from H3, H7, and H10 subtypes, and possibly H15 (42).
Two avian group 2 subtypes (H4 and H14) are bound by CR8020 with only moderate affinity (Table 1), and neutralization of an H4 virus was undetectable in vitro (43). The H4 and H14 isolates tested differ from HK68 at two contact positions (Glu15Gln and Gln34Thr), and each substitution confers a modest reduction (~10-fold) in CR8020 affinity for HK68 (Table 1)(44). Thus, reduced affinity of CR8020 for H4 and H14 can largely be accounted for by the combined effect of these mutations, but subtle structural differences in non-contact residues surrounding the epitope may also have significant effects on antibody binding.
Although the HA surface recognized by CR8020 is also relatively well conserved in group 1 viruses, no group 1 viruses tested were bound or neutralized (Table S7). Several key differences in group 1 HAs may account for lack of CR8020 reactivity. First, the Gln or Thr that predominates at position 34 in group 2 HAs is substituted by a bulkier Tyr in many group 1 subtypes, and would likely clash with HCDR3. CR8020 binding to a non-natural, Gln34Arg HK68 variant is reduced over 100-fold, further suggesting that larger residues cannot be accommodated. Second, group 1 HAs have a highly conserved N-linked glycosylation site at HA1:Asn21 (5801/5813 group 1 sequences analyzed), adjacent to CR8020 HCDR1 (Fig. S6). In most configurations, the glycan would conflict with CR8020 VH, and likely interfere with antibody binding (45). Finally, several individual substitutions in CR8020 contact residues that modestly reduce affinity (5–10 fold) are combined in group 1 isolates and may reduce binding below our detection threshold (~10uM) (Tables 1 and S7).
Mechanism of virus neutralization by CR8020
Unlike most HA antibodies, which block attachment, CR8020 has no detectable activity in hemagglutination-inhibition (HAI) assays and does not compete with antibodies against the head, consistent with CR8020 binding to the stalk region. With exception of HA1 residue 325, the CR8020 epitope maps entirely to HA2. Upon exposure to low pH, HA2 undergoes extensive conformational rearrangements, bringing the viral and target membranes into close proximity and triggering fusion (Fig. 4, D and E) (39, 46). Consequently, CR8020 is poised to inhibit these conformational changes, thereby blocking membrane fusion and viral entry. Whereas CR8020 binds readily to cell surface–expressed H3 HAs in both its uncleaved (HA0) and cleaved (HA) forms, it did not bind after HA exposure to a pH of 4.9 (Fig. 5A), in agreement with previous findings that the epitope structure is not maintained in the fusion-active conformation (46, 47). When CR8020 was added prior to the pH shift, it remained bound after the pH was lowered (Fig. 5B), indicating that the epitope remains intact at low pH and suggesting that CR8020 inhibits the pH-induced conformational changes in HA. Moreover, CR8020 prevented DTT-mediated dissociation of HA1 and HA2 at low pH (48), which would only be expected if the HA was retained in the pre-fusion state. Similar results were obtained with H7 and H10 HAs, indicating that the mechanism of inhibition is conserved (Fig. S7).
Fig. 5.

CR8020 inhibits the fusogenic conformational changes in HA and blocks proteolytic activation. (A) FACS binding of CR8020 (open bars) and a control mAb against the head (closed bars) to various conformations of surface-expressed H3 HAs from A/Hong Kong/1/1968 (1968), A/Hong Kong/24/1985 (1985) or A/Wisconsin/67/2005 (2005). Note that the control mAb has a narrow spectrum of neutralization and only binds A/Wisconsin/67/2005. The various conformations are depicted above the binding data and are as follows: uncleaved precursor (HA0); neutral pH, cleaved (HA); fusion pH, cleaved (fusion pH); trimeric HA2 (tHA2). Binding is expressed as the percentage of binding to untreated HA (HA0). Error bars represent SD of data obtained in 3 independent experiments. Ribbon diagrams are adapted from (57). (B) FACS binding of CR8020 (open bars) and a control mAb against the head (closed bars) to surface-expressed HA of A/Hong Kong/1/1968 (1968), A/Hong Kong/24/1985 (1985) or A/Wisconsin/67/2005 (2005) as above, except that mAb CR8020 was added before exposure of the cleaved HAs to a pH of 4.9. The anti-head mAb was added last to detect whether the HA1 heads remained associated with HA2. (C) SDS-PAGE results of the protease-susceptibility assay for H3 and H7 HAs. Exposure to low pH converts HAs to the protease-susceptible, post-fusion state (lane 3). Pre-treatment with CR8020 blocks the pH-induced conformational change, retaining HA in the protease-resistant, pre-fusion state (lane 7). (D) Immunoblot of uncleaved (HA0), recombinant soluble H3 HA after digestion with trypsin at pH 8.0. Digest reactions contained either HA alone, or HA pretreated with CR8020 or a control mAb against the head. Digestion was stopped at several time-points by adding 1% BSA. Uncleaved hemagglutinin (HA0) was detected using a polyclonal serum against H3.
