Abstract
Infections with highly pathogenic H5N1 avian (HPAI) and 1918 pandemic H1N1 influenza viruses cause uncontrolled local and systemic inflammation. The mechanism for this response is poorly understood, despite its importance as a determinant of virulence. Therefore we profiled cellular microRNAs of lung tissue from Cynomolgus macaques (Macaca fascicularis) infected with a HPAI and a less pathogenic 1918 H1N1 reassortant virus to understand microRNA contribution to host response. We identified 23 microRNAs associated with the extreme virulence of HPAI, with expression patterns inversely correlated with that of predicted gene targets. Pathway analyses confirmed that these targets were associated with aberrant and uncontrolled inflammatory responses and increased cell death. Importantly, similar microRNAs were associated with lethal 1918 pandemic virus infections in mice. This study suggests that virulence of highly pathogenic influenza viruses may be mediated in part by cellular microRNA through dysregulation of genes critical to the inflammatory process.
INTRODUCTION
The lethality of highly pathogenic H5N1 avian influenza viruses (HPAI) is extraordinary. As of July 2011, more than 560 laboratory–confirmed human cases of H5N1 virus infection have been reported with a high fatality rate of approximately 59% (www.who.int/csr/disease/avian_influenza/). This is considerably higher than that of the 1918 H1N1 influenza virus which caused the worst known influenza pandemic in history claiming at least 40 million lives. Concerns remain over the potential for a deadly HPAI pandemic due to the periodic emergence of HPAI cases and sporadic avian-to-human transmissions (Yuen et al., 1998). The virulence of HPAI has been attributed in part to the over induction of inflammatory cytokines and chemokines resulting in subsequent lung damage, as shown in a variety of animal models (Baskin et al., 2009; Maines et al., 2005; Szretter et al., 2007) and in human patients (de Jong et al., 2006; Hien et al., 2004). However, the molecular mechanisms causing this aberrant gene expression are largely unknown.
Cellular microRNAs were recently implicated in lethal infections of mice with a highly pathogenic 1918 pandemic H1N1 influenza virus (Li et al., 2010b). By inducing increased mRNA degradation and translational inhibition of their cellular targets, cellular microRNAs are known to play key roles in crucial physiologic and pathologic processes, particularly those involving inflammatory responses (Davidson-Moncada et al., 2010; O’Connell et al., 2010). Notably, miR-223 and miR-10a were shown to increase the activity of NF-κB, a principle regulator of inflammatory responses (Fang et al., 2010; Li et al., 2010a). Upon activation, NF-κB represses the expression of Let-7 causing an increased expression of Il-6, a direct target of Let-7 and key pro-inflammatory cytokine gene (Iliopoulos et al., 2009). Other cellular microRNAs regulate the inflammatory responses more directly. Acting as a fine-tuner of the inflammatory response, miR-223 was able to regulate the expression patterns of a large number of target genes related to inflammation (Johnnidis et al., 2008). Furthermore, over-expression of miR-21 resulted in elevated inflammation by repressing the expression of Il-12 (Lu et al., 2009), a cytokine that plays a critical role in restraining antigen-induced airway inflammation (Gavett et al., 1995). While the differential expression of cellular microRNAs was shown to contribute in part to the virulence of H1N1 pandemic influenza virus, it is not known if these cellular microRNAs also contribute to the virulence of other highly pathogenic influenza viruses, including HPAI.
To determine whether cellular microRNAs are associated with severe and fatal HPAI virus infection, we profiled 477 cellular microRNAs from lung tissues of Cynomolgus macaques infected by a human HPAI H5N1 virus (HPAI, most virulent), a reassortant H1N1 virus containing the hemagglutinin (HA) and neuraminidase (NA) surface proteins from the highly pathogenic 1918 pandemic strains (2:6) reassortant of intermediate virulence), and a human seasonal H1N1 virus strain (Tx/91, least virulent). We also attempted to identify a core group of microRNAs associated with high virulence across species and influenza subtypes by comparing the H5N1 data in macaques with data obtained from mice infected with the reconstructed 1918 pandemic (r1918) virus. This study sought to provide a better understanding of how microRNAs and their inversely regulated target genes affect cellular function during infections with highly pathogenic influenza viruses.
