Abstract
Optimizing the productivity of bioengineered strains requires balancing ATP generation and carbon atom conservation through fine-tuning cell respiration and metabolism. Traditional approaches manipulate cell respiration by altering air feeding, which are technically difficult especially in large bioreactors. An approach based on genetic regulation may better serve this purpose. With excess oxygen supply to the culture, we efficiently manipulated Escherichia coli cell respiration by adding different amount of coenzyme Q1 to strains lacking the ubiCA genes, which encode two critical enzymes for ubiquinone synthesis. As a proof-of-concept, the metabolic effect of the ubiCA gene knockout and coenzyme Q1 supplementation were characterized, and the metabolic profiles of the experimental strains showed clear correlations with coenzyme Q1 concentrations. Further proof-of-principle experiments were performed to illustrate that the approach can be used to optimize cell respiration for the production of chemicals of interest such as ethanol. This study showed that controlled respiration through genetic manipulation can be exploited to allow much larger operating windows for reduced product formation even under fully aerobic conditions.
Keywords: Escherichia coli, “controlled respiration”, ubiquinone, metabolic distribution, reduced product formation
Introduction
Escherichia coli coordinately adapt its respiration and metabolism to environmental changes, and metabolic profiles of the cells are intrinsically different under aerobic and anaerobic conditions (Böck and Sawers, 1996; Lynch and Lin, 1996, Gunsalus and Park, 1994; Salmon et al., 2003; 2005). With sufficient oxygen supply, cells produce ATP through respiration and grow rapidly, but a lot of carbon atoms are lost in the form of CO2. Under anaerobic conditions, E. coli cells convert most of the carbon source to several organic acids and biomass, but cells grow slower than in aerobic cultures.
To optimize the productivity of bioengineered strains, the ATP generation and carbon atom conservation has to be well balanced by fine-tuning the metabolism and respiration. Cell growth is slightly improved and the carbon loss from metabolism is marginally reduced when microaerobic growth is used as an alternative to anaerobic conditions. The microaerobic condition confines a partially active respiration to reoxidize NADH and generate ATP for cell growth and maintenance with limited oxygen supply, while the majority of carbon atoms are guided to desired products (Alexeeva et al., 2000; Hua et al., 1998; Jensen et al., 2001; Zeng and Deckwer, 1996; Zhu and Shimizu, 2005). We have previously studied metabolite patterns under defined low oxygen conditions (Shalel-Levanon et al., 2005a; Shalel-Levanon et al., 2005b), and have examined the effects of global regulators such as ArcA and Fnr and other genes such as yfiD, (Zhu et al., 2006, 2007) and creB (Nikel et al. 2009) on the pattern of metabolites under defined low oxygen conditions. However, it is technically difficult to maintain a constant microaerobic environment in real processes, especially in large bioreactors. Controlling respiration and redox through genetic modifications may serve as a promising alternative.
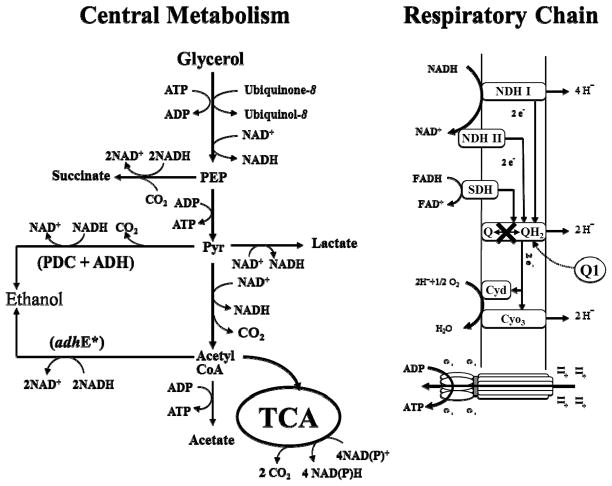
The respiratory system of E. coli has a modular design to facilitate system fine tuning according to the availability of oxygen or other electron acceptors (Gennis and Stewart, 1996). A diagram was drawn in light of Alexeeva et al. (2000) (Fig. 1). E. coli cells regenerate NAD+ and generate proton motive force for ATP production through the respiratory chain. Quinones are lipid-soluble molecules that mediate electron transfer between NADH or FADH dehydrogenases and cytochrome oxidases. Among several types of quinones synthesized in E. coli, ubiquinone is mainly used for oxygen respiration (Wallace and Young, 1977). The ubiC and ubiA genes encode the committed first and second reactions in the ubiquinone biosynthetic pathway of E. coli (Meganathan, 2001), and control of their expression are under complicated regulation by cell culture conditions (Kwon et al., 2005). The electron transport through ubiquinone affects the NADH and FADH levels in the cell and poses impacts on cell growth and related metabolic reactions through enzymes such as succinate dehydrogenase, lactate dehydrogenase and etc. (Hägerhäll, 1997; San et al., 2002). Besides mediating electron transport, quinones also serve as major regulation signals for global regulators such as ArcAB (Georgellis et al., 2001). The global regulators affect the transcription of genes encoding metabolic enzymes (Lynch and Lin, 1996; Matsuoka and Shimizu, 2011; Shalel-Levanon et al., 2005a; Shalel-Levanon et al., 2005b), and, therefore, regulate metabolic flux distributions in the cell (Alexeeva et al., 2003; Perrenoud and Sauer, 2005; Zhu et al., 2006).
Fig. 1.
Simplified central metabolic pathways of E. coli when using glycerol as a carbon source. The electron transfer through the respiratory chain was controlled by changing the concentration of coenzyme Q1 in the ubiCA− mutant strain, whose ubiquinone synthesis was disrupted. PEP: phosphoenolpyruvate; Pyr: pyruvate; Acetyl CoA: Acetyle coenzyme A; TCA: tricarboxylic acid cycle.
The regulation of the amount of NADH that is consumed by the respiratory chain vs the amount that is used for production of a reduced metabolite has been manipulated in several ways. If one wants to avoid reduced products, NADH oxidase can be introduced into the cells and in an aerobic culture would act to avoid formation of quantities of reduced products (Lopez de Felipe and Hugenholtz, 1999; Heux et al., 2006). On the other hand when reduced products are desired during aerobic or semi-aerobic growth, several strategies have been utilized. In our previous work we showed an effective strategy for producing reduced products (e.g. ethanol) in aerobic culture by employing expression of the C. boidinii FDH and supplying formate to generate extra NADH. During aerobic growth the cells produced ethanol. The amount of ethanol formed was dependent on the amount of formate addition to the FDH augmented strain (Berrios-Rivera et al., 2002). In this study, we use a different approach for redox control in order to channel NADH formed during glycolysis to reduced products under aerobic growth conditions by restricting the ability of the electron transport chain to oxidize the NADH.
