Abstract
Understanding in vivo regulation of photoautotrophic metabolism is important for identifying strategies to improve photosynthetic efficiency or re-route carbon fluxes to desirable end products. We have developed an approach to reconstruct comprehensive flux maps of photoautotrophic metabolism by computational analysis of dynamic isotope labeling measurements and have applied it to determine metabolic pathway fluxes in the cyanobacterium Synechocystis sp. PCC6803. Comparison to a theoretically predicted flux map revealed inefficiencies in photosynthesis due to oxidative pentose phosphate pathway and malic enzyme activity, despite negligible photorespiration. This approach has potential to fill important gaps in our understanding of how carbon and energy flows are systemically regulated in cyanobacteria, plants, and algae.
Keywords: metabolic flux analysis, cyanobacteria, isotopomer analysis, mass spectrometry, photoautotrophic metabolism, carbon fixation
1. INTRODUCTION
Photoautotrophic metabolism is the process by which plants, algae, and other photosynthetic organisms use light energy to fix carbon dioxide into complex organic molecules. This represents the primary source of all food on earth as well as raw materials for bio-based production of fuels and chemicals. To date, six different pathways of carbon fixation have been identified in nature (Berg et al., 2010), of which the Calvin-Benson-Bassham (CBB) cycle accounts for more than 99% of global primary biomass production (Overmann and Garcia-Pichel, 2006). Over one-third of this production is attributable to prokaryotic carbon fixation, mainly by marine cyanobacteria. Several groups have recently demonstrated the feasibility of engineering cyanobacteria to convert solar energy and atmospheric CO2 directly into biofuels (Atsumi et al., 2009; Dutta et al., 2005; Lindberg et al., 2010; Liu et al., 2011), thus providing a potential strategy to harness their vast photosynthetic capacity toward meeting global energy demands while transitioning to a carbon-neutral society. Despite these advances, the productivities achieved by cyanobacterial fermentations are currently too low for industrial feasibility (Sheehan, 2009), and few tools have been developed that specifically address the challenges of redirecting or enhancing metabolic flux in photosynthetic organisms. Furthermore, it has been estimated that less than 1% of the available solar energy flux is converted into chemical energy by photosynthetic processes (Overmann and Garcia-Pichel, 2006), and developing strategies to enhance the efficiency of photosynthetic carbon fixation is a key step toward solving food, energy, and environmental challenges of the future.
The ability to quantitatively map intracellular carbon fluxes using isotope tracers and metabolic flux analysis (MFA) is critical for identifying pathway bottlenecks and elucidating network regulation in biological systems, especially those that have been engineered to alter their native metabolic capacities (Sauer, 2006). Although 13C is the preferred isotope tracer for mapping central carbon metabolism in heterotrophic organisms, photoautotrophs assimilate carbon solely from CO2 and therefore produce a uniform steady-state 13C-labeling pattern that is insensitive to fluxes (Fig. 1). Thus, conventional steady-state 13C-MFA is incapable of quantifying autotrophic metabolic fluxes (Shastri and Morgan, 2007). As a result, prior 13C-MFA studies of plants (Schwender, 2008) and cyanobacteria (Yang et al., 2002) have been limited to heterotrophic (HT) or mixotrophic (MT) growth conditions, typically with sugar as the major carbon source. To overcome this limitation, we hypothesized that transient measurements of isotope incorporation following a step change from unlabeled to labeled CO2 could be used to map carbon fluxes under photoautotrophic (PA) growth conditions. This involves quantification of intracellular metabolic fluxes based upon computational analysis of dynamic isotope labeling trajectories, an approach that has been called isotopically nonstationary MFA (INST-MFA) (Wiechert and Noh, 2005). An underlying assumption of this approach is that metabolic fluxes and pool sizes remain constant throughout the labeling experiment and are not perturbed by the introduction of 13C tracer. If this assumption is valid, a single set of flux and pool size parameters (along with their associated uncertainties) can be estimated that is consistent with all transient isotopomer measurements obtained during the experiment. We have recently introduced computational routines that achieve more than 5000-fold speedup relative to previous INST-MFA algorithms (Young et al., 2008), which now makes this approach computationally feasible for autotrophic networks of realistic size. The aim of this contribution, therefore, was to apply these computational tools along with state-of-the-art mass spectrometry approaches to produce a comprehensive photoautotrophic flux map of a model photosynthetic organism.
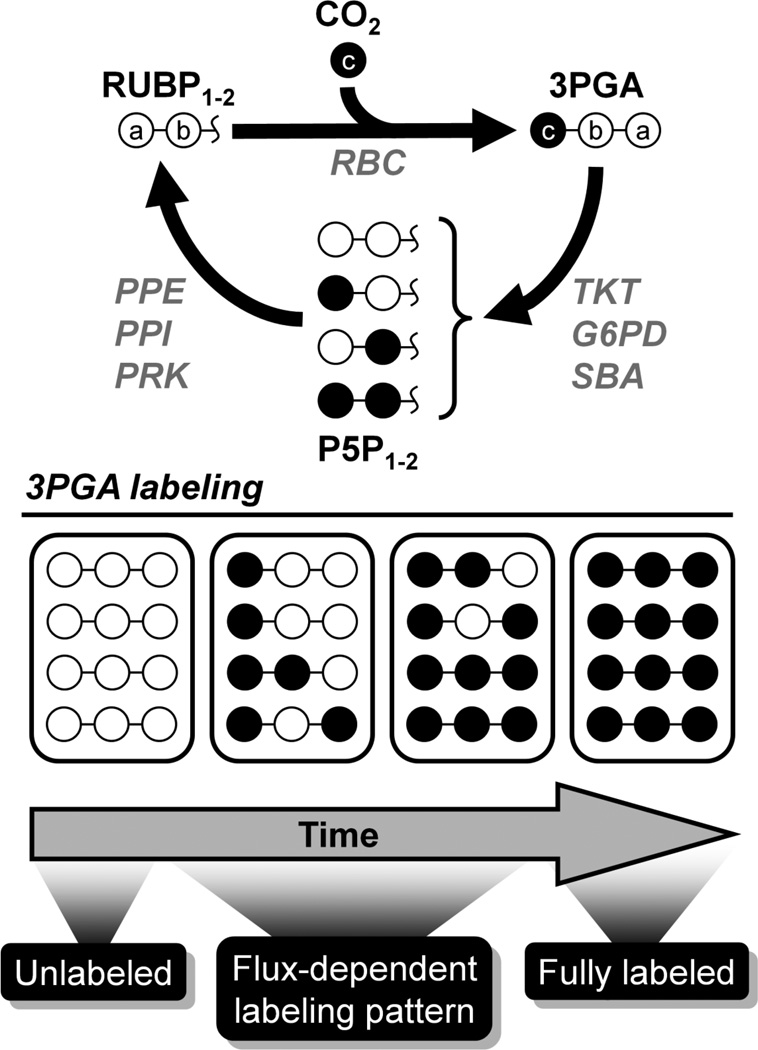
Figure 1. Simplified example of carbon labeling in a photoautotrophic system.
Following a switch from natural CO2 to 13CO2, intracellular metabolites become gradually labeled over time. 3-phosphoglycerate (3PGA) becomes 13C-labeled due to carbon fixation by RuBisCO (RBC), resulting in labeling of pentose-5-phosphate (P5P) intermediates of the Calvin-Benson-Bassham (CBB) cycle. Once steady-state labeling is achieved, all metabolites are uniformly 13C-labeled irrespective of fluxes and intracellular pool sizes. Labeling patterns observed during the isotopically transient period, however, can be computationally analyzed to determine fluxes.
2. MATERIALS AND METHODS
2.1. Strain and cultivation conditions
Synechocystis sp. PCC 6803 was obtained from ATCC (ATCC # 27150) and cultured in a 1.25 L bioreactor (Bioflo 3000, New Brunswick Scientific, NJ) on BG-11 medium buffered with 10 mM HEPES. Air was supplied at 1–1.5 L/min, and the reactor was stirred at 350 rpm. Temperature was controlled at 30°C and pH was maintained in the range 8.0–8.5 by automatic addition of 2 N H2SO4. Nine 23 W cool white fluorescent lights (Sylvania, MA) provided an average surface light flux of 400 µE/m2/s. Only 5 lights were turned on in the initial 12 hours of growth in order to prevent photoinhibition. The outlet gas CO2 concentration was monitored using a LI-820 CO2 gas analyzer with 14 cm optical bench (LI-COR Biosciences, NE).
2.2. Sample removal and metabolite extraction
The sideport of the standard reactor vessel was sealed with a thick autoclavable membrane. A two-way luer stopcock (Popper and Sons, NY) with an extremely small holdup volume (< 0.5 mL) was fitted with a gauge 16 needle and inserted into the reactor via the membrane. A short piece of tubing was attached to the end of the stopcock, which allowed the rapid fitting of a syringe. This enabled repeated withdrawal of 20 mL samples into 60 mL syringes (BD Biosciences, MD) containing 40 mL of quenching solution (60% methanol in water) precooled to −40°C or lower. Each sample took less than 3 seconds to withdraw. The contents of each syringe were rapidly transferred to 50 mL centrifuge tubes that were kept in a −20°C bath. Using this procedure, samples of 20 mL size could be withdrawn reliably every 20 seconds. Each quenched sample was centrifuged for 10 minutes at 8000g and −20°C. The supernatant was discarded and the cell pellet was extracted with 500 µL pure methanol, followed by two extractions with 500 µL of a 50/50 methanol-water solution. Each extraction was carried out for 30 minutes at a temperature below −20°C, and the extracts were pooled together and stored at −20°C.
