Abstract
Titanium dioxide nanoparticles (nanoTiO2) are used in various applications including in paints. NanoTiO2 inhalation may induce pulmonary toxicity and systemic effects. However, the underlying molecular mechanisms are poorly understood. In this study, the effects of inhaled surface-coated nanoTiO2 on pulmonary global messenger RNA (mRNA) and microRNA (miRNA) expression in mouse were characterized to provide insight into the molecular response. Female C57BL/6BomTac mice were exposed for 1 hr daily to 42.4 ± 2.9 (SEM) mg surface-coated nanoTiO2/m3 for 11 consecutive days by inhalation and were sacrificed 5 days following the last exposure. Physicochemical properties of the particles were determined. Pulmonary response to nanoTiO2 was characterized using DNA microarrays and pathway-specific PCR arrays and related to data on pulmonary inflammation from bronchial lavages. NanoTiO2 exposure resulted in increased levels of mRNA for acute phase markers serum amyloid A-1 (Saa1) and serum amyloid A-3 (Saa3), several C-X-C and C-C motif chemokines, and cytokine tumor necrosis factor genes. Protein analysis of Saa1 and 3 showed selective upregulation of Saa3 in lung tissues. Sixteen miRNAs were induced by more than 1.2-fold (adjusted P-value < 0.05) following exposure. Real time polymerase chain reaction confirmed the upregulation of miR-1, miR-449a and revealed dramatic induction of miR-135b (60-fold). Thus, inhalation of surface-coated nanoTiO2 results in changes in the expression of genes associated with acute phase, inflammation and immune response 5 days post exposure with concomitant changes in several miRNAs. The role of these miRNAs in pulmonary response to inhaled particles is unknown and warrants further research. Environ. Mol. Mutagen., 2011. © 2011 Wiley-Liss, Inc.†
Keywords: nanotitanium dioxide, gene expression, microRNA, inflammation
BACKGROUND
Rapid developments in nanotechnology are resulting in increased use and potential release of a variety of engineered nanomaterials into the workplace and environment. Nanoparticles (NPs) are particles with a diameter less than 100 nm along at least one dimension and exhibit new or enhanced size-associated properties compared with larger particles of the same material. The large surface area, unique surface chemistry, and reactivity of NPs pose a unique challenge in assessing their effects on biological systems. Consequently, there is little knowledge of the molecular mechanisms leading to toxic effects following exposure to the diverse types of NPs, and there is no accepted framework for risk assessment of NP exposure [Oberdorster et al.,2005a,b; Borm et al.,2006].
Titanium dioxide nano particles (nanoTiO2) are poorly soluble and were originally considered to be physiologically inert, posing little risk to human health. However, the International Agency for Research on Cancer has recently classified pigment-grade titanium dioxide as a group 2B carcinogen [IARC, Feb.2006]. NanoTiO2 has also been listed as a high priority NP by the Organization for Economic Cooperation and Development steering group for test guidelines. NanoTiO2 are widely used in paints, paper, plastics, ceramics, and cosmetic sunscreens. Thus, there is potential for human exposure to nanoTiO2 occupationally or through release from consumer products. As a result, there is an urgent need for experimental investigations on the biological effects of various types of NPs to increase our understanding of the mechanisms of NP action and establish safe regulatory guidelines for human exposure.
Studies have demonstrated that nanoTiO2 causes cellular damage in exposed rodents. For example, chronic exposure to high doses of nanoTiO2 by inhalation causes bronchoalveolar hyperplasia, metaplasia, pulmonary fibrosis, and tumor formation in rats [Lee et al.,1985; Heinrich et al.,1995], possibly resulting from impaired particle clearance [Warheit et al.,1997; Cullen et al.,2000]; lower doses cause lung inflammation in mice [Dankovic et al.,2007]. A single intratracheal dose of nanoTiO2 can disrupt alveolar septa and induce emphysema-like changes in mouse lungs [Chen et al.,2006]. Particle accumulation in lungs and interstitial pneumonia associated with alveolar septal thickening results following intraperitoneal injection of nanoTiO2 in mice [Chen et al.,2009]. A multispecies, subchronic, inhalation study demonstrated similar inflammatory responses, lung burdens, and pulmonary overload in both mice and rats exposed at high doses [Bermudez et al.,2004]. NanoTiO2 is also cytotoxic and causes DNA damage and mutation in various animal and human cell lines [Rahman et al.,2002; Bermudez et al.,2004; Sayes et al.,2006; Warheit et al.,2006; Wang et al.,2007].
To increase its stability in paints, lotion, or creams, nanoTiO2 in general are surface coated with silica, alumina, or other polymers. Surface-coated nanoTiO2 in sunscreen lotions is expected to reduce its photoactivity. However, surface coatings and functionalization play a major role in nanoTiO2-induced toxicity. For instance, mice challenged with nanoTiO2 coated with silica exhibited significantly larger inflammatory responses than mice challenged with uncoated nanoTiO2 or silica alone [Rossi et al.,2010]. Similarly, vanadium pentoxide-coated anatase nanoTiO2 particles induced more cytotoxicity and genotoxicity than natural anatase [Bhattacharya et al.,2008]. Therefore, nanoTiO2 toxicity is enhanced by surface modifications of the particles. However, it is unclear what changes arise at the molecular level that lead to differences in the toxicity induced by uncoated nanoTiO2 relative to particles that are surface modified.
Genomic analyses provide a means to analyze the entire transcriptome of cells or tissues [Mei et al.,2010]. Early changes in critical pathways may be used to predict eventual health outcomes as well as derive the molecular mechanisms leading to toxicity [Hanahan and Weinberg,2000; Bhattacharjee et al.,2001; Golpon et al.,2004; Ning et al.,2004; Spira et al.,2004; Granville and Dennis,2005]. However, mRNA expression does not always correlate with related protein abundance and activity, as mRNA and proteins may be post-transcriptionally regulated. Small noncoding RNAs known as microRNAs (miRNAs) have recently been found to play a central role in the regulation of gene expression and protein translation. MiRNAs are typically 21–25 nucleotides in length and function in translational repression or mRNA degradation via the RNA interference pathway [Kim et al.,2009; Winter et al.,2009]. MiRNAs are also thought to play an important role in the maintenance of chromatin structure and are therefore critical mediators of gene expression and genome stability [Guil and Esteller,2009]. MiRNAs are involved in numerous biological processes including apoptosis [Lynam-Lennon et al.,2009], cell cycle progression [Carleton et al.,2007; Bueno et al.,2008], development (reviewed in [Sun and Tsao,2008]), and immune response (reviewed in [Tsitsiou and Lindsay,2009]). Aberrant miRNA expression has been found in several human diseases (reviewed in [Wang et al.,2008]), and many cancers express unique miRNA signatures [Croce,2009; Garzon et al.,2009]. As such, disruption of miRNA expression is now being widely investigated and various techniques have been developed for this purpose (reviewed in [Kong et al.,2009]).
We have recently investigated the developmental and neurological effects of surface-coated nanoTiO2 particles [Hougaard et al.,2010]. We also reported pulmonary inflammation in nonpregnant adult female mice exposed by inhalation to a low, biologically relevant dose of rutile nanoTiO2 particles that were surface modified and coated [Hougaard et al.,2010]. In this study, we apply DNA microarrays, pathway-specific real-time polymerase chain reaction (RT-PCR) arrays, focussed RT-PCR, and protein analysis to characterize the molecular changes associated with the observed pulmonary inflammation in the same nonpregnant adult mice exposed to surface-coated nanoTiO2 particles from the above study.
MATERIALS AND METHODS
Material, Animal Handling, and Tissue Collection
This study used UV-titan L181 with particle size of 20 nm (Kemira, Pori, Finland) enriched in rutile and modified with amounts of zirconium, silicon, aluminum, and coated with polyalcohol.
The study was conducted in parallel with a developmental toxicity study and therefore the particle dose and the time point were selected accordingly. Detailed information about animals, exposure, and exposure monitoring is described by Hougaard et al. [2010]. Briefly, 45 time-mated, nulliparous, adult female C57BL/6BomTac mice were treated as described by Hougaard et al. [2008] by whole-body inhalation to 42.4 ± 2.9 (SEM) mg nanoTiO2/m3 or to filtered air for 1 hr/day for 11 days. Seventeen (nine controls and eight exposed) of these animals did not conceive (lack of pups and interuterine implantation sites) and were used in this study as exposed adult females.
Animals were sacrificed by cardiac puncture 5 days after the last exposure. Left lung and a section of the liver were sliced randomly into portions and used for (a) total RNA extraction; (b) total RNA extraction enriched with small RNAs; and (c) total protein extracts. Samples were stored at −80°C until analysis. All procedures complied with EC Directive 86/609/EEC and Danish laws regulating experiments on animals (permit 2006/561-1123).
Material Characterization
Details of sample preparation and particle analysis are described in the Supporting Information provided by Hougaard et al. [2010]. In brief, physical particle size, morphology, and general state of agglomeration/aggregation were determined by transmission electron microscope (TEM, Tecnai G20, FEI Company, Hillsboro, OR). Crystalline phases and crystallite sizes were determined by powder X-ray diffraction (XRD) with a Bruker D8 Advance diffractometer equipped with a Lynxeye CCD detector (Bruker AXS, Madison, WI 53711-5373). Results were obtained by Rietveld refinement of the X-ray diffractograms using Bruker 10 TOPAS V4.1 software. Specific surface area was determined on a Quantachrome Autosorp-1 (Quantachrome GmbH & Co. KG, Odelzhausen, Germany) using multipoint Brunauer, Emmett, and Teller (BET) nitrogen adsorption method after 1 hr degassing at 300°C. Analysis was completed according to DIN ISO 9277 as a commercial service by Quantachrome GmbH & Co. KG. Elemental composition was analyzed by X-ray Fluorescence analysis on a Philips PW-2400 spectrometer as a commercial service by the Department of Earth Sciences, University of Aarhus, Denmark.
Exposure Monitoring
The fine particle exposure (<600 nm) was monitored using a GRIMM Sequential (Stepping) Mobility Particle Sizer (SMPS, Model No. 5.521; Serial No. 5LP 10209) connected to a GRIMM Condensation Particle Counter (Model 5.400). The SMPS data sampling and calculations were completed using the GRIMM software 5.477/02 v. 1.34 and operated in the fast scan mode. Data were corrected for both Classifier and CPC efficiency using the available software options. Particles were neutralized using a 3.7 MBq Am-241 source (Model No. 5.521). Coarse particle exposure (0.75 to > 15 μm) was measured using a GRIMM Dust Monitor (Model 1.105) at a resolution of 6 sec. The Dust Monitor particle sizes were subsequently recalculated to geometric means assuming an upper channel cut point at 20 μm.
Titanium in Tissue
Approximately 25–75 mg of lung tissue was weighed and digested in concentrated nitric acid (PlasmaPure, SCP Science, Quebec, Canada) in a microwave oven (Multiwave, Anton Paar, Graz, Austria), and titanium content was determined by quadrupole-based inductively coupled plasma mass spectrometer (ICPMS 7500ce, Agilent Technologies, Tokyo, Japan) equipped with a collision/reaction cell (CRC). The limit of detection (LOD) for titanium in tissues, based on three times the standard deviation of repeated blank measurements, was estimated to be 0.2–5 mg/kg depending on sample intake and dilution. Additional details are described by Hougaard et al. [2010].
