Abstract
Two analyses of HIV-1 subtype C Gag quasispecies were performed in a prospective cohort of 42 acutely and recently infected individuals by SGA on viral RNA/proviral DNA templates. First, in vivo Gag substitutions were assessed in relation to the HIV-1C consensus sequence, which revealed that 29.3% of detected amino acid substitutions can be classified as reversions to subtype consensus, 61.3% as forward substitutions from subtype consensus, and 9.3% as polymorphisms not associated with the subtype consensus sequence. Second, the proportion, dynamics, and relationships within individual pools of viral quasispecies were analyzed. Among reverse substitutions, 16.1% were minor, 11.0% transient, 13.6% dominant, and 59.2% fixed. In contrast, 31.6% of forward substitutions were minor, 59.3% transient, 3.8% dominant, and 5.3% fixed. The distinct patterns in the spectrum and dynamics of reverse and forward Gag substitutions suggest that these differences should be considered in HIV-1 evolutionary studies and analyses of viral mutational pathways.
Introduction
A comprehensive assessment of viral evolution across HIV-1 genes is critical for the successful design of preventive and therapeutic interventions. Host-driven immune pressure involving both innate and adaptive components during the course of HIV infection results in a robust quasi-steady balance between emerging viral variants and virus fitness (Borghans et al., 2007; Crawford et al., 2007; Leslie et al., 2004; Liu et al., 2007; Martinez-Picado et al., 2006; Peyerl et al., 2004b). The first virus-mediated T cell response can rapidly select escape mutations concurrent with the peak decline in acute HIV infection (Goonetilleke et al., 2009). Successful viral escape from immune recognition can maintain viral replication (Feeney et al., 2004; Geels et al., 2006; Oxenius et al., 2004). However, if the escape mutation impairs virus replicative fitness, such escape might be associated with reduced viral load and better disease outcome in humans and rhesus macaques (Chopera et al., 2008; Crawford et al., 2007; Friedrich et al., 2004a; Friedrich et al., 2004b; Friedrich et al., 2004c; Goepfert et al., 2008; Martinez-Picado et al., 2006; Migueles et al., 2003; Peyerl et al., 2004a; Peyerl et al., 2004b; Schneidewind et al., 2007). For example, in subjects expressing HLA-B*57/5801, the virus mutates from Thr to Asn at position 242 in Gag which results in a reduction of viral replication due to the fitness cost of the T242N mutation (Brockman et al., 2007; Chopera et al., 2008; Crawford et al., 2007; Leslie et al., 2004; Martinez-Picado et al., 2006). Because host genetic factors play an important role in disease progression in HIV-infected individuals (Carrington et al., 1999; Fellay et al., 2007; Kaslow et al., 1996; Klein et al., 1998; O'Brien and Nelson, 2004), it is believed that the pathway of viral escape from immune recognition can be predicted by the profile of host MHC class I HLA alleles (Allen et al., 2005; Brumme et al., 2008a; Brumme et al., 2007; Brumme et al., 2008b; Rousseau et al., 2008).
Transmission of the viral escape variant to a new host may result in a rapid reversion to the wild type virus (which corresponds to the HIV-1 subtype consensus sequence in most epidemics), particularly in the absence of selective pressure in the new host due to a different profile of HLA alleles (Allen et al., 2004; Chopera et al., 2008; Crawford et al., 2007; Davenport et al., 2008; Duda et al., 2009; Friedrich et al., 2004a; Leslie et al., 2004; Li et al., 2007; Matthews et al., 2008; Rousseau et al., 2008). Reverse amino acid substitutions to the HIV-1 subtype consensus are critical for maintaining virus within the optimal evolutionary space in the local epidemic. The breadth of immune response plays an important role in disease outcome, and is likely to be associated with the breadth of viral mutational pathways. It is widely agreed that viral escape (or forward) and reverse amino acid subsitutions contribute significantly to HIV-1 evolution at both the intra-patient and population levels (Allen et al., 2005; Bhattacharya et al., 2007; Borrow et al., 1997; Brumme et al., 2008a; Friedrich et al., 2004a; Koenig et al., 1995; Leslie et al., 2005; Leslie et al., 2004; Li et al., 2007; Moore et al., 2002; Phillips et al., 1991; Price et al., 1997; Rousseau et al., 2008). While intrapatient evolution of HIV does not necessarily translate into evolution of HIV at the population level (Leslie et al., 2004), it is likely that the circulating variants of HIV-1 in the local epidemic represent equilibrium between cumulative immune pressure of the host population and viral fitness of locally transmitting viruses.
The HIV-1 Gag is able to induce potent virus-specific T cell responses that were shown to be associated with control of viral replication, lower viral set point, and better disease prognosis (Betts et al., 2001; Boaz et al., 2002; Edwards et al., 2002; Geldmacher et al., 2007; Kiepiela et al., 2007; Masemola et al., 2004; Ndongala et al., 2009; Novitsky et al., 2003; Novitsky et al., 2006; Ramduth et al., 2005; Rolland et al., 2008; Serwanga et al., 2009; Zuniga et al., 2006), and therefore represents an attractive target for an HIV-1 vaccine design. Better understanding of viral dynamics and the spectrum of in vivo Gag substitutions may advance the rational design and development of an HIV-1 vaccine.
Surprisingly, types of viral amino acid subsitutions have not been defined and used uniformly. While some types of substitutions are well defined (e.g., synonymous, nonsynonymous, fixed), terms for other viral substitutions are used inconsistently, leading to confusion or misunderstanding. For example, “reverse mutation” or “reverse substitution” may mean reversion either to the transmitted virus, or to subtype consensus. Using a detailed classification of terms, this study addressed and characterized the types and frequency of the in vivo Gag substitutions in the early stage of HIV-1 subtype C infection in a cohort of 42 subjects with estimated time of seroconversion. The frequency distributions of amino acid substitutions within the individual pools of viral quasispecies were analyzed longitudinally. Two temporal aspects in the evolution of in vivo Gag quasispecies were addressed. Amino acid substitutions across Gag were analyzed by their relation to the HIV-1 subtype C consensus, and by fixation and stability.
Methods
Study subjects
Longitudinal sets of gag sequences were analyzed in subjects enrolled in a primary HIV-1 subtype C infection cohort in Botswana (Novitsky et al., 2009c; Novitsky et al., 2009d; Novitsky et al., 2008) from April 2004 to April 2008. A total of 42 enrolled subjects included 8 detected with acute HIV-1 infection (Fiebig stage II) and 34 detected with “recent” infection (Fiebig stage IV or V). Details on estimating time of seroconversion are provided elsewhere (Novitsky et al., 2009b). A “zero” time corresponded to seroconversion, as a more accurate and measurable time point than estimation the time of HIV infection (Novitsky et al., 2010c). The time of HIV infection can be estimated by adding 14 to 21 days. The median (IQR) time elapsed between estimated time of seroconversion and first available sequence was 44.0 (10.8 to 54.0) days. There were 9 male (2 acute) and 33 female (6 acute) participants. The median age at enrollment was 27 years (IQR 25-32, range 20-56). All subjects were Botswana nationals, and all infections were HIV-1 subtype C (Novitsky et al., 2009a; Novitsky et al., 2009d). As described previously (Novitsky et al., 2009a; Novitsky et al., 2009d; Novitsky et al., 2008), viral load and CD4+ T cell counts were assessed during follow-up. Blood samples were analyzed at median (IRQ) of 5 (4-6) time points per subject. The follow up period was 415 (321-447) days post-seroconversion (p/s). Ten of 42 (24%) subjects initiated ART within the observed period of time due to a drop in CD4+ T cells (time of ART initiation, if any, is shown in Table S1). The study was approved by Institutional Review Boards in Botswana and the US. Written informed consent was obtained from each participant.
Single-genome amplification and sequencing
Previous evolutionary studies demonstrated that the HIV-1 gag and env sequences derived from PBMC are comparable to viruses circulating in plasma (Geels et al., 2003; Shankarappa et al., 1999). The similarities between viral sequences originating from alternative templates, viral RNA and proviral DNA, are particularly evident at the early phase of HIV infection (Novitsky V., unpublished data). In this study both viral RNA from plasma and cell-associated proviral DNA were used as templates for amplification of viral sequences. Single genome amplification and direct sequencing was performed as described previously (Novitsky et al., 2009c; Novitsky et al., 2010c). Sequence contigs were assembled by SeqScape v.2.6. Multiple sequence alignment was performed by HIV-align at the Los Alamos HIV Database site (http://www.hiv.lanl.gov/) using the hidden Markov model and the codon-alignment option followed by minor manual adjustments in BioEdit (Hall, 1999). The obtained sequences were tested by HYPERMUT v.2.0 (Rose and Korber, 2000) and hypermutated sequences were excluded from analysis. A total of 2,595 gag sequences included 827 sequences originating from the viral RNA template, and 1,768 sequences obtained from the proviral DNA template. The median (IQR) of analyzed gag sequences was 11.7 (8.8-13.9) per time point, and 61.5 (43.3-76.0) per subject. The accession numbers of the analyzed gag sequences are GQ275380–GQ277569, GQ375107–GQ375128, and GQ870874–GQ871183.