In the present study, as a proof of concept we supplemented the cultures of several ubiCA mutant strains with different amounts of coenzyme Q1, an unnatural analogue of ubiquinone. With sufficient oxygen supply to the cultures, variant limited respiration levels were achieved, and the metabolite profiles were affected accordingly. In order to demonstrate that the modified system can also be applied to the production of reduced chemicals normally produced under anaerobic conditions, plasmids carrying the Zymomonas mobilis pdc-adh genes or a mutated aerobic adhE* gene, respectively, were introduced into the ubiCA mutant host as a convenient means of assessing the proportion of NADH allotted to reduced products (i.e. ethanol production) under the experimental aerobic conditions. The experiments show that NADH (redox) partitioning between the electron transport chain to generate energy and the use of NADH for reduction of a metabolic intermediate could be precisely controlled by manipulating the level of ubiquinone. This study further suggests controlled respiration that simulates a semi-aerobic environment through genetic manipulations can be exploited to allow much larger operating windows for reduced product production even under fully aerobic culturing conditions.
Materials and Methods
Bacterial strains and plasmids
Strains and plasmids used in this study are listed in Table 1. The ubiCA gene deletion was transduced from RKP4152 (a kind gift from Dr. Poole, Soballe and Poole, 1998) to GJT001 using P1 phage transduction (Thomason et al., 2007). One colony, named AMS001, was isolated and selected for further study. YBS131 was constructed by insertion inactivation of the ldhA gene (Yang et al., 1999b). The ubiCA gene deletion was transduced from AMS001 to YBS131 using P1 phage transduction (Thomason et al., 2007). The mutant strain was screened on LB plates supplemented with coenzyme Q1 (Sigma Aldrich). One colony named AMS002 was isolated and selected for further study.
TABLE 1.
List of strains and plasmids used in this study
| Strain or plasmid | Relevant Characteristics | Reference or source |
|---|---|---|
| E. coli strain | ||
| GJT001 | laboratory wild type, Spontaneous cadR mutant of MC4100, SmR | (Tolentino et al., 1992) |
| YBS131 | GJT001 ldhA::Tn10, SmR, TcR | (Yang et al., 1999b) |
| AMS001 | GJT001 ΔubiCA, SmR, KmR | This work |
| AMS002 | YBS131ΔubiCA, SmR, TcR, KmR | This work |
| Plasmids | ||
| pLOI295 | Carries the pdc and adh genes from Zymomonas mobilis, ApR | (Ingram et al., 1987) |
| pCRADH2 | Carries the aerobic functional adhE* gene, ApR | (Holland-Staley et al., 2000) |
The plasmid pLOI295 carries Z. mobilis pdc and adh genes (Ingram et al., 1987), and pCRADH2 carries a mutated adhE* gene, which encodes an aerobically functional alcohol dehydrogenase (ADH) (Holland-Staley et al., 2000). The strains used in this study were transformed with either pLOI295 or pCRADH2 so that they can produce ethanol as an external sink of reducing power under aerobic conditions.
Medium and shake flask culture conditions
The aerobic culture medium used in this study was LB supplemented with glycerol as a carbon source with appropriate antibiotics. In the cultures of the ubiCAμ mutant strains, coenzyme Q1 was supplemented at different levels (1 nM, 10 nM, 100 nM, 1 μM, 5 μM, 10 μM, 20 μM, 40 μM, and 80 μM) to improve cell growth and change cell metabolism. In the cultures of the strains bearing the pLOI295 or pCRADH2 plasmid, 1 mM of IPTG was added to ensure full induction of gene expression. The strains were grown in 125 ml shake flask cultures with 15 ml of culture volume. The shaker (InnovaR44 New Brunswick Scientific, Edison, NJ) was set at 37 °C with a shaking rate at 250 rpm. The shake flask cultures in the current study were all performed under aerobic conditions if not further mentioned.
Analytical procedures
Cell densities were measured at 600 nm in a spectrophotometer (Bausch & Lomb Spectronic 1001). The measured optical densities (OD600) were correlated with dry cell weight using established OD600-dry cell weight correlation. Culture broth samples were centrifuged for one minute at 13,000×g in an AccuSpin™ micro centrifuge. The supernatant was filtered through a 0.2 μm syringe filter before HPLC measurements. The concentrations of glycerol and extracellular metabolites such as lactate, acetate, succinate, and ethanol were measured using an HPLC system (Shimadzu Scientific Instruments, Columbia, MD) equipped with a cation-exchange column (HPX-87 H, BioRad Labs, Hercules, CA), a differential refractive index detector (Waters, Milford, MA) and a UV-VIS detector (SPD-10A, Shimadzu Scientific Instruments, Columbia, MD). A mobile phase of 2.5 mM H2SO4 solution at a 0.6 ml/min flow rate was used and the column was operated at 55 °C (Sanchez et al., 2005; Yang et al., 1999a).
Results
Metabolic impact of coenzyme Q1 supplementation on AMS001 strain
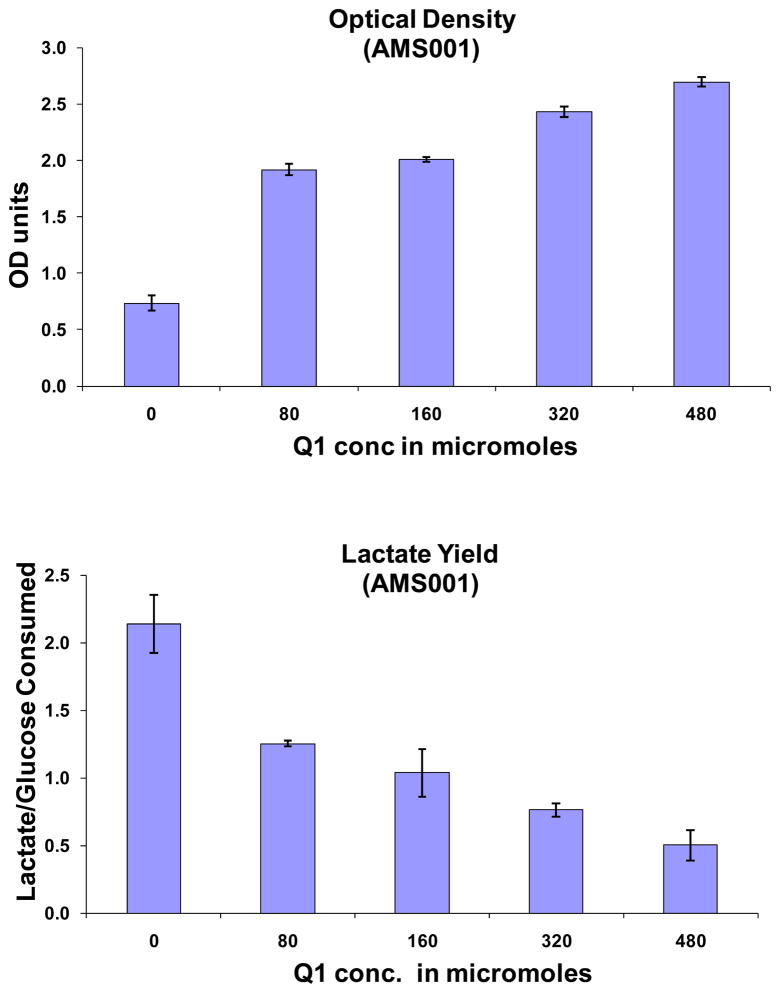
The effect of exogenous supplementation of ubiquinone-1 (Q-1) to an ubiCA mutant strain, AMS001, was first examined. This mutant cannot produce ubiquinone due to a disruption in its biosynthesis pathway. The aerobic experiments with this mutant strain were carried out in 125-ml shake flasks containing 10 ml of LB medium supplemented with 20 g/L of glucose in a rotary shaker at 37°C and 250 rpm. The final optical density and the specific lactate yields after 24 hours are summarized in Fig. 2. The mutant strain grew very poorly and produced a large amount of lactate (about 23 mM). The ubiCA mutant, due to the disruption in the electron chain, cannot use oxygen as the electron receptor. The cell opted to recycle the NADH to NAD+ through the formation of lactate (the ethanol pathway is not very active aerobically). In another words, the metabolic pattern of the mutant strain behaves very much like one that is grown anaerobically even in the presence of air.