Total RNA and MiRNA Extraction and Purification
Total RNA was isolated from the lung and the liver (n = 8/group) using TRIzol reagent (Invitrogen) and purified using RNeasy Mini Kit (Qiagen). The mirVana miRNA Isolation Kit (Ambion, Streetsville, ON, Canada) was used to prepare total RNA enriched with small RNA species from randomly selected left lung sections. RNA quality was confirmed by UV spectrophotometry and using an Agilent bioanalyzer (Agilent Technologies).
Microarray Hybridization
Individual total RNA (250 ng) samples from eight mice per treatment group (control or exposed) and universal reference total RNA (Stratagene) were used to synthesize double-stranded cDNA and cyanine labeled cRNA according to the manufacturer's instructions (Agilent Linear Amplification Kits, Agilent Technologies). Experimental samples were labeled with Cyanine 5-CTP and reference RNA with Cyanine 3-CTP (Perkin-Elmer Life Sciences). Cyanine-labeled cRNA targets were in vitro transcribed using T7 RNA polymerase, purified by RNeasy Mini Kit (Qiagen) and were hybridized to Agilent mouse 4 × 44 oligonucleotide microarrays (Agilent Technologies) at 60°C overnight. Arrays were washed and scanned on an Agilent G2505B scanner. Data were acquired using Agilent Feature Extraction software version 9.5.3.1.
MiRNA Expression Profiling
Freshly isolated individual total lung RNA samples from eight control and eight treated samples were labeled using the Agilent miRNA Complete Labeling and Hybridization Kit (Agilent Technologies). Briefly, 100 ng of total RNA was dephosphorylated by incubation with calf intestinal phosphatase at 37°C for 30 min, denatured using 100% DMSO at 100°C for 5 min, then labeled with pCp-Cy3 using T4 ligase by incubation at 16°C for 1 hr. The labeled RNA samples were hybridized to an individual array on 8 × 15K format Agilent mouse miRNA array slides. Hybridizations were performed in SureHyb chambers (Agilent) at 55°C for 20 hrs. Arrays were washed, scanned at a resolution of 5 μm using an Agilent G2505B scanner and data were acquired using Agilent Feature Extraction software version 9.5.3.1.
Statistical Analysis of Microarray Data
A reference design [Kerr,2003; Kerr and Churchill,2007] was used to analyze gene expression microarray data. Data were normalized using LOWESS in R [R-Development-Core-Team,2004] and differential expression was determined using MAANOVA [Wu et al.,2003]. The Fs statistic [Cui et al.,2005] was used to test for treatment effects, and P-values were estimated by the permutation method using residual shuffling, followed by adjustment for multiple comparisons using the false discovery rate (FDR) approach [Hochberg,1995]. Fold change calculations were based on the least-square means. Significant genes were identified as having an adjusted P-value < 0.05 for any individual contrast.
Agilent MiRNA Microarray Analysis
Nonbackground subtracted raw data were quantile normalized [Bolstad et al.,2003]. Present calls were determined as signals that were >3 trimmed SDs above the trimmed mean of the (−)3 × SLv1 probes on the array. Probes with technical replicates for a miRNA were averaged using the median signal intensity. Boxplots and cluster analyses were used to identify potential outliers (poor quality chips). This quality control check resulted in the elimination of six arrays from the analysis. Identification of differentially expressed miRNA was carried out at the probe level as well as the miRNA level. The MAANOVA model included the sample identity as a random effect and the gene specific variance estimate (F1 Test) was used to test for differences between the control and treated samples. In this analysis, parametric P-values were obtained and were then FDR corrected.
Real-Time Polymerase Chain Reaction
Microarray
Primers were designed using Beacon design 2.0 (Premier BioSoft International). Approximately 2.5 μg of total RNA per sample was reverse transcribed and RT-PCR was performed in duplicate using an iCycler IQ real-time detection system (Bio-Rad). Threshold cycle values were averaged. Gene expression levels were normalized to the GAPDH and Hprt gene, which were stable on the DNA microarray. PCR efficiency was examined using the standard curve for each gene. Primer specificity was assured by the melting curve for each gene. A Students' t-test was used for statistical evaluation. A minimum of five samples/treatment group were used.
MicroRNA
The Qiagen miScript PCR system was used. For each sample (n = 5 per group), 1 μg of total RNA enriched with small RNA species was polyadenylated and then converted to cDNA using an oligodT primer with a universal tag and miScript Reverse Transcription mix. Real-time PCR was performed in duplicate for each sample, using a primer complementary to the universal tag and a miScript primer (Qiagen) specific for each miRNA. Amplified product was detected using SYBR Green and a CFX real-time detection system (Bio-Rad). Expression levels of miRNAs were normalized to expression levels of small nuclear RNAs RNU1A1 and RNU5A1. A Students' t-test was used to statistically evaluate the data.
Pathway-Specific PCR Arrays
Approximately 800 ng of total RNA per sample (n = 6/group) was reverse transcribed using a RT2 first strand kit (SABiosciences™). Reverse transcription and real-time PCR reactions were carried out using RT2 SYBR Green PCR Master Mix on 96-well PCR arrays designed for the evaluation of mouse inflammatory cytokines and receptors (No. PAMM-011D, SABiosciences™) using a CFX real-time Detection System (BioRad). Threshold cycle values were averaged. Relative gene expression was determined according to the comparative Ct method and normalized to the Hprt and β-actin housekeeping genes. Fold changes were calculated using online PCR array data analysis software (SABiosciences™). Statistical significance was calculated using the REST method [Pfaffl et al.,2002].
Pathway Analysis
Gene ontology was used to assign genes to functional categories in DAVID [Huang da et al.,2009]. KEGG pathways were used to identify specific biological pathways associated with the differentially expressed genes. In addition, we also conducted pathway analyses using two programs we developed to perform gene set enrichment analysis in R. KEGG gene sets were obtained using the mgug4121a.db R library. The first method is a rank-based test that takes into accounts the magnitude of the intensity value as well as the correlation between genes within a specific pathway. The advantage of this test is that no assumptions on distribution or independence are made. Genes in a pathway are ranked within each observation and the average rank is calculated for each treatment. The distance between the two treatments is calculated. Permutation analysis is used to obtain a P-value for each pathway (Table I in the manuscript). The second method (Design-based test) utilizes Euclidean distance between the estimated mean vector of two treatment groups as the test statistic. The null distribution of the test statistic distance is obtained using bootstrapping as described by Kerr and Churchill [2001]. The advantage of this method is that the experimental design used in the study is accounted for.
Table I.
Gene Set Enrichment Analysis
| Probe | |||
|---|---|---|---|
| Pathway | P value | aTotal | bChanging |
| Rank-based test | |||
| Complement and coagulation cascades | 0.002 | 123 | 18 |
| Hematopoietic cell lineage | 0.002 | 253 | 13 |
| Olfactory transduction | 0.003 | 914 | 2 |
| Pantothenate and CoA biosynthesis | 0.008 | 26 | 1 |
| Systemic lupus erythematosus | 0.013 | 222 | 11 |
| Cytokine-cytokine receptor interaction | 0.018 | 652 | 33 |
| gamma-Hexachlorocyclohexane degradation | 0.021 | 27 | 3 |
| Neuro active ligand-receptor interaction | 0.041 | 421 | 3 |
| Toll-like receptor signaling pathway | 0.045 | 332 | 11 |
| Design-based test | |||
| Cytokine-cytokine receptor interaction | 0.001 | 652 | 33 |
| Pantothenate and CoA biosynthesis | 0.001 | 26 | 1 |
| Hematopoietic cell lineage | 0.006 | 253 | 13 |
| Complement and coagulation cascades | 0.011 | 123 | 18 |
| Systemic lupus erythematosus | 0.017 | 222 | 11 |
| ABC transporters | 0.018 | 87 | 2 |
| Natural killer cell mediated cytotoxicity | 0.018 | 334 | 13 |
| T cell receptor signaling pathway | 0.019 | 352 | 11 |
| gamma-Hexachlorocyclohexane degradation | 0.026 | 27 | 3 |
| Fc epsilon RI signaling pathway | 0.029 | 243 | 13 |
| Asthma | 0.032 | 87 | 3 |
| Jak-STAT signaling pathway | 0.032 | 390 | 10 |
| Linoleic acid metabolism | 0.041 | 53 | 6 |
Total indicates the total number of known probes in a given pathway.
Changing indicates the number of probes changing in a given pathway following the exposure.
Preparation of Tissue Protein Extracts and Western Blotting
A random section of the frozen left lung lobe was homogenized in lysis buffer (5 M HEPES, pH 7.5, 5 M NaCl, 10% Glycerol, 1% Triton X-100, 2 M EGTA, 1 M MgCl2, 0.5 M NaF, 0.2 M sodium pyrophosphate, protease inhibitor cocktail tablets (Roche Applied Science)) and centrifuged. The supernatant was quantified for protein content using a Bradford protein assay reagent kit (Bio-Rad).
Total lung protein extracts were immunoblotted on 14% SDS-PAGE gels and analyzed using antibodies against Saa3 (Santa Cruz Biotechnologies). Membranes were stripped and reprobed with anti-actin antibody for normalization. Band intensities were determined by averaging the densitometric readings from three biological replicates of control and treated samples from the same gel.
Total SAA Immunoassay
The Mouse SAA immunoassay (Invitrogen) was used to measure total Saa1 in lung tissue homogenates. The assay was conducted according to the manufacturer's instructions. Briefly, 100 μl of assay diluent and known quantities of controls (0–5 μg/ml mouse SAA supplied by the company) and individual samples (100 μg total tissue homogenates) in 50 μl of incubation buffer were loaded onto a microplate precoated with mouse Saa1 specific antibody followed by addition of biotinylated second monoclonal antibody. The plate was incubated at 37°C for 1 hr and unbound Saa1 was removed by washing. A streptavidin-HRP enzyme was then added to each well and incubated for 30 min at room temperature. Plates were washed to remove any unbound enzyme conjugate. One-hundred microliters of chromagen substrate solution was added to each well and incubated in the dark for 30 min at room temperature. The reaction was quenched by adding 100 μl of stop solution to individual wells. Optical Density for each well was determined at 450 nm using a microtiter plate spectrophotometer.
For analysis of Saa2, an Enzyme Immunoassay kit (Life Diagnostics) was used. One hundred micrograms of total protein from individual lung homogenates and controls (reference standard Saa2 0–500 ng/μl, supplied by the company) were loaded onto a plate precoated with anti-mouse Saa2 antibody along with horseradish peroxidase-conjugated polyclonal antibody. Plates were incubated for 1 hr at room temperature, wells were washed and 100 μl of chromagen was added to each well. Following incubation with chromagen for 20 min at room temperature, 100 μl of stop solution was added and the optical density was read at 450 nm in a microtiter plate reader. A minimum of 3 samples/treatment group were used.