RESULTS AND DISCUSSION
HPAI infections induced unique cellular microRNA expression profiles in macaque lungs
The potential of microRNA expression patterns as tools to predict clinical outcomes or response to treatment is intriguing as there is mounting evidence that specific microRNA signatures may be associated with a variety of human diseases (Bartels and Tsongalis, 2009). The identification of microRNAs associated with increased fatality or morbidity during influenza infection could be of significant benefit, particularly during pandemic outbreaks, for rapid diagnosis of potentially fatal cases. To identify microRNAs associated with HPAI virulence in the cynomolgus macaque model, we compared cellular microRNA expression profiles in HPAI-, 2:6- or Tx-infected lungs at matching time points using the cut off criteria of absolute fold change ≥ 1.5 and P ≤ 0.01. Overall, 67 cellular microRNAs were differentially expressed between HPAI- and 2:6- infected lung tissues at one or more time points (Figure 1a; Supplementary File 1) presumably reflecting the high and moderate pathogenicity of the viruses used (Baskin et al., 2009). The number of differentially expressed microRNAs in HPAI-infected tissues significantly increased on day 4 but the same increase did not occur until day 7 p.i. in 2:6-infected tissues (Figure 1b). Thus the HPAI infection had an earlier impact on cellular microRNA expression compared with the 2:6 infection. This impact was also a mix of up- and down-regulation, whereas infection with the 2:6 virus caused only increased expression of affected microRNAs. Taken together, our results suggest that influenza viruses of varying pathogenicity elicit distinct cellular microRNA expression patterns in the macaque model.
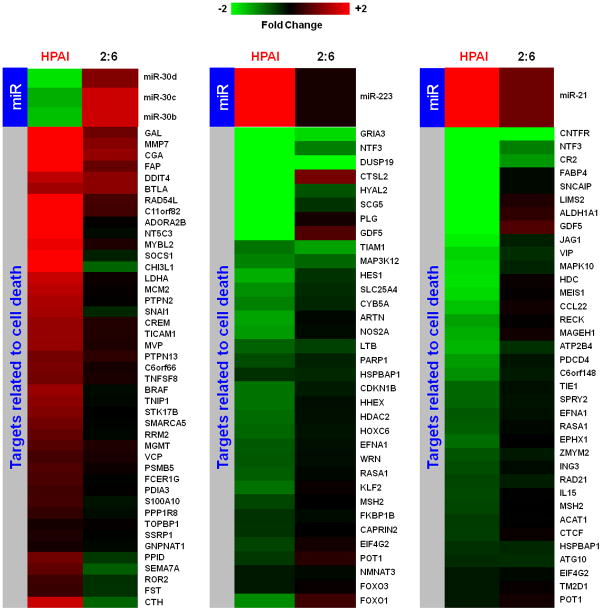
Figure 1. Infection with highly pathogenic H5N1 avian influenza virus (HPAI) and r1918 reassortant virus (2:6) of lesser pathogenicity induced distinct cellular microRNA expression patterns in macaques.
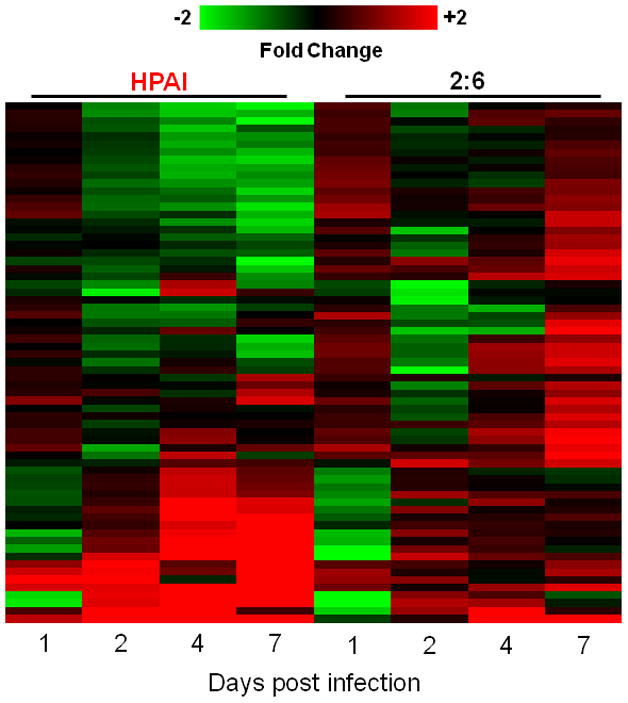
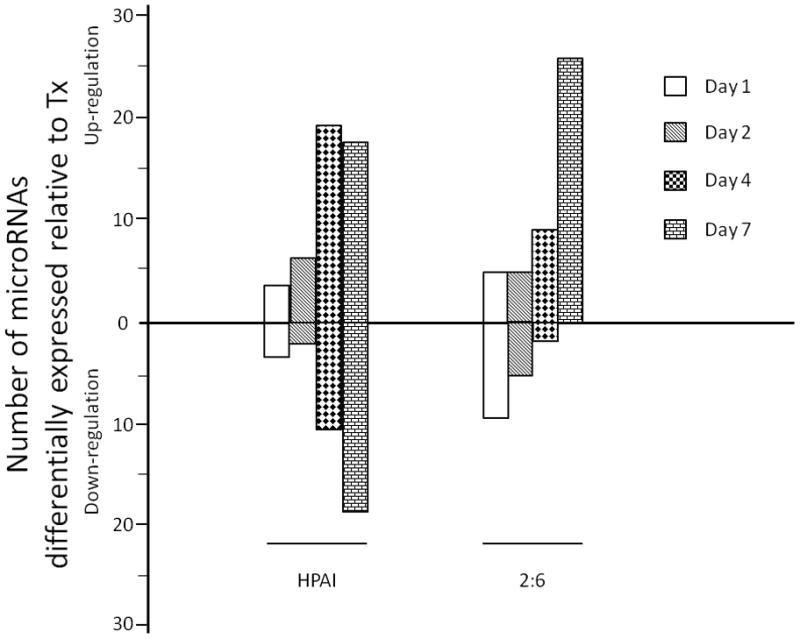
1a: Distinct cellular microRNA expression patterns in HPAI and 2:6 infected macaque lungs. The columns correspond to expression patterns of differentially expressed microRNAs between HPAI- and 2:6- infected macaque lungs relative to Tx-infected macaque lungs on days 1, 2, 4 and 7 post-infection. MicroRNAs satisfied a cut-off of ANOVA P ≤ 0.01 of direct comparison and absolute fold change between HPAI and Tx or between 2:6 and Tx ≥ 1.5. Red color represents microRNA with increased expression in HPAI- or 2:6-infected samples, relative to Tx-infected samples. Green color represents microRNA with decreased expression in HPAI- or 2:6-infected samples, relative to Tx-infected samples. 1b. The number of differentially expressed microRNAs in HPAI or 2:6 infected samples, relative to Tx-infected samples.