Fig. 2.
The effect of exogenous addition of ubiquinone-1 to an uibCA mutant strain, AMS001
Adding different quantities of ubiquinone Q1 to the culture was able to increase the final optical density and reduce the amount of lactate accumulated. The mutant strain responded to ubiquinone supplementation in a graded manner (Fig. 2). The reduction in lactate formation suggests that the function of the electron transfer chain is partially recovered and the cells are capable of using oxygen as the electron acceptor. However, the mutant strain did not fully recover even when the concentration was increased to 400 μM. It is possible that the low solubility of Q-1 in the medium limits the uptake of this molecule or that Q-1 does not act as efficiently as the normal E. coli ubiquinone. Another explanation might be that some intermediate products in the ubiquinone biosynthesis pathway are necessary for full recovery and restoration of the cell. These experiments, nevertheless, show that the cell could achieve some recovery by the exogenous addition of ubiquinone.
Metabolic impact of ubiCA gene knockout in the ethanol producing strains
To further examine the potential of exploiting the disruption of the respiration chain for reduced product formation besides lactate in the presence of air, we used the production of ethanol as the model system by introducing heterologous pathways which served as NADH sinks under the experimental aerobic conditions.
Four strains, GJT001 (parent strain), YBS131 (ldhA−), AMS001 (ubiCA−) and AMS002 (ubiCA−, ldhA−) carrying the pLOI295 and pCRADH2 plasmids were used in this study. These strains were cultured in LB medium supplemented with glycerol under aerobic conditions without coenzyme Q1. The metabolite concentrations and cell densities measured during the mid-exponential phases were used to calculate the biomass yields on glycerol and specific metabolite forming rates. The data are summarized in Table 2. Formic acid and succinic acid were not detected in any of the cultures, which indicate that the pyruvate formate lyase (PFL) was not functioning under the experimental conditions, and, therefore, the cultures were indeed operating under aerobic conditions.
TABLE 2.
Metabolite production in the strains using glycerol as a sole carbon source without coenzyme Q1 supplementation under aerobic conditions
| Strains | qmetabolite / qglycerola for:
|
Specific growth rate (h−1) | Biomass yield on glycerol (g/g) | ||
|---|---|---|---|---|---|
| Lactate | Acetate | Ethanol | |||
| GJT001 (pLOI295) | 0.00 ± 0.00 | 0.32 ± 0.18 | 0.52 ± 0.07 | 0.27 ± 0.03 | 1.85 ± 0.19 |
| GJT001(pCRADH2) | 0.04 ± 0.01 | 0.35 ± 0.01 | 0.20 ± 0.01 | 0.37 ± 0.05 | 0.91 ± 0.27 |
| YBS131(pLOI295) | 0.00 ± 0.00 | 0.29 ± 0.10 | 0.48 ± 0.04 | 0.33 ± 0.07 | 1.89 ± 0.17 |
| YBS131(pCRADH2) | 0.00 ± 0.00 | 0.42 ± 0.03 | 0.22 ± 0.01 | 0.42 ± 0.06 | 1.49 ± 0.39 |
| AMS001(pLOI295) | - b | - | - | - | - |
| AMS001(pCRADH2) | 0.64 ± 0.05 | 0.21 ± 0.12 | 0.17 ± 0.04 | 0.13 ± 0.01 | 0.16 ± 0.02 |
| AMS002(pLOI295) | 0.00 ± 0.00 | 0.17 ± 0.04 | 0.80 ± 0.06 | 0.07 ± 0.01 | 0.26 ± 0.01 |
| AMS002(pCRADH2) | 0.00 ± 0.00 | 0.53 ± 0.03 | 0.10 ± 0.02 | 0.05 ± 0.00 | 0.46 ± 0.03 |
qmetabolite denotes specific metabolite forming rates (mmol/ [g of dry weight]/h), and qglycerol denotes specific glycerol uptake rates (mmol/ [g of dry weight]/ h). These specific rates were calculated using the data during the mid-exponential phase of the cultures to qualify quasi-steady state assumption. The results represent the mean values ± standard deviations for measurements from at least three independent experiments.
The AMS001 (pLOI295) strain failed to grow without coenzyme Q1 supplementation under the experimental condition.
The ubiCA mutation resulted in less biomass formation and more secretion of reduced metabolites such as lactate and ethanol. The biomass yields on glycerol for ubiCA− mutant strains were about 14 – 31 percent of those for the ubiCA+ strains, and the pLOI295 plasmid had a more profound effect on cell growth in the ubiCA− mutant strains as compared to the ubiCA+ strains, in particular, the AMS001 (pLOI295) strain failed to grow in the culture without coenzyme Q1 supplementation. With lower biomass yield and acetate production compared to those in the cultures of GJT001 (pCRADH2), the AMS001 (pCRADH2) strain converted the majority of glycerol into lactate. The AMS002 (pLOI295) converted about 80% of the carbon atoms from glycerol into ethanol under the experimental aerobic condition as the ldhA gene was inactivated in this strain.
The strains carrying the pLOI295 plasmid converted more glycerol into ethanol than the strains bearing the pCRADH2 plasmid. In the cultures of the ubiCA+ strains with the pLOI295 plasmid, more than 60% of the secreted metabolites (on a molar basis) were ethanol, which indicate that the Z. mobilis pdc and adh genes are efficient in these cultures to compete pyruvate with pyruvate dehydrogenase. More significantly, ethanol production counted for 82% of the secreted metabolite molecules in the cultures of AMS002 (pLOI295). In comparison, ethanol accumulation in the strains carrying pCRADH2 were less than 45% of the metabolites accumulated in the culture medium. In particular, ethanol production only accounted for ca. 16% of the metabolites produced in AMS002 (pCRADH2). It seems that the flux partitioning at the acetyl coenzyme A (AcCoA) node favors acetate formation under the experimental conditions.