RESULTS
Exposure and Particle Characterization
Mature female mice (n = 9/controls, 8/treated) were exposed for 1 hr per day for 11 consecutive days to 42.4 ± 2.9 (SEM) mg/m3 nanoTiO2 particles. Physicochemical characteristics of the nanoTiO2 used in the study are presented in detail by Hougaard et al. [2010]. Briefly, the sample consisted of a rutile TiO2-based material, which accounted for 70.8 wt% TiO2. The other major constituents were modifiers of SiO2 > Al2O3 > ZrO2 and 5.2 wt% polyalcohol coating. Mass spectrometric analyses suggested that the polyalcohol consisted of a complex mixture with 4, 6, or 8 carbon chain length. BET measurements showed that the specific surface area was 107.7 m2/g, which is slightly higher than what was reported by the manufacturer (70 m2/g). Transmission electron microscopy showed that the powder particles consisted of aggregates and agglomerates of equidimensional to needle-shaped TiO2 crystals with particle size ranging from less than 10 nm to more than 100 nm diameter. Rietveld analysis of powder X-ray diffractograms revealed that the average crystallite size of the TiO2 was 20.6 ± 0.3 nm, but an elongation was also determined.
The mice were exposed to 42.4 ± 2.9 (SEM) mg nanoTiO2/m3 particles mass concentrations as indicated by periodic filter measurements. The number of particles in the exposure atmosphere was 1.70 ± 0.20 × 106/cm3 with major particle sizes of ∼100 nm and 4 μm. About 80% of the particles by number were between 40 and 200 nm with a maximum size of 12 μm. However, the mass–size distribution was dominated by micrometer-size particles. Sub-100 nm size particles made up less than 1% of the mass.
The concentration of TiO2 particles in tissues was analyzed by ICPMS and showed 38 mg TiO2/kg in the lungs of mice 5 days after the last exposure. The lungs of control mice did not contain TiO2. No TiO2 particles were detected in the liver tissues of the exposed or control mice [Hougaard et al.,2010].
Bronchoalveolar Lavage Fluid Analysis
To assess inflammatory response to nanoTiO2 exposure in mouse lungs, inflammatory cell counts were performed on the bronchoalveolar lavage fluid (BALF) as described by Hougaard et al. [2010] and the response was reported in detail by Hougaard et al. [2010]. Briefly, the total number of cells in BALF from mice exposed to nanoTiO2 increased by 21% compared with mice exposed to air only, a change that was not statistically significant. The percentage of neutrophils increased significantly from 2% in control (air exposed) mice to 43% in the nanoTiO2-treated group (a 19-fold increase). However, the total number of macrophages decreased from 87% in controls to 49% in the treated group. There was a 6-fold increase in lymphocytes in the treated group and no significant changes in the total number of eosinophils and epithelial cells [Hougaard et al.,2010].
Gene Expression Analysis
MAANOVA analysis revealed 353 transcripts that were differentially expressed (FDR—P-values < 0.05) compared with matched controls. Of these, 53 had fold changes >1.5 for exposed versus control (50 upregulated and 3 downregulated) (full list in Additional File 1; File 1: Supporting Information Table S1). Complete microarray data are available through the Gene Expression Omnibus at NCBI (http://www.ncbi.nlm.nih.gov/geo/), GSE 19487. A heatmap was generated using the Heatplus library [Ploner,2008] in R [R-Development-Core-Team,2009] using the log2 of the relative intensities. Any technical probe replicates were averaged using the mean. Outliers were removed based on the cluster analysis of the normalized log2 of the Cy3 and Cy5 channel. Hierarchical cluster analysis on differentially expressed genes revealed that samples within a treatment group were clustered together (Additional File 1; File 1: Supporting Information Fig. S1). Thus, a clear treatment effect was found as a result of exposure to nanoTiO2. The genes with increased mRNA levels included the following: serum amyloid A-3 (Saa3, 4.7-fold), chemokine (C-X-C motif) ligand 5 (cxcl5, 4.4-fold), lymphocyte antigen 6 complex, locus F (Ly6f, 4.3-fold), solute carrier family 26,member 4 (Slc26a4, 3.4-fold), lipocalin 2 (Lcn2, 3.3-fold), and NADPH oxidase organizer 1 (Noxo1, 3.0-fold). Among the genes with decreased mRNA levels, the largest decrease was observed for calcium-dependent phospholipid-binding protein, Copine5 (Cpne5, 2.1-fold). Real time RT-PCR confirmed the altered mRNA levels of serum amyloid A 1(Saa1), Saa3,Noxo1,NADPH oxidase activator 1 (Noxa1), matrix metallopeptidase 12 (Mmp12), and complement component 3 (C3) in the exposed mice (Fig. 1A).
Fig. 1.
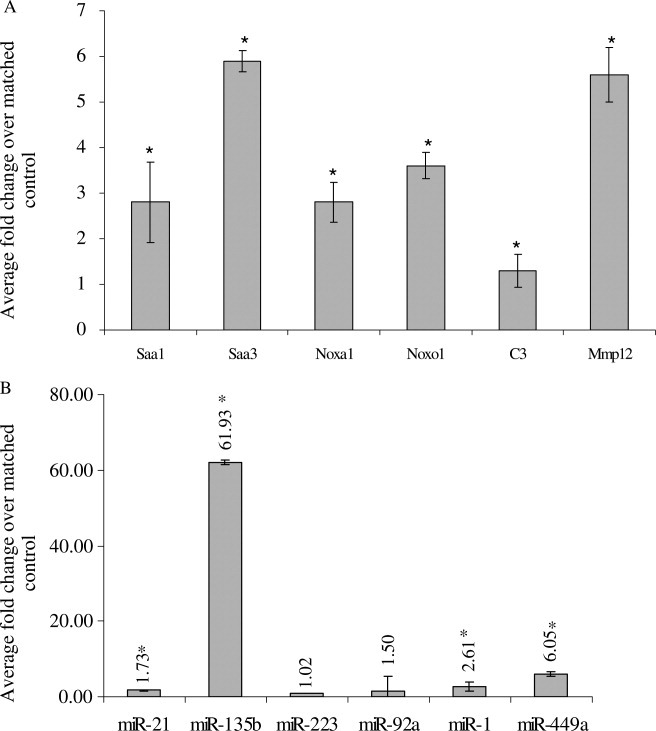
Real time quantitative PCR validation of array results. A: Microarray results. Data (n = 5 mice/group, ±SEM) are presented as average fold changes over matched controls (air exposed). Gene expression values were normalized to internal reference gene GAPDH and Hprt. B: MicroRNA array results. Data are presented as average fold changes over matched controls (air exposed). (n = 5 mice/group, ±SEM). Expression values were normalized to small nuclear RNA RNU1A1. Numbers indicate fold changes. * indicates significant by Students' t-test.
Gene ontology analysis was used to assign genes to functional categories in DAVID [Huang da et al.,2009]. KEGG pathways were used to identify specific biological pathways associated with the differentially expressed genes. Pathway enrichment analysis applied two programs using the mgug4121a.db R library (Table I, rank-based test [Alvo et al.,2010], and a design-based test). The major pathways that were identified remained the same regardless of the method applied. These included cytokine–cytokine receptor interaction, metabolism, complement and coagulation cascade, hematopoeitic cell lineage, biosynthesis of steroids, and systemic lupus erythematosus.
Induction of Acute Phase Response Reactants
NanoTiO2 exposure altered the expression of several acute phase response genes (Table II) including Saa1 and 3. Previous work has demonstrated that acute phase response genes including Saa are induced in response to air pollution and inhaled particulates [Pope et al.,2004; Ruckerl et al.,2006]. Protein analysis was conducted to elucidate the role of Saa genes in nanoTiO2-induced lung response. ELISA analysis of total lung tissue homogenates could not detect Saa2 in either treatment group (data not shown). A very modest decrease (0.86-fold) in total Saa1 protein was observed in the lung tissues of exposed mice (Additional File 1; File 1: Supporting Information Fig. S2A), in contrast to the gene expression results (Table II, 2.2-fold upregulated). This observation may have been due to outlying values from two control mice (expressing high amounts of Saa1 as indicated by the large error bars, Additional File 1; File 1: Supporting Information Fig. S2A.). Alternatively, Saa1 may be targeted post-transcriptionally by a miRNA that is also induced in response to nanoTiO2. Western blot analysis (no ELISA assay available) revealed a 2.2-fold increase in total Saa3 protein in the nanoTiO2-treated group relative to matched controls (Additional File 1; File 1: Supporting Information Fig. S2B).
Table II.
List of all Acute Phase Response Genes Showing Fold Changes Higher Than 1.2 in exposed mice
| Acute phase reactants | P value | Fold changea |
|---|---|---|
| Serum amyloid A1 | 0.00 | 2.24 |
| Serum amyloid A3 | 0.00 | 4.71 |
| Complement protein C3 | 0.00 | 1.37 |
| Complement component 1, s (C1s) | 0.00 | 1.28 |
| Complement component 3a receptor 1 (C3ar1) | 0.00 | 1.15 |
| Complement component 1, q beta polypeptide (C1qb) | 0.00 | 1.30 |
| Complement component 1, r subcomponent (C1r) | 0.00 | 1.31 |
| Complement component C1RB (C1rb) | 0.00 | 1.21 |
| Fibrinogen | 0.01 | 2.05 |
| Coagulation factor II (F2) | 0.01 | 1.72 |
| Mannose binding protein | 0.02 | 1.70 |
| Albumin | 0.01 | 1.79 |
| apoA1 | 0.01 | 1.51 |
| apoAII | 0.03 | 1.61 |
| alpha2-HS glycoprotein | 0.00 | 1.85 |
| S100A8 (calgranulin A) | 0.01 | −1.85 |
| Serpina3n | 0.00 | 1.37 |
Gene names in bold indicate FDR adjusted P value > 0.05.
Average fold change compared with matched controls.
Induction of Inflammatory Cytokines and Receptors
NanoTiO2 exposure resulted in increased mRNA levels of genes associated with cytokine–cytokine receptor signaling and chemokine signaling pathways. These include chemokine (C-X-C motif and C-C motif) ligands (cxcl5, cxcl1 and cxcl12, ccl7, ccl9, ccl2, ccl17, ccl12, and ccl19). We confirmed the altered mRNA levels of these genes using pathway-specific PCR arrays (mouse inflammatory cytokines and receptors; SABiosciences™) containing 70 different cytokines and chemokines. Six individual samples from the control and treatment groups were analyzed. Twenty one genes were statistically significantly differentially expressed (1.5-fold) by t-tests (data not shown) and 14 genes by the REST method [Pfaffl et al.,2002] (Table III and Additional File 1; File 1: Supporting Information Table S2). The analysis showed significant increase in levels of cxcl-5 (30.0-fold), cxcl1 (7.0-fold), ccl2 (4.2-fold), ccl22 (3.4-fold), ccl7 (3.8-fold), ccr4 (2.1-fold), and cytokine tumor necrosis factor (TNF, 1.8-fold) in lung tissues of mice treated with nanoTiO2. Expression of interleukin-6,interleukin 1 beta, and IFN gamma did not change. Overall, 14 transcripts that were significantly differentially expressed with FC > 1.5 as measured by the microarray were validated using the PCR arrays. Eleven of them are in agreement with the results of the microarray.
Table III.