Amino acid frequencies in the pool of viral quasispecies
Only non-synonymous substitutions were analyzed in this study. The frequencies of translated amino acid sequences in Gag were analyzed using MargFreq (Ray, 2008) per time point per subject. The frequency of each amino acid was expressed as a fraction of 1 in the pool of viral quasispecies at a given time point. Frequency changes in the pool of viral quasispecies at each amino acid position over time were tracked according the following criteria. If any change in the proportion of amino acid(s) in the pool of viral quasispecies was observed over time, the amino acid with increased frequency was the subject of frequency assessment. Sites with a single amino acid present in the pool of the earliest available quasispecies were examined for appearance of any alternative amino acid over the time of follow up, and the new amino acid was subject for evaluation. Examples of this approach were presented elsewhere (e.g., Figure 2 in (Novitsky et al., 2010c)). If two amino acids were present in the pool of the earliest quasispecies, the amino acid that showed an increase in frequency over time was a subject for analysis. Although multiple amino acids can be present in the pool of viral quasispecies, we observed no cases with more than two amino acids at the earliest time points in this study. Therefore, no sites with multiple increasing amino acids were detected.
Types of amino acid substitutions
By analogy with drug-resistance mutations, we define a mutational pathway as a series of sequential changes in the pool of viral quasispecies at a single or at multiple amino acid positions over time. Parameters quantified in this study include sequential frequency of observed amino acids over time in relation to the estimated time of seroconversion. At each position in amino acid alignment, the observed amino acid substitutions were classified based on their relation to subtype C consensus sequence, and fraction in the pool of viral quasispecies.
In analysis of relationships between the observed in vivo Gag substitutions and the subtype consensus, an amino acid substitution from the consensus amino acid to the non-consensus was treated as a forward substitution (a potential escape mutation that requires experimental confirmation of immune escape). In this study, an amino acid substitution from a non-consensus amino acid to the consensus-matched amino acid was considered a reverse substitution, or reversion (note: the alternative interpretation of reverse substitution is related to the transmitted virus, and was not used in this study). Viral substitutions that did not involve the most common HIV-1 subtype C amino acid were treated as non-subtype-consensus-related polymorphisms. Testing of immune responses was out of the scope of the study, resulting in a lack of evidence that observed viral substitutions are directly linked to immune responses.
Based on the frequency in the pool of viral quasispecies, we categorized amino acid substitutions as minor, dominant, fixed, and transient. A substitution was considered minor if its fraction was less than or equal to 50% in the pool of viral quasispecies during follow-up. A substitutions was treated as dominant if its fraction reached more than 50% in the pool of viral quasispecies but did not reach complete substitution of the original amino acid. A substitution was fixed if it reached 100% in the pool of viral quasispecies during the follow-up period. A substitution was considered transient if its frequency subsequently decreased below 1.0 for fixed substitutions, or below 0.5 for dominant substitutions, or if minor substitution disappeared over the observation period. Once amino acid substitutions in Gag reached the state of dominance or fixation, they rarely disappeared but rather shared their frequency in the pool of viral quasispecies with alternative amino acid(s) by balancing and frequency fluctuation over time. Thus, in this study transient substitutions are predominantly those that were minor and disappeared from the pool of viral quasispecies.
MHC class I HLA typing
High resolution HLA typing was performed for all study subjects as described previously (Novitsky et al., 2009c). Briefly, the AlleleSEQR HLA Sequencing-Based Typing kit (Celera, Alameda, CA) was used according to the manufacturer's instruction. Contig assembling and assignment of HLA alleles was implemented by Assign SBT ver. 3.5.1.42 (Conexio Genomics, Applecross, Australia). All ambiguous positions were resolved by re-sequencing. Polymorphisms outside the targeted exons that could not resolve heterozygote combinations were interpreted as two-digit HLA typing results.
CTL epitopes analysis
The Los Alamos National Laboratory HIV immunology database (http://www.hiv.lanl.gov/content/immunology) was screened for known human CTL epitopes identified in the context of HIV-1 subtype C infection and the MHC class I HLA alleles restriction. For each subject, the location of the observed amino acid substitutions in Gag was matched with the retrieved epitopes restricted by the corresponding class I HLA alleles. The number of matched amino acid substitutions within known CTL epitopes in the context of their class I HLA restriction was used for analysis the proportion of Gag substitutions within CTL epitopes.
Statistical analysis
Data are summarized with medians (IQR range for 25% and 75%). Frequencies of amino acid substitutions by type over time are presented.
Results
Intra-patient heterogeneity of HIV-1C Gag substitutions
Dynamics of Gag amino acid substitutions was analyzed in a cohort of 42 HIV-1 subtype C infected individuals with estimated time of seroconversion, which allowed us to synchronize the time of observed substitutions among patients. The actual distribution of HIV-1subtype C Gag substitutions is presented in supplementary Table S1. In the cumulative analysis of all types of Gag substitutions, the median (IQR) number of reverse substitutions was 7 (4-15.5), forward substitutions was 18 (15.3-25), and polymorphisms was 2 (1-4) per subject. The number of Gag substitutions per subject was considerably less for a subset of dominant and fixed substitutions: median (IQR) number of reverse amino acid substitutions was 5 (1.3-8.8); forward substitutions was 1 (0-3), and polymorphisms was 0 (0-1). The reverse dominant and fixed amino acid substitutions in HIV-1C Gag were found in 37 of 42 (88%) subjects, while forward dominant and fixed substitutions were detected in 27 of 42 (64%) subjects.