We identified a set of microRNAs that may be associated with high virulence across species and influenza viruses
We hypothesized that microRNAs affected by infection with the H5N1 influenza virus in macaques could be likewise regulated during infections with another highly pathogenic influenza virus, even in a different species. Therefore, we compared HPAI-associated microRNAs in macaques with those affected by the r1918 virus in mice (Li et al., 2010b). Although the timeline was not identical between these two independent studies, both used time-matched Tx-infections as controls for microRNA expression changes. Overall, we identified 23 cellular microRNAs whose expressions were affected by both the HPAI and r1918 viruses, suggesting that pathogenicity rather than subtype drove these changes. Among them, 22 microRNAs demonstrated similar expression patterns in both animal model systems at one or more time points, except for miR-188 which demonstrated opposite expression patterns in two models suggesting an unusual regulation on this particular microRNA. Remarkably, among these 23 microRNAs, none were expressed in similar patterns in 2:6-infected tissues as they were in HPAI-infected tissues except for miR-21, whose induction was twice as high in HPAI- than in 2:6-infected samples. Interestingly, a vast majority of these microRNAs had decreased expression in response to highly pathogenic viruses (Figure 2). Sequence alignments in humans, cynomolgus macaques and mice suggested a high degree of conservation of microRNA genes across these three species, especially in the putative target recognition sites or so called seed regions (Table 1). As microRNAs with critical functions are frequently evolutionarily conserved (Aravin et al., 2003), our finding suggests that these 23 microRNAs may play similar and important roles during infection across species.
Figure 2. A group of cellular microRNAs whose expression patterns are associated with the highly pathogenic influenza virus infection in both macaque and mouse models.
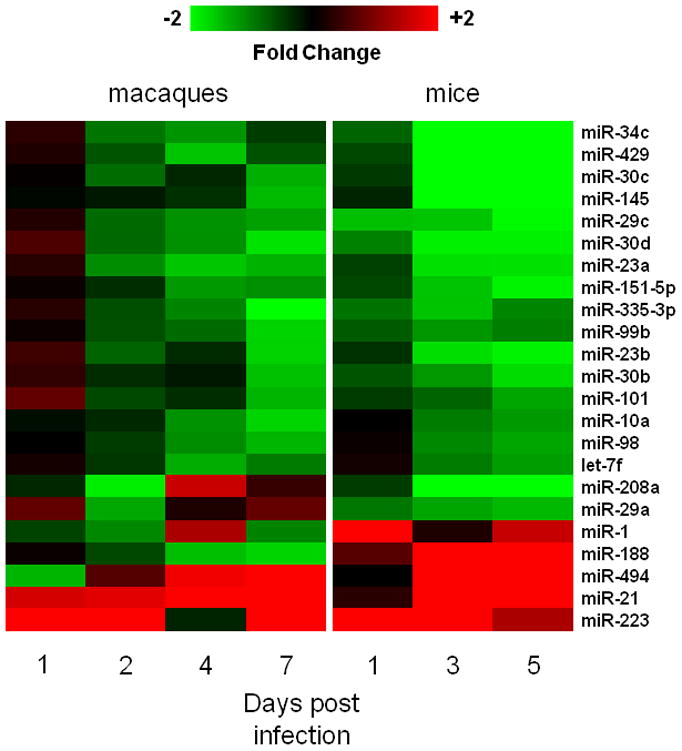
Red represents microRNA with increased expression in HPAI- or r1918-infected samples, relative to Tx-infected samples. Green represents microRNA with decreased expression in HPAI- or r1918- infected samples, relative to Tx-infected samples.