Results of Pathway-Specific PCR Array (Mouse Inflammatory Cytokines and Receptors)
| Gene name | P value | Fold changea |
|---|---|---|
| Chemokine (C-X-C motif) ligand 1 (Cxcl1) | 0.000 | 7.00 |
| Chemokine (C-C motif) ligand 2 (Ccl2) | 0.000 | 4.20 |
| Chemokine (C motif) receptor 1 (Xcr1) | 0.000 | 1.80 |
| Chemokine (C-X-C motif) ligand 5 (Cxcl5) | 0.010 | 30.00 |
| Secreted phosphoprotein 1 (Spp1) | 0.010 | 2.00 |
| Chemokine (C-C motif) ligand 6 (Ccl6) | 0.021 | 1.90 |
| Complement component 3 (C3) | 0.021 | 1.70 |
| Chemokine (C-C motif) ligand 22 (ccl22) | 0.021 | 3.40 |
| Chemokine (C-C motif) receptor 4 (Ccr4) | 0.021 | 2.20 |
| Chemokine (C-C motif) ligand 3 (ccl3) | 0.024 | 2.20 |
| Chemokine (C-C motif) ligand 12 (Ccl12) | 0.026 | 2.00 |
| Chemokine (C-C motif) ligand 9 (Ccl9) | 0.028 | 1.70 |
| Tumor necrosis factor (Tnf) | 0.041 | 1.80 |
| Chemokine (C-C motif) ligand 7 (Ccl7) | 0.048 | 4.00 |
Average fold change compared with matched controls.
Hepatic Gene Expression
We analyzed liver mRNA from the same mice and found differential expression of 11 genes (FDR P-value <0.05); however, these expression changes were modest; the largest change was a 1.6-fold downregulation of “similar to hepatocellular carcinoma-associated gene TD26.” The other 10 genes had fold changes below 1.2. The DNA microarray data are available through the NCBI (http://www.ncbi.nlm.nih.gov/geo/), GSE 19487. Overall ranking of all genes according to fold change demonstrated that gene expression was relatively unchanged in the livers.
Global MiRNA Expression Changes
Agilent arrays containing 567 mouse probes were used to examine changes in miRNA following exposure to nanoTiO2. The data are available through the NCBI (http://www.ncbi.nlm.nih.gov/geo/), GSE 19487. Fifty-five miRNAs were significantly altered by nanoTiO2 exposure following FDR-adjustment (Table IV). The levels of six miRNAs were increased by more than 1.5-fold; the only downregulated miRNAs were miR-92a (1.3-fold) and miR-223 (1.4-fold)). Real time RT-PCR analysis confirmed the upregulation of miR-449a, miR-1, and miR-135b (Fig. 1B).
Table IV.
MiRNA Results
| Probe | FDR P value | Fold changea |
|---|---|---|
| mmu-miR-449a | 0.000 | 2.81 |
| mmu-miR-1 | 0.000 | 1.92 |
| mmu-miR-135b | 0.000 | 1.83 |
| mmu-miR-133b | 0.004 | 1.59 |
| mmu-miR-144 | 0.008 | 1.53 |
| mmu-miR-21 | 0.000 | 1.52 |
| mmu-miR-133a | 0.018 | 1.39 |
| mmu-miR-34b-5p | 0.000 | 1.33 |
| mmu-miR-193 | 0.048 | 1.29 |
| mmu-miR-34c | 0.000 | 1.28 |
| mmu-miR-141 | 0.001 | 1.28 |
| mmu-miR-33 | 0.003 | 1.26 |
| mmu-miR-342-3p | 0.000 | −1.25 |
| mmu-miR-92a | 0.007 | −1.26 |
| mmu-miR-720 | 0.016 | −1.28 |
| mmu-miR-223 | 0.001 | −1.39 |
Average fold change compared with matched controls.
DISCUSSION
Short term or chronic exposure to nanoTiO2 particles by inhalation or instillation has revealed that nanoTiO2 particles induce a complex pulmonary response that potentially involves alterations in signaling cascades and associated expression of numerous genes and proteins [Lee et al.,1985; Bermudez et al.,2002; Bermudez et al.,2004; Warheit and Frame,2006]. Changes in gene expression are presumed to occur in response to exposure and in the early stages of disease development, and thus lead to the downstream biological outcome. MiRNAs are proposed to be one of the mechanisms regulating mRNA and protein levels following toxicant exposure [Hudder and Novak,2008; Taylor and Gant,2008], and thus are potentially important mediators of toxic outcome. Concurrent analysis of mRNAs and miRNAs is a powerful approach to explore the direct response of the genome to toxicant exposure.
We have recently reported the developmental and neurological effects of surface-coated nanoTiO2 particles [Hougaard et al.,2010]. Pregnant mice were exposed for 1 hr daily for 11 consecutive days to 42.4 ± 2.9 (SEM) mg/m3 suspended nanoTiO2 particles via inhalation from gestational day 8–18. The mice that failed to conceive were sacrificed 5 days following the exposure. These mice (nonpregnant adult) exhibited evidence of pulmonary inflammation by analysis of cell counts in BALF. In this study, we characterize the global mRNA and miRNA response in the lungs of the nonpregnant adult mice, to provide insight into the molecular mechanisms contributing to this effect. We also analyze global hepatic mRNA response to the inhaled nanoTiO2. We demonstrate alterations in pulmonary mRNA levels in pathways that are consistent with the observed and predicted effects of particle exposure and also in line with the observed phenotype. We also report perturbation of several miRNAs in the lungs of these mice. In contrast, gene expression was relatively unaffected in the livers of the same mice.
Pulmonary Inflammation
Analysis of different cell counts in BALF revealed an increase in the total number of neutrophils, supporting the presence of pulmonary inflammation, 5 days following nanoTiO2 exposure in mature female mice from this study [Hougaard et al.,2010]. Long-term pulmonary inflammation following inhalation of nanoTiO2 has previously been documented in both mouse and rat models [Bermudez et al.,2004; Ma-Hock et al.,2009; van Ravenzwaay et al.,2009]. For example, studies have demonstrated increased infiltration of neutrophils 2 weeks following inhalation of 100 mg/m3 mixed anatase and rutile nanoTiO2 for 6 hrs/day for 5 days [Ma-Hock et al.,2009; van Ravenzwaay et al.,2009]. Similarly, mice exposed to 10 mg/m3 nanoTiO2 for 6 hrs/day, 5 days/week for 13 weeks showed persistent increases in the number of neutrophils following a 52 day recovery period [Bermudez et al.,2004]. These studies, along with the data presented here, suggest that nanoTiO2-induced pulmonary inflammation is characterized by neutrophil infiltration.
This study also supports previous work demonstrating that surface-treated nanoTiO2 particles are inflammogenic. For example, intratracheal instillation of surface-treated (alumina or alumina and amorphous silica) nanoTiO2 caused pulmonary inflammation in rats [Warheit et al.,2005]. High doses of silanized hydrophobic ultrafine TiO2 instilled into rats resulted in increased lung toxicity [Pott et al.,1998]. Mild inflammatory changes were noted in rats instilled with low doses of silanized nanoTiO2 [Rehn et al.,2003]. Similar inflammogenic responses were observed in several other studies using ultrafine nonsurface-modifed nanoTiO2 both in vitro and in vivo [Baggs et al.,1997; Drumm et al.,1999; Bermudez et al.,2002; Bermudez et al.,2004; Renwick et al.,2004; de Haar et al.,2006]. The extent of inflammation observed in this study is in line with the overall calculated mass of nanoTiO2 deposited in the pulmonary region (72.5 μg with a surface area of 77.6 cm2). Differential cell count analysis of BALF revealed that 43% of the cells were neutrophils [Hougaard et al.,2010]. This finding is in close agreement with a previous experiment where mice exposed by instillation to nanoTiO2 with a surface area of 50 cm2 exhibited 45% neutrophils in BALF [Oberdorster et al.,2005b]. A linear correlation between the total number of neutrophils in BALF and particle surface area was noted in mice exposed for 24 hrs to five different types of nanoparticles, including the nanoTiO2 studied in this study (Saber et al., personal communication). Similar relationships between surface area and total number of neutrophils has also been reported by Tran et al. [Tran et al.,2000]. Taken together, these studies demonstrate that nanoTiO2 exposure promotes proinflammatory activities across a wide range of doses, necessitating an understanding of the molecular pathways involved in this response.
mRNA Expression Profiling
Global transcriptional profiling of lung samples from adult female mice exposed to nanoTiO2 revealed significant differential expression of several inflammation modulators. Inhalation exposure to 42.4 ± 2.9 (SEM) mg/m3 nanoTiO2 for 1 hr every day for 11 consecutive days induced significant changes in the mRNA levels of several chemokine genes. These include ccl2 (MCP-1 alpha), cxcl1 (keratinocyte cell-derived chemokine, KC), cxcl5 (epithelial cell-derived neutrophil activating peptide ENA78), ccl22 (macrophage-derived chemokine), ccl7 (monocyte chemotactic protein 3, MCP3), ccl9/MIP1 gamma, and ccl3 (monocyte inflammatory protein, MIP-1 alpha). These genes participate in the modulation of chemotaxis, infiltration of neutrophils, and epithelial proliferation. Because it is difficult to interpret the biological relevance of small fold changes of individual genes, we emphasize changes in pathways and groups of genes (i.e., pathways, and Gene Ontology molecular functions and biological processes) in our discussion below. When many genes that interact together to carry out a biological function are perturbed, this is more likely to be biologically relevant than an individual gene within that family. Thus, the gene expression changes we observed collectively provide molecular insight into the findings on inflammation described earlier.
Neutrophil sequesteration is a critical event during pulmonary inflammation and involves multiple steps including the following: Increased production of chemokines and cell adhesion molecules, and enhanced interactions between cells. Cxcl1, macrophage inflammatory protein −2 (MIP2), and cxcl5 are the major chemokines for neutrophil recruitment and accumulation in mouse lungs. Cxcl1 and MIP2 are produced predominantly by the infiltrating myeloid cells (macrophages and neutrophils) and increases in their levels in response to a stimulant are rapid and transient. In contrast, cxcl5 is produced by resident cells, such as alveolar epithelial type II cells. Elevated levels of cxcl5 persist for days during lung inflammation. Preferential and persistent induction of cxcl5 in resident cells may be important in establishing cellular communication between myeloid cells and resident cells, which is required for initiation, maintenance and resolution of an inflammatory process [Jeyaseelan et al.,2005]. We observed high levels of cxcl5 5 days following the last exposure in this study. This finding indicates epithelial cell activation and may suggest continued lung injury and persistent pulmonary inflammation.