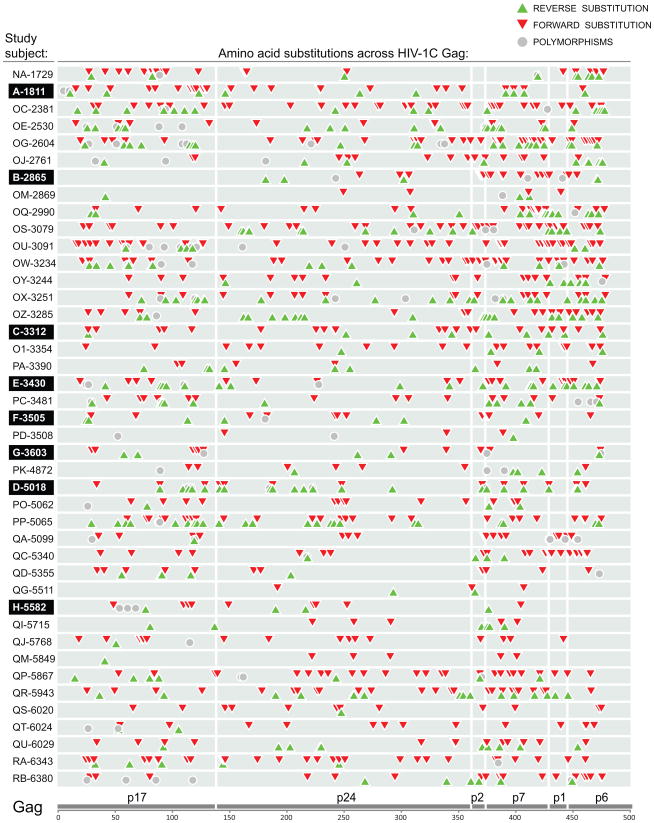
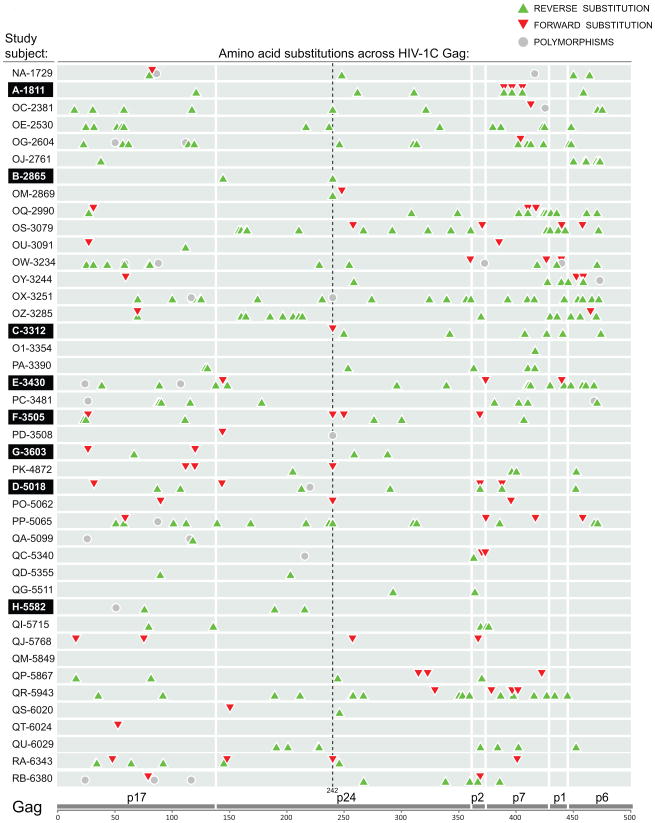
To address distribution of amino acid substitutions in HIV-1 subtype C Gag, the identified non-synonymous substitutions were plotted according to their location across Gag in each subject. The substitution map presented on Figure 1 shows distribution of three types of Gag substitutions – reverse (green up-triangles), forward (red down-triangles), and polymorphisms not associated with the subtype consensus sequence (gray circles) – that were accumulated over the follow-up period (up to 500 days p/s). The map on Figure 1 includes all minor, transient, dominant, and fixed substitutions in HIV-1C Gag. It is evident from this map that 1) all types of substitutions are scattered across Gag; 2) the number and type of Gag substitutions differ substantially between subjects, and 3) forward (red) substitutions dominate the map (Fig. 1). However, excluding of minor and transient substitutions revealed in a changed shape of the Gag substitution map (Fig. 2). The reverse substitutions (green) are prevailing on the map of dominant and fixed Gag substitutions, while forward substitutions are less common. The location and number of dominant and fixed substitutions differs considerably between subjects. Substitutions at Gag codon position 242 represented the most common pattern observed in 12 out of 42 subjects including 5 reverse, 5 forward, and 2 polymorphic substitutions, as described elsewhere (Novitsky et al., 2010c).
Figure 1.
Profile and location of total amino acid substitutions across Gag in a cohort of 42 HIV-1 subtype C-infected individuals. The cumulative non-synonymous substitutions identified by SGA during the time period from seroconversion to up to 500 days p/s are presented. The study subjects' code is shown at the left. Eight acutely infected subjects are highlighted. Three types of Gag substitutions are outlined as follows: green up triangle shows amino acid substitutions toward the HIV-1 subtype C consensus sequence – reverse substitutions, red down triangle denotes amino acid substitutions from the subtype C consensus – forward substitutions, and gray circle delineates viral amino acid polymorphisms not associated with the subtype C consensus sequence. Gray bars on the background denote HIV-1 Gag p17, p24, p2/p7p1/p6 for each subject.
Figure 2.
Profile and location of dominant and fixed amino acid substitutions across HIV-1C Gag. See legend to Figure 1. The dashed line highlights 12 amino acid substitutions observed at Gag codon position 242.
Overall, the analysis of viral amino acid substitutions by their relation to the subtype consensus sequence demonstrated the wide range and heterogeneity in the number and location of reverse and forward Gag substitutions among HIV-1 subtype C infected individuals.
Differential dynamics of Gag substitutions
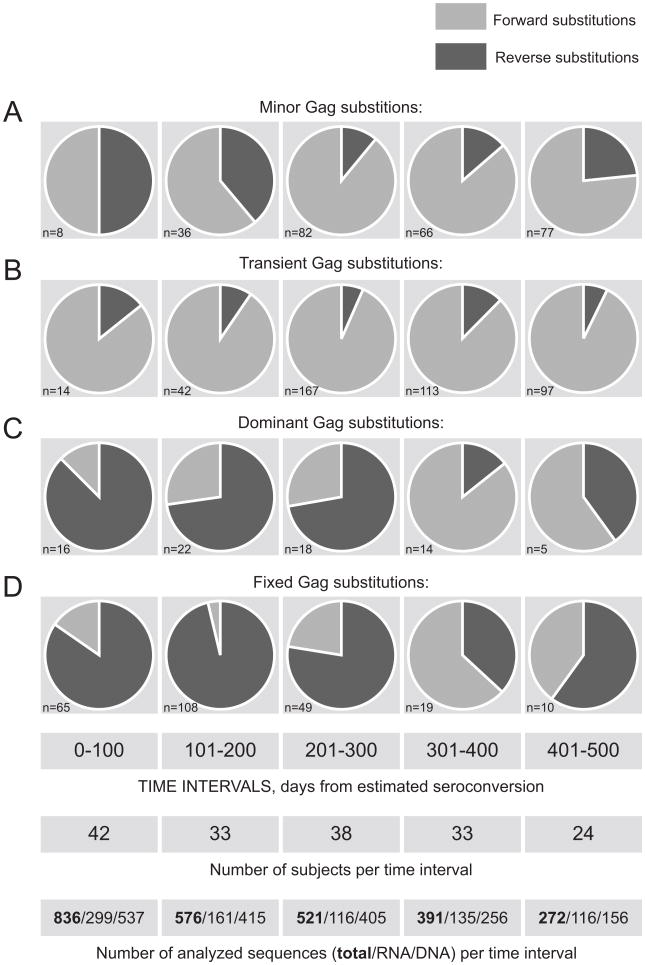
Temporal trends in kinetics of reverse and forward Gag substitutions in primary HIV-1 subtype C were analyzed over five 100-day intervals up to 500 days p/s (Fig. 3). Forward Gag substitutions were predominant among minor (Fig. 3A) and transient (Fig. 3B) substitutions, except even distribution of minor reverse and minor forward substitutions during the period 0 to 100 days p/s. The relationships between reverse and forward transient substitutions in Gag were remarkably constant over time (Fig. 3B). In contrast, the reverse substitutions outnumbered forward substitutions among dominant (Fig. 3C) and fixed (Fig. 3D) Gag substitutions during the period up to 300 days p/s. However a larger proportion of forward substitutions was evident during 301-400 days p/s with about equal number of forward and reverse dominant and fixed substitutions over the period 401-500 days p/s. This observation suggests a shift in the ratio of reverse/forward amino acid substitution in HIV-1 subtype C Gag around 300 days p/s. The Fisher exact test supported that the frequencies of reverse substitutions within total substitutions were changing over time (p-value for minor, transient, dominant, and fixed substitutions was equal 0.001, 0.36, <0.001, and <0.001, respectively). The observed shift in the ratio of reverse/forward substitutions after 300 days p/s seems to indicate fewer reversions that occur after the first year of HIV-1 infection.
Figure 3.
Relationships between reverse and forward substitutions in HIV-1C Gag over time. The number of identified amino acid substitutions is shown in the left bottom corner of each graph. Analyzed time intervals in days from estimated seroconversion, the number of subjects, and the number of analyzed sequences per time interval including total, RNA, and DNA are shown at the bottom. A: Minor Gag substitutions (did not reach dominance or fixation, and did not disappear over the follow up time). B: Transient Gag substitutions. C: Dominant Gag substitutions (did not reach fixation over the follow up time). D: Fixed substitutions.