Table 1.
Sequence alignment of microRNAs among human, macaque and mouse.

|
Note: The seed region, the 2–8 nucleotides from the 5′ end, was highlighted
To understand the possible biological implications of changes in microRNA expression after infections with highly pathogenic influenza viruses, we looked at publications experimentally validating the functions of these particular microRNAs. Interestingly, a majority, including Let-7f, miR-10a, miR-208a, miR-21, miR-223, miR-30b, miR-30c, miR-30d and miR-98 (Fang et al., 2010; Hu et al., 2009; Iliopoulos et al., 2009; Li et al., 2010a; Lu et al., 2009; Recchiuti et al.; Zhou et al., 2009) have been experimentally shown to affect key regulators in inflammatory response and cell death (Table 2).
Table 2.
Published examples of examples of experimentally validated functions for the microRNAs associated with the HPAI infections
| microRNA ID | Loci
|
Known function | Reference | ||
|---|---|---|---|---|---|
| Mouse | Macaque | Human | |||
| let-7f | 13 | 15 | 9 | Inhibiting IL-6 inflamation | Ibopoulos, cell 2009 |
| miR-1 | 2 | 10 | 18 | Inhibiting HDAC4 muscle proliferation | Chen, Nat Genetics 2006 |
| miR-101 | 4 | 1 | 1 | Inhibiting MPK in innate immune responce | Zhu, J Immunology 2010 |
| miR-10a | 11 | 16 | 17 | miR-10 knock-downleads to NF-κF activation | Fang, PNAS 2010 |
| miR-145 | 18 | 6 | 5 | targeting STAT1 in cokin cancer cells | Gregerson, Plos one 2010 |
| miR-151-5p | 15 | 5 | 5 | targeting RhoGDIA in tumor cell migration | Ding, Nature Cell Biology 2010 |
| miR-188 | X | X | X | unknown | |
| miR-208a | 14 | 7 | 14 | involved in regulating acute inflammation | Recchiufi. FASEB J 2010 |
| miR-21 | 11 | 16 | 17 | Targets i-12p35 in allergic airway inflammation | Lu. J Immunology 2009 |
| miR-223 | X | X | X | Modulate NF-κB pathway through targeting Iκk-α | Li. Nature Immunology 2010 |
| miR-23a | 8 | 19 | 19 | Involved in cell growth | Cherg, Nucleic Acids Res 2005 |
| miR-23b | 13 | 15 | 9 | Involved in cell growth | |
| miR-29a | 6 | 3 | 7 | Activates p53 by targeting CDC42 and p85α | Park, Nat Struct Mol Biol 2009 |
| miR-29c | 1 | 1 | 1 | Activates p53 by targeting CDC42 and p85α | Park, Nat Struct Mol Biol 2010 |
| miR-30b | 15 | 8 | 8 | Involved in NF-κb Meadiated immune responce | Zhou, Plos Pathogen 2009 |
| miR-30c | 4 | 1 | 1 | Involved in NF-κb Meadiated immune responce | Zhou, Plos Pathogen 2009 |
| miR-30d | 15 | 8 | 8 | Involved in NF-κb Meadiated immune responce | Zhou, Plos Pathogen 2009 |
| miR-335 | 6 | 3 | 7 | Targets Rb1 in p-53 meadiated stress responce | Scarola, Cancer Research 2010 |
| miR-34c | 9 | 14 | 11 | Targets Myc in responce to DNA | Cannel, PNAS 2010 |
| miR-429 | 4 | 1 | 1 | Involved in stem cell differentiation | Lin, EMBO J 2009 |
| miR-494 | 12 | 7 | 14 | unknown | |
| miR-98 | X | X | X | Targeting CIS, an NF-κB activator | Hu, J Immunology, 2009 |
| miR-99b | 17 | 19 | 19 | unknown | |
Predicted targets of microRNAs affected by infection with highly pathogenic influenza viruses were associated with inflammatory response pathways and cell death
To better understand the roles of the 23 microRNAs affected by infection with HPAI, we performed pathway analyses on predicted target genes, whose expression were inversely correlated with the microRNAs’. Predicting the targets of a microRNA and subsequently analyzing their functions by bioinformatics are proven strategies to identify the true biological functions of microRNAs (Olaru et al., 2011), by analyzing the potential functions of a large number of microRNAs simultaneously although with a relatively high false positive rate. By this strategy, 4890 predicted targets of these 23 microRNAs were identified in the HPAI-infected macaque lung samples (ANOVA P ≤ 0.01) (Supplementary File 2). This strategy could also be complemented by experimentally testing the functions of all identified microRNAs in tissue culture or transgenic animal systems. However, it is simply beyond the scope of this study and could not be accomplished in is a feasible time period.
