Abstract
Plants commonly respond to pathogen infection by increasing ethylene production, but it is not clear if this ethylene does more to promote disease susceptibility or disease resistance. Ethylene production and/or responsiveness can be altered by genetic manipulation. The present study used mutagenesis to identify soybean (Glycine max L. Merr.) lines with reduced sensitivity to ethylene. Two new genetic loci were identified, Etr1 and Etr2. Mutants were compared with isogenic wild-type parents for their response to different soybean pathogens. Plant lines with reduced ethylene sensitivity developed similar or less-severe disease symptoms in response to virulent Pseudomonas syringae pv glycinea and Phytophthora sojae, but some of the mutants developed similar or more-severe symptoms in response to Septoria glycines and Rhizoctonia solani. Gene-for-gene resistance against P. syringae expressing avrRpt2 remained effective, but Rps1-k-mediated resistance against P. sojae races 4 and 7 was disrupted in the strong ethylene-insensitive etr1-1 mutant. Rps1-k-mediated resistance against P. sojae race 1 remained effective, suggesting that the Rps1-k locus may encode more than one gene for disease resistance. Overall, our results suggest that reduced ethylene sensitivity can be beneficial against some pathogens but deleterious to resistance against other pathogens.
Ethylene is a growth hormone of relatively simple structure that influences many aspects of plant growth, development, and productivity (Matoo and Suttle, 1991; Abeles et al., 1992). Directed genetic manipulation of plant ethylene production or responsiveness has been demonstrated for plant species that include tomato, Arabidopsis, petunia, and tobacco (Bleecker et al., 1988; Hamilton et al., 1990; Klee et al., 1991; Oeller et al., 1991; Ecker, 1995; Wilkinson et al., 1997; Knoester et al., 1998). In many cases, these manipulations are carried out to alter economically important traits such as fruit ripening, floral senescence, or disease tolerance.
Ethylene signaling requires not only ethylene production, but also ethylene perception and response. Ethylene signal transduction pathways are currently the subject of extensive research. A number of ethylene-response mutants have been identified in Arabidopsis, and study of these mutants has been particularly revealing (Ecker, 1995; Kieber, 1997). Arabidopsis ETR1 and its homologs encode proteins with ethylene-binding activity that apparently function as the primary cellular receptors of ethylene. Mutations of ETR1 that cause insensitivity to ethylene are typically dominant to wild-type ETR1. Downstream signaling components of the ethylene-perception pathway have also been identified by mutational analysis, and the Arabidopsis CTR1, EIN2, and EIN3 genes have been isolated and characterized. Mutant alleles of these downstream genes are typically recessive. Ethylene apparently acts by disrupting negative regulation of ethylene-response pathways, and early stages of this pathway involve protein kinases with similarity to the Raf kinase family of signal transduction proteins. Ethylene-responsive promoter elements and transcription factors have also been characterized (Ecker, 1995; Deikman, 1997). Although many steps of the ethylene signal transduction pathway have been identified, it is likely that others remain to be discovered.
Ethylene is produced by plants in response to a wide variety of biotic or abiotic stresses (Matoo and Suttle, 1991; Abeles et al., 1992). The production of ethylene is one of the earliest responses of plants to pathogen attack, and ethylene production is observed in response to diverse viral, bacterial, and fungal plant pathogens (Pegg, 1976; Boller, 1991). Because ethylene formation occurs in many plant-pathogen interactions, the question of its role in disease resistance and disease susceptibility arises. Ethylene may be a stimulus for defense responses that lead to resistance, or conversely, it may play a role in disease symptom development and in the breakdown of endogenous resistance (Boller, 1991; Abeles et al., 1992; Lund et al., 1998).
Roles for ethylene in the activation of plant defense are suggested by the known ability of ethylene to induce the accumulation of pathogenesis-related proteins. Numerous workers have shown that ethylene will induce synthesis of PR-1, β-1,3-glucanase, chitinase, Phe ammonia-lyase, Hyp-rich glycoproteins, osmotin, and other defense-associated proteins (Boller, 1991; Deikman, 1997). Significantly, synthesis of many of these proteins can also be induced by ethylene-independent pathways (Dixon and Lamb, 1990). However, biological elicitation of some defense-associated proteins may require a functional ethylene response (e.g. Penninckx et al., 1996), and a rhizobacterium-stimulated “induced systemic resistance” response was disrupted in Arabidopsis etr1 mutants (Pieterse et al., 1998). Further evidence for a role of ethylene in resistance was provided using tobacco lines made ethylene insensitive by expression of a dominant mutant etr1 transgene from Arabidopsis (Knoester et al., 1998). These ethylene-insensitive tobacco plants displayed susceptibility to Pythium sylvaticum fungi that are usually not pathogenic on tobacco. However, ethylene-insensitive Arabidopsis and tomato lines have not shown excessive susceptibility to Pseudomonas, Xanthomonas, Peronospora, or Fusarium pathogens (Bent et al., 1992; Lawton et al., 1995; Lund et al., 1998).
Although ethylene is involved in the expression of a number of pathogenesis-related plant genes, it can also increase disease symptom severity. Ethylene increased disease severity of verticillium wilt of tomato and of gray mold on rose and carnation flowers and on detached leaves of tomato, pepper, bean, and cucumber (Boller, 1991). Similar correlations between ethylene production and infection-related chlorotic or necrotic symptom development have been demonstrated in other plant species (Gentile and Matta, 1975; Goto et al., 1980; Stall and Hall, 1984; Ben-David et al., 1986). More generally, ethylene is a well-known inducer of necrosis, ripening, chlorosis, and senescence in plants (Matoo and Suttle, 1991; Abeles et al., 1992). These findings imply that ethylene can promote disease symptom development and general disease susceptibility.
Because ethylene appears to increase disease severity, researchers have asked whether insensitivity to ethylene may increase plant tolerance to plant pathogens. Early evidence for this came from studies of a pepper line with enhanced sensitivity to ethylene that exhibited more rapid development of chlorosis in response to Xanthomonas campestris pv vesicatoria, the causal agent of bacterial spot disease (Stall and Hall, 1984). Studies with ethylene-insensitive mutants of Arabidopsis revealed enhanced disease tolerance to Pseudomonas syringae pv tomato or pv maculicola and Xanthomonas campestris pv campestris pathogens (Bent et al., 1992). Disease tolerance can be defined as quantitatively reduced symptom severity despite pathogen growth that is similar to that observed in disease-susceptible control plants. A simple role for ethylene insensitivity in disease tolerance remained uncertain in the studies cited above with Arabidopsis, because tolerance was observed only with ein2 mutants and not with etr1 (ein1) and ein3 mutants (Bent et al., 1992). The ein2 mutations may disrupt a specific branch of the ethylene-response pathway or provide an especially complete block in ethylene signaling. Alternatively, mutation of EIN2 may exert direct or indirect pleiotropic effects on other response pathways (e.g. Cary et al., 1995). Arabidopsis ethylene-insensitive mutants were also used to show that ethylene insensitivity does not disrupt gene-for-gene resistance (resistance gene/avirulence gene-dependent disease resistance) (Bent et al., 1992). In a separate study, systemic acquired resistance was not disrupted in these Arabidopsis ethylene-insensitive mutants (Lawton et al., 1995). These studies suggested that it may be possible to use a screen for ethylene insensitivity as a method to identify disease-tolerant lines in other plant species.
More recently, studies with the Never ripe mutant of tomato provided further evidence of enhanced disease tolerance in ethylene-insensitive plant lines (Lund et al., 1998). Enhanced tolerance was observed in response to both X. campestris pv vesicatoria and Fusarium oxysporum, an important fungal pathogen. It is interesting that the Never ripe mutation disrupts a tomato homolog of Arabidopsis ETR1 (Wilkinson et al., 1995), and in earlier studies the Arabidopsis etr1 mutants did not show enhanced tolerance (Bent et al., 1992). In tobacco lines made ethylene insensitive by expression of a dominant mutant etr1 transgene from Arabidopsis, gene-for-gene resistance against tobacco mosaic virus remained effective (Knoester et al., 1998). However, as mentioned above, these ethylene-insensitive tobacco plants displayed susceptibility to P. sylvaticum fungi that are not usually pathogenic on tobacco (Knoester et al., 1998). A limited number of pathogens were tested in the studies cited above, and different pathogens have very different modes of pathogenicity. In light of the known role of ethylene as an inducer of chlorosis and senescence, but also of pathogenesis-related gene expression, it has remained unclear if the ethylene-insensitivity trait could be used to confer enhanced disease tolerance to plants or if it might, on balance, be more disruptive of resistance.
To examine the potential contribution of ethylene insensitivity to disease tolerance and/or disease susceptibility in an important field crop, we identified ethylene-insensitive mutants of soybean (Glycine max). Soybean mutants displaying reduced ethylene sensitivity were then compared with their parental lines in tests with virulent Septoria glycines, Rhizoctonia solani, Pseudomonas syringae pv glycinea, and Phytophthora sojae pathogens chosen to represent different taxonomic groups and different primary sites of infection. In addition, gene-for-gene resistance responses were monitored using compatible and incompatible races of P. sojae and P. syringae pv glycinea. Mutations that reduce ethylene sensitivity altered plant resistance to some but not all of the soybean pathogens examined in this study, and altered gene-for-gene resistance against some but not all avirulent races of a single pathogen species.
MATERIALS AND METHODS
Plant Material
Mutagenized populations of soybean (Glycine max) derived from 13 experimental breeding lines, including A90-312022, were supplied as M3 seed by Dr. Walter Fehr (Iowa State University, Ames). These M3 seeds were in approximately 24,000 packets each derived from single M2 individuals, with packets bundled to group seed from 10 M2 individuals from each of approximately 2,400 M1 plants. M1 plants were from seed that had been mutagenized with either ethyl methanesulfonate, NMU, or nitrosoguanidine; A90-312022 lines were mutagenized with NMU. To sample the Fehr seed populations in the ethylene triple-response test, pools of 72 seeds were obtained by removing 12 M3 seeds from each of six packets from separate M2 plants from a given M1 plant. Additional M3 seed was furnished by Dr. James Specht (University of Nebraska, Lincoln). This seed was grouped in approximately 4,000 packets from individual M2 plants that came from 1,434 M1 individuals that had been derived from the soybean var Hobbit 87 (Cooper et al., 1991) by NMU mutagenesis. Twelve seeds from each Hobbit 87 M3 packet were used in the ethylene triple-response screen.
