Abstract
Background
Several sequence based genotyping schemes have been developed for Leptospira spp. The objective of this study was to genotype a collection of clinical and reference isolates using the two most commonly used schemes and compare and contrast the results.
Methods and Findings
A total of 48 isolates consisting of L. interrogans (n = 40) and L. kirschneri (n = 8) were typed by the 7 locus MLST scheme described by Thaipadungpanit et al., and the 6 locus genotyping scheme described by Ahmed et al., (termed 7L and 6L, respectively). Two L. interrogans isolates were not typed using 6L because of a deletion of three nucleotides in lipL32. The remaining 46 isolates were resolved into 21 sequence types (STs) by 7L, and 30 genotypes by 6L. Overall nucleotide diversity (based on concatenated sequence) was 3.6% and 2.3% for 7L and 6L, respectively. The D value (discriminatory ability) of 7L and 6L were comparable, i.e. 92.0 (95% CI 87.5–96.5) vs. 93.5 (95% CI 88.6–98.4). The dN/dS ratios calculated for each locus indicated that none were under positive selection. Neighbor joining trees were reconstructed based on the concatenated sequences for each scheme. Both trees showed two distinct groups corresponding to L. interrogans and L. kirschneri, and both identified two clones containing 10 and 7 clinical isolates, respectively. There were six instances in which 6L split single STs as defined by 7L into closely related clusters. We noted two discrepancies between the trees in which the genetic relatedness between two pairs of strains were more closely related by 7L than by 6L.
Conclusions
This genetic analysis indicates that the two schemes are comparable. We discuss their practical advantages and disadvantages.
Author Summary
Two independent multilocus sequence based genotyping schemes (denoted here as 7L and 6L for schemes with 7 and 6 loci, respectively) are in use for Leptospira spp., which has led to uncertainty as to which should be adopted by the scientific community. The purpose of this study was to apply the two schemes to a single collection of pathogenic Leptospira, evaluate their performance, and describe the practical advantages and disadvantages of each scheme. We used a variety of phylogenetic approaches to compare the output data and found that the two schemes gave very similar results. 7L has the advantage that it is a conventional multi-locus sequencing typing (MLST) scheme based on housekeeping genes and is supported by a publically accessible database by which genotypes can be readily assigned as known or new sequence types by any investigator, but is currently only applicable to L. interrogans and L. kirschneri. Conversely, 6L can be applied to all pathogenic Leptospira spp., but is not a conventional MLST scheme by design and is not available online. 6L sequences from 271 strains have been released into the public domain, and phylogenetic analysis of new sequences using this scheme requires their download and offline analysis.
Introduction
Leptospirosis is a common zoonotic disease worldwide, with a particularly high prevalence in warm humid countries [1]–[4]. About 350,000 severe cases of leptospirosis are estimated to occur annually, with case fatality reports up to 50% [5]–[7]. Reported cases are likely to be a gross under-estimate of global incidence rates, the result of a combination of factors including lack of surveillance, diagnostics and notification in those countries with the highest disease burden. Leptospirosis is currently considered a globally re-emerging disease, with frequent outbreaks in South East Asia (including Thailand, India, The Philippines and Sri Lanka) as well as in Latin America [3], [8]–[14]. International travel also leads to presentation of leptospirosis cases in settings where incidence is low and clinicians are unfamiliar with its clinical manifestations [7],[15].
Identification and typing of Leptospira species plays an important role in understanding disease epidemiology and pathogenicity, together with the development of diagnostic tools, effective vaccines and preventive strategies. During the last three decades many molecular typing methods have been proposed for Leptospira spp. These include DNA-DNA hybridization analysis [16]–[19], randomly amplified polymorphic DNA (RAPD) fingerprinting [20], arbitrarily primed PCR (AP-PCR) [21], [22], pulsed field gel electrophoresis (PFGE) [23], [24], restriction fragment length polymorphism (RFLP) analysis [25], [26], bacterial typing methods based on insertion sequences (IS) [27], detection of variable number of tandem repeats (VNTR) [28], [29], rrs sequencing [30]–[32], and sequencing of specific genes or gene fragments including rpoB, gyrB, secY and ligB [33]–[37].
Multilocus sequencing typing (MLST) has been widely adopted for the study of bacterial evolution and population biology of a large number of microbial species [38], and represents the leading molecular method for bacterial genotyping. MLST based on 7 housekeeping loci has been developed for Leptospira [39], and is supported by a publically accessible database by which genotypes can be readily assigned as known or new sequence types. An alternative sequence based genotyping scheme of 6 loci including housekeeping genes, a 16S rRNA gene and genes encoding surface-expressed proteins has also been developed and used by several groups. This has led to uncertainty as to which scheme should be adopted. The aim of the current study was to compare the two schemes in terms of their discriminatory ability, both within and between species, by generating data using both schemes for a single set of isolates. We also discuss the practical aspects relating to each scheme.
Materials and Methods
Leptospira isolates and DNA isolation
The Leptospira isolates used in this study and their providers are shown in Table 1. Genomic DNA was extracted from laboratory bacterial cultures as described previously [39], [40].