The ldhA mutation had very little effect on the metabolism of the ubiCA+ strains, but had significant effect on the ubiCA− strains. The lactate production levels were negligible through all the cultures of the GJT001 (pLOI295) and GJT001 (pCRADH2) strains in this study. The AMS001 (pCRADH2) accumulated significant amount of lactate with less acetate and biomass produced than GJT002 (pCRADH2). In the cultures of the ldhA−, ubiCA− double mutant strain, AMS002 (pCRADH2), instead of accumulating ethanol, another chemically reduced metabolite, the cells produced acetate as the major metabolite, and the biomass yield was partially restored as compared to AMS001(pCRADH2).
Metabolic impact of various levels of coenzyme Q1 supplementation
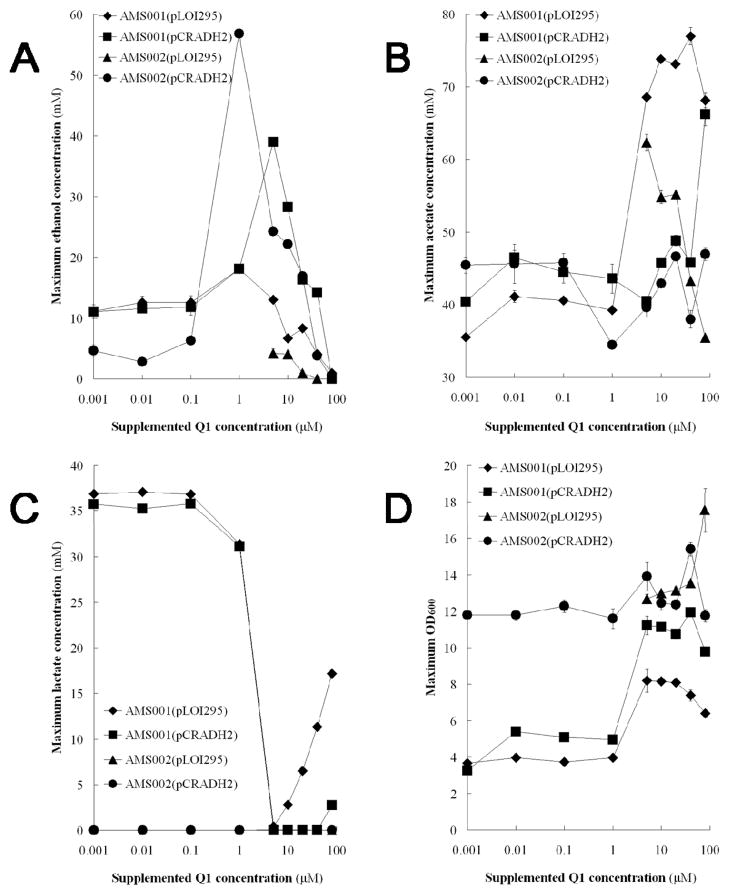
Several different levels of coenzyme Q1 (1 nM, 10 nM, 100 nM, 1 μM, 5 μM, 10 μM, 20 μM, 40 μM, and 80 μM) were supplemented into the cultures of the ubiCA− strains carrying pLOI295 or pCRADH2 plasmid, and the metabolic profiles are plotted in Fig. 3.
Fig. 3.
Metabolite and biomass accumulated with concentrations of coenzyme Q1 in the cultures of the ubiCA− mutant strains. The concentrations of (A) Ethanol, (B) Acetate, (C) lactate, and (D) cell density (OD600) were determined at the time point when glycerol is completely consumed. Symbols represent the mean values for each parameter ± standard deviation of measurements from at least three independent cultures.
The impact of Q1 supplementation was more profound when the Q1 concentration is higher than 1 μM. Supplementing 0.001 μM, 0.01 μM and 0.1 μM of coenzyme Q1 into the culture medium for the ubiCA− mutant strains had little effect on cell growth (Fig. 3D) and the production of metabolites such as ethanol (Fig. 2A), acetate (Fig. 2B), and lactate (Fig. 3C). With more than 1 μM or 5 μM of coenzyme Q1 supplemented in the culture medium, the production of metabolites and cell growth were significantly affected.
Ethanol production in the cultures of the ubiCA− mutant strains showed a clear correlation with the concentration of coenzyme Q1 (Fig. 3A). The ethanol production levels were the highest in the cultures of AMS002 (pCRADH2) and AMS001 (pLOI295) with 1 μM of coenzyme Q1 supplemented. While the cultures of AMS001 (pCRADH2) produced the highest amount of ethanol with 5 μM of coenzyme Q1 in the medium. Ethanol production decreased with coenzyme Q1 supplementation levels higher than 5 μM in the cultures of the ubiCA mutant strains. Since the ethanol and acetate production pathways compete for precursors of the reactions, the relationship between acetate production and Q1 supplementation was shown to be reverse to that between ethanol production and Q1 supplementation in the cultures of AMS001 (pLOI295), AMS001 (pCRADH2) and AMS002 (pCRADH2).
Lactate production in the cultures of AMS001 (pLOI295) and AMS001 (pCRADH2) had a clear correlation with Q1 supplementation in the medium. The production of lactate was diminished with 5 μM of coenzyme Q1 in the cultures (Fig. 3C). Correspondingly, cell growth densities of these two strains were significantly higher than in cultures with less coenzyme Q1 addition (Fig. 3D). By further increasing coenzyme Q1 concentration in the cultures of AMS001 (pLOI295), lactate production was revived and increased as coenzyme Q1 concentration increases. The cultures of AMS001 (pCRADH2) showed a similar relationship between lactate production and coenzyme Q1 concentrations. Thus, 5 μM of Q1 supplementation appeared to be a threshold value in the cultures of AMS001 (pLOI295) and AMS001 (pCRADH2).
The acetate production was not significantly correlated with coenzyme Q1 concentration in the cultures of AMS001 (pCRADH2) and AMS002 (pCRADH2) (Fig. 3B). However, in the cultures of AMS001 (pLOI295), acetate production was significantly higher with more than 5 μM of coenzyme Q1 in the medium compared with the cultures with less coenzyme Q1. Acetate production decreased as coenzyme Q1 concentration increases in the cultures of AMS002 (pLOI295), companying with higher biomass yields (Fig. 3D).
Culture profiles of experimental strains with 5 μM of coenzyme Q1
The correlations between metabolite production and coenzyme Q1 concentrations in the medium indicated that 5 μM is, or close to a threshold concentration to induce significant metabolic changes in the cultures. For instance, lactate productions were negligible in the cultures of AMS001 (pLOI295) and AMS001 (pCRADH2) with 5μM of coenzyme Q1 (Fig. 3C), although the ldhA gene was not disrupted in these strains, and the accumulation of ethanol in these cultures suggested that cells may need to regenerate NAD+ for continuous glycerol uptake.