Screen for Ethylene-Insensitive Soybean Lines
The method of Bleecker et al. (1988) was modified for use with soybean. Light-tight, gas-tight boxes (40 cm wide × 90 cm long × 25 cm tall) made from one-fourth-inch opaque gray sheets of polyvinyl chloride were constructed with welded seams and fitted with two brass gas ports. The removable top was fitted with a Neoprene gasket and latches. For each ethylene test the box was filled with autoclaved sand to a depth of approximately 2.5 cm, soybeans (typically approximately 1700 seeds) were planted at a depth of 0.5 to 1.0 cm, the sand was watered once, and the lid was secured. Compressed air with 10 μL L−1 (10 ppm) ethylene was bubbled through water and then into one of the two gas ports; the port at the other end released air through a second water trap to prevent inflow of room air. In later tests the flow-through system was replaced with a sealed system in which a small volume of pure ethylene was injected to achieve the desired ethylene concentration, typically 18 μL L−1. After 6 d the box was opened and seedlings were visually scored for the ethylene triple response (short hypocotyl length, exaggerated curl of the hypocotyl hook, and radial swelling of the hypocotyl). Any seedlings not exhibiting the wild-type ethylene triple response were measured, rescued, acclimated in a controlled environment, and then transplanted to 20-cm (diameter) pots and grown to maturity in a greenhouse. The remainder of the seedlings were transplanted in bulk to sand flats and moved to the greenhouse for other mutant screens.
Genetic Analysis
Candidate mutant lines were grown in the field and genetic crosses were performed by manual pollination. F1 and F2 seedlings from self-fertilized F1 plants were then tested for ethylene sensitivity using the ethylene triple-response assay described above. The female parent is listed first in the description of all crosses unless otherwise noted. Each ethylene sensitivity test of an F1 or F2 population typically included 10 or more individuals of both parents as the internal controls. Where overlap in the range of hypocotyl elongation phenotypes for wild-type and weak mutant parents was present, the cutoff values for phenotypic categories were set to equalize as much as possible the proportion of individuals from the parent data sets that fell into the category of the opposite parent.
Septoria glycines Tests
The S. glycines strain used was a purified, virulent strain isolated in Urbana, IL, in 1997 from a field-grown soybean plant. For inoculum preparation, S. glycines spores suspended in sterile distilled water were spread onto potato dextrose agar plates and grown for 1 week at room temperature (Dhingra and Sinclair, 1995). Plates were then flooded with distilled water and gently rubbed with a glass rod to dislodge spores; the liquid was then harvested for use. To compare ethylene-insensitive mutants with their respective parents, experiments were set up as completely randomized factorial designs with two treatments (inoculation with S. glycines or mock inoculation with water), four to nine soybean lines, and three or four replications, depending on the experiment. Five seeds were planted in an 8-cm pot for each replication. Plants were grown in a growth chamber set to a daylength of 16 h with a day temperature of 23.3°C stepped at nightfall to a night temperature of 18.8°C. Daybreak humidity was set to 68%, increasing to 80% 2 h later and for the remainder of the day, to 100% at nightfall, and then decreasing to 82% over a 4-h period and remaining at 82% until daybreak.
Spray-inoculation treatments were applied with a misting bottle when plants were 10 d old, a stage at which the first set of trifoliate leaves was expanded to near-full size. The S. glycines treatment contained S. glycines spores at a concentration of approximately 2 × 106 spores mL−1 in distilled water plus the surfactant Silwet L-77 (OSi Specialties, Danbury, CT) at 100 μL L−1. The control consisted of distilled water with L-77 but no spores. Immediately after spraying, plants were covered for 2 d with a tall, plastic dome and a shade cloth and placed back in the growth chamber. After 2 d the domes were removed and the plants were maintained under growth-chamber conditions for the duration of the experiment. S. glycines symptoms were evaluated 9 d after inoculation on a scale of 0 to 5: 0, no symptoms; 1, slight chlorotic flecking; 2, a few tiny necrotic lesions; 3, many smaller necrotic lesions; 4, a few large lesions (>1 mm in diameter); and 5, many large lesions. For the syringe-inoculation method, a plastic syringe (no needle) was used to lightly rub resuspended S. glycines spores or water alone (no surfactant in either case) over a small area of the underside of one fully expanded, unifoliate leaf on each plant. The rating scale for this technique also ranged from 0 to 5: 0, no symptoms; 1, very small lesions with no chlorosis; 2, small lesions with chlorosis; 3, lesions of intermediate size (1–2 mm); 4, large lesions with little chlorosis; and 5, large lesions with chlorosis.
Pseudomonas syringae pv glycinea Tests
P. syringae pv glycinea race 4 (Kobayashi et al., 1989) carrying pVSP61 (empty plasmid vector) or pV288 (pVSP61 + avrRpt2; Kunkel et al., 1993) were grown from frozen stocks at 28°C on NYG agar (Daniels et al., 1984) and then subcultured in NYG liquid, both containing rifampicin 50 μg/mL and kanamycin 25 μg/mL. Bacteria were harvested by centrifugation and resuspended in 10 mm MgCl2 to an optical density at 600 nm of 0.04 (approximately 4 × 107 colony-forming units/mL) in experiments 1 to 4, and an optical density at 600 nm of 0.01 for experiments 5 and 6. The surfactant Silwet L-77 was then added at a rate of 50 μL L−1. Experiments 1 to 5 consisted of three treatments (water/MgCl2/L-77 containing race 4 [pVSP61], race 4 [pV288], or no added bacteria), four to nine cultivars, and three or four replications, depending on the experiment. Experimental units (five seeds per 9-cm pot) were completely randomized within a particular experiment. Leaves were inoculated by vacuum infiltration when plants were approximately 2 weeks old and the first trifoliate leaf was partially expanded. Plants were then returned to the growth chamber and rated for symptom expression 1 week after infiltration. In experiments 1 to 4, a scale of 0 to 5 was used for rating unifoliate leaf symptoms: 0, healthy, no bacterial damage; 1, slight necrotic flecking; 2, more obvious necrotic lesions; 3, necrosis converging in small, dead sectors; 4, large, dead sectors; and 5, unifoliate leaves completely dead. Experiments 5 and 6 used a new 0 to 5 rating scale focused on both necrosis and chlorosis: 0, healthy, green leaves; 1, slight symptoms, leaves still very green; 2, more visible necrosis and/or slight chlorosis; 3, dispersed chlorosis and/or necrosis covering 20% to 50% of the leaf; 4, large, necrotic areas but leaves still partly green; and 5, leaves senesced. For chlorophyll assays leaf discs were removed from a standardized location on one unifoliate leaf from each of five plants per treatment, discs from each treatment were pooled, and chlorophyll measurements were performed according to the method of Lichtenthaler and Wellburn (1983).
Phytophthora sojae Tests
P. sojae strains of known race designation were obtained from Dr. Cecil Nickell (Urbana, IL) and maintained on lima bean agar plates at 4°C with minimal passaging. To foster rating of tolerance/partial-resistance phenotypes, an inoculum layer test method was used in which soybean seedlings were grown in vermiculite in containers carrying a P. soja-covered disc of V-8 agar at a depth 2 cm below the planted seeds (Schmitthenner and Bhat, 1994). Experimental units were completely randomized within a given experiment and each experiment consisted of three to five treatments (agar-only control plus various races of Phytophthora), the cultivars to be tested, and three to four replications each. Two weeks after planting, plants were rated for leaf/shoot symptoms and then removed from the vermiculite and rated for root rot symptoms. Shoots were rated on a scale of 0 to 6: 0, leaves appear healthy; 1, slight leaf damage; 2, plants of normal height with stunted leaves; 3, short, stunted plants; 4, tiny, unexpanded unifoliate leaves visible; 5, seed germinated but only cotyledons visible; and 6, seedling rotted, no leaves visible. Roots were rated on a scale of 0 to 7: 0, healthy roots; 1, slight root rot (browning at tips); 2, moderate root rot; 3, some healthy roots remain but severe rot; 4, many rotted roots present; 5, a few sick roots remaining; 6, all roots completely rotted; and 7, seedling rotted at emergence.
Rhizoctonia solani Tests
R. solani strain 2B-12, an isolate highly virulent on soybean, was obtained from the laboratory of Dr. James Sinclair (Urbana, IL) and maintained at 4°C with minimal passaging on potato dextrose agar plates. To compare the reaction to R. solani between soybean mutants and the parental lines from which they were derived, experiments were set up as completely randomized designs with two treatments (uninfested and R. solani-infested soil), six to nine cultivars, and three to four replications per experiment. R. solani inoculum was prepared from liquid cultures by placing plugs of mycelium from potato dextrose agar plates in a flask of potato dextrose broth on a shaker rotating at 80 rpm at a temperature of 28°C. After approximately 10 to 14 d a large mycelial mass developed that was harvested by centrifugation, blotted for less than 1 min until relatively dry, weighed, placed in distilled water, and ground into small pieces in a blender. This mycelial suspension was then added gradually and mixed thoroughly into a soil mixture of 1 part soil, 1 part sand, and 1 part vermiculite, to an inoculation density of 100 to 200 mg fresh weight kg−1 soil. Five seeds per pot were planted directly in the fungus-infested soil at a depth of approximately 1.5 cm and grown in the greenhouse. After 14 d the soil was gently washed from the roots, which were rated on a scale of 0 to 6: 0, roots completely healthy; 1, slight root rot; 2, moderate root rot; 3, severe root rot with some healthy roots remaining; 4, only rotted roots present; 5, root system completely rotted away; and 6, seedling completely rotted. Similarly, shoots were rated on a scale of 0 to 5: 0, healthy leaves; 1, normal plant height with stunted leaves; 2, mildly stunted plants; 3, small deformed plants; 4, seedling just emerged; and 5, seedling completely rotted at emergence.
For disease tests with all four pathogen species, disease severity was rated for randomized, numerically coded pots, and plant genotypes were then matched to the numerical code after disease scores had been recorded. Disease test data were analyzed using the GLM procedure in the SAS package (SAS, 1989). Within GLM, contrasts were written for each specific comparison between the parent and the mutant. In analysis of variance, treatments and cultivars were considered fixed and replications were considered random.