2.3. Carbon labeling experiment
The labeling experiment was initialized when the cell density reached an OD730 of 0.6. A sample corresponding to time zero (unlabeled) was withdrawn from the sideport using a syringe. The aeration was then stopped and gas inlets and outlets to the reactor were rapidly clamped to prevent unlabeled CO2 from entering the system. A 20 mL aliquot of 50% NaH13CO3 (Sigma, 98% isotopic purity) was injected swiftly into the system using a syringe, and 20 mL samples were withdrawn and rapidly quenched at time points of 20, 40, 60, 90, 130, 250, 480, and 610 seconds. Intracellular metabolites were obtained from these samples by solvent extraction of cell pellets. Additional samples were withdrawn without quenching and rapidly filtered through a syringe filter. These supernatants were used to identify metabolites secreted into the extracellular media. Metabolite samples were analyzed using a combination of GC-MS and LC-MS/MS to determine labeling and concentration of metabolites in both cell extracts and culture supernatants. Samples were also withdrawn for measurement of cell dry weight.
2.4. GC-MS measurement of metabolite labeling and concentration
The GC-MS method was adapted from Roessner et al. (2000) and was performed using an Agilent 6890N/5975B quadrupole GC-MS system. All samples were dried under vacuum at room temperature in a Centrivap (Labconco Corporation, MO). Ribitol was added as an internal standard to all the samples prior to the drying step to achieve a final concentration of 0.328 µM in the derivatized solution. The samples were subsequently methoximated with 50 µL of methoxyamine hydrochloride in pyridine (2mg/mL) for 90 minutes at 40°C. Next, 50µL BSTFA + 10% TMCS (Pierce Biotechnology, IL) was added to the reaction mixture and incubated at 40°C for 30 minutes. The derivatized samples remained for 6–12 hours at room temperature in a dessicator, to ensure complete reaction. All samples were run on the GC-MS within 30 hours of derivatization. An HP-5MS column of 180 µm diameter, 30 m length and 0.20 µm film thickness (Agilent Technologies, CA) was used with helium as the carrier gas. The total column flow was set to 1 mL/min, and 1 µL injections were made using a 230°C inlet temperature in splitless mode and a purge flow of 100 mL/min set to activate 1 min after injection. The column temperature was held at 70°C for 3 minutes, increased to 305°C at 3°C/min, and held at 305°C for 2 minutes. Spectra were recorded in the range of 50–600 m/z. Metabolites were identified by comparison to the retention time and characteristic ions of pure standards. Linear calibration curves were prepared with purchased standards and used to quantify the metabolites in the samples. All chemicals were purchased from Sigma Aldrich, USA.
2.5. LC-MS/MS measurement of metabolite labeling and concentration
An ion-pairing LC-MS/MS method was adapted from Oldiges et al. (2004) and was performed on a Shimadzu SCL10 LC system coupled to a linear ion-trap triple quadrupole MS/MS system (4000 Q TRAP, AB Sciex Instruments) at the Proteomics and Mass Spectrometry Facility, Donald Danforth Center, St. Louis, MO. A Synergi Hydro-RP column (150mm × 2 mm, Phenomenex Inc, CA) was used with gradient elution. The eluents used were 10 mM tributylamine + 15 mM acetic acid (A) and methanol (B). In order to dissolve tributylamine completely in water, the tributylamine and acetic acid were first mixed together in a dry flask before the requisite amount of ultrapure water was added. The final pH of eluent A was found to be 4.5–4.6. All chemicals were HPLC or LC-MS grade and purchased from Sigma Aldrich, USA. The eluents were filtered before use. An injection volume of 10 µL was used, and the column flow and temperature were constant at 0.3 mL/min and 25°C, respectively. The gradient profile was as follows: 0% B (0 min), 8% B (8 min), 22% B (18 min), 40% B (28 min), 60% B (32 min), 90% B (34 min), 90% B (37 min), 0% B (39 min), 0% B (49 min). The acquisition of labeling and concentration data was performed using negative electrospray ionization in the multiple reaction monitoring (MRM) mode. The final parameters used for isotopomer measurements are listed in Supplementary Table I. All data acquisition and analysis was performed on the Analyst 4.1.2 software (AB/MDS Sciex) supplied with the instrument.
2.6. Isotopically nonstationary metabolic flux analysis (INST-MFA)
We applied our previously developed INST-MFA approach (Young et al., 2008) to estimate intracellular metabolic fluxes and metabolite pool sizes based on the measured isotope labeling dynamics of intracellular metabolites. This approach relies upon an elementary metabolite unit (EMU) decomposition of the underlying isotopomer network to efficiently simulate the effects of varying fluxes on the labeling state of measurable metabolites (Antoniewicz et al., 2007). Metabolic fluxes and pool sizes were estimated by minimizing the lack-of-fit between experimentally measured and computationally simulated mass isotopomer distributions (MIDs) using least-squares regression. All isotopic measurements used for flux determination are listed in Table I. The flux and pool size parameters of the isotopomer network model were iteratively adjusted using a Levenberg-Marquardt algorithm until optimal agreement with experimental data was obtained (Madsen et al., 2004). All results were subjected to a chi-square statistical test to assess goodness-of-fit, and accurate 95% confidence intervals were computed for all estimated parameters by evaluating the sensitivity of the sum-of-squared residuals to parameter variations (Antoniewicz et al., 2006).
TABLE I. Isotope labeling measurements used for metabolic flux determination.
(a) LC-MS/MS ions and (b) GC-MS ions used to assess metabolite labeling. Standard errors of measurement (SEM) were determined based on the lack of agreement between measured and theoretically computed mass isotopomer distributions obtained from unlabeled cell extracts.
| (a) LC-MS/MS ions | |||||
|---|---|---|---|---|---|
| Metabolite | Mass | Carbons | Composition | SEM | Reference |
| 3PGA | 185 | 1-2-3 | C3H6O7P | 0.5 | (Luo et al., 2007) |
| DHAP | 169 | 1-2-3 | C3H6O6P | 1.3 | Ibid. |
| F6P | 259 | 1-2-3-4-5-6 | C6H12O9P | 1.7 | Ibid. |
| G6P | 259 | 1-2-3-4-5-6 | C6H12O9P | 1.4 | Ibid. |
| GAP | 169 | 1-2-3 | C3H6O6P | 1.3 | Ibid. |
| PEP | 167 | 1-2-3 | C3H4O6P | 0.5 | Ibid. |
| R5P | 229 | 1-2-3-4-5 | C5H10O8P | 1.5 | Ibid. |
| RUBP | 309 | 1-2-3-4-5 | C5H11O11P2 | 0.7 | Ibid. |
| S7P | 289 | 1-2-3-4-5-6-7 | C7H14O10P | 2.8 | Ibid. |
| RU5P | 229 | 1-2-3-4-5 | C5H10O8P | 1.3 | Ibid. |
| (b) GC-MS ions | |||||
| Metabolite | Mass | Carbons | Composition | SEM | Reference |
| 3PGA | 357 | 2-3 | C11H30O5PSi3 | 1.0 | (Kitson et al., 1996) |
| 3PGA | 459 | 1-2-3 | C14H36O7PSi4 | 1.0 | Ibid. |
| CIT | 273 | 1-2-3-4-5 | C11H21O4Si2 | 1.0 | (Tisza, 1994) |
| CIT | 363 | 1-2-3-4-5 | C14H31O5Si3 | 1.0 | Ibid. |
| CIT | 375 | 1-2-3-4-5-6 | C14H27O6Si3 | 1.6 | (Huege et al., 2007) |
| CIT | 465 | 1-2-3-4-5-6 | C17H37O7Si4 | 1.1 | Ibid. |
| FUM | 245 | 1-2-3-4 | C9H17O4Si2 | 1.0 | Ibid. |
| GA | 292 | 1-2 | C11H28O3Si3 | 1.0 | (Petersson, 1970) |
| GA | 307 | 1-2-3 | C11H27O4Si3 | 1.3 | (Huege et al., 2007) |
| MAL | 233 | 2-3-4 | C9H21O3Si2 | 1.0 | (Petersson, 1972) |
| SUC | 247 | 1-2-3-4 | C9H19O4Si2 | 1.0 | (Huege et al., 2007) |
A list of the reactions included in the isotopomer network model is provided in Supplementary Table II. (Refer to Supplementary Materials for a detailed description of the model formulation.) In total, the model was capable of fitting 921 independent mass isotopomer measurements using 224 adjustable parameters (34 free fluxes + 29 pool sizes + 161 measurement scaling factors). The measurement scaling factors were required to properly normalize the MIDs of each MS fragment ion (Mollney et al., 1999). The model required 559 ordinary differential equations (ODEs) to simulate the change in all measured MIDs over time. In addition, 35,217 ODEs were needed to compute sensitivities of these MIDs with respect to all adjustable parameters. Overall, the model consisted of 35,776 ODEs and required approximately 3 seconds to simulate using the best-fit parameters. To ensure that the final solution was the global optimum, flux estimation was repeated at least 50 times starting from random initial values. Supplementary Fig. 1 shows MID trajectories for all experimentally measured GC-MS and LC-MS/MS ions along with best-fit model simulations. The fit was statistically accepted based on a chi-square test of the sum-of-squared residuals (SSR), which was assessed at the 95% confidence level with 697 degrees of freedom (SSR = 684.7 versus the expected range [625.7, 772.1]). A full listing of the optimal parameter estimates can be found in Supplementary Tables III, IV and V.