Table 1. Leptospira isolates used in this study.
| Species | Serovar | Strain | ST (7 loci scheme)# | Origin | Source* |
| L. interrogans | Copenhageni | M 20 | 17 | Reference | Aus& KIT |
| L. interrogans | Guaratuba | An 7705 | 37 | Reference | Aus |
| L. interrogans | Hardjo | Hardjoprajitno | 20 | Reference | Aus& KIT |
| L. interrogans | Icterohaemorrhagiae | RGA | 17 | Reference | Aus& KIT |
| L. interrogans | Kenniwicki | LT1026 | 37 | Reference | KIT |
| L. interrogans | Kuwait | 136/2/2 | 26 | Reference | MORU |
| L. interrogans | Lai | Lai | 1 | Reference | GenBank† |
| L. interrogans | Pomona | Pomona | 37 | Reference | Aus& KIT |
| L. interrogans | Portlandvere | MY1039 | 37 | Reference | ND |
| L. interrogans | Schueffneri | Vleermuis90C | 3 | Reference | Aus |
| L. interrogans | Sumneri | Sumner | 7 | Reference | Aus& KIT |
| L. interrogans | Valbuzzi | Valbuzzi | 61 | Reference | Aus& KIT |
| L. interrogans | Autumanlis | 3 | 34 | Thailand | MORU |
| L. interrogans | Autumnalis | 86 | 34 | Thailand | MORU |
| L. interrogans | Autumnalis | L0020 | 34 | Thailand | MORU |
| L. interrogans | Autumnalis | L0661 | 34 | Thailand | MORU |
| L. interrogans | Autumnalis | L1151 | 34 | Thailand | MORU |
| L. interrogans | Autumnalis | UT227 | 34 | Thailand | MORU |
| L. interrogans | Autumnalis | 548 | 34 | Thailand | MORU |
| L. interrogans | Autumnalis | 729 | 34 | Thailand | MORU |
| L. interrogans | Autumnalis | LP101 | 22 | Thailand | MORU |
| L. interrogans | Bataviae | L1111 | 42 | Thailand | MORU |
| L. interrogans | Bataviae | UT229 | 46 | Thailand | MORU |
| L. interrogans | Bataviae | UT234 | 46 | Thailand | MORU |
| L. interrogans | Medanensis | L0448 | 46 | Thailand | MORU |
| L. interrogans | Medanensis | L0887 | 46 | Thailand | MORU |
| L. interrogans | Medanensis | L0941 | 46 | Thailand | MORU |
| L. interrogans | Pomona | UT364 | 38 | Thailand | MORU |
| L. interrogans | Pyrogenes | UD009 | 37 | Thailand | MORU |
| L. interrogans | Pyrogenes | L0443 | 49 | Thailand | MORU |
| L. interrogans | Pyrogenes | L0374 | 49 | Thailand | MORU |
| L. interrogans | Unknown | 654 | 33 | Thailand | MORU |
| L. interrogans | Unknown | M04 | 34 | Thailand | MORU |
| L. interrogans | Unknown | M08 | 34 | Thailand | MORU |
| L. interrogans | Unknown | UT126 | 40 | Thailand | MORU |
| L. interrogans | Unknown | L1085 | 42 | Thailand | MORU |
| L. interrogans | Unknown | L0996 | 46 | Thailand | MORU |
| L. interrogans | Unknown | UT053 | 46 | Thailand | MORU |
| L. interrogans | Unknown | M10 | 49 | Thailand | MORU |
| L. interrogans | Unknown | L1207 | 26 | Thailand | MORU |
| L. kirschneri | Grippotyphosa | Moskva V | 110 | Reference | KIT |
| L. kirschneri | Mozdok | 5621 | 117 | Reference | KIT |
| L. kirschneri | Ratnapura | Wumalasena | 116 | Reference | KIT |
| L. kirschneri | Tsaratsovo | B 81/7 | 115 | Reference | KIT |
| L. kirschneri | Vanderhoedeni | Kipod 179 | 110 | Reference | KIT |
| L. kirschneri | Grippotyphosa | UT130 | 68 | Thailand | MORU |
| L. kirschneri | Unknown | M06 | 68 | Thailand | MORU |
| L. kirschneri | Unknown | M07 | 71 | Thailand | MORU |
STs are not shown for the 6 loci scheme because this is not supported by a MLST website, and allelic numbers, profiles and STs have not been assigned to the sequence data.
*MORU, Mahidol-Oxford Tropical Medicine Research Unit, Bangkok, Thailand (MORU); KIT, KIT Biomedical Research, WHO/FAO/OIE Collaborating Center for Reference & Research on Leptospirosis, Amsterdam, Netherlands; Aus, WHO/FAO/OIE Collaborating Center for Reference & Research on Leptospirosis, Brisbane, Australia. Isolates from two different sources were identified using one of two MLST schemes only.
†: in silico analysis was performed on this isolate.