RESULTS
Characterization of the Ethylene Triple Response in Soybean
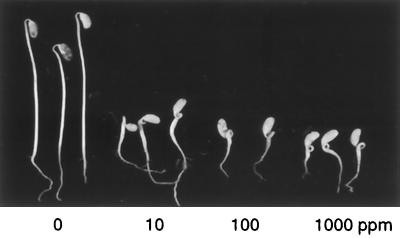
We adapted the ethylene triple-response assay of Bleecker et al. (1988) for use with soybean. When seedlings of most species are germinated in the dark in air, they become etiolated (tall, spindly, and chlorotic). In the ethylene triple-response assay, seedlings are germinated in the dark in an atmosphere containing ethylene. Plants with wild-type ethylene sensitivity typically remain quite short and exhibit thickening of the hypocotyl and excessive curling at the hypocotyl hook under these conditions (Knight et al., 1910; Ecker, 1995). Our initial experiments tested the ethylene triple response of wild-type soybean lines (Fig. 1). Soybean plants germinated in the dark in air typically grew to a hypocotyl length of 12 to 18 cm after 6 d in our experimental system. In contrast, plants germinated in the dark in 10 μL L−1 ethylene gave a classic triple response, with an average hypocotyl length of 2 cm, obvious radial thickening of the hypocotyl (diameter increased approximately 2-fold at thickest point), and exaggerated curvature of the hypocotyl hook to an angle exceeding 180o. As in other plant species, the response of soybean to ethylene in this assay was dose dependent. In establishing the lower limits of the response, we observed a 2- to 3-fold reduction in hypocotyl length in 0.5 μL L−1 ethylene but only a slight response in 0.1 μL L−1 ethylene (data not shown). The hypocotyl thickening and hook curling of wild-type soybeans became slightly more extreme as ethylene levels were increased up to 1000 μL L−1, but hypocotyl length was essentially saturated at 10 μL L−1 (Figs. 1 and 2).
Figure 1.
Ethylene triple response of etiolated soybean seedlings. Plants were germinated for 6 d in complete darkness in sealed vessels containing air or air supplemented with ethylene to the concentrations noted. The typical triple response to ethylene includes shortening of the hypocotyl, radial thickening of the central hypocotyl, and excessive curling of the hypocotyl hook.
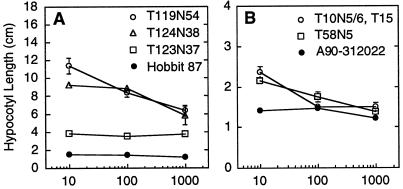
Figure 2.
Hypocotyl elongation of etiolated soybean seedlings germinated in ethylene. Mutants are presented with their respective nonmutagenized parent line Hobbit 87 (A) or A90-312022 (B). Seedlings were germinated for 6 d in complete darkness in sealed vessels containing air supplemented with ethylene to the concentrations noted. Values are means ± se. T10N5, T10N6, and T15 are sibling lines carrying the same mutation (see text). True-breeding progeny of originally identified mutants were used in these and subsequent studies.
Isolation of Ethylene-Insensitive Mutants of Soybean
In the absence of immediately available M2 soybean seed, mutant screens for plants displaying an altered ethylene triple response were initiated using M3 populations that were the kind gifts of Drs. Walter Fehr and James Specht. Approximately 142,000 M3 seeds representing approximately 1,985 M1 lines were tested from the Fehr seed populations, and approximately 48,000 M3 seeds representing 1,434 M1 lines were tested from the Specht seed population. Plants from 84 different M2 families were saved as possible ethylene-insensitive lines, although many were only slightly taller than their parents in the initial ethylene triple-response screen. Candidate mutants were grown to maturity and allowed to self-fertilize, and the progeny were tested for the ethylene triple response. Most of the lines resembled the wild type upon retesting, but seven lines showed reproducible reduction in ethylene sensitivity.
Figure 2 presents data from one of the many ethylene-sensitivity tests of the mutant lines that were used in further studies. The mutants presented in Figure 2 displayed full hypocotyl elongation when germinated in the dark in air with no added ethylene (data not shown; hypocotyl length typically 12–18 cm, depending on the experiment). When the “strong” ethylene-insensitive mutant T119N54 was germinated in 10 μL L−1 ethylene, hypocotyls extended to a length similar to that of etiolated seedlings grown in air (Fig. 2A). The “intermediate” mutants such as T123N37 or T124N38 displayed an obvious reduction in ethylene sensitivity relative to the parent line, but still exhibited some inhibition of average hypocotyl length when germinated under ethylene (Fig. 2A). The “weak” ethylene-insensitive mutants displayed a modest but reproducible reduction in their responsiveness to ethylene relative to the wild-type parent (Fig. 2B). Because average hypocotyl length varied somewhat from experiment to experiment, data comparisons focused on plants within a given experiment, and parental genotypes were routinely included as internal controls in these and subsequent ethylene triple-response tests.
All of the mutants retained some degree of ethylene sensitivity. Even in line T119N54, hypocotyls were more strongly shortened after germination at extremely high ethylene levels than they were at the 10 μL L−1 ethylene level that was saturating for the wild type (Fig. 2). At low ethylene levels the transition between ethylene-induced shortening and full hypocotyl elongation occurred at similar concentrations in the wild type and in the partial or weak ethylene-insensitive mutants. Partial loss of hypocotyl shortening was observed when ethylene concentrations decreased from 2.5 to 0.5 μL L−1, and essentially full hypocotyl elongation was observed in 0.1 μL L−1 (data not shown).
Genetic Analysis of Ethylene-Insensitive Mutants
Genetic studies of the inheritance and allelism of the mutations that cause ethylene insensitivity were performed, with a particular focus on mutants that were isolated earlier in the project. Line T119N54 was originally isolated from an M3 family in which three plants showed no triple response (hypocotyl approximately 15 cm), three were short (>3 cm), and six were intermediate (average approximately 10 cm). This 1:2:1 phenotypic ratio suggested the possibility of a semidominant trait. Subsequent retests of more than 200 progeny from the T119N54 family provided further support for this hypothesis. In ethylene triple-response tests the progeny from the tall phenotype plants were uniformly tall (12–19 cm), the progeny from the short plants were short (1.5–2.5 cm), and the progeny from the intermediate plants segregated for the short, intermediate, and tall phenotype in ratios resembling 1:2:1 (data not shown). Further genetic analyses focused on progeny from reciprocal crosses between a true-breeding ethylene-insensitive T119N54 line and its parent, Hobbit 87. F1 plants from these crosses exhibited an intermediate phenotype in the ethylene triple-response test (Table I). The F2 plants segregated in a manner consistent with a 1:2:1 ratio (Table II; Fig. 3). These data suggest that the ethylene-perception defect in line T119N54 is caused by mutation in a single nuclear locus and that the mutant allele confers ethylene insensitivity in a semidominant fashion. We propose the name Etr1 for this gene, and use etr1-1 to designate the mutant allele in line T119N54. True-breeding ethylene-insensitive T119N54 progeny are subsequently referred to as Hobbit 87 etr1-1, or simply etr1-1.
Table I.
Dominance and complementation tests for the ethylene-sensitivity trait among mutant and wild-type soybean lines
| Soybean Line | Hypocotyl Length |
|---|---|
| cm | |
| Parents | |
| Hobbit 87 | 2.4 ± 0.16 |
| T119N54 (etr1-1) | 13.3 ± 0.26 |
| A90-312022 | 2.2 ± 0.05 |
| T10N5/T10N6/T15N23 (“etr2-1 lines”) | 3.8 ± 0.05 |
| T58N5 | 3.4 ± 0.09 |
| F1 of crosses to wild type | |
| Hobbit 87 × T119N54 | 8.7 ± 0.32 |
| Different from Hobbit 87 and T119 (P < 0.001); semidominant | |
| A90-312022 × etr-1 lines | 2.3 ± 0.04 |
| Not different from A90-312022 (P > 0.05); recessive | |
| Different from etr2-1 lines (P < 0.001) | |
| A90-312022 × T58N5 | 2.8 ± 0.11 |
| Different from A90-312022 and T58N5 (P < 0.001); semidominant | |
| F1 for complementation tests | |
| T10N5 × T10N6 | 2.8 ± 0.15 |
| Different from parents (P < 0.001) | |
| T10N5 × T15N23 | 3.4 ± 0.11 |
| Not different from parents (P > 0.05) | |
| T10N6 × T15N23 | 4.0 ± 0.20 |
| Not different from parents (P < 0.05 and P = 0.049) | |
| T119N54 × T15N23 | 8.5 ± 0.67 |
| Different from parents (P < 0.001) (F2 data in Table II) | |
| T58N5 × etr2-1 lines | 3.2 ± 0.10 |
| Different from etr2-1 lines (P < 0.001) | |
| Not different from T58N5 (P > 0.05) | |
Seedlings were germinated in darkness for 6 d in 18 μL L−1 ethylene. For each designated cross, combined data for progeny of multiple reciprocal crosses are presented. Results of Student's t tests for similarity to parent lines (data at top of table) are presented below cross data, accept similarity if P > 0.05. Values are means ± se.
Table II.
Segregation of ethylene-sensitivity trait in F2 populations
| Soybean Line | No. of Plants within Hypocotyl Length Class | χ2, P | ||
|---|---|---|---|---|
| <4 | 4–12.4 | ≥12.5 | ||
| Hobbit 87 | 49 | 0 | 0 | |
| T119N54 (etr1-1) | 0 | 4 | 75 | |
| F2: Hobbit 87 × T119N54 | 28 | 70 | 30 | 1.19, P = 0.55 for 1:2:1 |
| F2: T119N54 × Hobbit 87 | 44 | 85 | 28 | 4.34, P = 0.11 for 1:2:1 |
| <1.8 | ≥1.8 | |||
| A90-312022 | 58 | 10 | ||
| T10N5 (etr2-1) | 1 | 74 | ||
| F2: A90 × T10N5 | 123 | 29 | 0.003, P = 0.96 for 3:1 | |
| F2: T10N5 × A90 | 80 | 27 | 2.82, P = 0.09 for 3:1 | |
| <1.7 | ≥1.7 | |||
| A90-312022 | 68 | 24 | ||
| T10N6 (etr2-1) | 13 | 66 | ||
| F2: A90 × T10N6 | 84 | 41 | 4.05, P = 0.04 for 3:1 | |
| F2: T10N6 × A90 | 107 | 35 | 0.003, P = 0.96 for 3:1 | |
| <2.2 | 2.2–2.8 | >2.8 | ||
| A90-312022 | 34 | 36 | 0 | |
| T58N5 | 3 | 22 | 38 | |
| F2: A90 × T58N5 | 30 | 57 | 21 | 1.68, P = 0.43 for 1:2:1 |
| F2: T58N5 × A90 | 24 | 70 | 26 | 3.40, P = 0.18 for 1:2:1 |
| <3 | 3–6 | >6 | ||
| Hobbit 87 | 11 | 2 | 0 | |
| T119N54 (etr1-1) | 0 | 0 | 15 | |
| A90-312022 | 15 | 0 | 0 | |
| T15N23 (etr2-1) | 2 | 13 | 0 | |
| F2: T119N54 × T15N23 | 15 | 8 | 77 | 1.29, P = 0.52 for 3:1:12 |
Seedlings were germinated in darkness for 6 d in 18 μL L−1 ethylene. Column subheadings define hypocotyl length ranges (cm) for the respective phenotypic classes (see Methods); integer values for each plant line are the number of individual plants within the phenotypic class. χ2 and P values are for χ2 tests of similarity of the data to the noted ratio; accept ratio if P > 0.05.