In order to determine the top biological functions associated with the identified targets, we performed functional analyses using the Ingenuity Pathway Analysis (IPA). The top five pathways we found induced by HPAI infections were HMGB1 signaling, the death receptor signaling, the TNFR1 signaling and the nitric oxide production pathways, all involved in inflammation and cell death (Karupiah et al., 1998; Kelliher et al., 1998; Wang et al., 1999). Neither the HMGB1 signaling pathway nor the nitric oxide production pathway were significantly altered during the 2:6 infection, with only temporary changes in the death receptor and TNFR1 signaling pathways as compared to the Tx infection (Table 2).
Thus, our data suggest that dysregulation of inflammation- and cell death-related pathways during the HPAI infections in macaques may be attributed in part to the microRNA-mediated expression changes of target genes, as we could not rule out the possibility that other mechanisms may be also in play to change the target expressions.
Synergistic regulation of inflammation and cell death by cellular microRNAs during the HPAI infection in macaques
We analyzed the role of individual microRNAs in regulating inflammatory responses and cell death during the HPAI infection. Using IPA, we interrogated the functions of inversely correlated targets of individual microRNAs. Inflammatory response and cell death were among the top functions of predicted targets regulated by miR-10a, miR-23b and miR-29c, and by miR-223, miR-21, miR-30b, miR-30c and miR-30d, respectively. The inversely correlated expression patterns of microRNAs and their targets associated with inflammatory or cell death were only observed in the animals infected with HPAI but not in animals infected with the 2:6 virus (Figure 3a and 4a).
Figure 3. Synergistic effect of cellular microRNAs on regulating inflammation during HPAI infection.

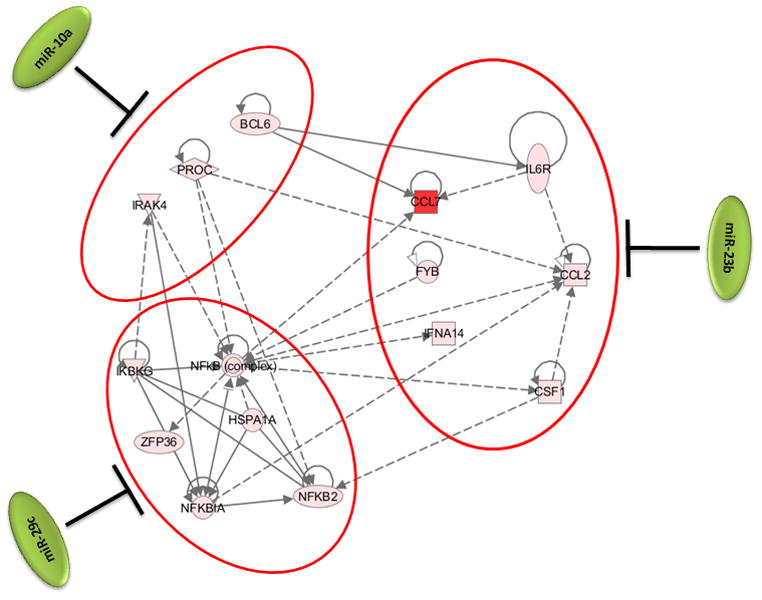
3a: Expression patterns of cellular microRNAs miR-10a, miR-29C and miR-23b and their inversely correlated target genes associated with the inflammation in HPAI infected lungs. Red color represents microRNA or target gene with increased expression in HPAI- or 2:6-infected samples, relative to Tx-infected samples. Green represents microRNA or target gene with decreased expression in HPAI- or 2:6- infected samples, relative to Tx-infected samples. 3b: Network demonstration of the functional connection between inversely correlated target genes associated with inflammation during the HPAI infection. Red color indicates increased expression in comparison with the Tx infected samples. Both figures represent the results from Day 7 post infection. HPAI: Highly pathogenic virus; 2:6: medium pathogenic virus.
Figure 4. Synergistic effect of cellular microRNAs on regulating cell death during HPAI infection.