3. RESULTS AND DISCUSSION
3.1. Carbon labeling experiment
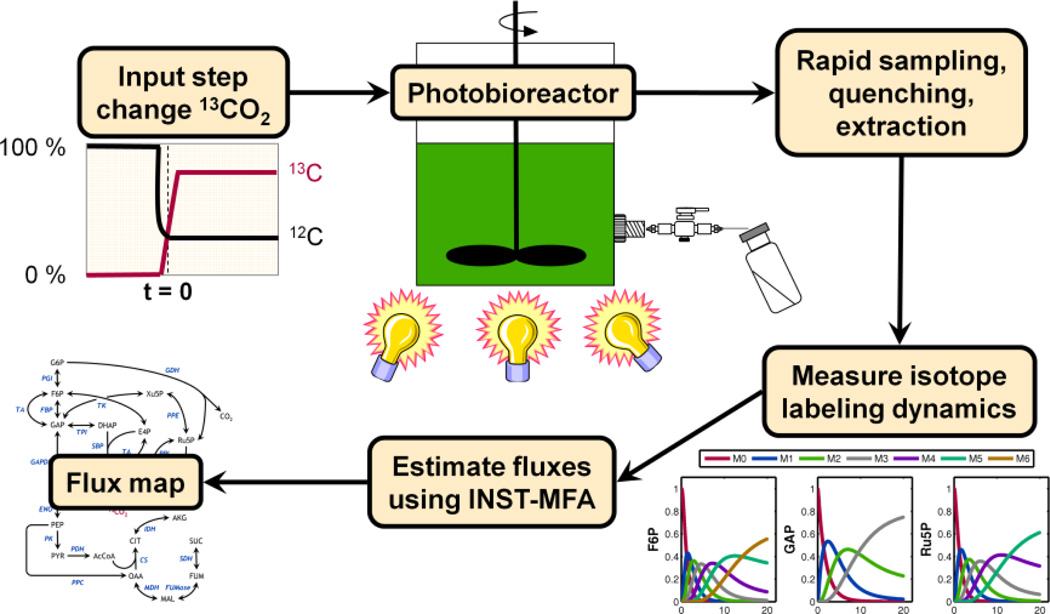
To assess the capability of 13C INST-MFA to quantify PA fluxes, we applied this approach to the cyanobacterium Synechocystis sp. PCC 6803 growing in a controlled photobioreactor environment with bicarbonate provided as the sole carbon source. (Bicarbonate equilibrates with dissolved CO2 in the culture medium and therefore provides a convenient route to administer CO2 to liquid cultures.) Following the introduction of 13C-labeled bicarbonate to the bioreactor, a time-series of metabolite samples was obtained by rapid quenching and extraction of cyanobacterial cells. Dynamic changes in isotope labeling patterns of central carbon metabolites were quantified using both gas chromatography-mass spectrometry (GC-MS) and liquid chromatography-tandem mass spectrometry (LC-MS/MS), followed by computational analysis of these trajectories to estimate metabolic pathway fluxes using INST-MFA (Fig. 2). This is the first time, to our knowledge, that a comprehensive flux analysis has been performed based on isotope labeling data obtained from a fully autotrophic system.
Figure 2. Overview of INST-MFA procedure.
Following introduction of 13C-labeled bicarbonate, a series of metabolite samples were obtained by rapid quenching and extraction of cyanobacterial cells. Dynamic changes in isotope labeling patterns were assessed using GC-MS and LC-MS/MS, followed by computational analysis of these trajectories to estimate metabolic pathway fluxes.
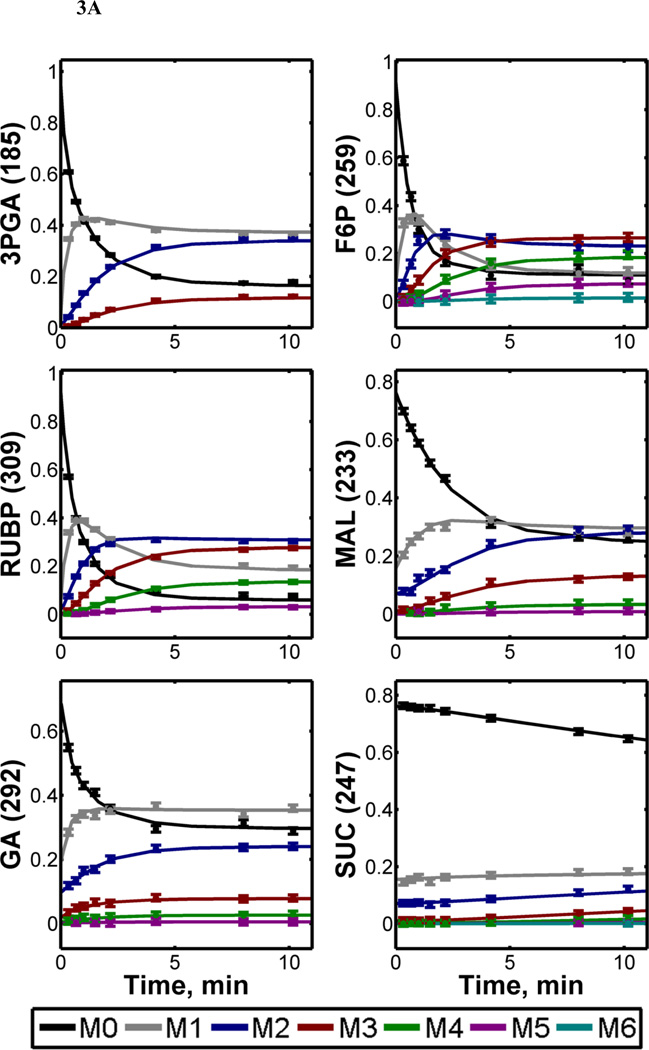
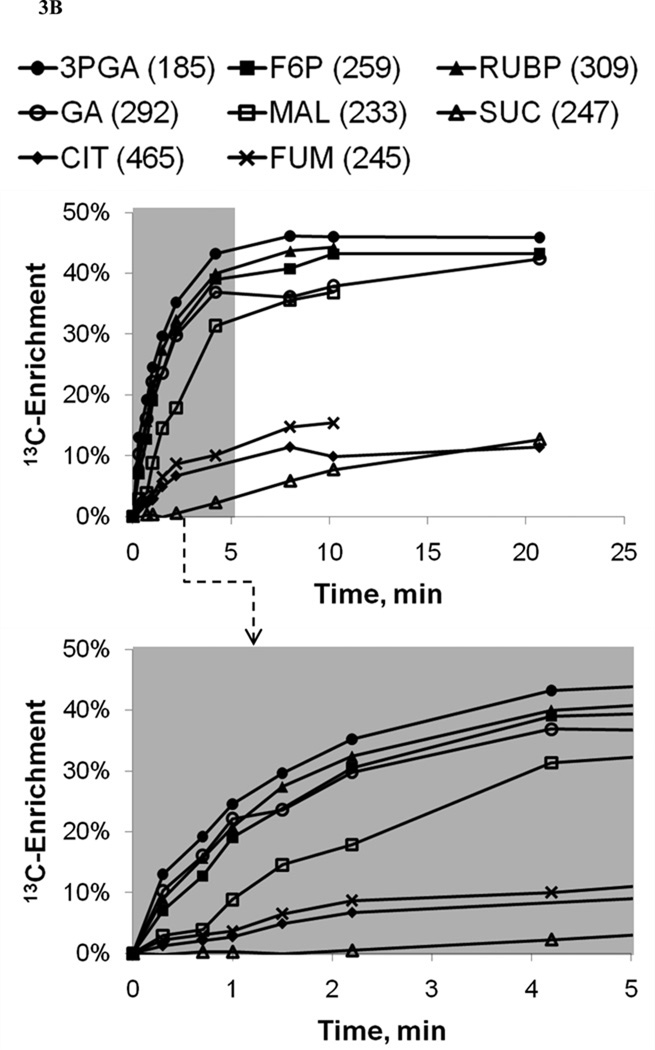
The labeling dynamics of 21 fragment ions derived from 15 different central metabolites were measured over a 20-minute time window following tracer administration. As shown in Fig. 3A, the relative abundances of unlabeled (M0) mass isotopomers dropped rapidly at the outset of the labeling period due to the emergence of 13C-enriched mass isotopomers (M1, M2, etc.). Initially, singly-labeled (M1) mass isotopomers accumulated due to incorporation of single 13C atoms into previously unlabeled metabolites. These M1 mass isotopomers were gradually supplanted by M2, M3, and higher mass isotopomers as further 13C atoms were incorporated, resulting in transient overshoots in M1 abundance. Note, however, that the average 13C-enrichment of each metabolite increased monotonically over time despite the rise and fall of M1 mass isotopomers (Fig. 3B). Steady-state labeling was typically obtained in less than 10 minutes, with the notable exception of tricarboxylic acid (TCA) pathway intermediates (e.g., succinate, fumarate, citrate) that were more slowly labeled.
Figure 3. 13C-Labeling trajectories of selected central carbon metabolites.