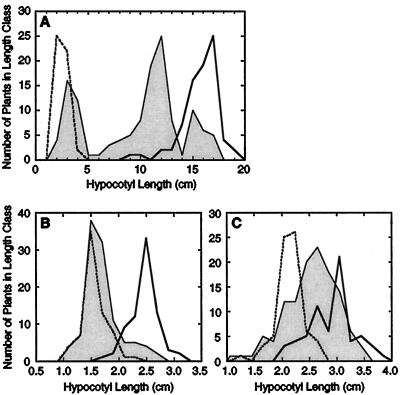
Figure 3.
Distribution of hypocotyl-length phenotypes in parents and segregating F2 populations germinated under etiolating conditions in 20 μL L−1 ethylene. Data on a given graph are from plants tested concurrently within the same chamber. A, Hobbit 87 (broken line), Hobbit 87etr1-1 (heavy, solid line), and F2 of Hobbit 87 × Hobbit 87etr1-1 (solid line with fill). B, A90-312022 (broken line), etr2-1 mutant T10N5 (heavy, solid line), and F2 of A90-312022 × T10N5 (solid line with fill). C, A90-312022 (broken line), T58N5 (heavy, solid line), and F2 of T58N5 × A90-312022 (solid line with fill).
The partially ethylene-insensitive mutants included T10N5, T10N6, and T15N23. All of these lines were derived from the experimental breeding line A90-312022 after mutagenesis with NMU. The mutants T10N5 and T10N6 were derived from two separate M2 lines derived from the same M1 parent, and T15N23 was a second line selected from the same M2 parent as T10N5. Thus, it was very likely that T10N5, T10N6, and T15N23 all carry the same mutant allele. Whereas traits of lines T10N5, T10N6, and T15N23 such as flower color, leaf shape, and pubescence color resembled those of A90-312022, seed derived from the M1 parent of T10N5, T10N6, and T15N23 also segregated for brown and black hilum color independent of the ethylene-insensitivity trait. The line from which these three ethylene-insensitive mutants was derived apparently carried a separate mutation affecting hilum color.
When lines T10N5, T10N6, and T15N23 were crossed reciprocally to the parental line A90-312022, F1 seedlings displayed wild-type ethylene sensitivity in the triple-response test (Table I), indicating that the mutant ethylene-insensitivity trait in these lines is recessive to the wild type. Tests were then performed using 10 different F2 populations from reciprocal crosses of the A90-312022 parental line and the mutants T10N5, T10N6, and T15N23. These F2 populations displayed the range of hypocotyl-elongation phenotypes that would be expected for a recessive mutation at a single locus, and segregation ratios consistent with a 3:1 hypothesis could be derived from the resulting F2 data sets (see examples in Table II and Fig. 3). However, in some tests the overlapping bell-shaped distributions of hypocotyl length for the controls (parental wild-type and weak ethylene-insensitive lines) required that a maximally differentiating but imperfect dividing value for phenotypic categories be chosen (see Methods). Figure 3 is presented to give a more precise example of the distribution of phenotypes obtained in these experiments, which were consistent with segregation of a single recessive gene.
T10N5, T10N6, and T15N23 were crossed reciprocally with each other for complementation tests to confirm allelism. Most of the F1 plants from these crosses exhibited the mutant ethylene-insensitivity phenotype, although some T10N5 × T10N6 plants were of a size intermediate between the parental line A90-312022 and the two mutants (Table I). The complementation data again suggest that these three closely related lines carry a mutation in the same gene. Complementation tests between the etr1-1 mutant T119N54 and T15N23 gave data that were inconsistent with allelism but consistent with the hypothesis that these lines carry mutations in separate genes. F1 plants from reciprocal crosses of etr1-1 and T15N23 exhibited strong ethylene insensitivity, as expected given the semidominant nature of the etr1-1 mutation. F2 plants from the etr1-1 × T15N23 cross segregated 12:1:3 for ethylene-sensitivity phenotypes resembling etr1-1 homozygotes or heterozygotes, T15N23, and the wild type, respectively (Table II and data not shown). No F2 plants would be expected in the small (wild type, <3 cm) category if etr1-1 and T15N23 carried allelic mutations. We propose, therefore, the name Etr2 for this second gene, and use etr2-1 to designate the mutant allele in lines T10N5, T10N6, and T15N23. In subsequent studies, data for T10N5, T10N6, and T15N23 were combined and presented as data for the etr2-1 lines.
A separate partially ethylene-insensitive mutant, T58N5, was also isolated from NMU-mutagenized parental line A90-312022 seed but was derived from a different M1 plant than the parent of the etr2-1 line. The F1 for a cross of T58N5 and the wild-type parent gave F1 individuals with ethylene-response phenotypes that were intermediate between the parents (Table I). The F2 segregation ratios were not consistent with a 3:1 ratio, but were consistent with a 1:2:1 ratio (Table II; Fig. 3). The mutant phenotype in line T58N5 is apparently conferred by a single locus that is semidominant with respect to the wild type. Complementation tests with the T58N5 line were complicated by the semidominant nature of the mutation in T58N5 and the overlap in the range of hypocotyl elongation phenotypes for homozygous mutant, heterozygote, and homozygous wild-type plants. Tests carried out with the F1 and F2 of crosses of T58N5 to etr2-1 (T10N5, T10N6, and T15N23) and etr1-1 (T119N54) suggested that this line carries a mutation in a third Etr gene (Table I and data not shown), but the overlap in hypocotyl phenotypes mentioned above reduced our level of confidence in such a conclusion.
Response to S. glycines
Previous work with other plant species has shown that some ethylene-insensitive lines exhibit enhanced disease tolerance against some pathogens, and we sought to examine the response of our ethylene-insensitive soybean mutants to four diverse soybean pathogens. S. glycines is a fungal pathogen that causes brown-spot disease, a foliar disease characterized by necrotic leaf lesions that turn brown with age and develop chlorosis around the margins (Sinclair and Backman, 1989; McGee, 1991). Under moderate to heavy S. glycines disease pressure, chlorotic areas merge and entire leaves senesce and drop from the plant. Despite extensive efforts, no strong genetic resistance to S. glycines has been identified in soybean (Sinclair and Backman, 1989; McGee, 1991). In the present study S. glycines tests were performed using two different methods. In the first method, plants were spray inoculated with S. glycines spore suspensions, maintained in a humid environment for 2 d, shifted to normal growth conditions, and then scored for disease development 9 d after inoculation. In test 1 of Table III, the highly ethylene-insensitive etr1-1 mutant did not appear to exhibit altered sensitivity to S. glycines infection. In contrast, the mutant T58N5 exhibited greater disease severity in this test than did the A90-312022 parent from which it was derived (Table III). Plants were scored at a second time point 14 d after inoculation, and similar results were obtained (data not shown). Mock-inoculated controls showed no significant differences between mutant and parental lines.
Table III.
Disease severity in ethylene-insensitive mutants in response to S. glycines
| Soybean Line | Spray
Inoculation
|
Syringe Inoculation
|
|
|---|---|---|---|
| 1 | 2a | 2b | |
| Hobbit 87 | 1.0 | 0.6 | 2.9 |
| etr1-1 | 1.2 | 0.4 | 4.3a,b |
| T123N37 | – | 0.1 | 3.5 |
| T124N38 | – | 0.9 | 4.2c,b |
| A90-312022 | 0.7 | 0.1 | 3.5 |
| etr2-1 | – | 0.2 | 3.4 |
| T58N5 | 2.3c,b | 1.9c,b | 4.1d,b |
Disease symptoms on unifoliate leaves, presented as mean disease severity on a scale ranging from 0 (no lesions) to 5 (many large lesions). –; Not tested. Test number is indicated at the top of the column. Statistical comparisons are between the mutant and its respective parent, Hobbit 87 or A90-312022.
Significant at P < 0.01.
Disease symptoms are more severe than parent.
Significant at P < 0.05.
Significant at P < 0.10.
The S. glycines spray-inoculation method gave reasonably reproducible results between replications within single experiments, but levels of plant infection varied substantially from experiment to experiment, so a different inoculation method was pursued. This second method was referred to as the syringe method for S. glycines application. Here a small area (approximately 0.5 × 2 cm) on the underside of one unifoliate leaf of each plant was rubbed gently with a plastic syringe (no needle) filled with a concentrated spore suspension. This treatment was designed to slightly wound the leaf epidermis so that spores might more easily become established in the leaf interior. Nine to fourteen days after inoculation, leaves of control plants (syringe inoculated with water only) remained green and healthy except for slight browning that did not extend past the abraded area. Leaves that had been inoculated with S. glycines typically developed severe necrosis that spread to double or triple the inoculated area. The rating scale used for syringe inoculation was based on lesion size and chlorosis. An experiment was conducted with the ethylene-insensitive lines in which the spray- and syringe-inoculation methods were used side by side. The two inoculation techniques produced overlapping but different results (Table III, tests 2a and 2b). The spray-inoculation technique once again produced symptoms that were more severe than those in the parent only for T58N5 and not for the other ethylene-insensitive lines. Syringe inoculation gave pronounced S. glycines lesions on all of the lines tested. The etr1-1 mutant and T124N38 exhibited significantly larger lesion size than the Hobbit 87 parent from which they were derived. T58N5 also developed slightly larger lesions than its parent when the syringe-inoculation method was used. Mock-inoculated controls showed no differences between mutants and parental lines for either inoculation method (data not shown). Thus, the weaker mutant allele in T58N5 and the stronger mutant alleles in etr1-1 and T124N38 each made the plant more susceptible to damage by S. glycines, but in an assay-dependent fashion.