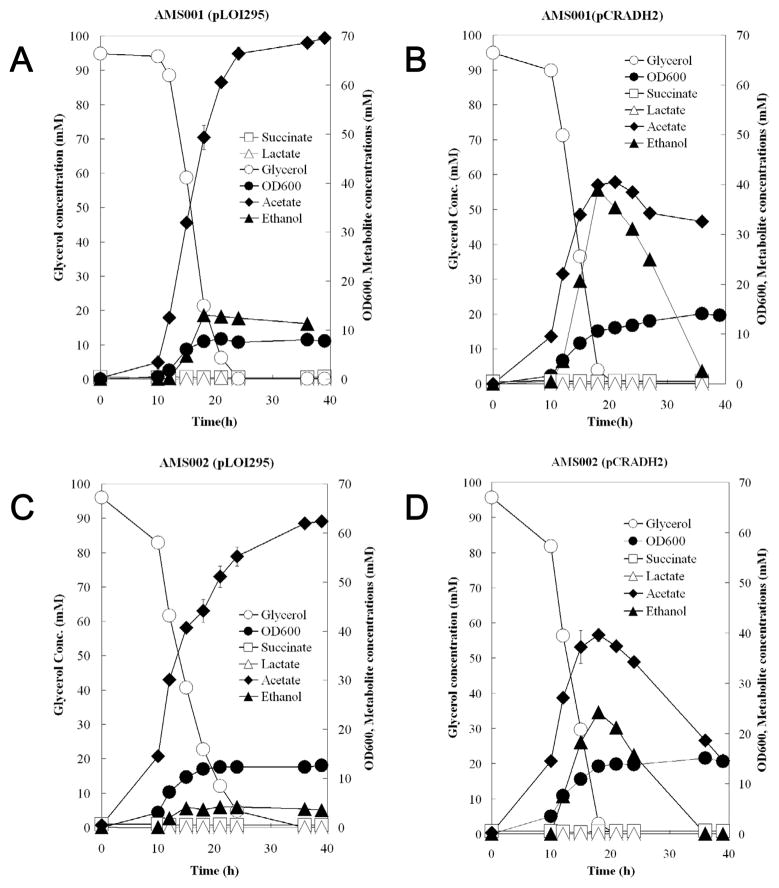
The culture profiles of the experimental strains with 5 μM of coenzyme Q1 are shown in Fig. 4. Ca. 96 mM of glycerol was completely consumed in about 20 hours in the cultures of the four experimental strains. Biomass, acetate, and ethanol were the major products of the cultures. The AMS001 (pLOI295) and AMS002 (pLOI295) strains produced ca. 66 mM and 55 mM of acetate as the major metabolic product, respectively, while the ethanol production of the cultures were only 12.5 and 4.2 mM, respectively (Fig. 4A and 4C). At the time when glycerol was depleted in the cultures, the amount of acetate and ethanol were comparable in the cultures of AMS001 and AMS002 strains carrying the pCRADH2 plasmid (Fig. 4B and 4D).
Fig. 4.
Shake flask cultures of E. coli (A) AMS001 (pLOI295), (B) AMS001 (pCRADH2), (C) AMS002 (pLOI295), and (D) AMS002 (pCRADH2) strains with 5 μM supplementation of coenzyme Q1. Symbols represent the mean values for each parameter ± standard deviation of measurements from at least three independent cultures.
The acetate and ethanol produced in the cultures of AMS001 (pCRADH2) and AMS002 (pCRADH2) were re-absorbed after glycerol was consumed. In contrast, the AMS001 (pLOI295) and AMS002 (pLOI295) strains did not metabolize acetate or ethanol until the end of the cultures.
Discussion
E. coli cells adapt to different levels of oxygen supply by changing respiratory intensity and metabolic profiles through complicated regulation systems (Lynch and Lin, 1996). Accurately controlling the oxygen levels to optimize cell metabolism for production of useful chemicals is usually very difficult in normal culture processes considering cell growth and medium composition changes during the process. A few attempts were made to accurately control the levels of oxygen supply in chemostat cultures (Alexeeva et al., 2002; Nikel et al., 2009; Shalel-Levanon et al., 2005a). However, the oxygen supply in batch cultures, which are more generally employed in industrial processes, is normally roughly manipulated with less accuracy, and this is particularly difficult under relatively low oxygen level conditions (Durnin et al., 2009; Zeng and Deckwer, 1996; Zhu and Shimizu, 2005). The current study aimed at manipulating cell respiration based on gene modification so that cell metabolism can be precisely manipulated via media additives accordingly. Although coenzyme Q1 is costly inhibiting its general use in industrial processes, the current strategy can be extended to controlling the expression levels of the ubiCA genes or other respiratory genes so that the process will be more economically viable. Such work is currently being pursued in our recent experiments.
There was no formic acid accumulation in the cultures of the experimental strains (Table 2, Fig. 3 and Fig. 4), as the glycyl radical in PFL is sensitive to oxygen (Wagner et al., 1992), which indicates that the cultures were made under aerobic conditions. Therefore, pyruvate was mainly converted into AcCoA through the pyruvate dehydrogenase (PDH) complex instead of PFL, which normally happens in the cultures under oxygen-limited conditions (Böck and Sawers, 1996). Ethanol accumulation in the cultures of AMS001 (pLOI295) and AMS002 (pLOI295) can be attributed to the pdc - adh pathway since the wild type ADH is mainly inactive under aerobic conditions (Holland-Staley et al., 2000; Leonardo et al., 1993).
Pyruvate and AcCoA are two of the major metabolic nodes in cell under the experimental conditions. The flux partitioning at these nodes are the results of cell regulations on different levels including gene transcription and metabolite balancing.
The concentration of coenzyme Q1 affects the flux distribution at the pyruvate node in the cultures of the ubiCA− mutant strains. Besides biomass synthesis, there are mainly three pathways that contribute to pyruvate metabolism, namely PDH, lactate dehydrogenase and pyruvate decarboxylase (PDC) introduced by the pLOI295 plasmid. Previous metabolic control analysis showed that the flux through PDH had significant control over the flux through LDH (Zhu and Shimizu, 2005). Since the expression of ldhA is roughly not changed by different oxygen levels in the wild type E. coli strains (Shalel-Levanon et al., 2005a), and the expression of PDC is controlled by the amount of IPTG inducer, the variant lactate production levels in the cultures were the result of changes on the PDH reaction rate, which corresponds to different concentrations of coenzyme Q1.