Genotyping
All isolates were evaluated using both genotyping schemes [39],[40]. The MLST scheme described by Thaipadungpanit et al. (2007), is based on pntA, sucA, pfkB, tpiA, mreA, glmU and fadD [39], and the scheme described by Ahmed et al. (2006) is based on adk, icdA, secY, rrs2, lipL41, and lipL32 [40]. The terms 7L and 6L have been adopted throughout to refer to the 7 and 6 gene schemes, respectively. No modifications were made to the published primers or cycling conditions of 7L. Table 2 lists the primer pairs used for 6L. Four of the 12 primers (adk-F, adk-R, secY-R and icdA-R) were modified compared with the published 6L scheme, and used in a repeat PCR reaction in the event that the original primers failed to generate an amplicon. Cycling conditions were as described previously for 6L, with the exception that reactions using the four new 6L primers had a reduced annealing temperature of 54°C. Sequence data were edited using SeqMan software contained within the DNASTAR package (DNASTAR Inc., Wisconsin, USA). The region of sequence used to define each locus of 7L was as described previously [39], but the region used to define each locus of 6L was altered as follows. Three loci (secY, lipL32 and lipL41) were changed because the published PCR product and the region of sequence used to define the locus were either identical (secY and lipL32) or different by just two bp [40]. This meant that we were unable to obtain high quality sequence traces for the first 10–20 bases of the amplicon, and so trimmed the sequence in frame by approximately 20 bp at either end for all three genes. The other 3 published loci of 6L (adk, icdA and rrs2), were trimmed by one or two bases to put them in frame, which simplifies the analysis of synonymous and non-synonymous substitutions. The sequence start and end points for the 6 loci of 6L are shown in Table 2.
Table 2. Primers for 6 locus genotyping scheme used during this study [39].
| Gene | Published primers (5′- 3′) | New primers (5′- 3′) | Location of sequence used to define MLST locus# | Size of MLST locus (bp) |
| adk | F-gggctggaaaaggtacacaa | F-acattatcttcatgggacctcc | 3458789–3458361 | 429 |
| R-acgcaagctccttttgaatc | R-ttacacaagctccctttgaat | |||
| icdA | F-gggacgagatgaccaggat | 3980926–3980372 | 555 | |
| R-ttttttgagatccgcagcttt | R-cttttttgagatctccggcttt | |||
| lipL32 | F-atctccgttgcactctttgc | 1667072–1666641 | 432 | |
| R-accatcatcatcatcgtcca | ||||
| lipL41 | F-taggaaattgcgcagctaca | 3603644–3604120 | 477 | |
| R-gcatcgagaggaattaacatca | ||||
| rrs2 | F-catgcaagtcaagcggagta | 1862535–1862984 | 450 | |
| R-agttgagcccgcagttttc | ||||
| secY | F-atgccgatcatttttgcttc | 3459402–3458902 | 501 | |
| R-ccgtcccttaattttagacttcttc | R-ccttcctttaattttagactttttc |
The alleles at each of the 7L loci were assigned and the sequence type (ST) defined using the publically accessible Leptospira MLST website (http://leptospira.mlst.net/). Allelic numbers, profiles and STs were not generated for the 6L data.
Sequence analysis
Sequence alignment, nucleotide diversity and reconstruction of phylogenetic trees were performed using Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0 [41]. Mean pairwise distances (p distance) were calculated using the Kimura Two Parameter nucleotide substitution model. Synonymous (dS) and non-synonymous (dN) nucleotide substitutions were calculated based on the Modified Nei-Gojobori method with Jukes Cantor correction using MEGA 4. Neighbor joining trees were reconstructed based on concatenated sequences of each scheme using the Kimura Two-Parameter substitution model. Gene order of the concatenated sequences were glmU, pntA, sucA, fadD, tpiA, pfkB, and mreA for 7L, and adk, icdA, lipL32, lipL41, rrs2, and secY for 6L. Discriminatory ability (D value) and 95% confidence intervals (CI) were estimated as described previously [42], [43]. These values were verified using the LIAN web tool housed on pubmlst.org [44]. A sliding window analysis of within- and between-species variation was carried out using DNAsp v. 5.0 [45]. An initial “window” of 400-bp was selected, as this is roughly equivalent to a single allele. The first window was thus from base 1 to base 400 of the concatenated sequence. From this we took each species in turn and calculated the average number of nucleotide differences per site over all pairwise comparisons (π), to give the within species polymorphism. Similarly, we calculated the number of fixed differences between species (substitutions) per site to gauge the divergence between L. interrogans and L. kirschneri. The window region was then moved along 50-bp and these parameters recalculated. GenBank accession numbers of 6L generated sequences are JF509178–JF509357.
Results
Discriminatory power of the two genotyping schemes
A total of 48 strains and isolates belonging to L. interrogans (n = 40) and L. kirschneri (n = 8) were included in this study, of which 17 were reference strains and 31 were clinical isolates – further referred to as strains (Table 1). Nine strains had been evaluated previously by both schemes [39], [40], and 39 strains typed previously by only one of the two schemes were typed by the other scheme during this study. Two strains (a Thai clinical isolate strain L1207 of unknown serovar and a reference strain of serovar Kuwait strain 136/2/2) could not be typed using 6L as both had a deletion of three nucleotides in the lipL32 sequence. These two strains were excluded from further analysis.