(A) Experimentally measured mass isotopomer abundances (data points) and INST-MFA model fits (solid lines). Error bars represent standard measurement errors. Raw mass isotopomer data are shown without correction for natural isotope abundance. (B) Average 13C-enrichments of selected ion fragments. Mass isotopomer distributions were corrected for natural isotope abundance using the method of Fernandez et al. (1996), and average 13C enrichment was calculated using the formula , where N is the number of carbon atoms in the metabolite and Mi is the fractional abundance of the ith mass isotopomer. The top axis shows the full labeling trajectory and the bottom axis shows an enlarged view of the highlighted region from 0–5 min. Ions shown are for 3-phosphoglycerate (3PGA), fructose-6-phosphate (F6P), ribulose-1,5-bisphosphate (RUBP), malate (MAL), glycerate (GA), succinate (SUC), citrate (CIT) and fumarate (FUM). Nominal masses of M0 mass isotopomers are shown in parentheses.
3.2. Secretion of organic metabolites
In addition to measurements of intracellular metabolite labeling, extracellular samples were collected for analysis of secreted metabolites. Glycolate was the only secreted metabolite that could be identified in the culture medium. The extracellular concentration of glycolate at the end of the 32-hour batch growth period was estimated to be less than 5µM. Based on this measurement and the final biomass concentration of 204 mg/L, we estimated that the carbon lost to glycolate was approximately 1000-fold less than the amount of carbon incorporated into biomass. Therefore, we concluded that secretion of organic metabolites by Synechocystis was essentially negligible under photoautotrophic conditions, and that all fixed carbon was either incorporated into biomass or released as CO2 due to oxidative processes. As a result, we were able to apply INST-MFA based solely on the intracellular metabolite labeling data while normalizing all flux values to the net rate of CO2 conversion into biomass (Fig. 4). This rate was estimated from the growth rate and carbon content of the culture, which was 0.09 h−1 × 41 mmol-C/g-DW = 3.7 mmol-CO2/g-DW/h.
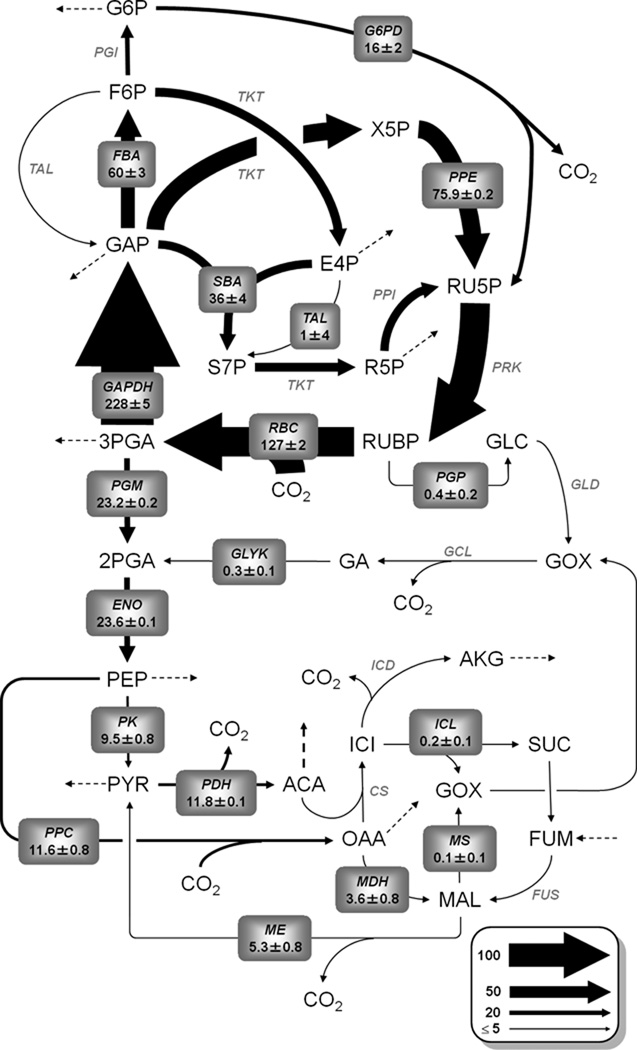
Figure 4. Synechocystis flux map determined under photoautotrophic growth conditions.
Net fluxes are shown normalized to a net CO2 uptake rate of 100. Values are represented as M ± SE, where M is the median of the 95% flux confidence interval and SE is the estimated standard error of M calculated as (UB95 – LB95)/3.92. (UB95 and LB95 are the upper and lower bounds of the 95% confidence interval, respectively.) Arrow thickness is scaled proportional to net flux. Dotted arrows indicate fluxes to biomass formation.
3.3. Comparison to optimal flux map
The resulting flux map was compared to a previously published linear programming (LP) solution that predicts the theoretical optimum flux profile that simultaneously maximizes biomass production and minimizes light utilization based on a specified CO2 uptake rate (Shastri and Morgan, 2005). Table II compares each reaction that exhibited a statistically significant deviation between MFA-determined and LP-predicted flux values. Overall, the INST-MFA results indicate that PA-grown Synechocystis cells exhibited suboptimal carbon efficiency, with significant loss of fixed carbon via the oxidative pentose phosphate (OPP) pathway. LP predicts that 111 moles of CO2 are fixed by RuBisCO (RBC) for every 100 moles of carbon converted to biomass, the difference being due to flux through pyruvate dehydrogenase (PDH), isocitrate dehydrogenase (ICD) and other CO2-releasing reactions that are necessary to generate biosynthetic precursors. INST-MFA finds that, in actuality, RBC fixed 127 ± 2 moles of CO2 for every 100 C-moles of biomass produced and released 16 ± 2 moles of fixed CO2 via the OPP pathway. A significant malic enzyme (ME) flux of 5.3 ± 0.8 moles per 100 C-moles biomass was also identified by INST-MFA, providing an alternate route of pyruvate formation (phosphoenolpyruvate → oxaloacetate → malate → pyruvate) that bypasses the ATP-generating pyruvate kinase (PK) reaction and closely resembles the pathway of carbon fixation found in C4 plants (Hatch, 1987). However, this cycle does not directly contribute to carbon loss since it involves concerted uptake and release of CO2 by PEP carboxylase (PPC) and ME enzymes acting in tandem, resulting in no net effect on the carbon balance.
TABLE II. Reactions that exhibit significant deviation between LP-predicted and MFA-determined fluxes.
All fluxes are relative to a net CO2 uptake rate of 100. The median, lower bound (LB), and upper bound (UB) of the 95% confidence interval is shown for each MFA-determined flux. Grouped fluxes are correlated due to pathway dependencies, and therefore deviations within each group can be attributed to the same underlying feature. Increased OPP pathway flux is responsible for deviations in Group I, while increased ME flux is responsible for deviations in Group II. The three rightmost columns show the net effect of these deviations on the balance of NADPH, NADH and ATP cofactors.
|
13C MFA |
Cofactor balance |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Flux | Reaction |
LP Predicted |
Median Value |
95% LB |
95% UB |
NADPH | NADH | ATP | |
| Group I | |||||||||
| PGI | F6P → G6P | 3 | 19 | 15 | 24 | ||||
| G6PD/6PGD | G6P → RU5P + CO2 + 2 NADPH | 0 | 16 | 12 | 21 | +32 | |||
| FBA/FBP | DHAP + GAP → F6P | 41 | 60 | 53 | 66 | ||||
| TPI | GAP → DHAP | 78 | 95 | 90 | 99 | ||||
| PGK/GAPDH | 3 PGA + ATP + NADPH → GAP | 196 | 228 | 219 | 237 | −32 | −32 | ||
| PRK | RU5P + ATP → RUBP | 111 | 127 | 123 | 132 | −16 | |||
| RBC | RUBP + CO2 → 2 3PGA | 111 | 127 | 123 | 132 | ||||
| Group II | |||||||||
| PK | PEP → PYR + ATP | 14.7 | 9.5 | 7.9 | 11.1 | −5.2 | |||
| MDH | OAA + NADH → MAL | −1.7 | 3.6 | 1.9 | 5.2 | −5.3 | |||
| ME | MAL → PYR + CO2 + NADPH | 0 | 5.3 | 3.7 | 6.9 | +5.3 | |||
| PPC | PEP + CO2 → OAA | 6.3 | 11.6 | 9.9 | 13.2 | ||||
| Net | +5.3 | −5.3 | −53 | ||||||
3.4. Oxidative pentose phosphate pathway flux
In most organisms, NADP-linked dehydrogenases such as those involved in the OPP pathway serve a predominantly anabolic role by providing reductant and intermediates for biosynthetic processes (Pelroy et al., 1972). However, cyanobacteria use NADPH as a key source of electrons for oxidative phosphorylation in the dark, which is necessary to produce ATP from NADP-linked oxidation of stored glycogen or exogenous glucose (Biggins, 1969). The coupling of NADPH production to oxidative phosphorylation represents a unique respiratory pathway found in few other organisms and is believed to compensate for the lack of an intact cyanobacterial TCA cycle (Pelroy et al., 1972). A previous MFA study reported that over 90% of glucose consumed by Synechocystis was oxidized through the OPP pathway under HT conditions (Yang et al., 2002). Our finding that residual levels of OPP pathway flux persist even under PA conditions was unexpected in view of the substantial amounts of NADPH produced by light-driven electron transport. This could indicate incomplete suppression of OPP flux by regulatory circuits that control the transition from dark to light metabolism or, alternatively, the presence of kinetic bottlenecks in the CBB cycle that are circumvented by OPP pathway activation under high-CO2 conditions.