Response to P. syringae pv glycinea
Soybean bacterial blight, caused by P. syringae pv glycinea, superficially resembles S. glycines brown spot in being a predominantly foliar disease characterized by necrotic lesions with chlorotic margins (Sinclair and Backman, 1989; McGee, 1991). However, the disease is caused by a bacterial pathogen with different virulence mechanisms and different responses to host defenses. Bacterial blight can be controlled using resistant cultivars, including cultivars carrying race-specific resistance genes (Sinclair and Backman, 1989; McGee, 1991). In the present study a number of tests were performed to compare mutants to parental lines for their reaction to virulent P. syringae pv glycinea. Additional studies examined reactions to an isogenic P. syringae pv glycinea strain that expresses the cloned avirulence gene avrRpt2 and is avirulent on the soybean lines used in this study. Tests concentrated on the etr1-1 and etr2-1 mutants. The mutants T58N5, T123N37, and T124N38 were identified at a later date and were included in later tests. In tests 1 to 4 of Table IV, mock-inoculated controls were included and no significant differences between mutants and parental lines were observed in these controls (data not shown).
Table IV.
Disease severity in ethylene-insensitive mutants in response to virulent P. syringae pv glycinea and to an isogenic avirulent strain that expresses avirulence gene avrRpt2
| Soybean Line | Virulent
Strain
|
Avirulent Strain
(avrRpt2+)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 1 | 2 | 3 | 4 | 5 | |
| Hobbit 87 | 4.7 | – | 3.5 | 3.4 | 4.8 | 2.1 | – | 1.5 | 2.6 | 2.5 |
| etr1-1 | 4.5 | – | 3.1 | 3.7 | 4.8 | 1.4a,b | – | 1.4 | 1.9c,b | 2.6 |
| T123N37 | – | – | – | – | 4.7 | – | – | – | – | 2.3 |
| T124N38 | – | – | – | – | 5.0 | – | – | – | – | 2.5 |
| A90-312022 | 4.8 | 4.1 | 4.1 | 3.6 | 4.9 | 2.1 | 2.0 | 1.3 | 2.4 | 2.0 |
| etr2-1 | 4.1a,b | 4.1 | 3.2a,b | 3.7 | 4.9 | 2.4c,d | 1.8 | 1.3 | 3.1e,d | 1.7a,b |
| T58N5 | – | – | – | – | 5.0 | – | – | – | – | 4.2a,d |
Data are from five separate tests; test number is indicated at top of the column. Data presented are mean disease severity scores for unifoliate leaves, rated on a scale ranging from 0 (no lesions) to 5 (leaves completely necrotic); –, not tested. Note that test 5 used a different rating scale based on necrosis and chlorosis (see text). Statistical comparisons are between the mutant and its respective parent, Hobbit 87 or A90-312022.
Significant at P < 0.01.
Disease symptoms less severe than parent.
Significant at P < 0.10.
Disease symptoms more severe than parent.
Significant at P < 0.05.
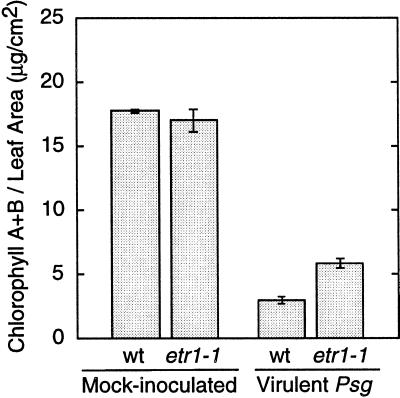
In studies with the virulent P. syringae pv glycinea strain, the more strongly ethylene-insensitive etr1-1 and T124N38 mutants were not significantly different from the parent Hobbit 87 in their disease-severity scores (Table IV). Infected leaves of etr1-1 often seemed to develop less chlorosis, but the rating scale used in tests 1 to 4 focused instead on necrosis. In test 3, leaf samples were taken from etr1-1 and Hobbit 87 and chlorosis was quantified by chlorophyll assay (Fig. 4). There was significantly less chlorosis in the ethylene-insensitive mutant. However, in test 5 a new rating scale was used that focused on leaf necrosis and chlorosis rather than on lesion size alone, and in that test no significant difference was observed between the strongly ethylene-insensitive etr1-1 line and Hobbit 87. In tests 1 to 5 the more weakly ethylene-insensitive lines gave disease-severity scores that often were not significantly different from those of the parent. However, in two of the five tests the etr2-1 mutants had significantly fewer disease lesions (Table IV). To summarize the results, most mutants performed as well as their parental lines, and in the etr1-1 and etr2-1 lines there was a trend toward reduced disease severity in response to virulent P. syringae pv glycinea.
Figure 4.
Chlorophyll content in leaves from mock-inoculated plants and plants infected by virulent P. syringae pv glycinea. Leaf discs were sampled 9 d after inoculation; values are means ± se for four separate groups of 5 plants (total of 20 plants per pathogen treatment). wt, Wild type.
P. syringae pv glycinea that express the avirulence gene avrRpt2 elicit a HR in resistant soybean hosts, and multiplication of such strains is restricted in these resistant hosts (Whalen et al., 1991; Innes et al., 1993; data not shown). The avrRpt2 gene also controls avirulence on Arabidopsis, and for that interaction the corresponding resistance gene RPS2 has been cloned and characterized (Bent et al., 1994; Mindrinos et al., 1994). For the experiments in this study, avirulent P. syringae pv glycinea bacteria were introduced uniformly into the mesophyll of entire leaves at relatively high initial population densities so that resistance-associated HR lesions developed in patches that were visible to the naked eye. Chlorosis developed around these lesions to a variable degree. Tests in Table IV that have the same number were run and rated simultaneously with virulent and avirulent P. syringae pv glycinea. The statistically significant resistance elicited by expression of avrRpt2 was evident when mean disease scores for a given plant genotype were compared for inoculations with the isogenic virulent and avirulent strains (Table IV, statistical data not shown).
The primary focus of Table IV is on a statistical comparison between the responses of parent and mutant plants to the same pathogen strain. The etr1-1 mutant exhibited lower HR severity than Hobbit 87 in response to avirulent P. syringae pv glycinea (avrRpt2+) in two of the four tests (Table IV). Overall, disease severity on the ethylene-insensitive Hobbit 87 mutants was similar to or less severe than disease severity on wild-type Hobbit 87 after infection with avirulent P. syringae pv glycinea (avrRpt2+). The mutations causing partial ethylene insensitivity in the etr2-1 line and in T58N5 also had a variable effect on the response to avirulent P. syringae pv glycinea (avrRpt2+). As mentioned above, avirulence gene-dependent resistance remained effective overall (Table IV; compare the reactions of a given mutant to the isogenic virulent and avirulent P. syringae strains). When reactions to avirulent P. syringae pv glycinea were compared between etr2-1 and its nonmutagenized parent, lesion scores for tests 1 to 4 (in which scoring focused on HR lesion size) revealed more extensive reactions in the etr2-1 mutant. However, the other two experiments gave no significant difference. In test 5 a severity scale was used that focused on overall leaf browning and chlorosis rather than on lesion size alone. In that case, as was observed for etr1-1 in the experiment reported in Figure 4, the etr2-1 line had significantly less chlorosis than the nonmutagenized parent. T58N5, on the other hand, had a more severe reaction than did the parent.
During the course of these studies five other mutant lines, T14N2B, T14N3A, T14N10, T35N20A, and T38N5, were originally selected as possible weak ethylene-insensitive mutants. In retests these lines failed to exhibit the ethylene-insensitive phenotype, but in the intervening period these lines were tested for their reaction to virulent and avirulent P. syringae pv glycinea. None of these mutants exhibited disease reactions that were significantly different from those of the parental lines from which they were derived (data not shown). Of the lines tested in these studies, only the ethylene-insensitive lines exhibited altered reactions to P. syringae pv glycinea infection.
Reactions to P. sojae
P. sojae is a soil-borne oomycete that is one of the most economically destructive soybean pathogens in the United States (Sinclair and Backman, 1989; Doupnik, 1993). Damage is caused primarily by rotting of the root and lower stem tissue, and disease control relies heavily on genetic strategies that involve both single race-specific resistance genes and multigenically controlled “field tolerance” (Sinclair and Backman, 1989; McGee, 1991). To test ethylene-insensitive mutants for their reaction to P. sojae, multiple races of Phytophthora were used. Hobbit 87 carries the Rps1-k resistance locus that conditions race-specific resistance to many common Phytophthora races, including races 1, 4, and 7, but not race 20 (Cooper et al., 1991; Kasuga et al., 1997). We found that race-specific resistance mediated by Rps1-k was partially compromised in the strongly ethylene-insensitive etr1-1 and T124N38 mutants (Table V). In particular, resistance against race 4 was significantly compromised and resistance against race 7 was also compromised in some of the tests. However, resistance against Phytophthora race 1 remained effective and was even improved. Hobbit 87 does not carry any other known Phytophthora resistance genes, and this differential effect of ethylene insensitivity on the reaction to different avirulent races suggests that races 1, 4, and 7 may stimulate different subsets of the plant defense response (see Discussion). Race 20 strains are virulent on Rps1-k lines such as Hobbit 87, and the reactions of the Hobbit 87 mutants etr1-1 and T124N38 to race 20 were not significantly different from those of the parental line (Table V).
Table V.
Disease severity in ethylene-insensitive mutants in response to races of P. sojae that are virulent or avirulent on the respective soybean genotypes
| Soybean Line | Race 1
|
Race
4
|
Race 7
|
Race 20
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 | 4 | 5 | 1 | 2 | 3 | 4 | 5 | 2 | 4 | 1 | 2 | 3 | 4 | |
| Hobbit 87 | 2.3 | 1.5 | – | 4.2 | 1.9 | 2.8 | 3.8 | – | 0.9 | 0.6 | 4.9 | 5.4 | 5.2 | 4.7 |
| T119N54 | 0.9a,b | 0.8 | – | 4.2 | 4.7a,c | 5.1a,c | 5.0a,c | – | 2.2d,c | 1.4e,c | 4.6 | 4.9 | 5.2 | 4.9 |
| T123N37 | – | 1.3 | – | – | – | – | 4.3 | – | – | 0.7 | – | – | – | 4.8 |
| T124N38 | – | 0.4d,b | – | – | – | – | 4.7d,c | – | – | 0.5 | – | – | – | 4.9 |
| A90-312022 | 5.5 | 5.0 | 4.2 | 4.4 | 4.2 | 5.1 | 5.6 | 3.8 | 4.9 | 4.0 | 4.6 | 3.9 | 4.9 | 5.3 |
| etr2-1 | 5.0 | 5.0 | – | 4.6 | 3.8 | 5.0 | 5.1 | – | 4.0e,b | 4.6 | 4.0d,b | 2.4a,b | 4.3e,b | 4.4d,b |
| T58N5 | – | 4.5 | 3.4e,b | 4.1 | 4.7 | – | 5.0 | 3.9 | 4.5 | 4.1 | 4.7 | 3.6 | – | 4.9 |
Disease symptom ratings for above ground plant tissues, presented as mean disease severity on a scale ranging from 0 (healthy plantlet) to 6 (seedling completely rotted); –, not tested. Data are presented from five separate tests; test number is indicated at the top of the column. Hobbit 87 and the three Hobbit 87 mutants carry Rsp1-k for resistance to races 1, 4, and 7 (data in bold); all other host-pathogen combinations involve compatible rather than gene-for-gene interactions. Statistical comparisons are between the mutant and its respective parent, Hobbit 87 or A90-312022.