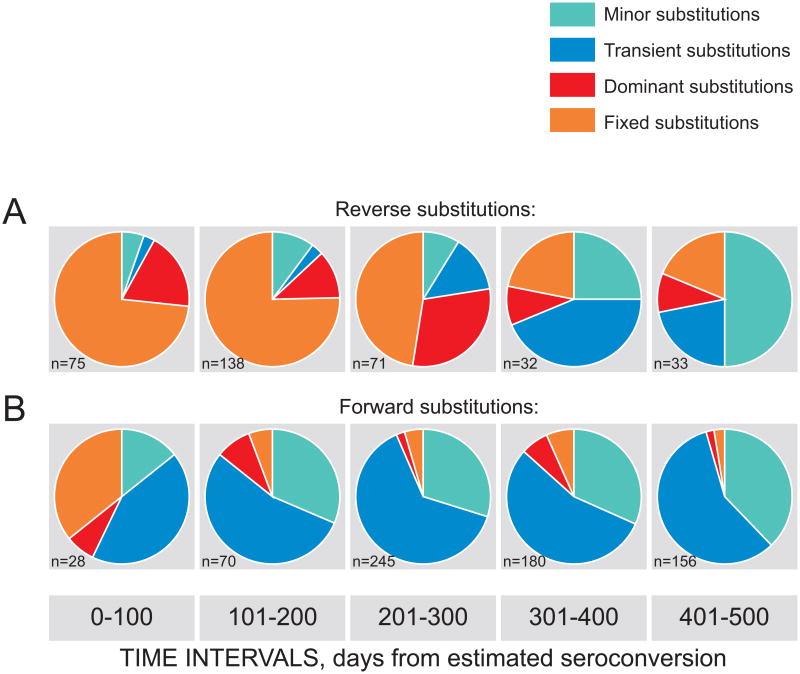
Changes in the spectrum of reverse and forward Gag substitutions were analyzed over five 100-day intervals (Fig. 4). A gradual shift over time was evident in the range of both reverse (Fig. 4A) and forward (Fig. 4B) substitutions. Among reverse Gag substitutions, gradual decrease of dominant and fixed substitutions was accompanied by increased proportion of minor and transient substitutions, which was particularly noticeable after 300 days p/s. Interestingly that among Gag forward substitutions, the fraction of complete substitutions was substantial (10 of 28) at the early stage of HIV-1 subtype C infection (Fig. 4B). However, the proportion of dominant and fixed amino acid substitutions among Gag forward substitutions diminished dramatically at later time points, e.g., after 100 days p/s. As a result, the majority of Gag forward substitutions after 100 days p/s were minor and transient.
Figure 4.
Relationships between different types of reverse and forward Gag substitutions over time. The number of identified substitutions is shown in the left bottom corner of each graph. Analyzed time intervals in days from estimated seroconversion are shown at the bottom. A: Reverse substitutions. B: Forward substitutions.
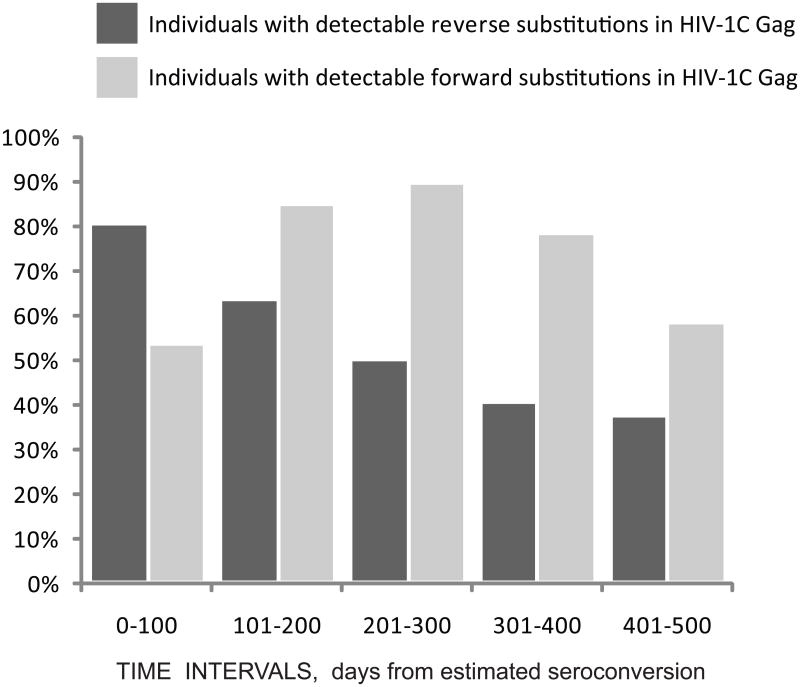
The proportion of individuals with detectable reverse or forward amino acid substitutions in Gag was analyzed over the same five 100-day intervals (Figure 5). A gradual decrease from about 80% to 40% of individuals with detectable reverse substitutions was evident over time. In contrast, the proportion of individuals with detectable forward substitutions was the highest during the 201-300 days p/s interval.
Figure 5.
Proportion of individuals with detectable amino acid substitutions in Gag over time. Individuals with detectable reverse substitutions are shown by dark bars. Individuals with detectable forward substitutions are shown by light bars. Analyzed time intervals in days from estimated seroconversion are shown at the bottom.
Time to fixation of amino acid substitutions in Gag was analyzed for different types of substitutions. The median (IQR) to fixation was 146 (100-185) days p/s for reverse substitutions, 268 (102-380) days p/s for forward substitutions, and 243 (190-440) days p/s for polymorphisms. The difference between time to fixation was statistically significant between reverse and forward substitutions (p<0.001), and between reverse substitutions and polymorphisms (p<0.001), but was similar between forward substitutions and polymorphisms not associated with subtype consensus sequence.
Relationships between types of amino acid substitutions in Gag
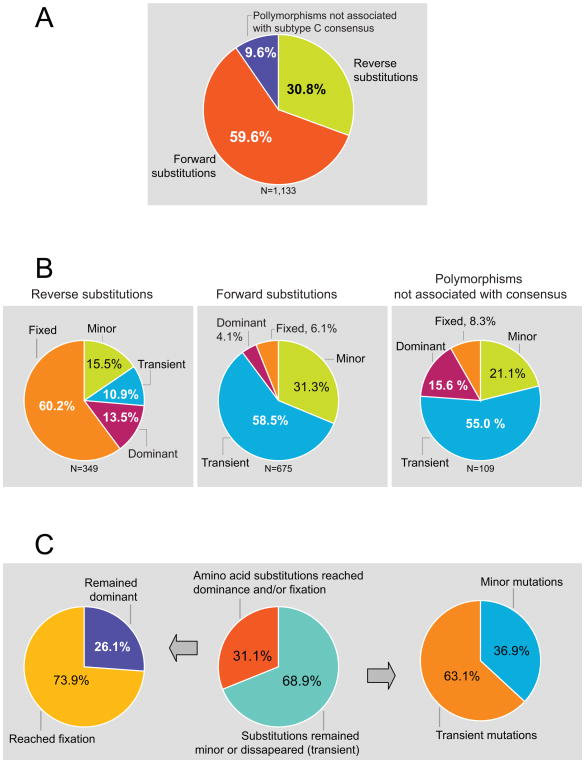
The quantitative relationships between different types of amino acid substitutions were analyzed. The proportions of total reverse substitutions, forward substitutions, and polymorphisms not directly associated with the subtype consensus in Gag were 30.8%, 59.6%, and 9.7%, respectively (Fig. 6A). The majority of reverse substitutions reached dominance (13.5%) and fixation (60.2%; Fig. 6B). In contrast, forward amino acid substitutions were predominantly minor (31.3%) or transient (58.5%), while only few reached dominance (4.1%) or fixation (6.1%) in the pool of viral quasispecies during the first year of HIV-1 subtype C infection (Fig. 6B). A large fraction of viral polymorphisms not associated with the subtype consensus sequence was represented by minor (21.1%) or transient (55.0%) substitutions, and only about a quarter of polymorphisms reaching dominance (15.6%) or fixation (8.3%) within a year following seroconversion (Fig. 6B). Overall, 68.9% of Gag amino acid substitutions did not exceed the level of 50% or were lost during the observation period (Fig. 6C). Of those, 36.9% were represented by minor amino acid substitutions, and 63.1% by transient substitutions. About one third of Gag substitutions (31.1%) reached dominance or fixation. Among these substitutions, 26.1% remained dominant, while 73.9% reached fixation (Fig. 6C).
Figure 6.
Relationships between different types of Gag amino acid substitutions. A: Overall relationships between reverse substitutions, forward substitutions, and viral polymorphisms not directly associate with the subtype consensus sequence. B: Relationships within reverse substitutions, forward substitutions, and viral polymorphisms not directly associate with the subtype C consensus sequence. C: Breakdown of viral substitutions in Gag that either reached or did not reach dominance and/or fixation.