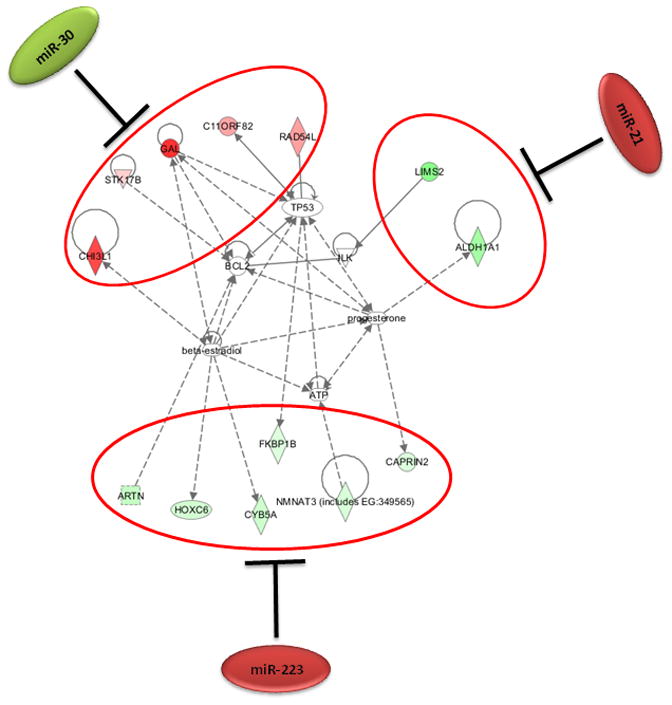
4a: Expression patterns of cellular microRNAs and their inversely correlated target genes associated with the cell death in HPAI infected lungs. Red represents microRNA or target gene with increased expression in HPAI- or 2:6-infected samples, relative to Tx-infected samples. Green represents microRNA or target gene with decreased expression in HPAI- or r1918- infected samples, relative to Tx-infected samples. 4b: Network demonstration of the functional connection between inversely correlated target genes associated with cell death during the HPAI infection. Red indicates increased expression in comparison with the Tx infected samples. Both figures represent the results from Day 7 post infection. HPAI: Highly pathogenic virus; 2:6 of intermediate virulence.
As shown in the networks of directly interacting inflammatory (Fig 3b) and cell death genes (Fig 4b), we observed that individual microRNAs were strongly anti-correlated with different subsets of genes in the same pathway. For example, among inflammatory target genes, miR-10a expression was most significantly inversely correlated with expression of signal transducer genes IRAK4 and PROC. We made identical observations for miR-29c and the expression of NF-κB and ZFP36, miR-23b and downstream genes including inflammatory cytokine genes CCL2 and CCL7 (Figure 3b). Similarly, miR-223, miR-21 and the miR-30 family members may regulate different subsets of genes in a network associated with cell death (Figure 4b). Notably, down-regulation of miR-30 family members in HPAI infections may cause up-regulation of a serine-threonine kinase STK17b, whose over-expression in transgenic mice will lead to increase apoptosis (Mao et al., 2006). On the other hand, up-regulation of miR-223 in HPAI infection may result in decreased expression of the transcriptional factor HOXC6, by which may lead to increased cell death (Ramachandran et al., 2005).
As demonstrated in this study, the infection of a highly pathogenic influenza virus induces the expression changes of certain microRNAs. These changes could in turn affect expression profiles of target genes, whose dysregulation may ultimately result in an uncontrolled inflammatory response and increased cell death. This was most significantly shown through an analysis of the pathway associated with the NF-κB mediated inflammatory response. Modulation of this pathway involves a number of microRNAs and the resulting expression changes were seen across several microRNA targets. Strongly down-regulated in HPAI infection, miR-10a has been shown to repress NF-κB mediated inflammation (Fang et al., 2010). This repression may involve the down-regulation of one of its predicted targets, IRAK4. Interestingly, the expressions of IRAK4 and miR-10a were inversely correlated likely due to a strong down-regulation of miR-10a during the HPAI infections (Figure 3a and 3b). This observation was particularly interesting as IRAK4 activity is required for induction of inflammatory responses mediated by the IL-1 receptor (Koziczak-Holbro et al., 2007). Known for its involvement in the p53 pathway and apoptosis (Park et al., 2009), miR-29c was strongly down-regulated during HPAI infection, which may suggest increased cell death. Down-regulation of miR-29c may also contribute to the uncontrolled inflammation by allowing up-regulation of its predicted targets, such as NF-κBIA and IκB in infections with HPAI but not with the less pathogenic 2:6 virus. We also saw decreased expression of miR-23b versus up-regulation of its predicted targets CCL2 and CCL7 as a result of HPAI infection. These results suggest that the presence of specific microRNAs may play an important role in controlling the level of inflammatory response.
One inherent caveat of this study was that infiltration of cells such as granulocytes may change the landscape of microRNA expressions in HPAI infected lungs. However, it may not be a major factor causing the differential microRNA expressions between the HPAI-infected and 2:6-infected lung tissues observed in this study. As shown in Figure 2, the vast majority of cellular microRNAs were down-regulated in the HPAI-infected but not in the 2:6-infected lungs. This was inconsistent with the significant influx of granulocytes in the macaque lungs (Baskin et al., 2009), as influx would only increase the abundance of microRNAs specific to granulocytes but not decrease the abundance of microRNAs specific to lung tissues. Therefore, cell infiltration alone cannot explain the significant down-regulation of cellular microRNAs during the HPAI infections.