7L resolved the 46 strains into 21 STs, shown in Table 1. 6L data were analysed off line, and the alleles at the six loci given arbitrary allelic numbers to construct an allelic profile and determine the number of genotypes. This demonstrated a total of 30 genotypes (data not shown). Overall levels of diversity (D) were comparable for the 7L and 6L schemes (92.0 (95% CI 87.5–96.5) and 93.5 (95% CI 88.6–98.4), respectively). The discriminatory ability per locus ranged from 59% (sucA) to 87% (glmU and mreA) for 7L and 66% (rrs2) to 92% (secY) for 6L (Table 3). All D values were verified using the LIAN web tool housed at pubmlst.org and found to be identical to the values shown. The majority of alleles of both schemes were species specific (that is, found in either L. interrogans or L. kirscheri but not both). There were three exceptions where alleles were found in both species, as follows: 7L, allele 1 of sucA; 6L, one allele of lipL32 and one allele of rrs2.
Table 3. Discriminatory ability of two genotyping schemes and their respective loci.
| Number of alleles | p-distance# | dN/dS* | Discriminatory ability (%) (95% confidence intervals) | |
| 7 loci scheme (21 STs) | 92.0 (87.5–96.5) | |||
| glmU | 11 | 2.30% | 0.073 | 86.9 (82.9–90.8) |
| pntA | 11 | 3.60% | 0.012 | 64.3 (49.0–79.5) |
| sucA | 7 | 4.70% | 0.007 | 59.3 (45.2–73.5) |
| fadD | 7 | 4.00% | 0.066 | 76.3 (69.1–83.5) |
| tpiA | 10 | 6.10% | 0.093 | 84.7 (79.1–90.4) |
| pfkB | 14 | 4.70% | 0.048 | 83.4 (76.0–90.7) |
| mreA | 12 | 4.20% | 0.007 | 86.9 (82.1–91.6) |
| Concatenated sequence of 6 loci (2,844 nt) | 3.60% | |||
| 6 loci scheme (30 genotypes) | 93.5 (88.6–98.4) | |||
| adk | 10 | 6.70% | 0.057 | 70.2 (57.2–83.2) |
| icdA | 12 | 2.50% | 0.022 | 74.8 (62.7–86.8) |
| lipL32 | 7 | 0.50% | 0.154 | 71.9 (62.3–81.5) |
| lipL41 | 7 | 2.70% | 0.01 | 81.9 (77.4–86.5) |
| rrs2 | 6 | 0.40% | ND | 66.3 (58.1–74.4) |
| secY | 20 | 5.50% | 0.019 | 91.8 (87.4–96.2) |
| Concatenated sequence of 7 loci (3,165 nt) | 2.30% |
p distances were estimated based on the Kimura Two Parameter nucleotide substitution model.
*dN/dS were estimated based on the Modified Nei-Gojobori Method with Jukes Cantor correction using MEGA 4. The values shown represent a combined value for L. interrogans and L. kirschneri. dN/dS was not estimated for rrs2 as this does not encode a protein.
Nucleotide diversity of genetic loci
Overall nucleotide diversity (based on concatenated sequences) for the 46 isolates was 3.6% and 2.3% for 7L and 6L, respectively (Table 3). The diversity within L. interrogans was lower than that within L. kirschneri (0.5% and 1.1% for 7L, and 0.4% and 0.8% for 6L, respectively). Table 3 also details the nucleotide diversity by locus. This ranged from 3.6% to 6.1% for 7L, and 0.5% to 6.7% for 6L. The lowest diversity was observed for lipL32 and rrs2 of 6L. The dN/dS ratios calculated for each locus indicated that none were under positive selection (that is, all values were lower than 1) (Table 3).
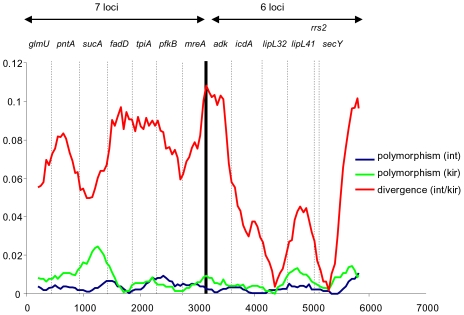
A sliding window analysis of the concatenated sequences was performed to provide a visual comparison of the degree of polymorphism within both species, and the level of divergence between them. This revealed a generally higher level of variation within L. kirschneri compared to L. interrogans, particularly at sucA (7L) and to a lesser extent lipL41 (6L), although the sample size for the former species was very small (n = 8) (Figure 1). This analysis confirmed that the degree of within species polymorphism showed very little difference between the 7L and 6L scheme. However, 7L tended to provide better resolution between species, which was largely accounted for by the low level of divergence for lipL32 and rrs2 of 6L.
Figure 1. Sliding window analysis of concatenated sequence of all 13 loci.