Amino acid substitutions in Gag within known CTL epitopes
To estimate the fraction of amino acid substitutions that are likely to be associated with immune pressure we quantified substitutions within known CTL epitopes restricted by individual MHC class I HLA alleles in each subject. A total of 462 of 1,243 (37.2%) observed amino acid substitutions in Gag were observed with known CTL epitopes restricted by individual class I HLA alleles. The restriction of involved epitopes was approximately even between HLA-A (31.2%), HLA-B (37.4%), and HLA-C (31.4%) loci. The proportion of different types of amino acid substitutions found within known CTL epitopes was similar to the overall distribution of substitutions in Gag: 29%, 63%, and 9% for reversions, forward substitutions, and polymorphisms, respectively. Interestingly, 48% of reverse substitutions and 39% of forward substitutions were found within known CTL epitopes restricted by more than one class I HLA allele. For example, a gradual substitution from Lys to Arg at position 26 in p17 was observed within known overlapping CTL epitope KIRLRPGGK restricted by HLA-A*03:01 and epitope RPGGKKRYM restricted by HLA-Cw*06:02 in recently infected subject OG-2604 expressing both HLA-A*03:01:01 and HLA-Cw*06:02. The median (IQR) of observed Gag substitutions within known CTL epitopes per patient was 9 (6; 14) ranging from 1 to 46.
Discussion
The study assessed the dynamic spectrum and inter-subject heterogeneity of the in vivo Gag substitutions during primary HIV-1 subtype C infection. Longitudinal analysis of viral evolution from seroconversion to up to 500 days p/s revealed the dynamic nature of the in vivo mutational pathways in HIV-1C Gag, and their complexity at selected amino acid residues. A high fraction of identified Gag substitutions, i.e., forward substitutions, were transient or minor. In contrast, the majority of Gag reverse substitutions were found to be fixed. A substantial inter-patient heterogeneity was evident in the patterns of appearance, dominance, and fixation of identified amino acid substitutions in HIV-1C Gag.
A considerable fraction of Gag reverse substitutions, 73.7%, reached dominance and/or fixation during the first year of HIV-1 subtype C infection, supporting the hypothesis that reverse substitutions in Gag are driven by viral fitness cost, e.g., ability of reverse substitutions to restore viral fitness. In contrast, only a small fraction of Gag forward substitutions, 10.2%, reached dominance or fixation. More than half of the detected forward substitutions, 58.5%, were transient. Given a relatively short period of detection of some transient substitutions even with frequent sampling, this is likely a conservative estimate. We propose that at least three mechanisms may account for a large fraction of transient forward substitutions in HIV-1 subtype C Gag. First, transient substitutions may represent mutational noise, although a relatively large overall number of transient substitutions might have a negative impact by diverting immune responses and exhausting the immune system. Second, assuming random appearance, transient substitutions are generated stochastically, they can dramatically reduce viral fitness, and therefore can be eliminated quickly from the pool of viral quasispecies. Third, hypothetically, transient substitutions might induce potent immune responses within a short period of time, and therefore become eliminated by immune pressure. If the last assumption is true, then certain transient substitutions could represent an attractive target for HIV vaccine design due to their potent immunogenicity. According to this scenario, a vaccine antigen with transient amino acid substitutions might rapidly induce a long lasting immune response of high magnitude that cannot be achieved in natural infection due to fast elimination of such viral variant by immune pressure. A high magnitude of the induced immune response might cause cross-reactivity toward viral variants without transient substitution, and if so, should have a protective effect. Presence of both fixed and transient viral substitutions indicates that the overall immune responses to HIV-1 Gag epitopes are substantial, but also provides evidence for a fast appearance of viral variants.
The reverse substitutions play an important role on the population level (Matthews et al., 2008) through their ability to restore the subtype consensus sequence. Upon virus transmission between HLA-distinct hosts and apparent shift in the specificities of HLA restriction, at least some forward substitutions that were generated in previous host revert to the subtype consensus. Thus, being amplified on a population level, the reverse substitutions maintain the subtype consensus sequence of virus in epidemic. Despite the increase in overall viral diversity, the stability of HIV-1C Gag consensus sequence in the epidemic over time is striking (Novitsky et al., 2010a; Novitsky et al., 2010b). A relatively low proportion of amino acid substitutions within known CTL epitopes might indicate that only fraction of CTL epitopes have been mapped in HIV-1C infection, and warrants further studies.
Utilizing the HIV-1 subtype consensus sequence might be biased to the extent of representative sampling in the epidemic, which is one of the study limitations. However our previous analyses demonstrated an extreme similarity between the HIV-1C consensus sequences originating from different geographic areas or collected at different time points (Novitsky et al., 2002; Novitsky et al., 2010b). Another study limitation is that some early reverse and forward substitutions could be missed due to the time of sampling. Samples with the first available sequence in Fiebig stage V might be particularly affected. Therefore, the current study had little to no power to address the earliest amino acid substitutions in Gag.
In summary, this study addressed the breadth and relationships of in vivo Gag substitutions in primary HIV-1 subtype C infection using a direct mapping of Gag amino acid substitutions along the time line of HIV-1 infection. The study contributes to better understanding of early events in HIV infection and might assist in facilitating the development of preventive strategies.
Supplementary Material
Acknowledgments
We are grateful to all participants and team members of the Tshedimoso study in Botswana. We thank Lendsey Melton for excellent editorial assistance. The primary HIV-1 subtype C infection study in Botswana, the Tshedimoso study, was supported and funded by NIH grant R01 AI057027 and R21 AI087493. This work was also supported in part by NIH grant R37 AI24643 (RW).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allen TM, Altfeld M, Geer SC, Kalife ET, Moore C, O'Sullivan KM, DeSouza I, Feeney ME, Eldridge RL, Maier EL, Kaufmann DE, Lahaie MP, Reyor L, Tanzi G, Johnston MN, Brander C, Draenert R, Rockstroh JK, Jessen H, Rosenberg ES, Mallal SA, Walker BD. Selective Escape from CD8+ T-Cell Responses Represents a Major Driving Force of Human Immunodeficiency Virus Type 1 (HIV-1) Sequence Diversity and Reveals Constraints on HIV-1 Evolution. J Virol. 2005;79:13239–13249. doi: 10.1128/JVI.79.21.13239-13249.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen TM, Altfeld M, Yu XG, O'Sullivan KM, Lichterfeld M, Le Gall S, John M, Mothe BR, Lee PK, Kalife ET, Cohen DE, Freedberg KA, Strick DA, Johnston MN, Sette A, Rosenberg ES, Mallal SA, Goulder PJR, Brander C, Walker BD. Selection, Transmission, and Reversion of an Antigen-Processing Cytotoxic T-Lymphocyte Escape Mutation in Human Immunodeficiency Virus Type 1 Infection. J Virol. 2004;78:7069–7078. doi: 10.1128/JVI.78.13.7069-7078.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts MR, Ambrozak DR, Douek DC, Bonhoeffer S, Brenchley JM, Casazza JP, Koup RA, Picker LJ. Analysis of Total Human Immunodeficiency Virus (HIV)-Specific CD4+ and CD8+ T-Cell Responses: Relationship to Viral Load in Untreated HIV Infection. J Virol. 2001;75:11983–11991. doi: 10.1128/JVI.75.24.11983-11991.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya T, Daniels M, Heckerman D, Foley B, Frahm N, Kadie C, Carlson J, Yusim K, McMahon B, Gaschen B, Mallal S, Mullins JI, Nickle DC, Herbeck J, Rousseau C, Learn GH, Miura T, Brander C, Walker B, Korber B. Founder effects in the assessment of HIV polymorphisms and HLA allele associations. Science. 2007;315:1583–1586. doi: 10.1126/science.1131528. [DOI] [PubMed] [Google Scholar]