Increased expression of ccl3, cxcl5, and cxcl1 has been shown following nanoTiO2 exposure in several in vivo mouse models; these findings were associated with the development of pulmonary inflammation, granulomas, emphysema, and pulmonary host defense [Driscoll et al.,1993; Park et al.,2010; Rossi et al.,2010]. Disregulation of inflammatory pathways following inhalation of nanoTiO2 has also been shown by Chen et al. using a DNA microarray approach [Chen et al.,2006]. These authors used small (∼6,000 clones) “home-made” cDNA microarrays to show that a single intratracheal instillation of 0.1 mg nanoTiO2 (rutile, uncoated pure particles of 19–21 nm in size) caused severe pulmonary inflammation with alterations in genes involved in several pathways that overlap with this work (chemokines and coagulation complement cascade). Chemotaxis and immune response pathways, and Th1 and 2 type cytokines were also differentially regulated in the BALF of mice treated with a single intratracheal dose of 20 mg/kg nanoTiO2 (P25, primary particle size 20 nm) 14 days after the last exposure [Park et al.,2009]. In many of the aforementioned studies, comparatively high doses of particles were directly deposited into the lungs of the animals by intratracheal instillation. In this study, mice were exposed using a biologically relevant exposure route to a relatively low dose of nanoTiO2 (mimicking the occupational scenario in terms of the total mass administered) and similar changes in gene expression profiles were noted despite differences in doses and time points. In contrast to our data and the studies described earlier, Rehn et al. [2003] reported no signs of inflammation in rats exposed to a range of occupationally relevant low doses of surface-treated nanoTiO2. Discrepancies between studies may be due to the targeted analysis of specific markers of inflammation and endpoints (e.g., total protein and BAL cells) applied by Rehn et al. [2003] compared with analysis of the entire transcriptome. Many nanotoxicological studies obtain data from a single endpoint representative of a particular mechanism of action, focusing on specific sets of genes or even a single pathway. The drawback of this approach is that the mechanism of action must be known beforehand; moreover, the expected phenotype or the physiological change may not be apparent in the very low dose range. Global gene expression profiles provide a snapshot of multiple cellular processes operating in a single experiment without prior knowledge of the mechanism of action. It is particularly suitable to identifying early molecular changes predictive of eventual pathological outcome. Thus, we demonstrate that global gene expression signatures support the observed phenotypes for exposure to nanoTiO2 that response can be measured in the low-dose range, and the results obtained can be used to provide insight into the molecular mechanisms operating in response.
Pulmonary response to nanoTiO2 also causes increase in levels of genes in the acute phase response pathway (Table II) including Saa1, Saa3, C3, complement components 1s (C1s), 3ar1 (C3ar1), 1qb (C1qb), 1r (C1r), and 1rb (C1rb). Moreover, several other acute phase proteins exhibited fold changes >1.5-fold with unadjusted P-values < 0.05. These include C- reactive protein,fibrinogen,coagulation factor II,mannose binding protein,albumin,apoAI,apoAII, and a2-hs-glycoprotein (regulator of SAA) (Table II). The complement cascade includes 30 proteins (some of which are enzymes), cofactors, inhibitors, and membrane-associated proteins. These molecules act in host defense by promoting phagocytosis and inflammation, which can activate the complement cascade resulting in chemotaxis and pulmonary inflammation. Acute-phase proteins have been used as biochemical markers of disease for numerous inflammatory processes. Induction of acute phase response has previously been observed in rodents following particle exposure. Saa is a generic name for a family of apolipoproteins [Meek and Benditt,1986] synthesized in response to activated monocyte/macrophage-released cytokines such as IL-1 and IL-6 [Ganapathi et al.,1988; Jiang et al.,1995; Jensen and Whitehead,1998]. Although the liver is the major site of synthesis of Saa [Morrow et al.,1981; Lowell et al.,1986], extrahepatic tissues including lung, spleen, adrenal gland, and others have been shown to express varying levels of individual members of Saa genes in response to different inflammatory stimuli [Meek and Benditt,1986]. In this study, Saa genes (1 and 3) were strongly induced in the lungs of mice exposed to nanoTiO2. The increase in Saa3 mRNA levels was much greater (5.9-fold) than Saa1 (2.8-fold). Corresponding protein levels of Saa3 were also changed in lung tissue homogenates of the treated group (Supporting Information Fig. S2B) but no change was observed for Saa1. Previous work has demonstrated that Saa genes are induced in mouse lung in response to dust and lipopolysaccharide (LPS) [Meek and Benditt,1986; Andre et al.,2006; Park et al.,2010]. In keeping with this study, while injection of LPS resulted in increased mRNA levels of all three Saa genes in liver, Saa3 was predominantly expressed in lungs. Similarly, the largest increase in Saa3 mRNA levels was noted in mouse lungs after ultrafine particle exposure [Andre et al.,2006].
Select upregulation of Saa3 and associated proteins, together with increased gene expression of a suite of cytokines, chemokines, and metalloproteases in our model suggests a critical role for Saa3 in nanoTiO2-induced pulmonary inflammation/immune response. In premetastatic lungs, Saa3 induction by chemoattractants such as S100A8 plays a role in myeloid cell accumulation and acts as a positive feedback regulator for chemoattractant secretion [Hiratsuka et al.,2008]. In this study, S100A8 mRNA levels were decreased compared with the controls (−1.9-fold; unadjusted P-value < 0.05) in the treatment group suggesting the involvement of a different regulatory mechanism for Saa3. Although the cytokines IL-1, IL-6, and TNF play a role in the expression of Saa genes, only TNF was marginally upregulated in our model. We also observed a marginal increase in TNF protein in BAL cells as well in lung tissue homogenates by protein arrays (data not shown), suggesting a possible role for TNF in Saa3 expression in response to nanoTiO2.
Several chemokines (cxcl5, cxcl1, and ccl3) and components of the acute phase pathway (Saa3, c3) measured in this study are rapidly induced in response to acute injury or stress. However, the increase in their expression levels is transient and falls rapidly once the stimulus is removed or the injury is repaired. Thus, high and persisting levels of these acute stressors 5 days after the last exposure may reflect the magnitude of the injury and indicate initiation of secondary pathological conditions.
In addition to confirming the response of these pathways, we also found a dramatic increase in polymeric Ig receptor (PIgr, 2.0-fold), solute carrier family 26, member 4 (Slc26a4, 3.4-fold), and lipocalin 2 (Lcn2, 3.3-fold). These genes are involved in mucosal immune defense. NADPH oxidase organizer 1 (Noxo1), a gene associated with antimicrobial defense [Flo et al.,2004; Schneeman et al.,2005; Nakao et al.,2008], was also upregulated by 3-fold. We also found upregulation of the proinflammatory genes Vanin-1 and 3 (Vnn-1,−3, 2.6- and 1.9-fold, respectively) in nanoTiO2-exposed lungs [Nitto et al.,2008]. Thus, our results confirm strong activation of pulmonary immune and inflammation in response to nanoTiO2 inhalation.
Despite significant upregulation of several genes associated with acute phase, inflammation and immune response in lung, the hepatic transcriptome of these mice was unaffected. These findings agree with our previous report showing few changes in hepatic transcription in mice exposed by inhalation to doses of carbon black and diesel exhaust that caused substantial pulmonary inflammation [Saber et al.,2009]. The results suggest that systemic effects in liver are secondary to pulmonary inflammation and may not require gene expression.
NanoTiO2 Exposure
Based on the exposure conditions, each animal in this study was exposed to ∼840 μg of nanoTiO2 (11 days × 1 hr/day × 40 mg/m3 × 0.0018 m3 inhaled/hr = 0.840 mg/animal). It is predicted that the majority of nanosized particles deposit in the lung once inhaled. The deposition of particles in the lungs depends on the size of the particles. In this study, the mass of the airborne fraction contained predominantly micrometer-sized particles. On the basis of the deposition model described by Jacobsen et al. [2009], we expected a deposition of 72.5 μg in the pulmonary region, 48 μg in the tracheobronchial region, 356 μg in the gastrointestinal tract, and 267 μg of nanoTiO2 in the skull. Hence, with an average lung weight of 274 mg, the estimated deposited pulmonary dose amounts to 265–439 mg/kg nanoTiO2 corresponding to 112–159 mg nanotitanium (Ti)/kg in the lung, depending on whether pulmonary or bronchopulmonary regions are considered. Detection in lung tissue using ICPMS revealed 38 mg/kg Ti in the lungs [Hougaard et al.,2010], indicating that although significant clearing of the lungs had taken place during the 16 days after the first exposure, considerable amounts of nanoTiO2 were still present 5 days postexposure. This corresponds to 24–34% of predicted nanoTiO2 deposition in lungs. Our results are in agreement with Rossi et al. [2010], who exposed mice by inhalation to 10 mg/m3 of surface-coated nanoTiO2 (40 nm) for a total of 32 hrs. They showed ∼60% of the deposited nanoTiO2 was cleared over a 4-week period [Rossi et al.,2010]. This suggests that the small particle size leads to alveolar deposition with subsequent slow clearing [Oberdorster,1995]. Depending on the particle size, some of the inhaled nanoTiO2 is also expected to be deposited in the nasal region. Approximately 90% of 1 nm, 20–30% of 5–10 nm, and less than 10% of 20 nm particles deposited in the nasopharyngeal region. A small fraction of the total particle mass deposited in the nasal region has previously been shown to be efficiently translocated to olfactory bulb in brain through axonal transport via olfactory neurons [Oberdorster et al.,2002,2004]. However, particles deposited in the nasal region of the respiratory system would be quickly cleared either by blowing the nose, sneezing, or by efficient mucociliary transport mechanism. As such, we have not assessed the deposition of particles in the upper respiratory tract and hence their uptake by the central nervous system. Since particles in the nasal region are rapidly cleared, and since we have measured the gene expression changes 5 days after the last exposure, we believe that the contribution of nasally deposited particles to overall outcome in the present model is likely to be minimal.
The dose administered in this study corresponds to the highest occupational exposure limit for dust (in terms of total mass administered, 5 mg/m3 during an 8 hr working day) in Denmark. Thus, this study used a relatively low, occupationally relevant exposure regime in combination with robust and conservative statistical analysis of a sufficiently large sample size of animals. However, one could argue that considering the slow clearance of the particles (as observed in this study), along with the dose used, lung overburden has occurred. Although the dose rate was high, the deposited dose is only a fraction of what would result in overload (i.e., slower motility of alveolar macrophages and impaired mucociliary clearance). Moreover, the particles in this study were surface coated with Si, Zr, and Al. It has been shown that surface-coated nanoTiO2 are more toxic than benign pure particles of nanoTiO2 [Rossi et al.,2010]. Although we did not test the possibility that surface coatings (Si, Al, and Zr) themselves contributed to the toxicity, it is documented in the literature that Si, Zr, and Al are less toxic when administered on their own [Rossi et al.,2010], suggesting that the slower clearance and retention of smaller size particles in the lung tissue could be the reason behind observed pulmonary response. Here, we confirm that nanoTiO2 inhalation results in changes in proinflammatory, immune response and complement cascade-related genes that persist for relatively long periods of time after the final exposure. The observed fold changes for any particular gene in our study were much lower than the other reports, which may be due to the lower doses used, the differences in exposure set up, or the types of particles used. However, a large portion of these changes were validated with quantitative RT-PCR and RT-PCR arrays. Thus, we argue that the pulmonary response observed in our study is likely due to the highly toxic nature of the particles and not lung overburden.
Although the mice in this study were exposed to doses near the occupational exposure limit for dust in Denmark in terms of total mass administered, it might be argued that the dose is high if the hours of exposure are considered (i.e., delivered in 1 hr vs. exposure over an 8 hr work day). Genomic tools have not been extensively used to understand nanoassociated toxicity. Using animals that exhibit an inflammatory response following the exposure, this work was initiated to establish proof of principle that genomic tools can be applied to characterize the effects of nanoparticles at relatively low doses and reveal the molecular perturbations that persist at this time point that contribute to the observed phenotype. However, the observed overall response at the transcriptional level must be investigated in studies incorporating multiple doses, time points, and different types of nanoTiO2, to truly dissect the mechanism of acute exposure and distinguish between immediate early effects and persistent effects. Thus, multiple time points and doses were not conducted in this study, but will be the subject of future work.