Further evidence that this mAb acts by inhibiting conversion from the pre-fusion to post-fusion conformation is illustrated by lack of protease susceptibility of the HA at low pH in the presence of CR8020. Exposure to low pH followed by trypsin digestion results in complete degradation of H3 and H7 HAs (Fig. 5C, lanes 1–4). In contrast, when the HA is pre-treated with CR8020, most HA is retained in a protease-resistant, pre-fusion form (Fig 5C, lanes 5–8). Furthermore, the crystals used for the structural studies were grown below pH 5.4, the fusion pH for HK68 HA (Fig. S8)(49). Nevertheless, the HA is retained in its pre-fusion state, even after extended exposure to low pH. However, the HA1 heads have opened up slightly and the B-loop adopts an alternate backbone conformation compared to neutral pH structures. The head opening is believed to be one of the first steps in the membrane fusion process and the B-loop must refold from an extended random coil to an α-helix to deliver the fusion peptide to the target membrane. Thus, our crystal structure appears to have captured an early fusion intermediate (50), trapped by the binding of CR8020. Taken together, our data suggest that CR8020 blocks fusion by sequestering the fusion peptide and preventing its release at low pH.
Furthermore, CR8020 may interfere with fusion by inhibiting activation of HA0 by host proteases. Whereas HA0 was rapidly cleaved by trypsin in vitro into HA1 and HA2, this cleavage was completely inhibited in the presence of CR8020, but not by a control, head-binding mAb (Fig. 5D). Thus, blocking HA0 maturation to HA1/HA2 may represent an additional mechanism by which mAbs can block viral entry.
Implications for therapy and vaccines
Influenza A viruses responsible for human pandemics have arisen from both group 1 (H1N1 and H2N2) and group 2 (H3N2) viruses (Fig. 1A). In addition, zoonotic viruses from both groups sporadically infect humans and have the potential to trigger future pandemics (including H5N1 and H9N2 from group 1 and H7N7 from group 2). Consequently, the ideal, universal therapy should provide protection against both group 1 and group 2 influenza viruses. Attempts to isolate broadly neutralizing antibodies against group 2 viruses from animals have generally yielded antibodies reported as non-neutralizing or of only modest potency or breadth (24, 51–55), and broadly neutralizing human antibodies against group 2 viruses have not been previously described. A cocktail of antibodies, such as CR6261 and CR8020, may protect against essentially all influenza A viruses implicated in human disease. Such a therapeutic cocktail would have undisputed benefits for high-risk groups, such as the elderly and immunocompromised, and for severe, life-threatening influenza infections. These antibodies also represent an ideal immunological solution to influenza infection, and could serve as a guide for development of vaccines that elicit broader, long-lasting immunity. The identification and characterization of CR6261-like antibodies has already sparked considerable advances, including 1) their detection in some individuals (21) and 2) design of immunization strategies that efficiently elicit stem antibodies in mice, ferrets, and monkeys (20, 25). Thus, the identification and characterization of CR8020 should facilitate similar advances for group 2 viruses, bringing us one step closer to the ultimate goal of a universal vaccine for influenza.