Significant at P < 0.01.
Disease symptoms less severe than parent.
Disease symptoms more severe than parent.
Significant at P < 0.05.
Significant at P < 0.1.
Races 1, 4, 7, and 20 are all virulent on plants of the A90-312022 genotype, which does not carry known genes for resistance to these Phytophthora races. The partially ethylene-insensitive lines derived from A90-312022 exhibited similar disease severity to the parent in most of the tests (Table V). However, in more than one-fourth of the tests disease severity was significantly reduced in the mutants, and there were no cases in which the mutants appeared worse than the parent. Mutation of the Etr2 locus in particular apparently caused a partial reduction in the extent of disease caused by virulent P. sojae.
Reactions to R. solani
R. solani is a genetically diverse species that causes root-rotting diseases on a wide variety of plant hosts (Agrios, 1997). As for many other root-rotting diseases, the effect is most pronounced on germinating seeds and young seedlings. Taxonomic subgroups within R. solani show a more limited host range, but single resistance genes against R. solani are not known and genetic resistance against this pathogen, where available, is multigenically controlled and only partially effective (Sinclair and Backman, 1989; McGee, 1991). Tests comparing the ethylene-insensitive mutants with their respective parental lines for their reaction to R. solani are reported in Table VI. Once again, a trend could be detected but statistical significance was not consistently observed. In one test etr2-1 and T58N5 exhibited more severe root rot than their parental line, A90-312022 (Table VI). In a separate test the shoots of etr2-1 and T58N5 were more severely affected by R. solani than their parental line, but differences in disease severity in roots were not significant. As for the other disease tests reported above, we present the full data set to illuminate the true variability observed in replicated experiments. The response of ethylene-insensitive soybean mutants to R. solani was variable, but more severe disease was observed in multiple instances.
Table VI.
Disease severity in ethylene-insensitive mutants in response to R. solani
| Soybean Line | Root/Hypocotyl
|
Shoot
|
|||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 2 | 3 | 4 | |
| Hobbit 87 | 1.5 | – | 4.5 | 4.7 | – | 3.5 | 3.6 |
| etr1-1 | 1.8 | – | 4.8 | 3.9a,b | – | 4.1 | 2.2c,b |
| T123N37 | – | – | – | 4.9 | – | – | 3.7 |
| T124N38 | – | – | – | 5.2 | – | – | 4.1 |
| A90-312022 | 0.8 | 3.6 | 4.1 | 4.9 | 0.0 | 2.0 | 4.0 |
| etr2-1 | 1.6a,d | 3.2 | 4.8 | 5.0 | 0.7 | 4.2c,d | 4.1 |
| T58N5 | 2.5c,d | 3.5 | 4.5 | 4.5 | 0.1 | 3.8a,d | 3.5 |
Disease symptom ratings for aboveground tissues or for root and hypocotyl tissues, presented as mean disease severity on a scale ranging from 0 (very healthy) to 6 (seedling completely rotted); –, not tested. Data are presented from four separate tests; test number is indicated at the top of the column. Statistical comparisons are between the mutant and its respective parent, Hobbit 87 or A90-312022.
Significant at P < 0.05.
Disease symptoms are less severe than parent.
Significant at P < 0.01.
Disease symptoms are more severe than parent.
DISCUSSION
The hormone ethylene modulates a number of traits that are relevant to plant growth and productivity. Plant lines can be generated with genetic modifications that alter ethylene production or responsiveness to ethylene. It is important to examine the effects of altered ethylene signaling on a wide variety of traits in these plants, including responses to infection by pathogens. The present work initiated such studies with soybean. Mutant plant lines with reduced ethylene sensitivity were identified, and the response of these lines to diverse virulent and avirulent pathogens was assessed. Reduction of ethylene sensitivity had a neutral or beneficial effect on the plant response to some pathogens, but a detrimental effect on others. Furthermore, ethylene insensitivity had differential effects on gene-for-gene resistance against different strains of the same pathogen species.
Ethylene-Insensitive Mutants
Strategies for the production of plants with reduced ethylene responses can target production of ethylene or responsiveness to ethylene (Bleecker et al., 1988; Hamilton et al., 1990; Klee et al., 1991; Oeller et al., 1991; Ecker, 1995; Wilkinson et al., 1997; Knoester et al., 1998). Either trait can be altered by mutation or through transgenic strategies. Variation within existing germplasm represents a third source of plants with reduced ethylene responses (Xie et al., 1996), but natural variants are less well suited for controlled comparisons because of the heterogeneity of genetic backgrounds among such plants. In light of the significant difficulty of soybean transformation and the regulatory and intellectual property issues that surround release of transgenic plant varieties to breeders and growers, we chose to pursue a mutational strategy. Characterization of the ethylene triple response of wild-type soybean seedlings revealed phenotypic behavior and dose-response relationships that are quite typical of previously studied dicotyledonous species. We were then able to adapt the screening method of Bleecker et al. (1988) and identify a number of mutant lines displaying reduced ethylene sensitivity. The spectrum of strong and weak ethylene-insensitive mutants that we identified resembles the outcome of similar screens performed with Arabidopsis (Ecker, 1995; Kieber, 1997).
Genetic studies were carried out on a subset of the identified mutants. Both semidominant and recessive mutations were observed and two new genetic loci of soybean, Etr1 and Etr2, were identified. Extensive work with Arabidopsis has shown that a very common class of ethylene-insensitive mutants contains dominant mutations in the ETR1 gene, which encodes an ethylene receptor (Ecker, 1995; Kieber, 1997). Mutations in ETR1 homologs can also be dominant (Hua et al., 1995; Sakai et al., 1998), whereas mutations in other ethylene-sensitivity loci are usually recessive (Ecker, 1995; Kieber, 1997). In tomato the Never ripe mutation confers semidominant ethylene insensitivity due to alteration of the Le-ETR3 gene, a homolog of Arabidopsis ethylene-receptor genes (Wilkinson et al., 1995). By analogy, the semidominance of the soybean etr1-1 mutation suggests that soybean Etr1 may encode a soybean ethylene receptor. The recessive etr2-1 mutation identified in the present study of soybean may be analogous to mutations in Arabidopsis EIN2, EIN3, or other genes. Three other lines with mutations that reduce sensitivity to ethylene were also identified and used in this study.
The incomplete ethylene insensitivity conferred by etr2-1 and by the unnamed mutations may be attributable to the retention of partial function in the mutant gene products, or to functional redundancy with other genes of soybean. In this regard it is interesting to note that modern soybeans, although diploid in overall genetic behavior, are believed to be descended from a diploidized tetraploid or from more complex polyploid ancestors (Shoemaker et al., 1996). Numerous recessive mutations that confer obvious phenotypes have been identified in soybean, but genetic redundancy remains a particularly strong possibility for any given gene in this species. Because of this, mutations with dominant behavior, such as the etr1-1 mutation, are particularly valuable. However, in previous work on responses to pathogen infection it was the recessive Arabidopsis ein2 mutants that displayed enhanced disease tolerance (Bent et al., 1992). In light of this fact, the other soybean mutants remained of interest. Genetic study of other more recently identified ethylene-insensitive soybean lines is being initiated in the present growing season. These newer studies may allow identification of additional soybean loci involved in responsiveness to ethylene.
Response of Ethylene-Insensitive Mutants to Virulent Pathogens
When the ethylene-insensitive soybean mutants were exposed to plant pathogens, altered responses were observed in many cases. Significantly, although the ethylene-insensitive mutants displayed reduced disease severity in response to some pathogens, more severe disease was observed in other plant-pathogen pairings. To enhance the reliability of the disease assays, pathogen tests were repeated on separate dates and plant samples within a test were typically replicated, numerically coded, and randomized to allow blinded scoring. Data analysis focused on the comparison of each ethylene-insensitive mutant line with its near-isogenic ethylene-sensitive parent. Although some host-pathogen combinations gave consistent differences between parent and mutant, in other cases significant differences were observed only in a subset of the repeats of similar tests.
Our results with S. glycines suggested that ethylene insensitivity, if anything, makes the plant more susceptible to this foliar fungal pathogen (Table III). The mutant allele in T58N5 may influence the host response to infection in a different manner than etr1-1 and the mutant allele in T124N38. T58N5 plants displayed greater symptom severity after inoculation by either of two methods, whereas the etr1-1 and T124N38 plants displayed enhanced disease severity only when the pathogen was applied to a gently wounded area.
In contrast to the results with S. glycines, the ethylene-insensitive mutants performed similarly to or better than their isogenic parents in response to virulent P. syringae pv glycinea (Table IV; Fig. 4). S. glycines and P. syringae, two very different pathogens, both infect foliar tissues and induce lesions surrounded by chlorosis. In previously published work, reduced symptom development was observed in the response of Arabidopsis ethylene-insensitive mutants to virulent P. syringae (Bent et al., 1992). However, in those studies tolerance was observed only in Arabidopsis ein2 mutants and not in ein3 or etr1 mutants. In the present study the data for experiments with virulent P. syringae pv glycinea were variable. In general, however, the ethylene mutants performed as well as their parental lines in response to virulent P. syringae pv glycinea, and the etr1-1 and etr2-1 mutant lines exhibited a trend toward reduced disease severity.
The ethylene-insensitive mutants also performed as well as or better than their parents in response to virulent P. sojae (Table V). No differences were observed with the strong ethylene-insensitive lines, but in 6 of 23 tests involving the weak ethylene-insensitive lines, the disease severity was significantly less severe on the ethylene-insensitive mutants. With R. solani a consistent theme was less prominent, but in four of the eight tests involving the weak ethylene-insensitive lines, root or shoot disease severity in response to R. solani was more severe on the mutant than on the parent (Table VI). Overall, for foliar pathogens (S. glycines and P. syringae) or for root/lower stem pathogens (P. sojae and R. solani), the direction in which ethylene insensitivity altered the reaction to virulent pathogens was not constant within a given type of plant tissue, but was instead dependent on the particular pathogen species. The plant reaction to virulent P. syringae (bacteria) and P. sojae (oomycete) was unaffected or modestly improved by mutations that reduce ethylene sensitivity. However, these same mutations led in many cases to more severe disease symptoms in response to the fungi S. glycines and R. solani.