Sliding window analysis of concatenated sequence of all 13 loci, carried out using DNAsp v 5 using a window size of 400-bp, a step size of 50-bp, and points based on the mid-point of each window (i.e. the first point is at position 200). The names of the individual loci are shown. Three plots are given to represent the level of polymorphism within each of the two species, and the level of diversity between them. In terms of the within species variation, there is little difference between the two schemes and both point to generally higher levels of variation within L. kirschneri than L. interrogans. However, there are two loci used in the 6L scheme that are highly conserved between species (lipL32 and rrs2), which means that in general the 7L scheme provides better between-species resolution.
Relatedness of Leptospira spp. inferred from the two genotyping schemes
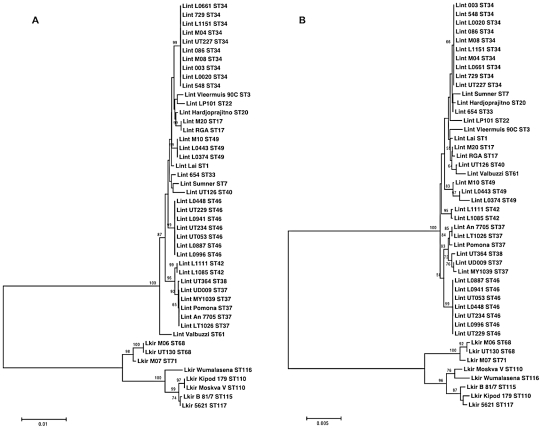
Neighbor joining trees were reconstructed for 7L and 6L based on the concatenated sequences of their respective loci (Figure 2). Both trees showed two distinct groups corresponding to L. interrogans and L. kirschneri. There were also several obvious similarities within L. interrogans between the two trees. For example, the clonal structure of ST34 and ST46 as defined by 7L was maintained by 6L. A common finding, however, was that 6L had a tendency to split single STs as defined by 7L into closely related clusters. For example, the three isolates designed as ST49 by 7L were split into three different genotypes by 6L. Further examples of splitting of a clone by the 6L scheme were 7L ST42, ST37, ST68 and ST17. A number of discrepancies were noted between the two trees. Two strains of L. kirschneri (strains Moskva V and Kipod 179) were designated by 7L as ST110, but these were resolved into different genotypes by 6L. These two strains differed by 9 nucleotides over 3 loci, with secY accounting for 7 of these. A difference was also noted for L. interrogans strain 654 (a Thai clinical isolate), which was closely related to L. interrogans strain Hardjoprajitno by 6L (differing by only 1 nucleotide), but was more distantly related by 7L (differing by 11 nucleotides over 6 loci).
Figure 2. Neighbor joining trees of the 7L scheme and the 6 loci scheme.
Neighbor joining trees reconstructed based on concatenated sequences of the 7L scheme (3,165 bp) (A), and the 6 loci scheme (2,844 bp) (B). Each bacterial strain is labeled by the following string: abbreviation of species name (Lint- L. interrogans, Lkir- L. kirschneri), strain name, and (for the 7L scheme only) sequence type (ST).
Discussion
The authors of this paper include representatives of the scientific groups that reported two independent genotyping schemes for Leptospira spp. Here, we provide the scientific community with the findings of a study that compared and contrasted the two schemes, together with a discussion of the practical aspects related to undertaking each.
The two schemes are unrelated and different by design. 7L was founded on a conventional strategy for MLST of selecting 7 housekeeping genes that were distributed around the genome and were not under positive selection. The design of 6L varied from this in that 6 loci were selected from different functional categories. For example, lipL41 and lipL32 encode surface expressed proteins that would be expected to be under positive selection as a result of being immunogenic and a target for the host response. At the other end of the spectrum, rrs2 is one of two 16S rRNA genes that would be predicted to be highly conserved.
Contrary to our expectations, we did not find that any of the 6L genes were under positive selection. More genotypes were resolved by 6L than by 7L, in part a function of the high number of alleles for secY. Analysis of genetic diversity indicated that there was little difference in within-species variation difference between the two schemes, both pointing to generally higher levels of variation within L. kirschneri than L. interrogans. The conserved nature of two loci used in 6L (lipL32 and rrs2), resulted in the finding on sliding window analysis that 7L provided better between-species resolution. Interestingly we noted that rrs2 of 6L showed a higher D value than the housekeeping gene sucA of 7L. Although this is an exception to the general rule that housekeeping metabolic genes provide more discrimination than conserved genes such as those encoding ribosomal RNA, such an observation is not unprecedented [46].
6L has been applied to six pathogenic Leptospira spp. [40], which compares favorably with 7L which was designed for the two closely related species L. interrogans and L. kirschneri. However, this disadvantage of 7L will be resolved within the next 12 months; the scheme has already been extended to L. borpetersenii (manuscript in preparation), and the laboratory work to extend this to all pathogenic species is now completed. These improvements will be made publicly available by the end of 2011.
Conversely, the 6L scheme does not conform to the original concept of MLST as it includes a non-housekeeping gene (rrs2), and genes that encode cell surface proteins. Furthermore, the sequence start and stop sites used to define the allele for each locus were not provided in the original description of 6L scheme and so could not be performed based on the published methodology alone, although these have been detailed in this study. Minor changes were necessary to the start and stop sites, but we think it unlikely that this led to a change in the performance of the scheme.