MiRNA Expression Profiling
Despite a dramatic increase in Saa1 gene transcript levels, we did not see a correlated increase in its protein synthesis. Similarly, in contrast to the ∼5-fold transcriptional increase of Saa3 mRNA, Saa3 protein was increased by only 2-fold. One mechanism that controls translation of proteins involves targeted translational repression by miRNAs. Thus, to address the observed discrepancies between the transcription and protein expression profiles and to explore the potential role of miRNAs in mitigating mRNA and translation in response to particle exposure, we analyzed global changes in pulmonary miRNA expression using DNA microarrays. Total RNA from lung tissues of control and exposed mice was used. The analysis revealed 55 statistically significant changes; 16 were altered by more than 1.2-fold and six had fold changes >1.5 (Table IV). Because an individual miRNA may target hundreds of mRNAs, a small change in a miRNA may have significant repercussions on transcription and translation. As such, we explored changes both above and below the 1.5-fold threshold.
Some of the miRNAs that are altered in response to nanoTiO2 have been implicated in inflammation and immune response. For example, upregulation of miR-21 and downregulation of miR-1 has recently been observed in IL-13 transgenic mice with allergic airway inflammation [Lu et al.,2009]. MiR-21 is expressed by inflammatory leukocytes in asthmatic lung [Jin et al.,2008], hematopoietic cells [Landgraf et al.,2007], and in macrophages and dendritic cells [Lu et al.,2009]. MiR-21 is induced in murine macrophages following LPS challenge [Lu et al.,2009], all suggesting a critical association for miR-21 in inflammation and immune response. MiR-1 levels increase during myocardial differentiation of mouse embryonic stem cells [Takaya et al.,2009]. Farraj et al. [2010] have shown that miR-1 and miR-21 are significantly downregulated in the myocardium of rats exposed to synthetic particulate matter. Other miRNAs affected by nanoTiO2 exposure have been linked to a cancer phenotype. For example, differential expression of miR-135b is found in human colon cancer samples [Sarver et al.,2009] and miR-449a is associated with antitumor activity in prostate cancer cells [Noonan et al.,2009].
Analysis by qRT-PCR confirmed the higher levels of miR-449a (fold six), miR-1 (2.6-fold), and miR-135b (60-fold) in this study (Fig. 1B) but not the suppression of miR-223 and miR-92a. Analysis of curated targets in TarBase (http://diana.cslab.ece.ntua.gr/tarbase/) did not reveal any known targets of miR-449a or miR-135b. As such, we searched for mRNA targets of miR-135b and miR-449a using the target-prediction softwares TargetScan and Pictar. Predicted targets for miR-135b (TargetScan or Pictar) that were differentially expressed (downregulated) included the following: checkpoint suppressor1,runt related transcription factor 2 (runx2), and phosphodiesterase 8b. Predicted targets of miR-449a that were differentially expressed included the following: lymphoid enhancer-binding factor 1 (lef1) and Kit ligand. Runx2 and lef1 are the downstream targets of WNT signaling suggesting that miR-135b and miR-449a may have a common function in regulating WNT signaling. However, it appears that the effect of the observed 6- and 60-fold induction of miR-449a and miR-135b on transcript levels of their predicted targets is negligible. There may be several reasons to explain the lack of changes in target mRNAs. First, the genes targeted by miR-449a and miR-135b in response to particle exposure may be different than the ones predicted in silico. Second, the majority of the genes changing in response to nanoTiO2 in our model are acutely induced and therefore the magnitude of their induction is expected to be 10–100-fold higher immediately following the exposure. Therefore, even if some of these genes are directly targeted by the miRNAs at the transcriptional level, their suppression may not be below those of control levels. Third, it is also possible that the predominant mode of action for these miRNAs is through translational repression.
Currently, the causes and consequences of miRNA response following exposure to nanoTiO2 are unknown. However, changes at the transcriptional level of several target genes are implicated in acute phase and inflammatory response. Thus, miR-1, miR-135b, and miR-449a may play a role in these biological processes. We have recently confirmed induction of miR-135b in other inflammatory models including particle-induced inflammation (Halappanavar et al., unpublished data). The induction of several inflammatory chemokines and acute phase components is well characterized in response to particle exposure, including nanoTiO2 (as described in detail earlier). However, the possibility that these molecular pathways could be under the direct control of miRNAs that play a role in particle-induced response is yet to be demonstrated. We hypothesize that some of the genes, including the Saa family and chemokines, are the direct targets of miR-135b and that miR-135b could be acting to resolve the inflammation process. A similar hypothesis was also proposed by Jardim et al. [2009]. These authors suggest that disruption in miRNA expression in response to diesel exhaust particles (DEP) in human airway epithelial cells could be implicated in modulation of the inflammatory process induced by DEP. Further characterization of miR-135b responsive genes and their role in particle-induced pulmonary response is being carried out in our laboratory to determine the biological relevance of these changes.
The miRNA field is still developing in parallel with more powerful tools to study their expression and activity. At present, a number of published studies describe changes in the expression of several miRNAs in diseases (reviewed in [Sun and Tsao,2008]) or in certain biological processes in vitro and in vivo [Bueno et al.,2008; Lynam-Lennon et al.,2009]. However, there are very few studies that have characterized changing mRNA and miRNA profiles in target tissues in vivo in response to toxic substances (reviewed in [Lema and Cunningham,2010]). To our knowledge, this is the first report on particle-induced pulmonary miRNA changes. Integration of gene and miRNA expression profiles with reports examining multiple endpoints at the molecular, cellular, tissue, and physiological levels will be very useful in understanding the toxicity of particles in the context of the whole organism. Further studies incorporating acute, subchronic, and chronic doses with different types of nano TiO2 particles and multiple time points are needed to validate our findings.
CONCLUSIONS
This work demonstrates the induction of acute phase reactants, chemoattractants, immune and host defense genes following exposure to occupationally relevant levels of surface-coated titanium dioxide nanoparticles via a biologically relevant exposure route. These findings are in keeping with the expected health outcome of chronic nanoparticle exposure, and the changes persist for up to 5 days following exposure. In parallel with perturbation of these genes, specific miRNAs are also induced in the lungs in response to nanoTiO2. The role of miRNAs in response to particle-induced lung injury is now under investigation in our laboratory.
Competing interests: There are no financial competing interests to disclose.
Acknowledgments
The authors like to acknowledge the expert technical assistance of Andrea Rowan-Carroll for carrying out the miRNA microarray experiments, Dongmei Wu for protein analysis, and Byron Kuo for analyzing miRNA targets.
Footnotes
Exclusive worldwide publication rights in the article have been transferred to Wiley-Liss, Inc.
Supplementary material
REFERENCES
- (IARC) IAfRoC. Feb. 2006. Titanium dioxide (IARC Group 2B). Summary of data reported.
- Alvo M, Liu Z, Williams A, Yauk C. Testing for mean and correlation changes in microarray experiments: An application for pathway analysis. BMC Bioinformatics. 2010;11:60. doi: 10.1186/1471-2105-11-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andre E, Stoeger T, Takenaka S, Bahnweg M, Ritter B, Karg E, Lentner B, Reinhard C, Schulz H, Wjst M. Inhalation of ultrafine carbon particles triggers biphasic pro-inflammatory response in the mouse lung. Eur Respir J. 2006;28:275–285. doi: 10.1183/09031936.06.00071205. [DOI] [PubMed] [Google Scholar]
- Baggs RB, Ferin J, Oberdorster G. Regression of pulmonary lesions produced by inhaled titanium dioxide in rats. Vet Pathol. 1997;34:592–597. doi: 10.1177/030098589703400607. [DOI] [PubMed] [Google Scholar]
- Bermudez E, Mangum JB, Asgharian B, Wong BA, Reverdy EE, Janszen DB, Hext PM, Warheit DB, Everitt JI. Long-term pulmonary responses of three laboratory rodent species to subchronic inhalation of pigmentary titanium dioxide particles. Toxicol Sci. 2002;70:86–97. doi: 10.1093/toxsci/70.1.86. [DOI] [PubMed] [Google Scholar]
- Bermudez E, Mangum JB, Wong BA, Asgharian B, Hext PM, Warheit DB, Everitt JI. Pulmonary responses of mice, rats, and hamsters to subchronic inhalation of ultrafine titanium dioxide particles. Toxicol Sci. 2004;77:347–357. doi: 10.1093/toxsci/kfh019. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, Loda M, Weber G, Mark EJ, Lander ES, Wong W, Johnson BE, Golub TR, Sugarbaker DJ, Meyerson M. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci USA. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya K, Cramer H, Albrecht C, Schins R, Rahman Q, Zimmermann U, Dopp E. Vanadium pentoxide-coated ultrafine titanium dioxide particles induce cellular damage and micronucleus formation in V79 cells. J Toxicol Environ Health A. 2008;71:976–980. doi: 10.1080/15287390801989218. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Borm PJ, Robbins D, Haubold S, Kuhlbusch T, Fissan H, Donaldson K, Schins R, Stone V, Kreyling W, Lademann J, Krutmann J, Warheit D, Oberdorster E. The potential risks of nanomaterials: A review carried out for ECETOC. Part Fibre Toxicol. 2006;3:11. doi: 10.1186/1743-8977-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno MJ, de Castro IP, Malumbres M. Control of cell proliferation pathways by microRNAs. Cell Cycle. 2008;7:3143–3148. doi: 10.4161/cc.7.20.6833. [DOI] [PubMed] [Google Scholar]