- Boaz MJ, Waters A, Murad S, Easterbrook PJ, Vyakarnam A. Presence of HIV-1 Gag-Specific IFN-gamma(+)IL-2(+) and CD28(+)IL-2(+) CD4 T Cell Responses is Associated with Nonprogression in HIV-1 Infection. J Immunol. 2002;169:6376–6385. doi: 10.4049/jimmunol.169.11.6376. [DOI] [PubMed] [Google Scholar]
- Borghans JAM, Mølgaard A, de Boer RJ, Keşmir C. HLA Alleles Associated with Slow Progression to AIDS Truly Prefer to Present HIV-1 p24. PLoS ONE. 2007;2:e920. doi: 10.1371/journal.pone.0000920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrow P, Lewicki H, Wei X, Horwitz MS, Peffer N, Meyers H, Nelson JA, Gairin JE, Hahn BH, Oldstone MB, Shaw GM. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat Med. 1997;3:205–211. doi: 10.1038/nm0297-205. [DOI] [PubMed] [Google Scholar]
- Brockman MA, Schneidewind A, Lahaie M, Schmidt A, Miura T, Desouza I, Ryvkin F, Derdeyn CA, Allen S, Hunter E, Mulenga J, Goepfert PA, Walker BD, Allen TM. Escape and compensation from early HLA-B57-mediated cytotoxic T-lymphocyte pressure on human immunodeficiency virus type 1 Gag alter capsid interactions with cyclophilin A. J Virol. 2007;81:12608–12618. doi: 10.1128/JVI.01369-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumme ZL, Brumme CJ, Carlson J, Streeck H, John M, Eichbaum Q, Block BL, Baker B, Kadie C, Markowitz M, Jessen H, Kelleher AD, Rosenberg E, Kaldor J, Yuki Y, Carrington M, Allen TM, Mallal S, Altfeld M, Heckerman D, Walker BD. Marked epitope- and allele-specific differences in rates of mutation in human immunodeficiency type 1 (HIV-1) Gag, Pol, and Nef cytotoxic T-lymphocyte epitopes in acute/early HIV-1 infection. J Virol. 2008a;82:9216–9227. doi: 10.1128/JVI.01041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumme ZL, Brumme CJ, Heckerman D, Korber BT, Daniels M, Carlson J, Kadie C, Bhattacharya T, Chui C, Szinger J, Mo T, Hogg RS, Montaner JS, Frahm N, Brander C, Walker BD, Harrigan PR. Evidence of differential HLA class I-mediated viral evolution in functional and accessory/regulatory genes of HIV-1. PLoS Pathog. 2007;3:e94. doi: 10.1371/journal.ppat.0030094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumme ZL, Tao I, Szeto S, Brumme CJ, Carlson JM, Chan D, Kadie C, Frahm N, Brander C, Walker B, Heckerman D, Harrigan PR. Human leukocyte antigen-specific polymorphisms in HIV-1 Gag and their association with viral load in chronic untreated infection. AIDS. 2008b;22:1277–1286. doi: 10.1097/QAD.0b013e3283021a8c. [DOI] [PubMed] [Google Scholar]
- Carrington M, Nelson GW, Martin MP, Kissner T, Vlahov D, Goedert JJ, Kaslow R, Buchbinder S, Hoots K, O'Brien SJ. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science. 1999;283:1748–1752. doi: 10.1126/science.283.5408.1748. [DOI] [PubMed] [Google Scholar]
- Chopera DR, Woodman Z, Mlisana K, Mlotshwa M, Martin DP, Seoighe C, Treurnicht F, de Rosa DA, Hide W, Karim SA, Gray CM, Williamson C. Transmission of HIV-1 CTL Escape Variants Provides HLA-Mismatched Recipients with a Survival Advantage. PLoS Pathogens. 2008;4:e1000033. doi: 10.1371/journal.ppat.1000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford H, Prado JG, Leslie A, Hue S, Honeyborne I, Reddy S, van der Stok M, Mncube Z, Brander C, Rousseau C, Mullins JI, Kaslow R, Goepfert P, Allen S, Hunter E, Mulenga J, Kiepiela P, Walker BD, Goulder PJ. Compensatory mutation partially restores fitness and delays reversion of escape mutation within the immunodominant HLA-B*5703-restricted Gag epitope in chronic human immunodeficiency virus type 1 infection. J Virol. 2007;81:8346–8351. doi: 10.1128/JVI.00465-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport MP, Loh L, Petravic J, Kent SJ. Rates of HIV immune escape and reversion: implications for vaccination. Trends Microbiol. 2008;16:561–566. doi: 10.1016/j.tim.2008.09.001. [DOI] [PubMed] [Google Scholar]
- Duda A, Lee-Turner L, Fox J, Robinson N, Dustan S, Kaye S, Fryer H, Carrington M, McClure M, McLean AR, Fidler S, Weber J, Phillips RE, Frater AJ. HLA-associated clinical progression correlates with epitope reversion rates in early human immunodeficiency virus infection. J Virol. 2009;83:1228–1239. doi: 10.1128/JVI.01545-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards BH, Bansal A, Sabbaj S, Bakari J, Mulligan MJ, Goepfert PA. Magnitude of Functional CD8+ T-Cell Responses to the Gag Protein of Human Immunodeficiency Virus Type 1 Correlates Inversely with Viral Load in Plasma. J Virol. 2002;76:2298–2305. doi: 10.1128/jvi.76.5.2298-2305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeney ME, Tang Y, Roosevelt KA, Leslie AJ, McIntosh K, Karthas N, Walker BD, Goulder PJR. Immune Escape Precedes Breakthrough Human Immunodeficiency Virus Type 1 Viremia and Broadening of the Cytotoxic T-Lymphocyte Response in an HLA-B27-Positive Long-Term- Nonprogressing Child. J Virol. 2004;78:8927–8930. doi: 10.1128/JVI.78.16.8927-8930.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellay J, Shianna KV, Ge D, Colombo S, Ledergerber B, Weale M, Zhang K, Gumbs C, Castagna A, Cossarizza A, Cozzi-Lepri A, De Luca A, Easterbrook P, Francioli P, Mallal S, Martinez-Picado J, Miro JM, Obel N, Smith JP, Wyniger J, Descombes P, Antonarakis SE, Letvin NL, McMichael AJ, Haynes BF, Telenti A, Goldstein DB. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–947. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich TC, Dodds EJ, Yant LJ, Vojnov L, Rudersdorf R, Cullen C, Evans DT, Desrosiers RC, Mothe BR, Sidney J, Sette A, Kunstman K, Wolinsky S, Piatak M, Lifson J, Hughes AL, Wilson N, O'Connor DH, Watkins DI. Reversion of CTL escape-variant immunodeficiency viruses in vivo. Nat Med. 2004a;10:275–281. doi: 10.1038/nm998. [DOI] [PubMed] [Google Scholar]
- Friedrich TC, Frye CA, Yant LJ, O'Connor DH, Kriewaldt NA, Benson M, Vojnov L, Dodds EJ, Cullen C, Rudersdorf R, Hughes AL, Wilson N, Watkins DI. Extraepitopic compensatory substitutions partially restore fitness to simian immunodeficiency virus variants that escape from an immunodominant cytotoxic-T-lymphocyte response. J Virol. 2004b;78:2581–2585. doi: 10.1128/JVI.78.5.2581-2585.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich TC, McDermott AB, Reynolds MR, Piaskowski S, Fuenger S, de Souza IP, Rudersdorf R, Cullen C, Yant LJ, Vojnov L, Stephany J, Martin S, O'Connor DH, Wilson N, Watkins DI. Consequences of Cytotoxic T-Lymphocyte Escape: Common Escape Mutations in Simian Immunodeficiency Virus Are Poorly Recognized in Naive Hosts. J Virol. 2004c;78:10064–10073. doi: 10.1128/JVI.78.18.10064-10073.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geels MJ, Cornelissen M, Schuitemaker H, Anderson K, Kwa D, Maas J, Dekker JT, Baan E, Zorgdrager F, van den Burg R, van Beelen M, Lukashov VV, Fu TM, Paxton WA, van der Hoek L, Dubey SA, Shiver JW, Goudsmit J. Identification of sequential viral escape mutants associated with altered T-cell responses in a human immunodeficiency virus type 1-infected individual. J Virol. 