Knockout mutants of the cyanobacterium species Synechococcus sp. PCC 7942 lacking either of the two OPP pathway enzymes glucose-6-phosphate dehydrogenase (G6PD) or 6-phosphogluconate (6PGD) exhibit no significant decrease in growth rate under PA conditions, indicating that OPP pathway activity is dispensable in the light (Broedel and Wolf, 1990; Scanlan et al., 1995). However, prior studies have reported no reduction in enzymatic activity of G6PD or 6PGD in Synechocystis cell extracts collected under PA conditions relative to MT or HT conditions (Knowles and Plaxton, 2003) or after shifting cells from light to dark (Singh and Sherman, 2005), suggesting that these key OPP pathway enzymes are highly expressed under PA conditions and are primarily regulated at the posttranslational level. Indeed, it is well known that G6PD controls carbon entry into the OPP pathway and is competitively inhibited by NADPH and allosterically deactivated by reduced thioredoxin in the light (Buchanan, 1991; Grossman and McGowan, 1975; Pelroy et al., 1976a). However, this response is apparently incomplete under the conditions of our study, which leads to a small but significant CO2 loss amounting to ~13% of total fixed carbon and a concomitant reduction in photosynthetic efficiency.
3.5. Malic enzyme flux
Our results are in agreement with a previous 13C-MFA study that also reported substantial ME flux in Synechocystis under HT and MT growth conditions (Yang et al., 2002). Interestingly, a previously reported Synechocystis knockout mutant lacking NADP-dependent malic enzyme grew about 15 times slower than its wild-type parent when grown in continuous light, which provides indirect evidence that ME is expressed and active under the conditions of our study and that ME flux provides a functional benefit to PA growth (Bricker et al., 2004). Because supplementation of pyruvate in the culture medium was able to completely restore PA growth of this knockout strain, the authors concluded that ME flux is required to supply pyruvate for biosynthetic processes due to down-regulation of PK activity in the light. This hypothesis might also explain the presence of ME flux in our system, which effectively bypasses the PK reaction step. Alternatively, Yang et al.(2002) have suggested that this pathway could be involved in the capture and concentration of intracellular CO2 by cyanobacteria via a process analogous to that observed in C4 plants.
3.6. Cofactor balance
In addition to the direct impact on intracellular carbon flows, the presence of nonzero OPP and ME pathway fluxes also alters the balance of reduced NADPH and NADH cofactors and increases the ATP demand relative to the LP prediction (Table II). Both OPP and ME pathways are coupled to production of the reduced cofactor NADPH in Synechocystis (Bricker et al., 2004; Knowles and Plaxton, 2003), although NADPH production in the OPP pathway is completely offset by its consumption in the CBB cycle and therefore has no net effect on NADPH availability. OPP pathway activity indirectly causes a substantial increase in ATP demand due to increased CBB cycling through the ATP-consuming reactions phosphoglycerate kinase (PGK) and phosphoribulokinase (PRK) reactions. This ATP drain is further compounded by ME pathway activity, which converts NADH to NADPH at the expense of ATP. The increased demands for NADPH and ATP are ultimately met by photosynthetic electron transport. Based on the reaction network and assumptions used by Shastri and Morgan (2005) to compute their LP solution, the minimum photosynthetic flux increases from 1297 to 1419 photons per C-mole biomass formed due to the presence of nonzero OPP and ME pathway activities, which represents a 9% loss of energetic efficiency.
3.7. Photorespiration
In contrast to the significant carbon and energetic losses attributable to OPP and ME pathway activities, the INST-MFA results indicate negligible photorespiration. The oxygenase side reaction of RuBisCO exhibited a net flux of 0.4 ± 0.2 moles of 2-phosphoglycolate (2PG) produced per 100 C-moles biomass formed, which was subsequently converted to 3-phosphoglycerate (3PGA) via photorespiration (Fig. 4). This meager flux confirms the presence of an efficient carbon concentrating mechanism in cyanobacteria, which functions to elevate the local concentration of CO2 in close proximity to RuBisCO and thereby compensate for its low affinity for CO2 (Kaplan and Reinhold, 1999). In addition, the high concentration of bicarbonate supplied during the carbon labeling experiment would have further suppressed photorespiration under the conditions of our study (Huege et al., 2011). Eisenhut et al. (2008) have shown that blocking photorespiration in Synechocystis leads to a high-CO2-requiring phenotype, and that these mutants accumulated intracellular glycolate even in the presence of 5% CO2. Our work demonstrates that flux through photorespiration is quantitatively very small compared to the total rate of CO2 fixation under high-CO2 conditions. This result is not necessarily inconsistent with the findings of Eisenhut et al., however, since complete removal of photorespiratory pathways would be expected to cause gradual accumulation of glycolate, even in the presence of low oxygenase activity.
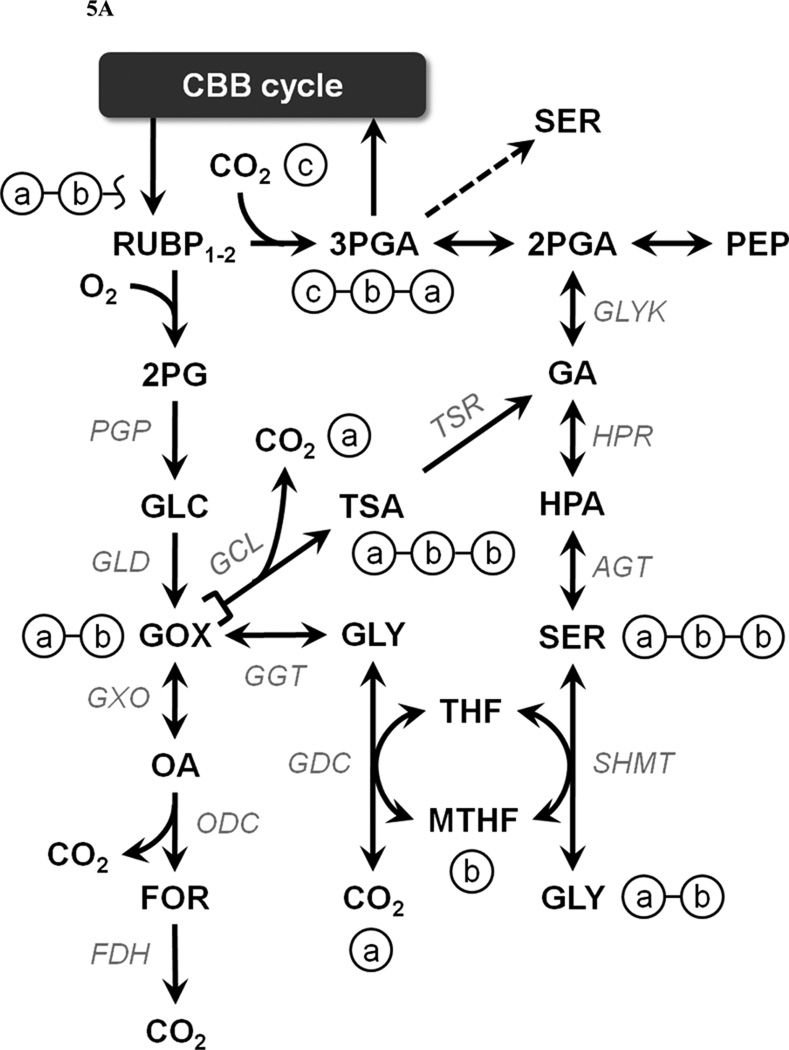
Photorespiratory 2PG metabolism comprises three alternative pathways in Synechocystis: (i) a plant-like C2 cycle, (ii) a bacterial glycerate pathway, and (iii) complete decarboxylation of glyoxylate via oxalate (Eisenhut et al., 2008; Hagemann et al., 2010). Flux carried by either of the first two pathways is readily detectable in the measured labeling patterns of glycerate, 3PGA, and PEP (Fig. 5A). However, the C2 cycle and glycerate pathways cannot be distinguished based on these measurements alone, since both produce identical carbon rearrangements as flux proceeds through the non-overlapping reaction steps connecting glyoxylate to glycerate. Although we analyzed isotope labeling in serine and glycine (which are intermediates in the C2 cycle but not the glycerate pathway), these measurements were not precise enough to be included in the flux analysis due to low signal intensities. Therefore, pathways (i) and (ii) were lumped into a single photorespiratory pathway that represents carbon recycling from 2PG to 3PGA and had a net flux of 0.3 ± 0.1 moles per 100 C-moles biomass under the conditions of our study (Fig. 4). The decarboxylation pathway flux could not be accurately quantified using the available isotopomer measurements and was therefore neglected. However, we found that oxalate achieved a maximum average 13C enrichment of approximately 10% even after 20+ minutes of labeling (not shown). Therefore, we concluded that decarboxylation of glyoxylate was likely insignificant under the conditions of our study.
Figure 5. Isotope labeling in photorepiratory pathways.