The 6L scheme is not associated with a publically accessible website that allows an investigator to compare new data with existing sequence data. 6L has recently been applied to an extended set of strains and isolates (n = 271) encompassing a wide diversity of hosts and geographic regions [47], providing a rich source of sequence data that has been released into the public domain (GenBank). Comparative phylogenetic analysis by individual investigators will require downloading and storage of these data. In contrast, a website for 7L was launched at the time of publication and is regularly maintained and curated. At least one representative of each ST is recorded in a downloadable spreadsheet, providing a mechanism by which a picture of global bacterial diversity can be developed over time. This is easy to use, provides tools for comparison of a given strain with all of the other strains in the database, is more suited to investigators with limited phylogenetic training and experience, and so has the power to reach a wider audience.
In conclusion, we have provided detailed comparisons of two major genotyping schemes for Leptospira spp., and have described their advantages and disadvantages. 7L complies with the philosophy of MLST (housekeeping genes only supported by website), but will not be ready for use for the study of all pathogenic Leptospira spp. until the end of 2011. In the meantime, a bioinformatics analysis of the discriminatory power of 4 genes (three of which are not present in either scheme) as well as a new scheme with 7 loci both limited to L. interrogans and L. kirschneri have been reported [48], [49], adding further diversity to the tools available for the phylogenetic study of Leptospira spp. There is a pressing need for consensus within the leptospirosis community as to the preferred genotyping scheme, an essential step if the wealth of knowledge gathered for other bacterial species based on detailed analysis within a single scheme is to be replicated for Leptospira spp. Both schemes contain highly discriminative and less discriminative loci. While it is feasible to formulate a consensus MLST combining the most discriminative housekeeping genes from both schemes, we have resisted the temptation of presenting an interim scheme that has not been extensively validated. Instead, we aim to expedite the release of the 7L MLST scheme for all the major pathogenic species, and recommend its use for the study of the global epidemiology of pathogenic Leptospira spp.
Footnotes
The authors have declared that no competing interests exist.
This study was funded by the Wellcome Trust. SJP is supported by the National Institute for Health Research (NIHR) Cambridge Biomedical Research Centre. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.World Health Organization. Human leptospirosis: Guidance for diagnosis, surveillance and control. Geneva: World Health Organization & International Leptospirosis Society; 2003. [Google Scholar]
- 2.Faine SB, Adler B, Bolin C, Perolat P. Leptospira and leptospirosis. Melbourne, Australia: MediSci; 1999. [Google Scholar]
- 3.Bharti AR, Nally JE, Ricaldi JN, Matthias MA, Diaz MM, et al. Leptospirosis: a zoonotic disease of global importance. Lancet Infect Dis. 2003;3:757–771. doi: 10.1016/s1473-3099(03)00830-2. [DOI] [PubMed] [Google Scholar]
- 4.Levett PN. Leptospirosis. Clin Microbiol Rev. 2001;14:296–326. doi: 10.1128/CMR.14.2.296-326.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. World Health Organization (WHO) and International Leptospirosis Society (ILS), 2006; online at www.leptonet.net.
- 6.McBride AJ, Athanazio DA, Reis MG, Ko AI. Leptospirosis. Curr Opin Infect Dis. 2005;18:376–386. doi: 10.1097/01.qco.0000178824.05715.2c. [DOI] [PubMed] [Google Scholar]
- 7.Hartskeerl RA. Leptospirosis: current status and future trends. Indian J Med Microbiol. 2006;24:309. doi: 10.4103/0255-0857.29404. [DOI] [PubMed] [Google Scholar]
- 8.Agampodi S, Peacock SJ, Thevanesam V. The potential emergence of leptospirosis in Sri Lanka. Lancet Infect Dis. 2009;9:524–526. doi: 10.1016/S1473-3099(09)70211-7. [DOI] [PubMed] [Google Scholar]
- 9.Koizumi N, Gamage CD, Muto M, Kularatne SA, Budagoda BD, et al. Serological and genetic analysis of leptospirosis in patients with acute febrile illness in Kandy, Sri lanka. Jpn J Infect Dis. 2009;62:474–475. [PubMed] [Google Scholar]
- 10.McCurry J. Philippines struggles to recover from typhoons. Lancet. 2009;374:1489. doi: 10.1016/s0140-6736(09)61888-2. [DOI] [PubMed] [Google Scholar]
- 11.World Health Organization. Leptospirosis, India: report of the investigation of a post-cyclone outbreak in Orissa, November 1999. Wkly Epidemiol Rec. 2000;75:217–23. [PubMed] [Google Scholar]