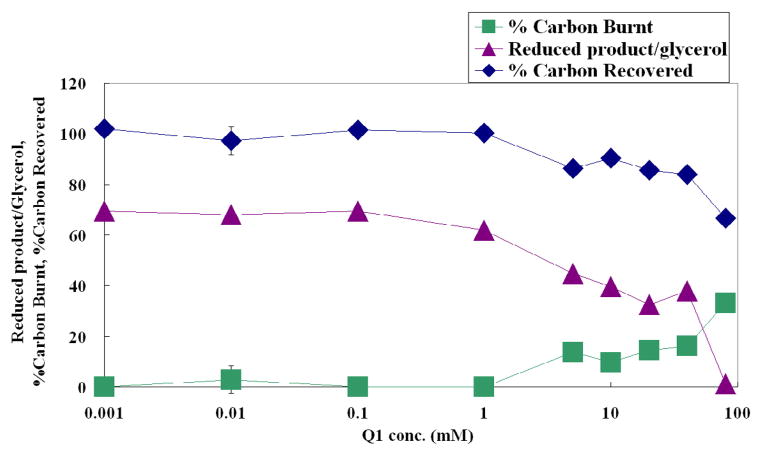
To further clarify the impact of coenzyme Q1 amount on the flux portioning at the pyruvate node, the flask culture data of AMS001 (pCRADH2) were reorganized to roughly evaluate the balance of reducing equivalents (Fig. 5). To simplify the estimation, we have to assume that biomass was derived from the LB medium, and the glycerol supplemented was mainly used for energy generation and metabolite productions such as lactate, acetate, ethanol, succinate and carbon dioxide. As can be seen on Fig. 5, there is almost no carbon was released as CO2 when coenzyme Q1 supplement is less than 1 μM. There are two main pathways for carbon dioxide generation in E. coli, namely tricarboxylic acid (TCA) cycle and the oxidative pentose phosphate (OPP) pathway. Under the above conditions, the electron transport through succinate dehydrogenase is largely blocked as a consequence of low quinone concentration (Tran et al., 2006), and therefore the CO2 generation through TCA cycle is very low. Since glycerol and rich medium were used for the current study, and the OPP pathway flux is low under oxygen-limited conditions (Zhu et al., 2006), little CO2 can be produced through the OPP pathway. Thus we believe that the reducing power generated by glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was oxidized by excreting reducing products such as lactate, ethanol and succinate. Under low oxygen conditions, the cell has high intracellular NADH concentration (Zhu and Shimizu, 2004, 2005) repressing the activity of PDH (Snoep et al., 1990), and resulted lactate accumulation is the major pathway for pyruvate metabolism. As the supplementation of coenzyme Q1 increases, some NADH will be oxidized through the respiratory chain. Therefore, more carbons from glycerol can go through the PDH and citrate synthase reactions, and” burnt” by the partially functional TCA cycle.
Fig. 5.
Impact of coenzyme Q1 supplementation on the reducing product accumulation in AMS001 (pCRADH2). % Carbon Burnt refers to the difference between glycerol consumption and metabolite accumulation, which can be attributed to CO2 releasing from the culture. Reduced product refers to lactate, ethanol and succinate which consumes NADH during the product synthesis.
An interesting observation is that the lactate production is reduced to almost zero in the cultures of AMS001 (pLOI295) and AMS001 (pCRADH2) with 5 μM of coenzyme Q1, while lactate production was partially restored when more coenzyme Q1 was added into the medium. Since the lactate flux is largely controlled by the PDH activity as discussed above, the lactate accumulation may be a result of the changes on PDH expression and activity levels. Shalel-Levanon et al. have shown that the transcription level of aceE gene, which encodes a subunit of the PDH complex, increases as the oxygen level increases when the head-space oxygen concentration is between 1– 5%, while with higher oxygen level, the aceE transcription level dropped (Shalel-Levanon et al., 2005a). Alexeeva et al. also proved that the flux through PDH increases as aerobiosis increases from 0 to about 40% (defined by the oxygen conditions related with the amount of acetate produced in chemostat cultures), and above that the flux through PDH decreases with higher aerobiosis even without PFL flux (Alexeeva et al., 2000). At the pyruvate node, higher PDH converts more pyruvate into AcCoA, which leads to less lactate accumulation, and vice versa. Therefore, our observation is consistent with these previous works, and the 1 – 5 μM of coenzyme Q1 supplementation in the culture suggests similar electron flux in the respiratory chain to previous chemostat cultures made with about 40% aerobiosis (Alexeeva et al., 2000), and 5 % head-space oxygen concentration (Shalel-Levanon et al., 2005a). Since PDH provides AcCoA as the precursor of Ack-pta pathway, which is used for acetate production, the increase of acetate accumulation in the cultures of AMS001 (pLOI295) and AMS002 (pLOI295) with more than 5 μM of coenzyme Q1 also suggested that the PDH flux was higher than the cultures with less Q1 (Fig. 3B).
Ethanol and acetate production are also indicators of the flux distribution at AcCoA node. Ethanol was produced through ADH in the cultures of AMS001 (pCRADH2) and AMS002 (pCRADH2). The highest ethanol accumulation in this study was observed in the cultures supplemented with 1 – 5 μM of coenzyme Q1. Ethanol production in the cultures of AMS001 (pCRADH2) and AMS002 (pCRADH2) decreased (Fig. 3A) as the concentration of coenzyme Q1 increases. This observation is consistent with the discussion about PDH flux. Firstly, the reaction through PDH provides AcCoA as the substrate of ADH and Ack-pta reactions. The peak of ethanol production can be an indication of peak PDH reaction assuming the flux partitioning at the AcCoA node is relatively stable. Secondly, the flux distribution at the AcCoA node is also affected by the reaction through citrate synthase, which competes for AcCoA as a precursor. As the coenzyme Q1 concentration increases, the cell metabolism should be similar to those microaerobic cultures with increasing oxygen supplies. In the study made by Shalel-Levanon et al., the transcription level of gltA, which encodes citrate synthase, increases as the oxygen level increases (Shalel-Levanon et al., 2005b). Thirdly, higher concentration of coenzyme Q1 will result in higher electron flux through the respiratory chain, and the cell can re-oxidize more NADH through oxidative phosphorylation for ATP production. Since one mole of ethanol production through ADH requires two moles of NADH as a metabolic cofactor, lack of NADH may also contribute to the drop of ethanol production in these cultures.
The re-absorption of ethanol in cultures of AMS001 (pCRADH2) and AMS002 (pCRADH2) after glycerol was used up suggested that the ADH enzyme was functional on both reaction directions, for ethanol production and absorption under the experimental conditions (Fig. 3B and 3D). It’s not clear why the ubiCA− mutant strains carrying the pCRADH2 plasmid re-utilized acetate after glycerol was used up, while those strains carrying the pLOI295 plasmid did not. The depletion of the pyruvate pool caused by the pdc - adh reactions could be one of the reasons, but the detailed regulation mechanism remains unclear.
The expression levels of cytochrome bd and cytochrome bo oxidases in E. coli are regulated according to oxygen levels in the culture (Lynch and Lin 1996). Since cytochrome bd and bo oxidases have different oxygen affinity and turnover rates (Gunsalus 1992), the electron flux partition between the two oxidases results at different overall P/O ratio and affects the metabolic flux distribution in the central metabolic network. Alexeeva et al. (2002) have studied the flux distribution in the cytochrome components under different oxygen levels in E. coli. It will provide a better picture of the cell regulation and metabolism to measure the electron flux distribution in the respiratory chain in future studies.