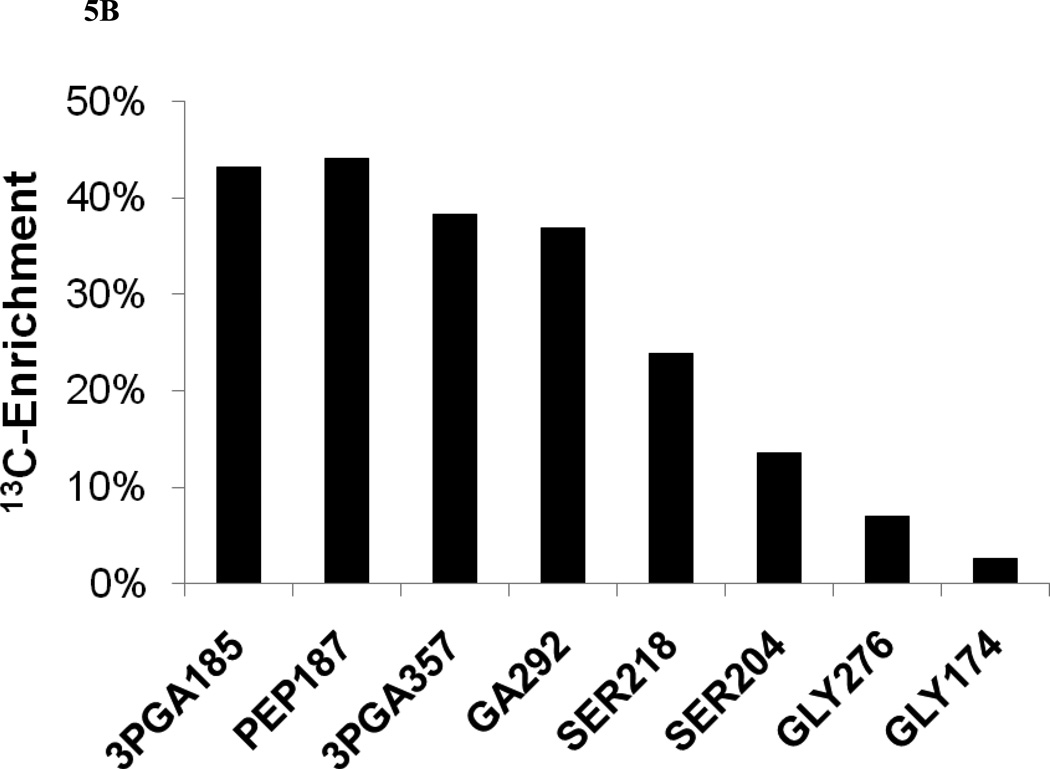
(A) Schematic diagram of the complete 2PG metabolism of Synechocystis showing the fate of the first two carbon atoms of ribulose-1,5-bisphosphate. The three photorespiratory pathways diverge at glyoxylate into the plant-like C2 cycle (via glycine and serine), the bacterial-like glycerate pathway (via tartronic semialdehyde), and the decarboxylation branch (via oxalate). The first two pathways re-converge at glycerate. (B) 13C enrichments of photorespiratory pathway intermediates after 4.2 minutes of 13CO2 labeling. Average enrichments were calculated as described in the caption to Figure 3B. The compositions of selected 3-phosphoglycerate (3PGA), phosphoenolpyruvate (PEP), and glycerate (GA) fragment ions are listed in Table I. The carbon atoms of serine (SER) and glycine (GLY) contained in the selected fragment ions were taken from Huege et al. (2007): SER218 (C1-C2), SER204 (C2-C3), GLY276 (C1-C2), GLY174 (C2).
Based on their large-scale in silico reconstruction of the Synechocystis metabolic network, Knoop et al. (2010) predicted that a photorespiratory flux of approximately 4% of the total RuBisCO activity is necessary to achieve optimal PA growth. However, this prediction was based on the assumption that Synechocystis does not possess a pathway to synthesize serine directly from 3PGA and must therefore use glyoxylate as a precursor for glycine, serine, and cysteine biosynthesis. A corollary of this assumption is that all serine carbon would be derived from glycine, with the amino acid backbone contributed directly from glycine and the hydroxyl carbon indirectly from glycine (C2) via methylene tetrahydrofolate (Fig. 5A). Although the serine and glycine labeling measurements were not considered precise enough to be included in the flux analysis, the 13C enrichment of serine was nonetheless substantially higher than glycine at all sample time points, indicating that serine was synthesized directly from 3PGA rather than by photorespiration via glycine. Fig. 5B depicts the labeling of several CBB and C2 cycle intermediates after approximately 4 minutes of 13CO2 labeling. At this time point, both serine fragment ions exhibited considerably higher 13C enrichments than was found in glycine fragment ions. This is consistent with a recent paper by Huege et al. (2011) and would appear to contradict the assumption of Knoop et al.
3.8 Glyoxylate shunt
The INST-MFA flux map indicates absence of glyoxylate shunt activity under PA conditions. Although Yang et al. (2002) reported substantial flux through the glyoxylate shunt under HT and MT conditions, Knoop et al. (2010) could find no biochemical or genetic evidence to support the inclusion of the pathway in their large-scale reconstruction of the Synechocystis metabolic network. Shastri and Morgan (2005) included the glyoxylate shunt in their earlier flux balance model, but the resulting LP solution predicted that the pathway should be completely inactivated to achieve optimal PA growth. We also included the glyoxylate shunt in our isotopomer model to facilitate comparisons to the earlier work of Yang et al. (2002) and Shastri and Morgan (2005). However, our results confirm that this pathway is either altogether missing from Synechocystis or strongly repressed under PA conditions.
3.9 Pool size estimation
Most prior INST-MFA studies have incorporated direct metabolite pool size measurements into the SSR objective function (Noh et al., 2007; Schaub et al., 2008). This was considered necessary to maximize flux identifiability, due to the fact that the pool sizes appear as additional adjustable parameters in the isotopomer balance equations and must be fitted during the flux estimation procedure (Noh and Wiechert, 2006). However, our results indicate that precise flux determination can be achieved using INST-MFA even in the absence of direct pool size measurements. This provides a distinct advantage, since intracellular concentration measurements are easily corrupted by metabolite losses that can occur during sample quenching and extraction. These same losses, however, would not be expected to impact the labeling measurements so long as sufficient material remains for mass isotopomer analysis. We hypothesize that the reason we can achieve good flux identifiability without measuring pool sizes is because the feasible labeling patterns that can occur at convergent nodes in the metabolic network are mostly determined by the atom rearrangements and relative fluxes of the tributary pathways that feed into those nodes. Therefore, by simultaneously fitting mass isotopomer measurements derived from metabolites both upstream and downstream of such nodes, one can adequately constrain the resulting flux estimates to narrow confidence regions. While the pool size parameters serve to match the labeling dynamics of adjacent pools and account for any lags in the network, they cannot drastically alter the isotopomer patterns that emerge within the network once an acceptable flux distribution has been identified.
On the other hand, it is clear from our results that most pool size parameters remain unidentifiable without including direct pool size measurements in the fitting routine (Supplementary Table V). This is to be expected, since many of the pools are close to quasi-equilibrium with respect to surrounding metabolites in the network, and therefore only an upper bound can be estimated for these pool sizes. Also, if there are several intermediate pools separating measurable upstream and downstream metabolites, a delay in the dynamic response of the downstream metabolites could be attributable to any one of the intervening pools. This redundancy will inevitably lead to poor identifiability of pool sizes. As a result of these issues, only a handful of pool sizes could be estimated with both finite upper and nonzero lower bounds (citrate, fructose-1,6-bisphosphate, fumarate, malate, PEP, and succinate). While the estimated values for these pool sizes are within the correct order of magnitude based on prior measurements (Shastri and Morgan, 2007), it is important to note that if a metabolite is in rapid exchange with other metabolites not represented in the isotopomer model, the estimated values will reflect the size of the combined pool. This complicates direct comparisons to experimental concentration measurements.
3.10 Metabolic channeling
Although the steady-state isotope labeling measurements are not directly useful for flux analysis, they do provide information on subcellular compartmentation and metabolic channeling. After correcting for natural isotope abundances, PEP and 3PGA exhibited the highest steady-state 13C-enrichments of 47% and 46%, respectively. Huege et al. (2011) reported significantly higher PEP enrichments compared to 3PGA throughout their labeling period, which they attributed to metabolic channeling of carboxysomal 3PGA into PEP. We also noted slightly higher PEP enrichments in all samples collected after 1 minute of 13CO2 labeling, but the differences were typically less than 1 mol% and were not substantial enough to warrant special treatment in our isotopomer model. On the other hand, the CBB cycle intermediates fructose-6-phosphate (F6P), glyceraldehyde-3-phosphate (GAP), dihydroxyacetone phosphate (DHAP), and ribose-5-phosphate (R5P) were significantly less labeled than their downstream products sedoheptulose-7-phosphate (S7P), ribulose-5-phosphate (RU5P), and ribulose-1,5-bisphosphate (RUBP) (Fig. 6). This implies the presence of metabolic channeling that can arise due to the formation of multienzyme complexes that transfer intermediates directly between catalytic sites without diffusion into the bulk phase of the cell. We estimated the contribution of metabolic channeling by including “dilution parameters” for F6P, GAP, DHAP, and R5P in our isotopomer model. The values obtained from INST-MFA suggest that approximately 10% of the total intracellular concentrations of these intermediates were metabolically inactive and therefore remained unlabeled at steady state. Although the organization of CBB cycle enzymes into multienzyme complexes has been previously reported to occur in plants and green algae (Gontero et al., 1988; Winkel, 2004), our results along with those of Huege et al. appear to provide the first evidence of metabolic channeling in the CBB cycle of cyanobacteria.
Figure 6. Average steady-state 13C enrichment of CBB cycle intermediates.
Average enrichments were calculated as described in the caption to Figure 3B. The labeling of all CBB intermediates achieved steady state at times greater than 8 minutes after tracer administration. Averages of at least two steady-state time points are shown. Error bars indicate standard errors of the mean values.