Supplementary Material
Acknowledgments
We thank H. Tien and D. Marciano of the Robotics Core at the JCSG for automated crystal screening, T. Doukov and the staff of the SSRL BL9-2 for beamline support, X. Dai and R. Stanfield for excellent assistance with data collection, processing, and analyses, R. Lerner, J. Paulson, and D. Burton for valuable comments and insightful discussion, E. Geelen, D. Spek, and V. Klaren for excellent assistance and advice, K. Hegmans, A. Lourbakos, J. Meijer, and A. Apetri and their teams for producing the mAbs, C.Y.H. Leung for providing the A/WF/Hong Kong/MPA892/06 virus, E. de Boer-Luijtze and technicians in the groups of P. van Rossum and S. Riemersma for assistance with the animal experiments, E. Brown from Ottawa University, Canada for the mouse-adapted A/Hong Kong/1/68 strain, and A. Dingemans for critical review of the manuscript. This project has been funded in part by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, USA, under contract No. HHSN272200900060C; the Area of Excellence Scheme of the University Grants Committee, Hong Kong (Grant AoE/M-12/06); a predoctoral fellowship from the Achievement Rewards for College Scientists Foundation (D.C.E.), Grant GM080209 from the NIH Molecular Evolution Training Program (D.C.E.), and the Skaggs Institute (I.A.W.). Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences. This is publication #20951 from the Scripps Research Institute. Coordinates and structure factors will be deposited in the Protein Data Bank (PDB code 3SDY). Nucelotide sequences for the CR8020 variable regions have been deposited in GenBank (accession numbers JN093122-JN093123). A patent application relating to antibody CR8020 has been filed (International Publication Number WO2010/130636). Sharing will be subject to standard MTAs.
References and Notes
- 1.Beigel J, Bray M. Current and future antiviral therapy of severe seasonal and avian influenza. Antiviral Res. 2008;78:91. doi: 10.1016/j.antiviral.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bright RA, Shay DK, Shu B, Cox NJ, Klimov AI. Adamantane resistance among influenza A viruses isolated early during the 2005–2006 influenza season in the United States. JAMA. 2006;295:891. doi: 10.1001/jama.295.8.joc60020. [DOI] [PubMed] [Google Scholar]
- 3.Kiso M, et al. Resistant influenza A viruses in children treated with oseltamivir: descriptive study. Lancet. 2004;364:759. doi: 10.1016/S0140-6736(04)16934-1. [DOI] [PubMed] [Google Scholar]
- 4.Lowen AC, Palese P. Influenza virus transmission: basic science and implications for the use of antiviral drugs during a pandemic. Infect Disord Drug Targets. 2007;7:318. doi: 10.2174/187152607783018736. [DOI] [PubMed] [Google Scholar]
- 5.The most common vaccine formulations include influenza A H1N1 and H3N2 and influenza B components.
- 6.Salzberg S. The contents of the syringe. Nature. 2008;454:160. doi: 10.1038/454160a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Interim within-season estimate of the effectiveness of trivalent inactivated influenza vaccine--Marshfield, Wisconsin, 2007–08 influenza season. MMWR Morb Mortal Wkly Rep. 2008;57:393. [PubMed] [Google Scholar]
- 8.Fleury D, Daniels RS, Skehel JJ, Knossow M, Bizebard T. Structural evidence for recognition of a single epitope by two distinct antibodies. Proteins. 2000;40:572. [PubMed] [Google Scholar]
- 9.Barbey-Martin C, et al. An antibody that prevents the hemagglutinin low pH fusogenic transition. Virology. 2002;294:70. doi: 10.1006/viro.2001.1320. [DOI] [PubMed] [Google Scholar]
- 10.Fleury D, et al. A complex of influenza hemagglutinin with a neutralizing antibody that binds outside the virus receptor binding site. Nat Struct Biol. 1999;6:530. doi: 10.1038/9299. [DOI] [PubMed] [Google Scholar]
- 11.Fleury D, Wharton SA, Skehel JJ, Knossow M, Bizebard T. Antigen distortion allows influenza virus to escape neutralization. Nat Struct Biol. 1998;5:119. doi: 10.1038/nsb0298-119. [DOI] [PubMed] [Google Scholar]
- 12.Xu R, et al. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science. 2010;328:357. doi: 10.1126/science.1186430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ekiert DC, et al. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Throsby M, et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS One. 2008;3:e3942. doi: 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hemagglutinins cluster into two distinct groups on the basis of their primary sequence. Group 1 HAs include 10 of the 16 subtypes: H1, H2, H5, H6, H8, H9, H11, H12, H13, and H16. Group 2 HAs account for the remaining 6 subtypes: H3, H4, H7, H10, H14, and H15.