E. coli adjusts its metabolism according to environmental changes including oxygen supplies. As a proof of concept, instead of changing/controlling the oxygen levels in the culture, we have explored adding different amounts of coenzyme Q1 in the cultures of ubiCA− mutant strains and showed that it coordinately changed metabolite production under aerobic conditions. This work demonstrated the potential of manipulating the ubiquinone level in E. coli cultures to optimize cell metabolism for the production of chemicals of interest under easily controlled fermentation conditions. This study further illustrated that controlled respiration that simulated a semi-aerobic environment through genetic manipulation can be exploited to allow much larger operating windows for reduced product production even under fully aerobic conditions. Other potential methods need to be tested to control respiration by altering cytochrome oxidase levels or the level of other ETS components.
Highlights.
Manipulated Escherichia coli cell respiration
Novel strategy to increase metabolically engineered product formation through “controlled” respiration
Manipulating ubiquinone level in E. coli can be used to control cell metabolism
Acknowledgments
We gratefully acknowledge Mary Harrison for her excellent technical assistance. The work was supported in part by grants from the National Institutes of Health (NIH GM090152) and NSF (CBET-0828516).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexeeva S, de Kort B, Sawers G, Hellingwerf KJ, de Mattos MJ. Effects of limited aeration and of the ArcAB system on intermediary pyruvate catabolism in Escherichia coli. Journal of Bacteriology. 2000;182:4934–4940. doi: 10.1128/jb.182.17.4934-4940.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexeeva S, Hellingwerf KJ, Teixeira de Mattos MJ. Quantitative Assessment of Oxygen Availability: Perceived Aerobiosis and Its Effect on Flux Distribution in the Respiratory Chain of Escherichia coli. Journal of Bacteriology. 2002;184:1402–1406. doi: 10.1128/JB.184.5.1402-1406.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexeeva S, Hellingwerf KJ, Teixeira de Mattos MJ. Requirement of ArcA for redox regulation in Escherichia coli under microaerobic but not anaerobic or aerobic conditions. Journal of Bacteriology. 2003;185:204–209. doi: 10.1128/JB.185.1.204-209.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böck A, Sawers G. Fermentation. In: Neidhardt FC, Curtiss R, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella: Cellular and Molecular Biology. ASM Press; Washington, D.C: 1996. pp. 262–282. [Google Scholar]
- Berrios-Rivera SJ, Bennett GN, San K-Y. Metabolic engineering of Escherichia coli: increase of NADH availability by overexpressing an NAD+-dependent formate dehydrogenase. Metab Eng. 2002;4:217–229. doi: 10.1006/mben.2002.0227. [DOI] [PubMed] [Google Scholar]
- Durnin G, Clomburg J, Yeates Z, Alvarez PJJ, Zygourakis K, Campbell P, Gonzalez R. Understanding and harnessing the microaerobic metabolism of glycerol in Escherichia coli. Biotechnol Bioeng. 2009;103:148–161. doi: 10.1002/bit.22246. [DOI] [PubMed] [Google Scholar]
- Gennis RB, Stewart V. Respiration. In: Neidhardt FC, Curtiss R, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella: Cellular and Molecular Biology. ASM Press; Washington, D.C: 1996. pp. 217–261. [Google Scholar]
- Georgellis D, Kwon O, Lin ECC. Quinones as the redox signal for the Arc two-component system of bacteria. Science. 2001;292:2314–2316. doi: 10.1126/science.1059361. [DOI] [PubMed] [Google Scholar]
- Gunsalus RP. Control of electron flow in Escherichia coli: coordinated transcription of respiratory pathway genes. Journal of Bacteriology. 1992;174(22):7069–74. doi: 10.1128/jb.174.22.7069-7074.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunsalus RP, Park SJ. Aerobic-anaerobic gene regulation in Escherichia coli: control by the ArcAB and Fnr regulons. Res Microbiol. 1994;145:437–50. doi: 10.1016/0923-2508(94)90092-2. [DOI] [PubMed] [Google Scholar]
- Hägerhäll C. Succinate: quinone oxidoreductases: Variations on a conserved theme. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 1997;1320(2):107–141. doi: 10.1016/s0005-2728(97)00019-4. [DOI] [PubMed] [Google Scholar]
- Heux S, Cachon R, Dequin S. Cofactor engineering in Saccharomyces cerevisiae: Expression of a H2O-forming NADH oxidase and impact on redox metabolism. Metab Eng. 2006;8:303–14. doi: 10.1016/j.ymben.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Holland-Staley CA, Lee K, Clark DP, Cunningham PR. Aerobic activity of Escherichia coli alcohol dehydrogenase is determined by a single amino acid. Journal of Bacteriology. 2000;182:6049–6054. doi: 10.1128/jb.182.21.6049-6054.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Q, Fu PC, Yang C, Shimizu K. Microaerobic lysine fermentations and metabolic flux analysis. Biochemical Engineering Journal. 1998;2:89–100. [Google Scholar]
- Ingram LO, Conway T, Clark DP, Sewell GW, Preston JF. Genetic engineering of ethanol production in Escherichia coli. Appl Environ Microbiol. 1987;53:2420–2425. doi: 10.1128/aem.53.10.2420-2425.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen NBS, Melchiorsen CR, Jokumsen KV, Villadsen J. Metabolic behavior of Lactococcus lactis MG1363 in microaerobic continuous cultivation at a low dilution rate. Appl Environ Microbiol. 2001;67:2677–2682. doi: 10.1128/AEM.67.6.2677-2682.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon O, Druce-Hoffman M, Meganathan R. Regulation of the Ubiquinone (Coenzyme Q) Biosynthetic Genes ubiCA in Escherichia coli. Current Microbiology. 2005;50:180–189. doi: 10.1007/s00284-004-4417-1. [DOI] [PubMed] [Google Scholar]
- Leonardo MR, Cunningham PR, Clark DP. Anaerobic regulation of the adhE gene, encoding the fermentative alcohol dehydrogenase of Escherichia coli. Journal of Bacteriology. 1993;175:870–878. doi: 10.1128/jb.175.3.870-878.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez de Felipe F, Hugenholtz J. Pyruvate flux distribution in NADH-oxidase- overproducing Lactococcus lactis strain as a function of culture conditions. FEMS Microbiology Letters. 1999;179:461–6. doi: 10.1111/j.1574-6968.1999.tb08763.x. [DOI] [PubMed] [Google Scholar]