In summary, we identified a group of cellular microRNAs with expression signatures that were associated with the extreme virulence of HPAI H5N1 virus infection in cynomolgus macaques and the reconstructed 1918-pandemic influenza virus infection in mice. Functional analysis of microRNA-targeted genes, which were disrupted by HPAI infection, indicated that these microRNAs may work coordinately with other cellular factors to mediate the inflammatory response and cell death following infection. MicroRNA function can be experimentally altered in vivo by using mimics or antagomirs to offset expression changes induced by viruses, so further studies may ultimately lead to the identification of unique microRNA expression signatures associated with pathogenicity of an emerging influenza virus.
MATERIAL AND METHODS
Viruses
The human isolate of the H5N1 HPAI virus, A/Vietnam/1203/2004 (HPAI), was obtained from the World Health Organization (WHO) influenza collaborating laboratory at the Centers for Disease Control (CDC), Atlanta, GA. The seasonal flu virus A/Texas/36/1991 (Tx) and the reassortant A/Texas/36/1991 influenza viruses (2:6) possessing the A/South Carolina/1/18 HA (GenBank AF117241) and A/Brevig Mission/1/1918 NA (GenBank AF250356) were generated and characterized previously (Baskin et al., 2009; Tumpey et al., 2004; Tumpey et al., 2005).
Macaque experiments
Influenza virus infections in Macaca fascicularis (cynomolgus macaques) were conducted previously (Baskin et al., 2009). Animals in this study were matched for age, weight, and sex and were inoculated by intratracheal, intranasal, tonsillar, and conjunctival routes with a total of 107 pfu of HPAI, 2:6 or Tx. Two animals per treatment group were sacrificed on days 1, 2, 4, and 7 post-inoculation and lung tissue collected. Lung samples from 7 uninfected cynomolgus macaques were also collected. All animal work was performed in a ABSL-3AG facility with appropriate personal protection equipment and standard operating procedures, as approved by the Battelle Bio-safety Committee, and performed following the guidelines of the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC).
RNA Isolation
For total RNA extraction, influenza virus infected macaque lung samples were harvested on days 1, 2, 4 and 7 post-infection (p.i.). Lung samples from 7 uninfected cynomolgus macaques were also collected. Lung tissue was homogenized in QIAzol lysis reagent (Qiagen, CA). Total RNA was isolated with the miRNeasy kit (Qiagen, CA) according to the manufacturer’s protocol which retains small RNAs. Total RNA integrity was confirmed by Bioanalyzer 2100 (Agilent Technologies, CA).
MicroRNA microarray analysis
MicroRNA expression profiling was carried out using the miRCURY LNA™ microRNA array, v.11.0-other species (Exiqon, MA). The dual channel miRCURY LNA™ microRNA array was designed based on the Sanger miRBase release v.14 and contains probes for 477 rhesus macaque (Macaca mulatta) specific microRNAs. The sequence for microRNA of rhesus macaque was used as that of cynomolgus has not been made available. We expected that the expression profiles of a few microRNAs may be underrepresented on the array output due to slight sequence changes between rhesus and cynomolgus macaques and therefore non-optimized hybridization. 500 ng of total RNA was used to make microRNA probes labeled with Hy3 (pooled mock RNA from 7 animals) or Hy5 (RNA from individual animals infected with influenza virus), according to the manufacturer’s protocol (miRCURY LNA™ microRNA array Power Labeling kit, Instruction manual v2.0). Probes were hybridized at 56°C for 16 hours. The slides were then washed according to manufacturer’s protocol. After being washed, the slides were scanned using the Agilent Microarray scanner (Model#: G2505C; Agilent, CA).
The microRNA microarray results were extracted using Agilent Feature Extraction software v9.5. The total microRNA signal from the GeneView result files, which summarized the fluorescence intensity measurements of two channels (Hy3 and Hy5) for all probes for each microRNA on an array, was used in the analyses. Expression data were normalized across arrays using a median-centered approach. The expression difference of microRNA between HPAI-infected and Tx-infected samples or between 2:6-infected and Tx-infected samples was calculated using fluorescence intensities. Differential expression of microRNAs between two groups of samples was assessed by one-way analysis of variance (ANOVA). The average expression change (Fold change) of a microRNA in two HPAI-infected or two 2:6-infected samples from the same groups compared to that in the time-matched Tx-infected sample was calculated. A cutoff ANOVA (P ≤ 0.01) and an absolute fold change of ≥1.5 between infection groups were used to select the differentially expressed microRNAs. A fold change of 1.5 was chosen because a microRNA expression change of 1.5 fold was enough to induce a significant biological impact on the cellular functions (Hu et al., 2008).
mRNA microarray analysis
The mRNA microarray experiments were conducted in a previous study (Baskin et al., 2009). The data was reprocessed for a direct comparison of the cellular gene expression profiles between HPAI- and Tx- infected lungs and between 2:6- and Tx- infected lungs using Resolver 7.2 (Rosetta Biosoftware, WA). The gene expression profiles of Tx- infected lungs from two animals at each time point were in silico pooled and used as a time-matched reference. The cellular gene expressions in lungs from two HPAI- or 2:6- infected animals at each time point were compared to the time-matched reference to assess relative expression changes by one-way analysis of variance (ANOVA). The average Fold change was used to indicate the gene expression change upon HPAI- or 2:6-infection relative to the time-matched Tx-infection.