- 12.Sanders EJ, Rigau-Perez JG, Smits HL, Deseda CC, Vorndam VA, et al. Increase of leptospirosis in dengue-negative patients after a hurricane in Puerto Rica in 1996. Am J Trop Med Hyg. 1999;61:935–941. doi: 10.4269/ajtmh.1999.61.399. [DOI] [PubMed] [Google Scholar]
- 13.Ko AI, Galvao RM, Ribeiro Dourado CM, Johnson WD, Jr, Riley LW. Urban epidemic of severe leptospirosis in Brazil. Salvador Leptospirosis Study Group. Lancet. 1999;354:820–825. doi: 10.1016/s0140-6736(99)80012-9. [DOI] [PubMed] [Google Scholar]
- 14.Zaki SR, Shieh WJ. Leptospirosis associated with outbreak of acute febrile illness and pulmonary haemorrhage, Nicaragua, 1995. The Epidemic Working Group at Ministry of Health in Nicaragua. Lancet. 1996;347:535–536. doi: 10.1016/s0140-6736(96)91167-8. [DOI] [PubMed] [Google Scholar]
- 15.Lau C, Smythe L, Weinstein P. Leptospirosis: an emerging disease in travellers. Travel Med Infect Dis. 2010;8:33–39. doi: 10.1016/j.tmaid.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 16.Yasuda PH, Steigerwalt AG, Sulzer KR, Kaufmann AF, Rogers FC, et al. Deoxyribonucleic acid relatedness between serogroups and serovars in the Family Leptospiraceae with proposals for seven new Leptospira species. Int J Syst Bacteriol. 1987;37:407–415. doi: 10.1099/00207713-49-2-839. [DOI] [PubMed] [Google Scholar]
- 17.Ramadass P, Jarvis BD, Corner RJ, Penny D, Marshall RB. Genetic characterization of pathogenic Leptospira species by DNA hybridization. Int J Syst Bacteriol. 1992;42:215–219. doi: 10.1099/00207713-42-2-215. [DOI] [PubMed] [Google Scholar]
- 18.Brenner DJ, Kaufmann AF, Sulzer KR, Steigerwalt AG, Rogers FC, et al. Further determination of DNA relatedness between serogroups and serovars in the family Leptospiraceae with a proposal for Leptospira alexanderi sp. nov. and four new Leptospira genomospecies. Int J Syst Bacteriol. 1999;49 Pt 2:839–858. doi: 10.1099/00207713-49-2-839. [DOI] [PubMed] [Google Scholar]
- 19.Perolat P, Chappel RJ, Adler B, Baranton G, Bulach DM, et al. Leptospira fainei sp. nov., isolated from pigs in Australia. Int J Syst Bacteriol. 1998;48 Pt 3:851–858. doi: 10.1099/00207713-48-3-851. [DOI] [PubMed] [Google Scholar]
- 20.Corney BG, Colley J, Djordjevic SP, Whittington R, Graham GC. Rapid identification of some Leptospira isolates from cattle by random amplified polymorphic DNA fingerprinting. J Clin Microbiol. 1993;31:2927–2932. doi: 10.1128/jcm.31.11.2927-2932.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ralph D, McClelland M, Welsh J, Baranton G, Perolat P. Leptospira species categorized by arbitrarily primed polymerase chain reaction (PCR) and by mapped restriction polymorphisms in PCR-amplified rRNA genes. J Bacteriol. 1993;175:973–981. doi: 10.1128/jb.175.4.973-981.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perolat P, Merien F, Ellis WA, Baranton G. Characterization of Leptospira isolates from serovar Hardjo by ribotyping, arbitrarily primed PCR, and mapped restriction site polymorphisms. J Clin Microbiol. 1994;32:1949–1957. doi: 10.1128/jcm.32.8.1949-1957.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herrmann JL, Baril C, Bellenger E, Perolat P, Baranton G, et al. Genome conservation in isolates of Leptospira interrogans. J Bacteriol. 1991;173:7582–7588. doi: 10.1128/jb.173.23.7582-7588.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herrmann JL, Bellenger E, Perolat P, Baranton G, Saint Girons I. Pulsed-field gel electrophoresis of NotI digests of leptospiral DNA: a new rapid method of serovar identification. J Clin Microbiol. 1992;30:1696–1702. doi: 10.1128/jcm.30.7.1696-1702.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuerner RL, Bolin CA. Nucleic acid probe characterizes Leptospira interrogans serovars by restriction fragment length polymorphisms. Vet Microbiol. 1990;24:355–366. doi: 10.1016/0378-1135(90)90183-v. [DOI] [PubMed] [Google Scholar]
- 26.Zuerner RL, Ellis WA, Bolin CA, Montgomery JM. Restriction fragment length polymorphisms distinguish Leptospira borgpetersenii serovar Hardjo type Hardjo-bovis isolates from different geographical locations. J Clin Microbiol. 1993;31:578–583. doi: 10.1128/jcm.31.3.578-583.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zuerner RL, Alt D, Bolin CA. IS1533-based PCR assay for identification of Leptospira interrogans sensu lato serovars. J Clin Microbiol. 1995;33:3284–3289. doi: 10.1128/jcm.33.12.3284-3289.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Majed Z, Bellenger E, Postic D, Pourcel C, Baranton G, et al. Identification of variable-number tandem-repeat loci in Leptospira interrogans sensu stricto. J Clin Microbiol. 2005;43:539–545. doi: 10.1128/JCM.43.2.539-545.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slack A, Symonds M, Dohnt M, Smythe L. An improved multiple-locus variable number of tandem repeats analysis for Leptospira interrogans serovar Australis: a comparison with fluorescent amplified fragment length polymorphism analysis and its use to redefine the molecular epidemiology of this serovar in Queensland, Australia. J Med Microbiol. 2006;55:1549–1557. doi: 10.1099/jmm.0.46779-0. [DOI] [PubMed] [Google Scholar]