- 16.Kashyap AK, et al. Combinatorial antibody libraries from survivors of the Turkish H5N1 avian influenza outbreak reveal virus neutralization strategies. Proc Natl Acad Sci U S A. 2008;105:5986. doi: 10.1073/pnas.0801367105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sui J, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol. 2009;16:265. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen GL, Subbarao K. Attacking the flu: neutralizing antibodies may lead to ‘universal’ vaccine. Nat Med. 2009;15:1251. doi: 10.1038/nm1109-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang TT, Palese P. Universal epitopes of influenza virus hemagglutinins? Nat Struct Mol Biol. 2009;16:233. doi: 10.1038/nsmb.1574. [DOI] [PubMed] [Google Scholar]
- 20.Bommakanti G, et al. Design of an HA2-based Escherichia coli expressed influenza immunogen that protects mice from pathogenic challenge. Proc Natl Acad Sci U S A. 2010;107:13701. doi: 10.1073/pnas.1007465107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corti D, et al. Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. J Clin Invest. 2010;120:1663. doi: 10.1172/JCI41902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steel J, et al. Influenza virus vaccine based on the conserved hemagglutinin stalk domain. MBio. 2010;1 doi: 10.1128/mBio.00018-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang TT, et al. Vaccination with a synthetic peptide from the influenza virus hemagglutinin provides protection against distinct viral subtypes. Proc Natl Acad Sci U S A. 2010;107:18979. doi: 10.1073/pnas.1013387107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang TT, et al. Broadly protective monoclonal antibodies against H3 influenza viruses following sequential immunization with different hemagglutinins. PLoS Pathog. 2010;6:e1000796. doi: 10.1371/journal.ppat.1000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei CJ, et al. Induction of broadly neutralizing H1N1 influenza antibodies by vaccination. Science. 2010;329:1060. doi: 10.1126/science.1192517. [DOI] [PubMed] [Google Scholar]
- 26.Wrammert J, et al. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J Exp Med. 2011;208:181. doi: 10.1084/jem.20101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Materials and methods are available as supporting material on Science Online
- 28.Although CR8020 binds to H4 and H14, the affinity is probably too low to result in in vitro neutralization, at least at the concentration tested here. Neutralization of H14 and H15 viruses was not tested.
- 29.All increases in body weight were statistically significant (p 0.018), except for the H7N7 challenge group treated with 3mg/kg CR8020 (p=0.168).
- 30.Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981;289:366. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 31.The final model includes HA1 residues 11–328, HA2 residues 1–175, CR8020 heavy chain residues 2–222 and light chain residues 1–212. The asymmetric unit contains one HA protomer bound to on CR8020 Fab, with the additional protomers in the trimer generated by crystallographic symmetry operations.
- 32.In this regard, CR8020 can be thought of as being analogous to the anti-HIV envelope antibodies 2F5, 4E10, and Z13. These antibodies recognize the membrane-proximal external region (MPER), a short helical peptide from the gp41 subunit that is closely associated with the viral envelope.
- 33.CR8020 also contacts: 1) HA1 residue 325, near the C-terminus of the chain, 2) HA2 residue 25, in the fourth strand out in the small -sheet, 3) HA2 residue 38, at the bottom of helix A, and 4) HA2 residues 146 and 150, in a short helix adjacent to the sheet. It is worth noting that, in addition to protein-protein interactions, the Fab also makes contacts with the core fucose of a universally conserved glycan linked to Asn154 in HA2, although the extent of the contribution of this interaction to the overall binding energy is unclear.
- 34.Bao Y, et al. The influenza virus resource at the National Center for Biotechnology Information. J Virol. 2008;82:596. doi: 10.1128/JVI.02005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Details of the dataset and analysis can be found in the supporting material on Science Online
- 36.In this analysis, residues are considered conserved if they fall within one of the following groups: 1) Asp/Asn/Glu/Gln; 2) Phe/Tyr; 3) Ile/Leu/Val/Met; 4) Lys/Arg; or 5) Ser/Thr.
- 37.Lorieau JL, Louis JM, Bax A. The complete influenza hemagglutinin fusion domain adopts a tight helical hairpin arrangement at the lipid:water interface. Proc Natl Acad Sci U S A. 2010;107:11341. doi: 10.1073/pnas.1006142107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Twenty of the first 23 positions in HA2 are well-conserved across all subtypes (See Fig. S3). Two of the remaining positions (HA2 positions 12 and 15) have differing, group-specific residues. The final position, HA2 residue 19 is also conserved as D or N across most subtypes from both groups. However, we regard this substitution as non-conservative in the context of CR8020, since D19N mutations escape virus neutralization.