Alternative interpretations of these results must also be considered. For example, although numerically unlikely, it is possible that the lines carrying mutations that cause ethylene insensitivity also carry separate mutations that are the cause of altered interaction with pathogens. This possibility could be tested using extensively backcrossed lines, or using a population of homozygous etr1/etr1-1 and homozygous Etr1/Etr1 lines derived from F2 individuals segregating for the ethylene-insensitivity trait.
A separate issue concerns the possible pleiotropic effects of mutations that cause ethylene insensitivity. It must be emphasized that alterations in the response to pathogens in these lines do not necessarily imply a direct role for ethylene signaling in a particular pathogen-response pathway. However, even if the effect is indirect, it remains relevant that genetic alterations that cause ethylene insensitivity can alter the response of plants to pathogens.
In considering how ethylene and ethylene insensitivity might influence disease resistance, it is interesting to note the involvement of ethylene in jasmonic acid-mediated defense signaling. Multiple studies have provided evidence that signaling through jasmonic acid and salicylic acid pathways can show a degree of interaction and a tendency toward mutual exclusivity (for review, see Dong, 1998; see also Pieterse et al., 1998).
Response of Ethylene-Insensitive Mutants to Avirulent Pathogens
Soybean plants exhibiting reduced ethylene sensitivity were altered in their response to avirulent pathogens. When the strong ethylene-insensitive mutants were tested for their Rps1-k-mediated resistance against Phytophthora races 1, 4, and 7, two very interesting results were obtained. First, in contrast to previous findings with other species (Bent et al., 1992; Knoester et al., 1998), some gene-for-gene resistance interactions were significantly hindered by mutations causing strong ethylene insensitivity in soybean. Second, resistance based on the Rps1-k resistance locus was disrupted only for some and not all of the avirulent races against which Rps1-k is effective. The response of soybean to an avirulent strain of P. syringae pv glycinea expressing avrRpt2 was less obviously affected by ethylene insensitivity. Alterations in the HR to P. syringae pv glycinea were observed, but overall, avrRpt2-specific resistance remained effective.
The finding that ethylene insensitivity can inhibit gene-for-gene resistance suggests that ethylene signaling can influence some gene-for-gene defense-signaling pathways. It seems very unlikely that ethylene is globally required for gene-for-gene signaling given that ethylene insensitivity did not perturb resistance in Arabidopsis-Pseudomonas interactions mediated by resistance genes RPS2 and RPM1, or in the N-gene-mediated reaction of tobacco to tobacco mosaic virus (Bent et al., 1992; Knoester et al., 1998). Even an essential role for ethylene in all Rps1-k-mediated resistance of soybean against P. sojae is unlikely given the successful resistance to Phytophthora race 1 in the ethylene-insensitive mutants. Instead, we more narrowly conclude that ethylene signaling modulates plant processes that are also modulated in Rps1-k-mediated resistance to Phytophthora races 4 and 7.
The bifurcation of resistance responses controlled by the single Rps1-k locus might be explained by the production of different Rps1-k-recognized avirulence factors in race 1, as opposed to races 4 and 7. A clear example of this is provided by RPM1-mediated resistance in Arabidopsis. RPM1 provides resistance against P. syringae pathogens that express either avrB or avrRpm1, two genes that encode very different protein avirulence factors (Bisgrove et al., 1994). However, in the case of RPM1 it is known that the same RPM1 gene controls both responses (Grant et al., 1995). No substantial differences have been identified in the defense signaling that is activated downstream of RPM1 after stimulation by these two different avirulent pathogens.
The differential effect of etr1-1 on Rps1-k-mediated resistance to different P. sojae races might arise if the Rps1-k locus carries multiple tightly clustered but distinct resistance genes that confer separate pathogen specificities. The tomato-P. syringae pv tomato interaction involving the resistance genes Pto, Fen, and Prf and elicitation by AvrPto or fenthion provides an excellent model for this type of finding (Bent, 1996; Hammond-Kosack and Jones, 1997). The Pto/Fen/Prf gene cluster was treated previously as a single resistance gene. Prf is a resistance gene encoding an NBS-LRR protein that mediates responses to both AvrPto and fenthion (Salmeron et al., 1996). Pto and Fen encode two closely related but distinct protein kinases; Pto is apparently the primary binding site for the AvrPto ligand, whereas Fen controls the plant response to fenthion (Martin et al., 1994; Zhou et al., 1995; Scofield et al., 1996; Tang et al., 1996). Overlapping but distinct downstream plant responses are activated, depending on which of these two Prf-dependent kinases is activated. Other resistance genes also occur in clusters with related homologs (Parniske et al., 1997, and refs. therein). If the soybean Rps1-k locus is composed of multiple genes, different Rps1-k genes may be responsible for the response to different Phytophthora races and these genes may activate defense pathways that are differentially affected by ethylene insensitivity.
As a distinct alternative, the Phytophthora race 1, 4, and 7 strains used in this work may all induce similar plant responses. If the soybean etr1-1 mutant activates only a subset of these defenses, then differential resistance could be explained if the race 1 strain exhibited much greater sensitivity than races 4 or 7 to the subset of Rps1-k-dependent plant defenses that can still be induced in the etr1-1 mutant.
SUMMARY
The present report outlines the initial isolation, genetic characterization, and disease testing of ethylene-insensitive mutants of soybean. A variety of plant mutants with different alterations in ethylene sensitivity were obtained. The disease tests involved multiple pathogen species, and our results suggest that the response of ethylene-insensitive plant lines, although unchanged or improved in response to some pathogens, may be hindered in response to other pathogens or in specific gene-for-gene interactions. These results bear out the prediction of Boller and others that the effects of ethylene on plant responses to pathogens might be variable in different plant-pathogen interactions (e.g. Boller, 1991; Bent et al., 1992; Knoester et al., 1998). Given the negative effect of ethylene insensitivity on resistance to some pathogens, caution is advisable in the engineering of ethylene insensitivity and in the cultivation of ethylene-insensitive plant lines.
Further study of the responses to plant pathogens in these ethylene-insensitive lines will be of interest. Interactions with beneficial microbes are also of interest, and we have investigated the interaction of these soybean lines with symbiotic nitrogen-fixing Bradyrhizobium japonicum bacteria (Schmidt, et al., 1999). The ethylene-insensitive soybean mutants and their near-isogenic parental lines are available for other laboratory and field studies. Such studies could investigate the role of ethylene and ethylene insensitivity in seedling emergence, response to nematodes and insects, flooding tolerance, late-season senescence, plant dry-down, yield, and many other relevant phenotypic traits.
ACKNOWLEDGMENTS
We thank Jim Specht and Walter Fehr for mutagenized populations, Hilary Heustis for her extensive assistance with the mutant screen, Glen Hartman and Fritz Schmitthenner for advice on pathogen assays, Phue Vanchiasong and Steve Damon for their assistance with mutant characterization, Charlie Smyth for help with data analysis, and Cecil Nickell and colleagues for their generous assistance with the field component of this work.
Abbreviations:
- HR
hypersensitive response
- NMU
nitrosomethyl urea
Footnotes
This research was supported by the Illinois Soybean Program Operating Board.
LITERATURE CITED
- Abeles GB, Morgan PW, Saltveit ME. Ethylene in Plant Biology. San Diego, CA: Academic Press; 1992. [Google Scholar]
- Agrios GN. Plant Pathology. San Diego, CA: Academic Press; 1997. [Google Scholar]
- Ben-David A, Bashan Y, Okon Y. Ethylene production in pepper (Capsicum annuum) leaves infected with Xanthomonas campestris pv. vesicatoria. Physiol Mol Plant Pathol. 1986;29:305–316. [Google Scholar]
- Bent A, Innes R, Ecker J, Staskawicz B. Disease development in ethylene-insensitive Arabidopsis thaliana infected with virulent and avirulent Pseudomonas and Xanthomonas pathogens. Mol Plant Microbe Interact. 1992;5:372–378. doi: 10.1094/mpmi-5-372. [DOI] [PubMed] [Google Scholar]
- Bent AF. Plant disease resistance genes: function meets structure. Plant Cell. 1996;8:1757–1771. doi: 10.1105/tpc.8.10.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent AF, Kunkel BN, Dahlbeck D, Brown KL, Schmidt RL, Giraudat J, Leung JL, Staskawicz BJ. RPS2 of Arabidopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science. 1994;265:1856–1860. doi: 10.1126/science.8091210. [DOI] [PubMed] [Google Scholar]
- Bisgrove SR, Simonich MT, Smith NM, Sattler A, Innes RW. A disease resistance gene in Arabidopsis with specificity for two different pathogen avirulence genes. Plant Cell. 1994;6:927–933. doi: 10.1105/tpc.6.7.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleecker AB, Estelle MA, Somerville C, Kende H. Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis thaliana. Science. 1988;241:1086–1089. doi: 10.1126/science.241.4869.1086. [DOI] [PubMed] [Google Scholar]
- Boller T. Ethylene in pathogenesis and disease resistance. In: Matoo AK, Suttle JC, editors. The Plant Hormone Ethylene. Boca Raton, FL: CRC Press; 1991. pp. 293–314. [Google Scholar]
- Cary AJ, Liu WN, Howell SH. Cytokinin action is coupled to ethylene in its effects on the inhibition of root and hypocotyl elongation in Arabidopsis thaliana seedlings. Plant Physiol. 1995;107:1075–1082. doi: 10.1104/pp.107.4.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper RL, Martin RJ, Schmitthenner AF, McBlain BA, Fioritto RJ, St. Martin SK, Calip-DuBois A. Registration of ‘Hobbit 87′ soybean. Crop Sci. 1991;31:1093. [Google Scholar]