4. CONCLUSION
Overall, we were able to precisely quantify the rates of all CBB cycle reactions, as well as several “wasteful” side reactions including G6PD, ME, and photorespiratory fluxes which contribute to suboptimal PA growth of Synechocystis. Although earlier studies have applied dynamic isotope labeling experiments to obtain important information about in vivo kinetics of CBB cycle reactions (Pelroy et al., 1976b) and photorespiration (Huege et al., 2011) in cyanobacteria, these approaches were not capable of integrating numerous isotopic measurements into a comprehensive flux map that encompasses all major pathways of central carbon metabolism. Furthermore, because these studies did not apply the INST-MFA approach, it was impossible to estimate fluxes without simultaneously measuring the intracellular pool sizes of the labeled intermediates. We have shown that application of INST-MFA in combination with GC-MS and LC-MS/MS analysis of isotope labeling trajectories can effectively quantify photoautotrophic fluxes in cultured cyanobacteria to a high degree of precision. This is possible even in the absence of direct measurements of intracellular pool sizes, which are treated as additional adjustable parameters in the isotopomer model.
Our study provides an example of how flux analysis can be used to identify pathways responsible for reduced cyanobacterial productivity, by pinpointing reactions that contribute to carbon and energetic losses. By determining how these losses are altered by genetic manipulations or by modulation of environmental conditions, we expect that INST-MFA can be a powerful approach for identifying metabolic engineering strategies to improve photosynthetic efficiency or re-route metabolic fluxes to desirable end products in these hosts. Furthermore, INST-MFA is not limited to photosynthetic bacteria, but can be extended to algae, plants, and other microorganisms that grow exclusively on C1 compounds. As a result, the integration of quantitative INST-MFA flux maps with gene expression and proteomic datasets is expected to fill a critical gap in our ability to assess network-wide regulation of carbon and energy flows in photosynthetic organisms.
HIGHLIGHTS.
Isotopically nonstationary metabolic flux analysis (INST-MFA) was applied to map intracellular fluxes in a photoautotroph (Synechocystis sp. PCC6803) for the first time.
Inefficiencies in photosynthesis were discovered due to oxidative pentose phosphate pathway and malic enzyme activity.
Recycling of 2-phosphoglycolate to 3-phosphoglycerate was negligible under the high-CO2 conditions of our study.
This approach can be applied to map fluxes in cyanobacteria, algae, and plants.
Supplementary Material
ACKNOWLEDGEMENT
This work was supported by NSF BES 0348458 (to JAM), NIH R01 DK075850 (to GS) and NIH F32 DK072856 (to JDY).
ABBREVIATIONS
- 2PG
2-phosphoglycolate
- 2PGA
2-phosphoglycerate
- 3PGA
3-phosphoglycerate
- 6PGD
6-phosphogluconate dehydrogenase
- ACA
acetyl-CoA
- AGT
alanine/glyoxylate aminotransferase
- AKG
alpha-ketoglutarate
- CIT
citrate
- CS
citrate synthase
- DHAP
dihydroxyacetone phosphate
- E4P
erythrose 4-phosphate
- EC2
transketolase-bound 2-carbon fragment
- EC3
transaldolase-bound 3-carbon fragment
- EMU
elementary metabolite unit
- ENO
enolase
- F6P
fructose-6-phosphate
- FBA
fructose bisphosphate aldolase
- FBP
fructose 1,6-bisphosphate
- FDH
formate dehydrogenase
- FOR
formate
- FUM
fumarate
- FUS
fumarase
- G6P
glucose 6-phosphate
- G6PD
glucose-6-phosphate dehydrogenase
- GA
glycerate
- GAP
glyceraldehyde 3-phosphate
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GCL
glyoxylate carboligase
- GC-MS
gas chromatography-mass spectrometry
- GDC
glycine decarboxylase
- GGT
glycine/glutamate aminotransferase
- GLC
glycolate
- GLD
glycolate dehydrogenase
- GLY
glycine
- GLYC
glycogen
- GLYK
glycerate kinase
- GOX
glyoxylate
- GXO
glyoxylate oxidase
- HPA
hydroxypyruvate
- HPR
hydroxypyruvate reductase
- ICD
isocitrate dehydrogenase
- ICI
isocitrate
- ICL
isocitrate lyase
- INST-MFA
isotopically nonstationary metabolic flux analysis
- LB
lower bound
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- LP
linear programming
- MAL
malate
- MDH
malate dehydrogenase
- MFA
metabolic flux analysis
- ME
malic enzyme
- MID
mass isotopomer distribution
- MRM
multiple reaction monitoring
- MS
malate synthase
- MTHF
5,10-methylenetetrahydrofolate
- OAA
oxaloacetate
- ODC
oxalate decarboxylase
- ODE
ordinary differential equation
- OPP
oxidative pentose phosphate
- P5P
pentose-5-phosphate
- PDH
pyruvate dehydrogenase
- PEP
phosphoenolpyruvate
- PGI
phosphoglucose isomerase
- PGK
phosphoglycerate kinase
- PGM
phosphoglycerate mutase
- PGP
phosphoglycolate phosphatase
- PK
pyruvate kinase
- PPC
phosphoenolpyruvate carboxylase
- PPE
phosphopentose epimerase
- PPI
phosphopentose isomerase
- PRK
phosphoribulokinase
- PYR
pyruvate
- R5P
ribose-5-phosphate
- RBC
ribulose-1,5-bisphosphate carboxylase oxygenase (RuBisCO)
- RU5P
ribulose-5-phosphate
- RUBP
ribulose-1,5-bisphosphate
- S7P
sedoheptulose-7-phosphate
- SBA
sedoheptulose-1,7-bisphosphate aldolase
- SBP
sedoheptulose-1,7-bisphosphate
- SER
serine
- SHMT
serine hydroxymethyltransferase
- SSR
sum-of-squared residuals
- SUC
succinate
- TAL
transaldolase
- TCA
tricarboxylic acid
- THF
tetrahydrofolate
- TKT
transketolase
- TSA
tartronic semialdehyde
- TSR
tartronic semialdehyde reductase
- UB
upper bound
- X5P
xylulose 5-phosphate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
JDY designed the experiment and flux calculation method, analyzed the data and wrote the manuscript. AAS designed and performed the experiment, analyzed the data and wrote the manuscript. GS designed the flux calculation method and wrote the manuscript. JAM designed the experiment, analyzed the data and wrote the manuscript.
REFERENCES
- Antoniewicz MR, Kelleher JK, Stephanopoulos G. Determination of confidence intervals of metabolic fluxes estimated from stable isotope measurements. Metab Eng. 2006;8:324–337. doi: 10.1016/j.ymben.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Antoniewicz MR, Kelleher JK, Stephanopoulos G. Elementary metabolite units (EMU): a novel framework for modeling isotopic distributions. Metab Eng. 2007;9:68–86. doi: 10.1016/j.ymben.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsumi S, Higashide W, Liao JC. Direct photosynthetic recycling of carbon dioxide to isobutyraldehyde. Nat Biotechnol. 2009;27:1177–1180. doi: 10.1038/nbt.1586. [DOI] [PubMed] [Google Scholar]
- Berg IA, Kockelkorn D, Ramos-Vera WH, Say RF, Zarzycki J, Hugler M, Alber BE, Fuchs G. Autotrophic carbon fixation in archaea. Nat Rev Microbiol. 2010;8:447–460. doi: 10.1038/nrmicro2365. [DOI] [PubMed] [Google Scholar]
- Biggins J. Respiration in blue-green algae. Journal of Bacteriology. 1969;99:570–575. doi: 10.1128/jb.99.2.570-575.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricker TM, Zhang S, Laborde SM, Mayer PR, 3rd, Frankel LK, Moroney JV. The malic enzyme is required for optimal photoautotrophic growth of Synechocystis sp. strain PCC 6803 under continuous light but not under a diurnal light regimen. Journal of Bacteriology. 2004;186:8144–8148. doi: 10.1128/JB.186.23.8144-8148.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broedel SE, Jr, Wolf RE., Jr Genetic tagging, cloning, and DNA sequence of the Synechococcus sp. strain PCC 7942 gene (gnd) encoding 6-phosphogluconate dehydrogenase. Journal of Bacteriology. 1990;172:4023–4031. doi: 10.1128/jb.172.7.4023-4031.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan BB. Regulation of CO2 assimilation in oxygenic photosynthesis: the ferredoxin/thioredoxin system. Perspective on its discovery, present status, and future development. Archives of Biochemistry and Biophysics. 1991;288:1–9. doi: 10.1016/0003-9861(91)90157-e. [DOI] [PubMed] [Google Scholar]
- Dutta D, De D, Chaudhuri S, Bhattacharya SK. Hydrogen production by Cyanobacteria. Microbial Cell Factories. 2005;4 doi: 10.1186/1475-2859-4-36. -. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhut M, Ruth W, Haimovich M, Bauwe H, Kaplan A, Hagemann M. The photorespiratory glycolate metabolism is essential for cyanobacteria and might have been conveyed endosymbiontically to plants. Proc Natl Acad Sci U S A. 2008;105:17199–17204. doi: 10.1073/pnas.0807043105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez CA, Des Rosiers C, Previs SF, David F, Brunengraber H. Correction of 13C mass isotopomer distributions for natural stable isotope abundance. J Mass Spectrom. 1996;31:255–262. doi: 10.1002/(SICI)1096-9888(199603)31:3<255::AID-JMS290>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Gontero B, Cardenas ML, Ricard J. A functional five-enzyme complex of chloroplasts involved in the Calvin cycle. Eur J Biochem. 1988;173:437–443. doi: 10.1111/j.1432-1033.1988.tb14018.x. [DOI] [PubMed] [Google Scholar]