- 39.Chen J, Skehel JJ, Wiley DC. N- and C-terminal residues combine in the fusion-pH influenza hemagglutinin HA(2) subunit to form an N cap that terminates the triple-stranded coiled coil. Proc Natl Acad Sci U S A. 1999;96:8967. doi: 10.1073/pnas.96.16.8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.While there is clear density for the VL:Arg53 side chain placing the guanidinium moiety in close proximity to HA2:Asp19, the preferred rotamer for the Arg side chain cannot be assigned unambiguously. Various rotamers consistent with the observed electron density result in charged atom contact distances of ~4–4.5Å. Although somewhat longer than expected for a salt bridge that would make a major contribution to antibody binding, this discrepancy may be due in part to shielding by a nearby sulfate from the crystallization solution, which is sandwiched between the VL:Arg53–Arg54 side chains.
- 41.Most of the group 2 HAs with Asp19Asn are restricted to a single lineage of the H7 subtype, and disproportionate sampling in birds in this region may exaggerate the prevalence of Asp19Asn substitutions in the H7 population. See discussion in the SOM on Science Online.
- 42.Neutralization of H15 has not been tested, but the KD for CR8020 binding is comparable to that of H7, which is neutralized.
- 43.Neutralization of H14 viruses has not been tested, but comparable binding of CR8020 to H4 and H14 suggests that H14 will not be neutralized.
- 44.H14 also has a Glu325G mutation in HA1, which has a negligible effect on CR8020 in a HK68 background.
- 45.This scenario is reminiscent of the group 1 restriction of CR6261, which cannot interact with group 2 viruses, such as H3 and H7, at least in part due to a conserved glycan at HA1:Asn38, and the more general and well-documented use of glycans to mask and unmask surfaces to evade immune recognition, such as vividly illustrated in the evolution of human H1N1 viruses (12, 56)
- 46.Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 47.In contrast, a control mAb against the HA1 head bound to A/Wisconsin/67/2005 HA in all three conformations and binding was only lost after DTT treatment, which dissociates HA1 from HA2 in the post-fusion state.
- 48.As evidenced by the fact that in this case treatment with DTT did not diminish CR8057 binding to A/Wisconsin/67/2005 HA. In line with its narrow spectrum of neutralization (Table S1), CR8057 did not bind to any conformation of the HAs of A/Hong Kong/1/1968 or A/Hong Kong/24/1985.
- 49.Since the initial crystals only appeared between 3 and 7 days after the start of the experiment, CR8020 must be capable of retaining HA in the pre-fusion state for several days at low pH.
- 50.Xu R, Wilson IA. Structural characterization of an early fusion intermediate of influenza virus hemagglutinin. J Virol. 2011;85:5172. doi: 10.1128/JVI.02430-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Russ G, Polakova K, Kostolansky F, Styk B, Vancikova M. Monoclonal antibodies to glycopolypeptides HA1 and HA2 of influenza virus haemagglutinin. Acta Virol. 1987;31:374. [PubMed] [Google Scholar]
- 52.Vareckova E, Mucha V, Wharton SA, Kostolansky F. Inhibition of fusion activity of influenza A haemagglutinin mediated by HA2-specific monoclonal antibodies. Arch Virol. 2003;148:469. doi: 10.1007/s00705-002-0932-1. [DOI] [PubMed] [Google Scholar]
- 53.Yoshida R, et al. Cross-protective potential of a novel monoclonal antibody directed against antigenic site B of the hemagglutinin of influenza A viruses. PLoS Pathog. 2009;5:e1000350. doi: 10.1371/journal.ppat.1000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hashem AM, et al. Universal antibodies against the highly conserved influenza fusion peptide cross-neutralize several subtypes of influenza A virus. Biochem Biophys Res Commun. 2010;403:247. doi: 10.1016/j.bbrc.2010.11.030. [DOI] [PubMed] [Google Scholar]
- 55.Stropkovska A, et al. Broadly cross-reactive monoclonal antibodies against HA2 glycopeptide of Influenza A virus hemagglutinin of H3 subtype reduce replication of influenza A viruses of human and avian origin. Acta Virol. 2009;53:15. [PubMed] [Google Scholar]
- 56.Wei CJ, et al. Cross-neutralization of 1918 and 2009 influenza viruses: role of glycans in viral evolution and vaccine design. Sci Transl Med. 2010;2:24ra21 . doi: 10.1126/scitranslmed.3000799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Skehel JJ, Wiley DC. Influenza haemagglutinin. Vaccine. 2002;20(Suppl 2):S51. doi: 10.1016/s0264-410x(02)00131-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