- Lynch AS, Lin ECC. Responses to molecular oxygen. In: Neidhardt FC, Curtiss R, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella Cellular and Molecular Biology. ASM Press; Washington, D.C: 1996. pp. 262–282. [Google Scholar]
- Matsuoka Y, Shimizu K. Metabolic regulation in Escherichia coli in response to culture environments via global regulators. Biotechnol J. 2011 doi: 10.1002/biot.201000447. [DOI] [PubMed] [Google Scholar]
- Meganathan R. Ubiquinone biosynthesis in microorganisms. FEMS Microbiology Letters. 2001;203:131–139. doi: 10.1111/j.1574-6968.2001.tb10831.x. [DOI] [PubMed] [Google Scholar]
- Nikel PI, Zhu JF, San K-Y, Mendez BS, Bennett GN. Metabolic Flux Analysis of Escherichia coli creB and arcA Mutants Reveals Shared Control of Carbon Catabolism under Microaerobic Growth Conditions. Journal of Bacteriology. 2009;191:5538–5548. doi: 10.1128/JB.00174-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrenoud A, Sauer U. Impact of global transcriptional regulation by ArcA, ArcB, Cra, Crp, Cya, Fnr, and Mlc on glucose catabolism in Escherichia coli. Journal of Bacteriology. 2005;187:3171–3179. doi: 10.1128/JB.187.9.3171-3179.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San KY, Bennett GN, Berrios-Rivera SJ, Vadali RV, Yang YT, Horton E, Rudolph FB, Sariyar B, Blackwood K. Metabolic engineering through cofactor manipulation and its effects on metabolic flux redistribution in Escherichia coli. Metab Eng. 2002;4(2):182–192. doi: 10.1006/mben.2001.0220. [DOI] [PubMed] [Google Scholar]
- San KY, Bennett GN. Reduced activity of UbiCA in E. coli. Rice University; 20100009403 U S Patent application. 2010
- Sanchez AM, Bennett GN, San K-Y. Novel pathway engineering design of the anaerobic central metabolic pathway in Escherichia coli to increase succinate yield and productivity. Metab Eng. 2005;7:229–239. doi: 10.1016/j.ymben.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Shalel-Levanon S, San K-Y, Bennett GN. Effect of ArcA and FNR on the expression of genes related to the oxygen regulation and the glycolysis pathway in Escherichia coli under microaerobic growth conditions. Biotechnol Bioeng. 2005;92:147–159. doi: 10.1002/bit.20583. [DOI] [PubMed] [Google Scholar]
- Shalel-Levanon S, San K-Y, Bennett GN. Effect of oxygen, and ArcA and FNR regulators on the expression of genes related to the electron transfer chain and the TCA cycle in Escherichia coli. Metab Eng. 2005;7:364–374. doi: 10.1016/j.ymben.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Snoep J, Teixeira de Mattos M, Postma P, Neijssel O. Involvement of pyruvate dehydrogenase in product formation in pyruvate-limited anaerobic chemostat cultures of Enterococcus faecalis NCTC 775. Arch Microbiol. 1990;154(1):50–55. doi: 10.1007/BF00249177. [DOI] [PubMed] [Google Scholar]
- Søballe B, Poole RK. Requirement for ubiquinone downstream of cytochrome(s) b in the oxygen-terminated respiratory chains of Escherichia coli K-12 revealed using a null mutant allele of ubiCA. Microbiology. 1998;144:361–73. doi: 10.1099/00221287-144-2-361. [DOI] [PubMed] [Google Scholar]
- Salmon K, Hung SP, Mekjian K, Baldi P, Hatfield GW, Gunsalus RP. Global gene expression profiling in Escherichia coli K12. The effects of oxygen availability and FNR. J Biol Chem. 2003;278:29837–55. doi: 10.1074/jbc.M213060200. [DOI] [PubMed] [Google Scholar]
- Salmon KA, Hung SP, Steffen NR, Krupp R, Baldi P, Hatfield GW, Gunsalus RP. Global gene expression profiling in Escherichia coli K12: effects of oxygen availability and ArcA. J Biol Chem. 2005;280:15084–96. doi: 10.1074/jbc.M414030200. [DOI] [PubMed] [Google Scholar]
- Thomason LC, Costantino N, Court DL. Curr Protoc Mol Biol. Unit 1. Chapter 1. 2007. E. coli genome manipulation by P1 transduction; p. 17. [DOI] [PubMed] [Google Scholar]
- Tolentino GJ, Meng SY, Bennett GN, San K-Y. A Ph-Regulated Promoter for the Expression of Recombinant Proteins in Escherichia coli. Biotechnology Letters. 1992;14:157–162. [Google Scholar]
- Tran QM, Rothery RA, Maklashina E, Cecchini G, Weiner JH. The Quinone Binding Site in Escherichia coli Succinate Dehydrogenase Is Required for Electron Transfer to the Heme b. Journal of Biological Chemistry. 2006;281(43):32310–32317. doi: 10.1074/jbc.M607476200. [DOI] [PubMed] [Google Scholar]
- Wagner AF, Frey M, Neugebauer FA, Schafer W, Knappe J. The free radical in pyruvate formate-lyase is located on glycine-734. Proc Natl Acad Sci. 1992;89:996–1000. doi: 10.1073/pnas.89.3.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace BJ, Young IG. Role of Quinones in Electron-Transport to Oxygen and Nitrate in Escherichia coli Studies with a ubiA- menA- Double Quinone Mutant. Biochimica Et Biophysica Acta. 1977;461:84–100. doi: 10.1016/0005-2728(77)90071-8. [DOI] [PubMed] [Google Scholar]
- Yang YT, San K-Y, Bennett GN. Redistribution of metabolic fluxes in Escherichia coli with fermentative lactate dehydrogenase overexpression and deletion. Metab Eng. 1999;1:141–152. doi: 10.1006/mben.1998.0111. [DOI] [PubMed] [Google Scholar]
- Yang YT, Bennett GN, San K-Y. Effect of inactivation of nuo and ackA-pta on redistribution of metabolic fluxes in Escherichia coli. Biotechnol Bioeng. 1999;65:291–297. [PubMed] [Google Scholar]
- Zeng AP, Deckwer WD. Bioreaction techniques under microaerobic conditions: From molecular level to pilot plant reactors. Chemical Engineering Science. 1996;51:2305–2314. [Google Scholar]
- Zhu J, Shalel-Levanon S, Bennett GN, San K-Y. Effect of the global redox sensing/regulation networks on Escherichia coli and metabolic flux distribution based on C-13 labeling experiments. Metab Eng. 2006;8:619–627. doi: 10.1016/j.ymben.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Zhu J, Shimizu K. The effect of pfl gene knockout on the metabolism for optically pure D-lactate production by Escherichia coli. Appl Microbiol Biotech. 2004;64(3):367–375. doi: 10.1007/s00253-003-1499-9. [DOI] [PubMed] [Google Scholar]
- Zhu J, Shimizu K. Effect of a single-gene knockout on the metabolic regulation in Escherichia coli for D-lactate production under microaerobic condition. Metab Eng. 2005;7:104–115. doi: 10.1016/j.ymben.2004.10.004. [DOI] [PubMed] [Google Scholar]