- Daniels MJ, Barber CE, Turner DC, Cleary SG, Sawzyc MK. Isolation of mutants of Xanthomonas campestris pv. campestris showing altered pathogenicity. J Gen Microbiol. 1984;130:2447–2455. [Google Scholar]
- Deikman J. Molecular mechanisms of ethylene regulation of gene transcription. Physiol Plant. 1997;100:561–566. [Google Scholar]
- Dhingra OD, Sinclair JB. Basic Plant Pathology Methods. Boca Raton, FL: CRC Press; 1995. [Google Scholar]
- Dixon RA, Lamb CJ. Molecular communication in interactions between plants and microbial pathogens. Annu Rev Plant Physiol Plant Mol Biol. 1990;41:339–367. [Google Scholar]
- Dong X. SA, JA, ethylene, and disease resistance in plants. Curr Opin Plant Biol. 1998;1:316–323. doi: 10.1016/1369-5266(88)80053-0. [DOI] [PubMed] [Google Scholar]
- Doupnik B. Soybean production and disease loss estimates for north central United States from 1989 to 1991. Plant Dis. 1993;77:1170–1171. [Google Scholar]
- Ecker JR. The ethylene signal transduction pathway in plants. Science. 1995;268:667–674. doi: 10.1126/science.7732375. [DOI] [PubMed] [Google Scholar]
- Gentile IC, Matta C. Production of and some effects of ethylene in relation to Fusarium wilt of tomato. Physiol Plant Pathol. 1975;5:27–35. [Google Scholar]
- Goto M, Yaguchi Y, Hyodo H. Ethylene production in citrus leaves infected with Xanthomonas citri and its relation to defoliation. Physiol Plant Pathol. 1980;16:343–350. [Google Scholar]
- Grant MR, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes RW, Dangl JL. Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science. 1995;269:843–846. doi: 10.1126/science.7638602. [DOI] [PubMed] [Google Scholar]
- Hamilton AJ, Lycett GW, Grierson D. Antisense gene that inhibits synthesis of the hormone ethylene in transgenic plants. Nature. 1990;346:284–287. [Google Scholar]
- Hammond-Kosack KE, Jones JDG. Plant disease resistance genes. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:575–607. doi: 10.1146/annurev.arplant.48.1.575. [DOI] [PubMed] [Google Scholar]
- Hua J, Chang C, Sun Q, Meyerowitz EM. Ethylene insensitivity conferred by Arabidopsis ERS gene. Science. 1995;269:1712–1714. doi: 10.1126/science.7569898. [DOI] [PubMed] [Google Scholar]
- Innes RW, Bent AF, Kunkel BN, Bisgrove SR, Staskawicz BJ. Molecular characterization of avirulence gene avrRpt2 and identification of a putative regulatory sequence common to all known Pseudomonas syringae avirulence genes. J Bacteriol. 1993;175:4859–4869. doi: 10.1128/jb.175.15.4859-4869.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga T, Salimath SS, Shi JR, Gijzen M, Buzzell RI, Bhattacharyya MK. High resolution genetic and physical mapping of molecular markers linked to the Phytophthora resistance gene Rps1-k in soybean. Mol Plant Microbe Interact. 1997;10:1035–1044. [Google Scholar]
- Kieber J. The ethylene response pathway in Arabidopsis. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:277–296. doi: 10.1146/annurev.arplant.48.1.277. [DOI] [PubMed] [Google Scholar]
- Klee H, Hayford M, Kretzmer K, Barry G, Kishore G. Control of ethylene synthesis by expression of a bacterial enzyme in transgenic tomato plants. Plant Cell. 1991;3:1187–1193. doi: 10.1105/tpc.3.11.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight LI, Rose RC, Crocker W. Effects of various gases and vapors upon etiolated seedlings of the sweet pea. Science. 1910;31:635–636. [Google Scholar]
- Knoester M, van Loon LC, van den Heuvel J, Hennig J, Bol JF, Linthorst HJM. Ethylene-insensitive tobacco lacks non-host resistance against soil-borne fungi. Proc Natl Acad Sci USA. 1998;95:1933–1937. doi: 10.1073/pnas.95.4.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi DY, Tamaki SJ, Keen NT. Cloned avirulence genes from the tomato pathogen Pseudomonas syringae pv. tomato confer cultivar specificity on soybean. Proc Natl Acad Sci USA. 1989;86:157–161. doi: 10.1073/pnas.86.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel BN, Bent AF, Dahlbeck D, Innes RW, Staskawicz BJ. RPS2, an Arabidopsis disease resistance locus specifying recognition of Pseudomonas syringae strains expressing the avirulence gene avrRpt2. Plant Cell. 1993;5:865–875. doi: 10.1105/tpc.5.8.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawton K, Weymann K, Friedrich L, Vernooij B, Uknes S, Ryals J. Systemic acquired resistance in Arabidopsis requires salicylic acid but not ethylene. Mol Plant Microbe Interact. 1995;8:863–870. doi: 10.1094/mpmi-8-0863. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK, Wellburn AR. Determination of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem Soc Trans. 1983;11:591–592. [Google Scholar]
- Lund ST, Stall RE, Klee HJ. Ethylene regulates the susceptible response to pathogen infection in tomato. Plant Cell. 1998;10:371–382. doi: 10.1105/tpc.10.3.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GB, Frary A, Wu T, Brommonschenkel S, Chunwongse J, Earle ED, Tanksley SD. A member of the tomato Pto gene family confers sensitivity to fenthion resulting in rapid cell death. Plant Cell. 1994;6:1543–1552. doi: 10.1105/tpc.6.11.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoo AK, Suttle JC. The Plant Hormone Ethylene. Boca Raton, FL: CRC Press; 1991. [Google Scholar]
- McGee DC (1991) Soybean Diseases: A Reference Source for Seed Technologists. APS Press, St. Paul, MN
- Mindrinos M, Katagiri F, Yu G-L, Ausubel FM. The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell. 1994;78:1089–1099. doi: 10.1016/0092-8674(94)90282-8. [DOI] [PubMed] [Google Scholar]
- Oeller PW, Min-Wong L, Taylor LP, Pike DA, Theologis A. Reversible inhibition of tomato fruit senescence by antisense RNA. Science. 1991;254:437–439. doi: 10.1126/science.1925603. [DOI] [PubMed] [Google Scholar]
- Parniske M, Hammond-Kosack KE, Golstein C, Thomas CM, Jones DA, Harrison K, Wulff BBH, Jones JDG. Novel disease resistance specificities result from sequence exchange between tandemly repeated genes at the Cf-4/9 locus of tomato. Cell. 1997;91:821–832. doi: 10.1016/s0092-8674(00)80470-5. [DOI] [PubMed] [Google Scholar]
- Pegg GF. The involvement of ethylene in plant pathogenesis. In: Heitfuss R, Williams PH, editors. Encyclopedia of Plant Pathology, New Series. Heidelberg, Germany: Springer-Verlag; 1976. pp. 582–591. [Google Scholar]
- Penninckx IAM, Eggermont K, Terras FRG, Thomma BPHJ, De Samlanx GW, Buchala A, Metraux J-P, Manners JM, Broekaert WF. Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell. 1996;8:2309–2323. doi: 10.1105/tpc.8.12.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CMJ, van Wees SCM, van Pelt JA, Knoester M, Laan R, Gerrits H, Weisbeek PJ, van Loon LC. A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell. 1998;10:1571–1580. doi: 10.1105/tpc.10.9.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H, Hua J, Chen QG, Chang C, Medrano LJ, Bleecker AB, Meyerowitz EM. ETR2 is an ETR1-like gene involved in ethylene signaling in Arabidopsis. Proc Natl Acad Sci USA. 1998;95:5812–5817. doi: 10.1073/pnas.95.10.5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmeron JM, Oldroyd GED, Rommens CMT, Scofield SR, Kim HS, Lavelle DT, Dahlbeck D, Staskawicz BJ. Tomato Prf is a member of the leucine-rich repeat class of plant disease resistance genes and lies embedded within the Pto kinase gene cluster. Cell. 1996;86:123–133. doi: 10.1016/s0092-8674(00)80083-5. [DOI] [PubMed] [Google Scholar]
- SAS (1989) SAS/STAT User’s Guide, Version 6. SAS Institute, Cary, NC
- Schmidt JS, Harper JE, Hoffman TK, Bent AF. Regulation of soybean nodulation independent of ethylene signaling. Plant Physiol. 1999;119:951–959. doi: 10.1104/pp.119.3.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitthenner AF, Bhat RG (1994) Useful Methods for Studying Phytophthora in the Laboratory. Ohio Agricultural Research and Development Center, Wooster
- Scofield SR, Tobias CM, Rathjen JP, Chang JH, Lavelle DT, Michelmore RW, Staskawicz BJ. Molecular basis of gene-for-gene specificity in bacterial speck disease of tomato. Science. 1996;274:2063–2065. doi: 10.1126/science.274.5295.2063. [DOI] [PubMed] [Google Scholar]
- Shoemaker RC, Polzin K, Labate J, Specht J, Brummer EC, Olson T, Young N, Concibido V, Wilcox J, Tamulonis JP and others. Genome duplication in soybean (Glycine subgenus soja) Genetics. 1996;144:329–338. doi: 10.1093/genetics/144.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair JB, Backman PA (1989) Compendium of Soybean Diseases. APS Press, St. Paul, MN
- Stall RE, Hall CB. Chlorosis and ethylene production in pepper leaves infected by Xanthomonas campestris pv. vesicatoria. Phytopathology. 1984;74:373–375. [Google Scholar]
- Tang X, Frederick RD, Zhou J, Halterman DA, Jia Y, Martin GB. Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science. 1996;274:2060–2063. doi: 10.1126/science.274.5295.2060. [DOI] [PubMed] [Google Scholar]
- Whalen M, Innes R, Bent A, Staskawicz B. Identification of Pseudomonas syringae pathogens of Arabidopsis thaliana and a bacterial gene determining avirulence on both Arabidopsis and soybean. Plant Cell. 1991;3:49–59. doi: 10.1105/tpc.3.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson JQ, Lanahan MB, Clark DG, Bleecker AB, Chang C, Meyerowitz EM, Klee HJ. A dominant mutant receptor from Arabidopsis confers ethylene insensitivity in heterologous plants. Nature Biotechnol. 1997;15:444–447. doi: 10.1038/nbt0597-444. [DOI] [PubMed] [Google Scholar]
- Wilkinson JQ, Lanahan MB, Yen H-C, Giovannoni JJ, Klee HJ. An ethylene-inducible component of signal transduction encoded by Never ripe. Science. 1995;270:1807–1809. doi: 10.1126/science.270.5243.1807. [DOI] [PubMed] [Google Scholar]
- Xie Z-P, Staehelin C, Wiemken A, Boller T. Ethylene responsiveness of soybean cultivars characterized by leaf senescence, chitinase induction and nodulation. J Plant Physiol. 1996;149:690–694. [Google Scholar]
- Zhou J, Loh Y-T, Bressan RA, Martin GB. The tomato gene Pti1 encodes a serine/threonine kinase that is phosphorylated by Pto and is involved in the hypersensitive response. Cell. 1995;83:925–935. doi: 10.1016/0092-8674(95)90208-2. [DOI] [PubMed] [Google Scholar]