2003;77:12430–12440. doi: 10.1128/JVI.77.23.12430-12440.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geels MJ, Jansen CA, Baan E, De Cuyper IM, van Schijndel GJ, Schuitemaker H, Goudsmit J, Pollakis G, Miedema F, Paxton WA, van Baarle D. CTL escape and increased viremia irrespective of HIV-specific CD4+ T-helper responses in two HIV-infected individuals. Virology. 2006;345:209–219. doi: 10.1016/j.virol.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Geldmacher C, Currier JR, Herrmann E, Haule A, Kuta E, McCutchan F, Njovu L, Geis S, Hoffmann O, Maboko L, Williamson C, Birx D, Meyerhans A, Cox J, Hoelscher M. CD8 T-cell recognition of multiple epitopes within specific Gag regions is associated with maintenance of a low steady-state viremia in human immunodeficiency virus type 1-seropositive patients. J Virol. 2007;81:2440–2448. doi: 10.1128/JVI.01847-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goepfert PA, Lumm W, Farmer P, Matthews P, Prendergast A, Carlson JM, Derdeyn CA, Tang J, Kaslow RA, Bansal A, Yusim K, Heckerman D, Mulenga J, Allen S, Goulder PJ, Hunter E. Transmission of HIV-1 Gag immune escape mutations is associated with reduced viral load in linked recipients. J Exp Med. 2008;205:1009–1017. doi: 10.1084/jem.20072457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goonetilleke N, Liu MK, Salazar-Gonzalez JF, Ferrari G, Giorgi E, Ganusov VV, Keele BF, Learn GH, Turnbull EL, Salazar MG, Weinhold KJ, Moore S, Letvin N, Haynes BF, Cohen MS, Hraber P, Bhattacharya T, Borrow P, Perelson AS, Hahn BH, Shaw GM, Korber BT, McMichael AJ. The first T cell response to transmitted/founder virus contributes to the control of acute viremia in HIV-1 infection. J Exp Med. 2009;206:1253–1272. doi: 10.1084/jem.20090365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Kaslow RA, Carrington M, Apple R, Park L, Munoz A, Saah AJ, Goedert JJ, Winkler C, O'Brien SJ, Rinaldo C, Detels R, Blattner W, Phair J, Erlich H, Mann DL. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat Med. 1996;2:405–411. doi: 10.1038/nm0496-405. [DOI] [PubMed] [Google Scholar]
- Kiepiela P, Ngumbela K, Thobakgale C, Ramduth D, Honeyborne I, Moodley E, Reddy S, de Pierres C, Mncube Z, Mkhwanazi N, Bishop K, van der Stok M, Nair K, Khan N, Crawford H, Payne R, Leslie A, Prado J, Prendergast A, Frater J, McCarthy N, Brander C, Learn GH, Nickle D, Rousseau C, Coovadia H, Mullins JI, Heckerman D, Walker BD, Goulder P. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat Med. 2007;13:46–53. doi: 10.1038/nm1520. [DOI] [PubMed] [Google Scholar]
- Klein MR, van der Burg SH, Hovenkamp E, Holwerda AM, Drijfhout JW, Melief CJ, Miedema F. Characterization of HLA-B57-restricted human immunodeficiency virus type 1 Gag- and RT-specific cytotoxic T lymphocyte responses. J Gen Virol. 1998;79( Pt 9):2191–2201. doi: 10.1099/0022-1317-79-9-2191. [DOI] [PubMed] [Google Scholar]
- Koenig S, Conley AJ, Brewah YA, Jones GM, Leath S, Boots LJ, Davey V, Pantaleo G, Demarest JF, Carter C, al, e Transfer of HIV-1-specific cytotoxic T lymphocytes to an AIDS patient leads to selection for mutant HIV variants and subsequent disease progression. Nat Med. 1995;1:330–336. doi: 10.1038/nm0495-330. [DOI] [PubMed] [Google Scholar]
- Leslie A, Kavanagh D, Honeyborne I, Pfafferott K, Edwards C, Pillay T, Hilton L, Thobakgale C, Ramduth D, Draenert R, Le Gall S, Luzzi G, Edwards A, Brander C, Sewell AK, Moore S, Mullins J, Moore C, Mallal S, Bhardwaj N, Yusim K, Phillips R, Klenerman P, Korber B, Kiepiela P, Walker B, Goulder P. Transmission and accumulation of CTL escape variants drive negative associations between HIV polymorphisms and HLA. J Exp Med. 2005;201:891–902. doi: 10.1084/jem.20041455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie AJ, Pfafferott KJ, Chetty P, Draenert R, Addo MM, Feeney M, Tang Y, Holmes EC, Allen T, Prado JG, Altfeld M, Brander C, Dixon C, Ramduth D, Jeena P, Thomas SA, St John A, Roach TA, Kupfer B, Luzzi G, Edwards A, Taylor G, Lyall H, Tudor-Williams G, Novelli V, Martinez-Picado J, Kiepiela P, Walker BD, Goulder PJ. HIV evolution: CTL escape mutation and reversion after transmission. Nat Med. 2004;10:282–289. doi: 10.1038/nm992. [DOI] [PubMed] [Google Scholar]
- Li B, Gladden AD, Altfeld M, Kaldor JM, Cooper DA, Kelleher AD, Allen TM. Rapid Reversion of Sequence Polymorphisms Dominates Early Human Immunodeficiency Virus Type 1 Evolution. J Virol. 2007;81:193–201. doi: 10.1128/JVI.01231-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, McNevin J, Zhao H, Tebit DM, Troyer RM, McSweyn M, Ghosh AK, Shriner D, Arts EJ, McElrath MJ, Mullins JI. Evolution of human immunodeficiency virus type 1 cytotoxic T-lymphocyte epitopes: fitness-balanced escape. J Virol. 2007;81:12179–12188. doi: 10.1128/JVI.01277-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Picado J, Prado JG, Fry EE, Pfafferott K, Leslie A, Chetty S, Thobakgale C, Honeyborne I, Crawford H, Matthews P, Pillay T, Rousseau C, Mullins JI, Brander C, Walker BD, Stuart DI, Kiepiela P, Goulder P. Fitness Cost of Escape Mutations in p24 Gag in Association with Control of Human Immunodeficiency Virus Type 1. J Virol. 2006;80:3617–3623. doi: 10.1128/JVI.80.7.3617-3623.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masemola A, Mashishi T, Khoury G, Mohube P, Mokgotho P, Vardas E, Colvin M, Zijenah L, Katzenstein D, Musonda R, Allen S, Kumwenda N, Taha T, Gray G, McIntyre J, Karim SA, Sheppard HW, Gray CM. Hierarchical targeting of subtype C human immunodeficiency virus type 1 proteins by CD8+ T cells: correlation with viral load. J Virol. 2004;78:3233–3243. doi: 10.1128/JVI.78.7.3233-3243.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews PC, Prendergast A, Leslie A, Crawford H, Payne R, Rousseau C, Rolland M, Honeyborne I, Carlson J, Kadie C, Brander C, Bishop K, Mlotshwa N, Mullins JI, Coovadia H, Ndung'u T, Walker BD, Heckerman D, Goulder PJ. Central role of reverting mutations in HLA associations with human immunodeficiency virus set point. J Virol. 2008;82:8548–8559. doi: 10.1128/JVI.00580-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migueles SA, Laborico AC, Imamichi H, Shupert WL, Royce C, McLaughlin M, Ehler L, Metcalf J, Liu S, Hallahan CW, Connors M. The differential ability of HLA B*5701+ long-term nonprogressors and progressors to restrict human immunodeficiency virus replication is not caused by loss of recognition of autologous viral gag sequences. J Virol. 2003;77:6889–6898. doi: 10.1128/JVI.77.12.6889-6898.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore CB, John M, James IR, Christiansen FT, Witt CS, Mallal SA. Evidence of HIV-1 Adaptation to HLA-Restricted Immune Responses at a Population Level. Science. 2002;296:1439–1443. doi: 10.1126/science.1069660. [DOI] [PubMed] [Google Scholar]
- Ndongala ML, Peretz Y, Boulet S, Doroudchi M, Yassine-Diab B, Boulassel MR, Rouleau D, Tremblay C, Leblanc R, Routy JP, Sekaly RP, Bernard NF. HIV Gag p24 specific responses secreting IFN-gamma and/or IL-2 in treatment-naive individuals in acute infection early disease (AIED) are associated with low viral load. Clin Immunol. 2009 doi: 10.1016/j.clim.2008.11.006. [DOI] [PubMed] [Google Scholar]