- Grossman A, McGowan RE. Regulation of glucose 6-phosphate dehydrogenase in blue-green algae. Plant Physiology. 1975;55:658–662. doi: 10.1104/pp.55.4.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann M, Eisenhut M, Hackenberg C, Bauwe H. Pathway and importance of photorespiratory 2-phosphoglycolate metabolism in cyanobacteria. Adv Exp Med Biol. 2010;675:91–108. doi: 10.1007/978-1-4419-1528-3_6. [DOI] [PubMed] [Google Scholar]
- Hatch MD. C4 photosynthesis: a unique elend of modified biochemistry, anatomy and ultrastructure. Biochimica et Biophysica Acta (BBA) - Reviews on Bioenergetics. 1987;895:81–106. [Google Scholar]
- Huege J, Goetze J, Schwarz D, Bauwe H, Hagemann M, Kopka J. Modulation of the major paths of carbon in photorespiratory mutants of synechocystis. PLoS One. 2011;6:e16278. doi: 10.1371/journal.pone.0016278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huege J, Sulpice R, Gibon Y, Lisec J, Koehl K, Kopka J. GC-EI-TOF-MS analysis of in vivo carbon-partitioning into soluble metabolite pools of higher plants by monitoring isotope dilution after 13CO2 labelling. Phytochemistry. 2007;68:2258–2272. doi: 10.1016/j.phytochem.2007.03.026. [DOI] [PubMed] [Google Scholar]
- Kaplan A, Reinhold L. CO2 Concentrating Mechanisms in Photosynthetic Microorganisms. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:539–570. doi: 10.1146/annurev.arplant.50.1.539. [DOI] [PubMed] [Google Scholar]
- Kitson FG, Larsen BS, McEwen CN. Gas chromatography and mass spectrometry : a practical guide. San Diego: Academic Press; 1996. [Google Scholar]
- Knoop H, Zilliges Y, Lockau W, Steuer R. The metabolic network of Synechocystis sp. PCC 6803: systemic properties of autotrophic growth. Plant Physiology. 2010;154:410–422. doi: 10.1104/pp.110.157198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles VL, Plaxton WC. From genome to enzyme: analysis of key glycolytic and oxidative pentose-phosphate pathway enzymes in the cyanobacterium Synechocystis sp. PCC 6803. Plant and Cell Physiology. 2003;44:758–763. doi: 10.1093/pcp/pcg086. [DOI] [PubMed] [Google Scholar]
- Lindberg P, Park S, Melis A. Engineering a platform for photosynthetic isoprene production in cyanobacteria, using Synechocystis as the model organism. Metab Eng. 2010;12:70–79. doi: 10.1016/j.ymben.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Liu X, Sheng J, Curtiss R., Iii Fatty acid production in genetically modified cyanobacteria. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1103014108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo B, Groenke K, Takors R, Wandrey C, Oldiges M. Simultaneous determination of multiple intracellular metabolites in glycolysis, pentose phosphate pathway and tricarboxylic acid cycle by liquid chromatography-mass spectrometry. J Chromatogr A. 2007;1147:153–164. doi: 10.1016/j.chroma.2007.02.034. [DOI] [PubMed] [Google Scholar]
- Madsen K, Nielsen HB, Tingleff O. Methods for non-linear least squares problems. 2nd edition. Lecture notes, Technical University of Denmark; 2004. [Google Scholar]
- Mollney M, Wiechert W, Kownatzki D, de Graaf AA. Bidirectional reaction steps in metabolic networks: IV. Optimal design of isotopomer labeling experiments. Biotechnol Bioeng. 1999;66:86–103. doi: 10.1002/(sici)1097-0290(1999)66:2<86::aid-bit2>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Noh K, Gronke K, Luo B, Takors R, Oldiges M, Wiechert W. Metabolic flux analysis at ultra short time scale: isotopically non-stationary 13C labeling experiments. Journal of Biotechnology. 2007;129:249–267. doi: 10.1016/j.jbiotec.2006.11.015. [DOI] [PubMed] [Google Scholar]
- Noh K, Wiechert W. Experimental design principles for isotopically instationary 13C labeling experiments. Biotechnology and Bioengineering. 2006;94:234–251. doi: 10.1002/bit.20803. [DOI] [PubMed] [Google Scholar]
- Oldiges M, Kunze M, Degenring D, Sprenger GA, Takors R. Stimulation, monitoring, and analysis of pathway dynamics by metabolic profiling in the aromatic amino acid pathway. Biotechnology Progress. 2004;20:1623–1633. doi: 10.1021/bp0498746. [DOI] [PubMed] [Google Scholar]
- Overmann J, Garcia-Pichel F. The Phototrophic Way of Life. In: Dworkin M, Falkow S, editors. The prokaryotes: A handbook on the biology of bacteria. vol. 2. New York ; London: Springer; 2006. pp. 32–85. [Google Scholar]
- Pelroy RA, Kirk MR, Bassham JA. Photosystem II regulation of macromolecule synthesis in the blue-green alga Aphanocapsa 6714. Journal of Bacteriology. 1976a;128:623–632. doi: 10.1128/jb.128.2.623-632.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelroy RA, Levine GA, Bassham JA. Kinetics of light-dark CO2 fixation and glucose assimilation by Aphanocapsa 6714. Journal of Bacteriology. 1976b;128:633–643. doi: 10.1128/jb.128.2.633-643.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelroy RA, Rippka R, Stanier RY. Metabolism of glucose by unicellular blue-green algae. Arch Mikrobiol. 1972;87:303–322. doi: 10.1007/BF00409131. [DOI] [PubMed] [Google Scholar]
- Petersson G. Mass spectrometry of aldonic and deoxyaldonic acids as trimethylsilyl derivatives. Tetrahedron. 1970;26:16. [Google Scholar]
- Petersson G. Mass spectrometry of hydroxy dicarboxylic acids as trimethylsilyl derivatives. Rearrangement fragmentations. Organic Mass Spectrometry. 1972;6:12. [Google Scholar]
- Roessner U, Wagner C, Kopka J, Trethewey RN, Willmitzer L. Simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. Plant Journal. 2000;23:131–142. doi: 10.1046/j.1365-313x.2000.00774.x. [DOI] [PubMed] [Google Scholar]
- Sauer U. Metabolic networks in motion: 13C-based flux analysis. Mol Syst Biol. 2006;2:62. doi: 10.1038/msb4100109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlan DJ, Sundaram S, Newman J, Mann NH, Carr NG. Characterization of a zwf mutant of Synechococcus sp. strain PCC 7942. Journal of Bacteriology. 1995;177:2550–2553. doi: 10.1128/jb.177.9.2550-2553.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub J, Mauch K, Reuss M. Metabolic flux analysis in Escherichia coli by integrating isotopic dynamic and isotopic stationary 13C labeling data. Biotechnol Bioeng. 2008;99:1170–1185. doi: 10.1002/bit.21675. [DOI] [PubMed] [Google Scholar]
- Schwender J. Metabolic flux analysis as a tool in metabolic engineering of plants. Curr Opin Biotechnol. 2008;19:131–137. doi: 10.1016/j.copbio.2008.02.006. [DOI] [PubMed] [Google Scholar]
- Shastri AA, Morgan JA. Flux balance analysis of photoautotrophic metabolism. Biotechnology Progress. 2005;21:1617–1626. doi: 10.1021/bp050246d. [DOI] [PubMed] [Google Scholar]
- Shastri AA, Morgan JA. A transient isotopic labeling methodology for 13C metabolic flux analysis of photoautotrophic microorganisms. Phytochemistry. 2007;68:2302–2312. doi: 10.1016/j.phytochem.2007.03.042. [DOI] [PubMed] [Google Scholar]
- Sheehan J. Engineering direct conversion of CO2 to biofuel. Nature Biotechnology. 2009;27:1128–1129. doi: 10.1038/nbt1209-1128. [DOI] [PubMed] [Google Scholar]
- Singh AK, Sherman LA. Pleiotropic effect of a histidine kinase on carbohydrate metabolism in Synechocystis sp. strain PCC 6803 and its requirement for heterotrophic growth. Journal of Bacteriology. 2005;187:2368–2376. doi: 10.1128/JB.187.7.2368-2376.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisza SaM-P, Ibolya GC-MS quantitation of isocitric acid in the presence of a large excess of citric acid. Journal of High Resolution Chromatography. 1994;17:4. [Google Scholar]
- Wiechert W, Noh K. From stationary to instationary metabolic flux analysis. Adv Biochem Eng Biotechnol. 2005;92:145–172. doi: 10.1007/b98921. [DOI] [PubMed] [Google Scholar]
- Winkel BS. Metabolic channeling in plants. Annu Rev Plant Biol. 2004;55:85–107. doi: 10.1146/annurev.arplant.55.031903.141714. [DOI] [PubMed] [Google Scholar]
- Yang C, Hua Q, Shimizu K. Metabolic flux analysis in Synechocystis using isotope distribution from 13C-labeled glucose. Metab Eng. 2002;4:202–216. doi: 10.1006/mben.2002.0226. [DOI] [PubMed] [Google Scholar]
- Young JD, Walther JL, Antoniewicz MR, Yoo H, Stephanopoulos G. An elementary metabolite unit (EMU) based method of isotopically nonstationary flux analysis. Biotechnol Bioeng. 2008;99:686–699. doi: 10.1002/bit.21632. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.