- Carleton M, Cleary MA, Linsley PS. MicroRNAs and cell cycle regulation. Cell Cycle. 2007;6:2127–2132. doi: 10.4161/cc.6.17.4641. [DOI] [PubMed] [Google Scholar]
- Chen HW, Su SF, Chien CT, Lin WH, Yu SL, Chou CC, Chen JJ, Yang PC. Titanium dioxide nanoparticles induce emphysema-like lung injury in mice. FASEB J. 2006;20:2393–2395. doi: 10.1096/fj.06-6485fje. [DOI] [PubMed] [Google Scholar]
- Chen J, Dong X, Zhao J, Tang G. In vivo acute toxicity of titanium dioxide nanoparticles to mice after intraperitioneal injection. J Appl Toxicol. 2009;29:330–337. doi: 10.1002/jat.1414. [DOI] [PubMed] [Google Scholar]
- Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Hwang JT, Qiu J, Blades NJ, Churchill GA. Improved statistical tests for differential gene expression by shrinking variance components estimates. Biostatistics. 2005;6:59–75. doi: 10.1093/biostatistics/kxh018. [DOI] [PubMed] [Google Scholar]
- Cullen RT, Tran CL, Buchanan D, Davis JM, Searl A, Jones AD, Donaldson K. Inhalation of poorly soluble particles. I. Differences in inflammatory response and clearance during exposure. Inhal Toxicol. 2000;12:1089–1111. doi: 10.1080/08958370050166787. [DOI] [PubMed] [Google Scholar]
- Dankovic D, Kuempel E, Wheeler M. An approach to risk assessment for TiO2. Inhal Toxicol. 2007;19(Suppl 1):205–212. doi: 10.1080/08958370701497754. [DOI] [PubMed] [Google Scholar]
- de Haar C, Hassing I, Bol M, Bleumink R, Pieters R. Ultrafine but not fine particulate matter causes airway inflammation and allergic airway sensitization to co-administered antigen in mice. Clin Exp Allergy. 2006;36:1469–1479. doi: 10.1111/j.1365-2222.2006.02586.x. [DOI] [PubMed] [Google Scholar]
- Driscoll KE, Hassenbein DG, Carter J, Poynter J, Asquith TN, Grant RA, Whitten J, Purdon MP, Takigiku R. Macrophage inflammatory proteins 1 and 2: Expression by rat alveolar macrophages, fibroblasts, and epithelial cells and in rat lung after mineral dust exposure. Am J Respir Cell Mol Biol. 1993;8:311–318. doi: 10.1165/ajrcmb/8.3.311. [DOI] [PubMed] [Google Scholar]
- Drumm K, Schindler H, Buhl R, Kustner E, Smolarski R, Kienast K. Indoor air pollutants stimulate interleukin-8-specific mRNA expression and protein secretion of alveolar macrophages. Lung. 1999;177:9–19. doi: 10.1007/pl00007628. [DOI] [PubMed] [Google Scholar]
- Farraj AK, Hazari MS, Haykal-Coates N, Lamb C, Winsett DW, Ge Y, Ledbetter AD, Carll AP, Bruno M, Ghio A, Costa DL. ST depression, arrhythmia, vagal dominance, and reduced cardiac microRNA in particulate-exposed rats. Am J Respir Cell Mol Biol. 2010 doi: 10.1165/rcmb.2009-0456OC. (in press) [DOI] [PubMed] [Google Scholar]
- Flo TH, Smith KD, Sato S, Rodriguez DJ, Holmes MA, Strong RK, Akira S, Aderem A. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature. 2004;432:917–921. doi: 10.1038/nature03104. [DOI] [PubMed] [Google Scholar]
- Ganapathi MK, May LT, Schultz D, Brabenec A, Weinstein J, Sehgal PB, Kushner I. Role of interleukin-6 in regulating synthesis of C-reactive protein and serum amyloid A in human hepatoma cell lines. Biochem Biophys Res Commun. 1988;157:271–277. doi: 10.1016/s0006-291x(88)80043-3. [DOI] [PubMed] [Google Scholar]
- Garzon R, Calin GA, Croce CM. MicroRNAs in cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- Golpon HA, Coldren CD, Zamora MR, Cosgrove GP, Moore MD, Tuder RM, Geraci MW, Voelkel NF. Emphysema lung tissue gene expression profiling. Am J Respir Cell Mol Biol. 2004;31:595–600. doi: 10.1165/rcmb.2004-0008OC. [DOI] [PubMed] [Google Scholar]
- Granville CA, Dennis PA. An overview of lung cancer genomics and proteomics. Am J Respir Cell Mol Biol. 2005;32:169–176. doi: 10.1165/rcmb.F290. [DOI] [PubMed] [Google Scholar]
- Guil S, Esteller M. DNA methylomes, histone codes and miRNAs: Tying it all together. Int J Biochem Cell Biol. 2009;41:87–95. doi: 10.1016/j.biocel.2008.09.005. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Heinrich U, Fuhst R, Rittinghausen S, Creutzenberg O, Bellmann B, Koch W, Levesen K. Chronic inhalation exposure of wistar rats and two different strains of mice to diesel engine exhaust, carbon black, and titanium dioxide. Inhal Toxicol. 1995;7:533–556. [Google Scholar]
- Hiratsuka S, Watanabe A, Sakurai Y, Akashi-Takamura S, Ishibashi S, Miyake K, Shibuya M, Akira S, Aburatani H, Maru Y. The S100A8-serum amyloid A3-TLR4 paracrine cascade establishes a pre-metastatic phase. Nat Cell Biol. 2008;10:1349–1355. doi: 10.1038/ncb1794. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B Stat Methodol. 1995;57:289–300. [Google Scholar]
- Hougaard KS, Jackson P, Jensen KA, Sloth JJ, Loschner K, Larsen EH, Birkedal RK, Vibenholt A, Boisen AM, Wallin H, Vogel U. Effects of prenatal exposure to surface-coated nanosized titanium dioxide (UV-Titan). A study in mice. Part Fibre Toxicol. 2010;7:16. doi: 10.1186/1743-8977-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hougaard KS, Jensen KA, Nordly P, Taxvig C, Vogel U, Saber AT, Wallin H. Effects of prenatal exposure to diesel exhaust particles on postnatal development, behavior, genotoxicity and inflammation in mice. Part Fibre Toxicol. 2008;5:3. doi: 10.1186/1743-8977-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hudder A, Novak RF. miRNAs: Effectors of environmental influences on gene expression and disease. Toxicol Sci. 2008;103:228–240. doi: 10.1093/toxsci/kfn033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen NR, Moller P, Jensen KA, Vogel U, Ladefoged O, Loft S, Wallin H. Lung inflammation and genotoxicity following pulmonary exposure to nanoparticles in ApoE-/- mice. Part Fibre Toxicol. 2009;6:2. doi: 10.1186/1743-8977-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardim MJ, Fry RC, Jaspers I, Dailey L, Diaz-Sanchez D. Disruption of microRNA expression in human airway cells by diesel exhaust particles is linked to tumorigenesis-associated pathways. Environ Health Perspect. 2009;117:1745–1751. doi: 10.1289/ehp.0900756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen LE, Whitehead AS. Regulation of serum amyloid A protein expression during the acute-phase response. Biochem J. 1998;334(Part 3):489–503. doi: 10.1042/bj3340489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyaseelan S, Manzer R, Young SK, Yamamoto M, Akira S, Mason RJ, Worthen GS. Induction of CXCL5 during inflammation in the rodent lung involves activation of alveolar epithelium. Am J Respir Cell Mol Biol. 2005;32:531–539. doi: 10.1165/rcmb.2005-0063OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang SL, Lozanski G, Samols D, Kushner I. Induction of human serum amyloid A in Hep 3B cells by IL-6 and IL-1 beta involves both transcriptional and post-transcriptional mechanisms. J Immunol. 1995;154:825–831. [PubMed] [Google Scholar]
- Jin P, Wang E, Ren J, Childs R, Shin JW, Khuu H, Marincola FM, Stroncek DF. Differentiation of two types of mobilized peripheral blood stem cells by microRNA and cDNA expression analysis. J Transl Med. 2008;6:39. doi: 10.1186/1479-5876-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr MK. Design considerations for efficient and effective microarray studies. Biometrics. 2003;59:822–828. doi: 10.1111/j.0006-341x.2003.00096.x. [DOI] [PubMed] [Google Scholar]
- Kerr MK, Churchill GA. Bootstrapping cluster analysis: Assessing the reliability of conclusions from microarray experiments. Proc Natl Acad Sci USA. 2001;98:8961–8965. doi: 10.1073/pnas.161273698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr MK, Churchill GA. Statistical design and the analysis of gene expression microarray data. Genet Res. 2007;89:509–514. doi: 10.1017/S0016672308009713. [DOI] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Kong W, Zhao JJ, He L, Cheng JQ. Strategies for profiling microRNA expression. J Cell Physiol. 2009;218:22–25. doi: 10.1002/jcp.21577. [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foa R, Schliwka J, Fuchs U, Novosel A, Muller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, hoksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KP, Trochimowicz HJ, Reinhardt CF. Pulmonary response of rats exposed to titanium dioxide (TiO2) by inhalation for two years. Toxicol Appl Pharmacol. 1985;79:179–192. doi: 10.1016/0041-008x(85)90339-4. [DOI] [PubMed] [Google Scholar]
- Lema C, Cunningham MJ. MicroRNAs and their implications in toxicological research. Toxicol Lett. 2010;198:100–105. doi: 10.1016/j.toxlet.2010.06.019. [DOI] [PubMed] [Google Scholar]
- Lowell CA, Stearman RS, Morrow JF. Transcriptional regulation of serum amyloid A gene expression. J Biol Chem. 1986;261:8453–8461. [PubMed] [Google Scholar]
- Lu TX, Munitz A, Rothenberg ME. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J Immunol. 2009;182:4994–5002. doi: 10.4049/jimmunol.0803560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynam-Lennon N, Maher SG, Reynolds JV. The roles of microRNA in cancer and apoptosis. Biol Rev Camb Philos Soc. 2009;84:55–71. doi: 10.1111/j.1469-185X.2008.00061.x. [DOI] [PubMed] [Google Scholar]
- Ma-Hock L, Burkhardt S, Strauss V, Gamer AO, Wiench K, van Ravenzwaay B, Landsiedel R. Development of a short-term inhalation test in the rat using nano-titanium dioxide as a model substance. Inhal Toxicol. 2009;21:102–118. doi: 10.1080/08958370802361057. [DOI] [PubMed] [Google Scholar]
- Meek RL, Benditt EP. Amyloid A gene family expression in different mouse tissues. J Exp Med. 1986;164:2006–2017. doi: 10.1084/jem.164.6.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei N, Fuscoe JC, Lobenhofer EK, Guo L. Application of microarray-based analysis of gene expression in the field of toxicogenomics. Methods Mol Biol. 2010;597:227–241. doi: 10.1007/978-1-60327-389-3_16. [DOI] [PubMed] [Google Scholar]
- Morrow JF, Stearman RS, Peltzman CG, Potter DA. Induction of hepatic synthesis of serum amyloid A protein and actin. Proc Natl Acad Sci USA. 1981;78:4718–4722. doi: 10.1073/pnas.78.8.4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao I, Kanaji S, Ohta S, Matsushita H, Arima K, Yuyama N, Yamaya M, Nakayama K, Kubo H, Watanabe M, Sagara H, Sugiyama K, Tanaka H, Toda S, Hayashi H, Inoue H, Hoshino T, Shiraki A, Inoue M, Suzuki K, Aizawa H, Okinami S, Nagai H, Hasegawa M, Fukuda T, Green ED, Izuhara K. Identification of pendrin as a common mediator for mucus production in bronchial asthma and chronic obstructive pulmonary disease. J Immunol. 2008;180:6262–6269. doi: 10.4049/jimmunol.180.9.6262. [DOI] [PubMed] [Google Scholar]