- 30.Hookey JV, Bryden J, Gatehouse L. The use of 16S rDNA sequence analysis to investigate the phylogeny of Leptospiraceae and related spirochaetes. J Gen Microbiol. 1993;139:2585–2590. doi: 10.1099/00221287-139-11-2585. [DOI] [PubMed] [Google Scholar]
- 31.Morey RE, Galloway RL, Bragg SL, Steigerwalt AG, Mayer LW, et al. Species-specific identification of Leptospiraceae by 16S rRNA gene sequencing. J Clin Microbiol. 2006;44:3510–3516. doi: 10.1128/JCM.00670-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cerqueira GM, McBride AJ, Queiroz A, Pinto LS, Silva EF, et al. Monitoring Leptospira strain collections: the need for quality control. Am J Trop Med Hyg. 2010;82:83–87. doi: 10.4269/ajtmh.2010.09-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.La SB, Bui LT, Baranton G, Khamis A, Raoult D. Partial rpoB gene sequencing for identification of Leptospira species. FEMS Microbiol Lett. 2006;263:142–147. doi: 10.1111/j.1574-6968.2006.00377.x. [DOI] [PubMed] [Google Scholar]
- 34.Slack AT, Symonds ML, Dohnt MF, Smythe LD. Identification of pathogenic Leptospira species by conventional or real-time PCR and sequencing of the DNA gyrase subunit B encoding gene. BMC Microbiol. 2006;6:95. doi: 10.1186/1471-2180-6-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Victoria B, Ahmed A, Zuerner RL, Ahmed N, Bulach DM, et al. Conservation of the S10-spc-alpha locus within otherwise highly plastic genomes provides phylogenetic insight into the genus Leptospira. PLoS One. 2008;3:e2752. doi: 10.1371/journal.pone.0002752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahmed A, Engelberts MF, Boer KR, Ahmed N, Hartskeerl RA. Development and validation of a real-time PCR for detection of pathogenic Leptospira species in clinical materials. PLoS One. 2009;4:e7093. doi: 10.1371/journal.pone.0007093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cerqueira GM, McBride AJ, Picardeau M, Ribeiro SG, Moreira AN, et al. Distribution of the leptospiral immunoglobulin-like (lig) genes in pathogenic Leptospira species and application of ligB to typing leptospiral isolates. J Med Microbiol. 2009;58:1173–1181. doi: 10.1099/jmm.0.009175-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maiden MC. Multilocus sequence typing of bacteria. Annu Rev Microbiol. 2006;60:561–588. doi: 10.1146/annurev.micro.59.030804.121325. [DOI] [PubMed] [Google Scholar]
- 39.Thaipadungpanit J, Wuthiekanun V, Chierakul W, Smythe LD, Petkanchanapong W, et al. A dominant clone of Leptospira interrogans associated with an outbreak of human leptospirosis in Thailand. PLoS Negl Trop Dis. 2007;1:e56. 10. doi: 10.1371/journal.pntd.0000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahmed N, Devi SM, Valverde ML, Vijayachari P, Machang'u RS, et al. Multilocus sequence typing method for identification and genotypic classification of pathogenic Leptospira species. Ann Clin Microbiol Antimicrob. 2006;5:28. doi: 10.1186/1476-0711-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 42.Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grundmann H, Hori S, Tanner G. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J Clin Microbiol. 2001;39:4190–4192. doi: 10.1128/JCM.39.11.4190-4192.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haubold B, Hudson RR. LIAN 3.0: detecting linkage disequilibrium in multilocus data. Linkage Analysis. Bioinformatics. 2000;16:847–848. doi: 10.1093/bioinformatics/16.9.847. [DOI] [PubMed] [Google Scholar]
- 45.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 46.Cooper JE, Feil EJ. The phylogeny of Staphylococcus aureus - which genes make the best intra-species markers? Microbiology. 2006;152:1297–1305. doi: 10.1099/mic.0.28620-0. [DOI] [PubMed] [Google Scholar]
- 47.Nalam K, Ahmed A, Devi SM, Francalacci P, Baig M, et al. Genetic affinities within a large global collection of pathogenic Leptospira: implications for strain identification and molecular epidemiology. PLoS One. 2010;5:e12637. doi: 10.1371/journal.pone.0012637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cerqueira GM, McBride AJ, Hartskeerl RA, Ahmed N, Dellagostin OA, et al. Bioinformatics describes novel Loci for high resolution discrimination of Leptospira isolates. PLoS One. 2010;5:e15335. doi: 10.1371/journal.pone.0015335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leon A, Pronost S, Fortier G, Andre-Fontaine G, Leclercq R. Multilocus sequence analysis for typing Leptospira interrogans and Leptospira kirschneri. J Clin Microbiol. 2009;48:581–585. doi: 10.1128/JCM.00543-09. [DOI] [PMC free article] [PubMed] [Google Scholar]