- Novitsky V, Gilbert P, Peter T, McLane MF, Gaolekwe S, Rybak N, Thior I, Ndung'u T, Marlink R, Lee TH, Essex M. Association between virus-specific T-cell responses and plasma viral load in HIV-1 subtype C infection. J Virol. 2003;77:882–890. doi: 10.1128/JVI.77.2.882-890.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitsky V, Lagakos S, Herzig M, Bonney C, Kebaabetswe L, Rossenkhan R, Nkwe D, Margolin L, Musonda R, Moyo S, Woldegabriel E, van Widenfelt E, Makhema J, Essex M. Evolution of proviral gp120 over the first year of HIV-1 subtype C infection. NIHMSID # 79286. Virology. 2009a;383:47–59. doi: 10.1016/j.virol.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitsky V, Smith UR, Gilbert P, McLane MF, Chigwedere P, Williamson C, Ndung'u T, Klein I, Chang SY, Peter T, Thior I, Foley BT, Gaolekwe S, Rybak N, Gaseitsiwe S, Vannberg F, Marlink R, Lee TH, Essex M. HIV-1 subtype C molecular phylogeny: consensus sequence for an AIDS vaccine design? J Virol. 2002;76:5435–5451. doi: 10.1128/JVI.76.11.5435-5451.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitsky V, Wang R, Essex M. Abstract 49, 17th International HIV Dynamics & Evolution. University of California, San Diego, Asilomar Conference Grounds - Monterey, CA: 2010a. Phylodynamics of HIV-1 Subtype C Gag in the Global Epidemic. [Google Scholar]
- Novitsky V, Wang R, Kebaabetswe L, Greenwald J, Rossenkhan R, Moyo S, Musonda R, Woldegabriel E, Lagakos S, Essex M. Better control of early viral replication is associated with slower rate of elicited antiviral antibodies in the detuned enzyme immunoassay during primary HIV-1C infection. J Acquir Immune Defic Syndr. 2009b;52:265–272. doi: 10.1097/QAI.0b013e3181ab6ef0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitsky V, Wang R, Lagakos S, Essex M. HIV-1 Subtype C Phylodynamics in the Global Epidemic. Viruses. 2010b;2:33–54. doi: 10.3390/v2010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitsky V, Wang R, Margolin L, Baca J, Kebaabetswe L, Rossenkhan R, Bonney C, Herzig M, Nkwe D, Moyo S, Musonda R, Woldegabriel E, van Widenfelt E, Makhema J, Lagakos S, Essex M. Timing constraints of in vivo gag mutations during primary HIV-1 subtype C infection. PLoS One. 2009c;4:e7727. doi: 10.1371/journal.pone.0007727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitsky V, Wang R, Margolin L, Baca J, Moyo S, Musonda R, Essex M. Dynamics and timing of in vivo mutations at Gag residue 242 during primary HIV-1 subtype C infection. Virology. 2010c;403:37–46. doi: 10.1016/j.virol.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitsky V, Woldegabriel E, Kebaabetswe L, Rossenkhan R, Mlotshwa B, Bonney C, Finucane M, Musonda R, Moyo S, Wester C, van Widenfelt E, Makhema J, Lagakos S, Essex M. Viral Load and CD4+ T Cell Dynamics in Primary HIV-1 Subtype C Infection. J Acquir Immune Defic Syndr. 2009d;50:65–76. doi: 10.1097/QAI.0b013e3181900141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitsky V, Woldegabriel E, Wester C, McDonald E, Rossenkhan R, Ketunuti M, Makhema J, Seage GR, Essex M., 3rd Identification of primary HIV-1C infection in Botswana. NIHMSID # 79283. AIDS Care. 2008;20:806–811. doi: 10.1080/09540120701694055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitsky VA, Gilbert PB, Shea K, McLane MF, Rybak N, Klein I, Thior I, Ndung'u T, Lee TH, Essex ME. Interactive association of proviral load and IFN-gamma-secreting T cell responses in HIV-1C infection. Virology. 2006;349:142–155. doi: 10.1016/j.virol.2006.02.006. [DOI] [PubMed] [Google Scholar]
- O'Brien SJ, Nelson GW. Human genes that limit AIDS. Nat Genet. 2004;36:565–574. doi: 10.1038/ng1369. [DOI] [PubMed] [Google Scholar]
- Oxenius A, Price DA, Trkola A, Edwards C, Gostick E, Zhang HT, Easterbrook PJ, Tun T, Johnson A, Waters A, Holmes EC, Phillips RE. Loss of viral control in early HIV-1 infection is temporally associated with sequential escape from CD8+ T cell responses and decrease in HIV-1-specific CD4+ and CD8+ T cell frequencies. J Infect Dis. 2004;190:713–721. doi: 10.1086/422760. [DOI] [PubMed] [Google Scholar]
- Peyerl FW, Barouch DH, Letvin NL. Structural constraints on viral escape from HIV- and SIV-specific cytotoxic T-lymphocytes. Viral Immunol. 2004a;17:144–151. doi: 10.1089/0882824041310658. [DOI] [PubMed] [Google Scholar]
- Peyerl FW, Bazick HS, Newberg MH, Barouch DH, Sodroski J, Letvin NL. Fitness Costs Limit Viral Escape from Cytotoxic T Lymphocytes at a Structurally Constrained Epitope. J Virol. 2004b;78:13901–13910. doi: 10.1128/JVI.78.24.13901-13910.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips RE, Rowland-Jones S, Nixon DF, Gotch FM, Edwards JP, Ogunlesi AO, Elvin JG, Rothbard JA, Bangham CR, Rizza CR, McMichael AJ. Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature. 1991;354:453–459. doi: 10.1038/354453a0. [DOI] [PubMed] [Google Scholar]
- Price DA, Goulder PJ, Klenerman P, Sewell AK, Easterbrook PJ, Troop M, Bangham CR, Phillips RE. Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc Natl Acad Sci U S A. 1997;94:1890–1895. doi: 10.1073/pnas.94.5.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramduth D, Chetty P, Mngquandaniso NC, Nene N, Harlow JD, Honeyborne I, Ntumba N, Gappoo S, Henry C, Jeena P, Addo MM, Altfeld M, Brander C, Day C, Coovadia H, Kiepiela P, Goulder P, Walker B. Differential immunogenicity of HIV-1 clade C proteins in eliciting CD8+ and CD4+ cell responses. J Infect Dis. 2005;192:1588–1596. doi: 10.1086/496894. [DOI] [PubMed] [Google Scholar]
- Ray SC. MargFreq. SCRoftware; 2008. http://sray.med.som.jhmi.edu/SCRoftware/MargFreq/ [Google Scholar]
- Rolland M, Heckerman D, Deng W, Rousseau CM, Coovadia H, Bishop K, Goulder PJ, Walker BD, Brander C, Mullins JI. Broad and Gag-biased HIV-1 epitope repertoires are associated with lower viral loads. PLoS ONE. 2008;3:e1424. doi: 10.1371/journal.pone.0001424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose PP, Korber BT. Detecting hypermutations in viral sequences with an emphasis on G --> A hypermutation. Bioinformatics. 2000;16:400–401. doi: 10.1093/bioinformatics/16.4.400. [DOI] [PubMed] [Google Scholar]
- Rousseau CM, Daniels MG, Carlson JM, Kadie C, Crawford H, Prendergast A, Matthews P, Payne R, Rolland M, Raugi DN, Maust BS, Learn GH, Nickle DC, Coovadia H, Ndung'u T, Frahm N, Brander C, Walker BD, Goulder PJ, Bhattacharya T, Heckerman DE, Korber BT, Mullins JI. HLA class I-driven evolution of human immunodeficiency virus type 1 subtype c proteome: immune escape and viral load. J Virol. 2008;82:6434–6446. doi: 10.1128/JVI.02455-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidewind A, Brockman MA, Yang R, Adam RI, Li B, Le Gall S, Rinaldo CR, Craggs SL, Allgaier RL, Power KA, Kuntzen T, Tung CS, LaBute MX, Mueller SM, Harrer T, McMichael AJ, Goulder PJ, Aiken C, Brander C, Kelleher AD, Allen TM. Escape from the dominant HLA-B27-restricted cytotoxic T-lymphocyte response in Gag is associated with a dramatic reduction in human immunodeficiency virus type 1 replication. J Virol. 2007;81:12382–12393. doi: 10.1128/JVI.01543-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serwanga J, Shafer LA, Pimego E, Auma B, Watera C, Rowland S, Yirrell D, Pala P, Grosskurth H, Whitworth J, Gotch F, Kaleebu P. Host HLA B*allele-associated multi-clade Gag T-cell recognition correlates with slow HIV-1 disease progression in antiretroviral therapy-naive Ugandans. PLoS ONE. 2009;4:e4188. doi: 10.1371/journal.pone.0004188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankarappa R, Margolick JB, Gange SJ, Rodrigo AG, Upchurch D, Farzadegan H, Gupta P, Rinaldo CR, Learn GH, He X, Huang XL, Mullins JI. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol. 1999;73:10489–10502. doi: 10.1128/jvi.73.12.10489-10502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga R, Lucchetti A, Galvan P, Sanchez S, Sanchez C, Hernandez A, Sanchez H, Frahm N, Linde CH, Hewitt HS, Hildebrand W, Altfeld M, Allen TM, Walker BD, Korber BT, Leitner T, Sanchez J, Brander C. Relative Dominance of Gag p24-Specific Cytotoxic T Lymphocytes Is Associated with Human Immunodeficiency Virus Control. J Virol. 2006;80:3122–3125. doi: 10.1128/JVI.80.6.3122-3125.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.