- Ning W, Li CJ, Kaminski N, Feghali-Bostwick CA, Alber SM, Di YP, Otterbein SL, Song R, Hayashi S, Zhou Z, Pinsky DJ, Watkins SC, Pilewski JM, Sciurba FC, Peters DG, Hogg JC, Choi AM. Comprehensive gene expression profiles reveal pathways related to the pathogenesis of chronic obstructive pulmonary disease. Proc Natl Acad Sci USA. 2004;101:14895–14900. doi: 10.1073/pnas.0401168101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitto T, Inoue T, Node K. Alternative spliced variants in the pantetheinase family of genes expressed in human neutrophils. Gene. 2008;426:57–64. doi: 10.1016/j.gene.2008.08.019. [DOI] [PubMed] [Google Scholar]
- Noonan EJ, Place RF, Pookot D, Basak S, Whitson JM, Hirata H, Giardina C, Dahiya R. miR-449a targets HDAC-1 and induces growth arrest in prostate cancer. Oncogene. 2009;28:1714–1724. doi: 10.1038/onc.2009.19. [DOI] [PubMed] [Google Scholar]
- Oberdorster G. Lung particle overload: Implications for occupational exposures to particles. Regul Toxicol Pharmacol. 1995;21:123–135. doi: 10.1006/rtph.1995.1017. [DOI] [PubMed] [Google Scholar]
- Oberdorster G, Maynard A, Donaldson K, Castranova V, Fitzpatrick J, Ausman K, Carter J, Karn B, Kreyling W, Lai D, Olin S, Monteiro-Riviere N, Warheit D, Yang H. Principles for characterizing the potential human health effects from exposure to nanomaterials: Elements of a screening strategy. Part Fibre Toxicol. 2005a;2:8. doi: 10.1186/1743-8977-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberdorster G, Oberdorster E, Oberdorster J. Nanotoxicology: An emerging discipline evolving from studies of ultrafine particles. Environ Health Perspect. 2005b;113:823–839. doi: 10.1289/ehp.7339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberdorster G, Sharp Z, Atudorei V, Elder A, Gelein R, Kreyling W, Cox C. Translocation of inhaled ultrafine particles to the brain. Inhal Toxicol. 2004;16:437–445. doi: 10.1080/08958370490439597. [DOI] [PubMed] [Google Scholar]
- Oberdorster G, Sharp Z, Atudorei V, Elder A, Gelein R, Lunts A, Kreyling W, Cox C. Extrapulmonary translocation of ultrafine carbon particles following whole-body inhalation exposure of rats. J Toxicol Environ Health A. 2002;65:1531–1543. doi: 10.1080/00984100290071658. [DOI] [PubMed] [Google Scholar]
- Park EJ, Kim H, Kim Y, Yi J, Choi K, Park K. Inflammatory responses may be induced by a single intratracheal instillation of iron nanoparticles in mice. Toxicology. 2010;275:65–71. doi: 10.1016/j.tox.2010.06.002. [DOI] [PubMed] [Google Scholar]
- Park EJ, Yoon J, Choi K, Yi J, Park K. Induction of chronic inflammation in mice treated with titanium dioxide nanoparticles by intratracheal instillation. Toxicology. 2009;260:37–46. doi: 10.1016/j.tox.2009.03.005. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploner A. Heatplus: A heat map displaying covariates and coloring clusters. R package Version 1.14.0. 2008 [Google Scholar]
- Pope CA, III, Hansen ML, Long RW, Nielsen KR, Eatough NL, Wilson WE, Eatough DJ. Ambient particulate air pollution, heart rate variability, and blood markers of inflammation in a panel of elderly subjects. Environ Health Perspect. 2004;112:339–345. doi: 10.1289/ehp.6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pott F, Althoff G-H, Roller M, Hohr D, Friemann J. High acute toxicity of hydrophobic ultrafine titanium dioxide in an intratracheal study with several dusts in rats. In: Mohr U, Dungworth DL, editors. ILSI Monographs Relationship Between Respiratory Disease and Exposure to Air Pollution. Washington, DC: ILSI Press; 1998. pp. 270–277. [Google Scholar]
- R-Development-Core-Team. R: A Language and Environment for Statistical Computing. TRDC. 2004 [Google Scholar]
- R-Development-Core-Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2009. [Google Scholar]
- Rahman Q, Lohani M, Dopp E, Pemsel H, Jonas L, Weiss DG, Schiffmann D. Evidence that ultrafine titanium dioxide induces micronuclei and apoptosis in Syrian hamster embryo fibroblasts. Environ Health Perspect. 2002;110:797–800. doi: 10.1289/ehp.02110797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehn B, Seiler F, Rehn S, Bruch J, Maier M. Investigations on the inflammatory and genotoxic lung effects of two types of titanium dioxide: Untreated and surface treated. Toxicol Appl Pharmacol. 2003;189:84–95. doi: 10.1016/s0041-008x(03)00092-9. [DOI] [PubMed] [Google Scholar]
- Renwick LC, Brown D, Clouter A, Donaldson K. Increased inflammation and altered macrophage chemotactic responses caused by two ultrafine particle types. Occup Environ Med. 2004;61:442–447. doi: 10.1136/oem.2003.008227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi EM, Pylkkanen L, Koivisto AJ, Vippola M, Jensen KA, Miettinen M, Sirola K, Nykasenoja H, Karisola P, Stjernvall T, Vanhala E, Kiilunen M, Pasanen P, Makinen M, Hameri K, Joutsensaari J, Tuomi T, Jokiniemi J, Wolff H, Savolainen K, Matikainen S, Alenius H. Airway exposure to silica-coated TiO2 nanoparticles induces pulmonary neutrophilia in mice. Toxicol Sci. 2010;113:422–433. doi: 10.1093/toxsci/kfp254. [DOI] [PubMed] [Google Scholar]
- Ruckerl R, Ibald-Mulli A, Koenig W, Schneider A, Woelke G, Cyrys J, Heinrich J, Marder V, Frampton M, Wichmann HE, Peters A. Air pollution and markers of inflammation and coagulation in patients with coronary heart disease. Am J Respir Crit Care Med. 2006;173:432–441. doi: 10.1164/rccm.200507-1123OC. [DOI] [PubMed] [Google Scholar]
- Saber AT, Halappanavar S, Folkmann JK, Bornholdt J, Boisen AM, Moller P, Williams A, Yauk C, Vogel U, Loft S, Wallin H. Lack of acute phase response in the livers of mice exposed to diesel exhaust particles or carbon black by inhalation. Part Fibre Toxicol. 2009;6:12. doi: 10.1186/1743-8977-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarver AL, French AJ, Borralho PM, Thayanithy V, Oberg AL, Silverstein KA, Morlan BW, Riska SM, Boardman LA, Cunningham JM, Subramanian S, Wang L, Smyrk TC, Rodrigues CM, Thibodeau SN, Steer CJ. Human colon cancer profiles show differential microRNA expression depending on mismatch repair status and are characteristic of undifferentiated proliferative states. BMC Cancer. 2009;9:401. doi: 10.1186/1471-2407-9-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayes CM, Wahi R, Kurian PA, Liu Y, West JL, Ausman KD, Warheit DB, Colvin VL. Correlating nanoscale titania structure with toxicity: A cytotoxicity and inflammatory response study with human dermal fibroblasts and human lung epithelial cells. Toxicol Sci. 2006;92:174–185. doi: 10.1093/toxsci/kfj197. [DOI] [PubMed] [Google Scholar]
- Schneeman TA, Bruno ME, Schjerven H, Johansen FE, Chady L, Kaetzel CS. Regulation of the polymeric Ig receptor by signaling through TLRs 3 and 4: Linking innate and adaptive immune responses. J Immunol. 2005;175:376–384. doi: 10.4049/jimmunol.175.1.376. [DOI] [PubMed] [Google Scholar]
- Spira A, Beane J, Pinto-Plata V, Kadar A, Liu G, Shah V, Celli B, Brody JS. Gene expression profiling of human lung tissue from smokers with severe emphysema. Am J Respir Cell Mol Biol. 2004;31:601–610. doi: 10.1165/rcmb.2004-0273OC. [DOI] [PubMed] [Google Scholar]
- Sun BK, Tsao H. Small RNAs in development and disease. J Am Acad Dermatol. 2008;59:725–737. doi: 10.1016/j.jaad.2008.08.017. quiz 738–740. [DOI] [PubMed] [Google Scholar]
- Takaya T, Ono K, Kawamura T, Takanabe R, Kaichi S, Morimoto T, Wada H, Kita T, Shimatsu A, Hasegawa K. MicroRNA-1 and microRNA-133 in spontaneous myocardial differentiation of mouse embryonic stem cells. Circ J. 2009;73:1492–1497. doi: 10.1253/circj.cj-08-1032. [DOI] [PubMed] [Google Scholar]
- Taylor EL, Gant TW. Emerging fundamental roles for non-coding RNA species in toxicology. Toxicology. 2008;246:34–39. doi: 10.1016/j.tox.2007.12.030. [DOI] [PubMed] [Google Scholar]
- Tran CL, Buchanan D, Cullen RT, Searl A, Jones AD, Donaldson K. Inhalation of poorly soluble particles. II. Influence of particle surface area on inflammation and clearance. Inhal Toxicol. 2000;12:1113–1126. doi: 10.1080/08958370050166796. [DOI] [PubMed] [Google Scholar]
- Tsitsiou E, Lindsay MA. microRNAs and the immune response. Curr Opin Pharmacol. 2009;9:514–520. doi: 10.1016/j.coph.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ravenzwaay B, Landsiedel R, Fabian E, Burkhardt S, Strauss V, Ma-Hock L. Comparing fate and effects of three particles of different surface properties: Nano-TiO(2), pigmentary TiO(2) and quartz. Toxicol Lett. 2009;186:152–159. doi: 10.1016/j.toxlet.2008.11.020. [DOI] [PubMed] [Google Scholar]
- Wang JJ, Sanderson BJ, Wang H. Cyto- and genotoxicity of ultrafine TiO2 particles in cultured human lymphoblastoid cells. Mutat Res. 2007;628:99–106. doi: 10.1016/j.mrgentox.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Wang Y, Liang Y, Lu Q. MicroRNA epigenetic alterations: Predicting biomarkers and therapeutic targets in human diseases. Clin Genet. 2008;74:307–315. doi: 10.1111/j.1399-0004.2008.01075.x. [DOI] [PubMed] [Google Scholar]
- Warheit DB, Brock WJ, Lee KP, Webb TR, Reed KL. Comparative pulmonary toxicity inhalation and instillation studies with different TiO2 particle formulations: Impact of surface treatments on particle toxicity. Toxicol Sci. 2005;88:514–524. doi: 10.1093/toxsci/kfi331. [DOI] [PubMed] [Google Scholar]
- Warheit DB, Frame SR. Characterization and reclassification of titanium dioxide-related pulmonary lesions. J Occup Environ Med. 2006;48:1308–1313. doi: 10.1097/01.jom.0000215385.71548.b0. [DOI] [PubMed] [Google Scholar]
- Warheit DB, Hansen JF, Yuen IS, Kelly DP, Snajdr SI, Hartsky MA. Inhalation of high concentrations of low toxicity dusts in rats results in impaired pulmonary clearance mechanisms and persistent inflammation. Toxicol Appl Pharmacol. 1997;145:10–22. doi: 10.1006/taap.1997.8102. [DOI] [PubMed] [Google Scholar]
- Warheit DB, Webb TR, Sayes CM, Colvin VL, Reed KL. Pulmonary instillation studies with nanoscale TiO2 rods and dots in rats: Toxicity is not dependent upon particle size and surface area. Toxicol Sci. 2006;91:227–236. doi: 10.1093/toxsci/kfj140. [DOI] [PubMed] [Google Scholar]
- Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: MicroRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- Wu H, Kerr M, Cui X, Churchill G. Maanova: A Software Package for the Analysis of Spotted cDNA Microarray Experiments. New York: Springer; 2003. pp. 313–341. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