MicroRNA target database
The predicted targets of human microRNAs were used in this study because the macaque microRNA target prediction was unavailable when the analyses were performed. The identifiers of predicted human microRNA targets were downloaded from the Sanger Institute (http://microrna.sanger.ac.uk/targets/v5/). To make the gene identifiers in the target database consistent with the gene identifiers assigned by the Agilent Feature Extraction Software to the Agilent microarray, the ENSEMBL gene IDs in the Sanger human target database were translated into Entrez Gene ID by BioMart (www.ensembl.org/biomart).
Statistical analysis
To assess the relationship between microRNAs and their predicted target genes, the correlation coefficients for the mean values of the differentially expressed microRNAs and their potential target genes at days 1, 2, 4 and 7 post-infection were calculated in the R environment (http://www.r-project.org/). The differentially expressed microRNA target genes between the HPAI- and the 2:6-infected samples were identified via direct comparisons using a single selection criterion of a P ≤ 0.01. The microRNA-target pairs identified with inversely correlated expression patterns were then selected for further analyses. The probability of enrichment of an inversely correlated target by chance was assessed by hypergeometric (HG) tests that considered the following factors: the number of genes on the arrays, the number of targets on the arrays, the number of differentially expressed genes on the arrays, and the number of inversely correlated targets on the arrays. The calculation for HG tests was performed in the R environment. An HG test, with P≤ 0.05, was used as the cutoff for statistically significant enrichment.
Bioinformatics analysis
Functional analysis of statistically significant gene expression changes was performed with Ingenuity Pathways Analysis (IPA; Ingenuity Systems). This software analyzes RNA expression data in the context of known biological response, regulatory networks, and higher-order response pathways. For all analyses, a Benjamini-Hochberg test correction was applied to the IPA-generated P-value to determine the probability that each biological function assigned to that data set was due to chance alone. In the functional networks, genes are represented as nodes, and the biological relationship between two nodes is represented as an edge (line). All biological relationships used to determine the edges are supported by at least one published reference stored in the Ingenuity Pathways Knowledge Base.
Supplementary Material
Table 3.
Top pathways associated with the inversely correlated targets of analyzed microRNAs.
| HPAI
|
Top Pathways | 2:6
|
||||||
|---|---|---|---|---|---|---|---|---|
| Number of genes | Number of genes | |||||||
| Day 1 | Day 2 | Day 4 | Day 7 | Day 1 | Day 2 | Day 4 | Day 7 | |
| 36 | 20 | 20 | 34 | Hepatic fibrosis | 27 | 16 | * | * |
| 24 | 24 | * | * | HMGB 1 signaling | * | * | * | * |
| 14 | 16 | * | 12 | TNFR 1 signaling | 11 | 8 | * | * |
| 16 | 19 | * | 16 | Death receptor signaling | 13 | 11 | * | * |
| 33 | 36 | * | 40 | Production of nitric oxide | * | * | * | * |
p-value > 0.05
Acknowledgments
We thank Janine Bryan and Marcus Korth for critical reading of the manuscript. We also thank Sean Proll, Lynn Law, and Glenn Zhang for their helpful discussion. This work was supported by NIH Grant P51 RR00166 and U54 AI081680. The findings and conclusions in this report are those of the author(s) and do not necessarily represent the views of the funding agency.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. The Small RNA Profile during Drosophila melanogaster Development. Developmental Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- Bartels CL, Tsongalis GJ. MicroRNAs: Novel Biomarkers for Human Cancer. Clin Chem. 2009;55:623–631. doi: 10.1373/clinchem.2008.112805. [DOI] [PubMed] [Google Scholar]
- Baskin CR, Bielefeldt-Ohmann H, Tumpey TM, Sabourin PJ, Long JP, García-Sastre A, Tolnay AE, Albrecht R, Pyles JA, Olson PH, Aicher LD, Rosenzweig ER, Murali-Krishna K, Clark EA, Kotur MS, Fornek JL, Proll S, Palermo RE, Sabourin CL, Katze MG. Early and sustained innate immune response defines pathology and death in nonhuman primates infected by highly pathogenic influenza virus. Proceedings of the National Academy of Sciences. 2009;106:3455–3460. doi: 10.1073/pnas.0813234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson-Moncada J, Papavasiliou FN, Tam W. MicroRNAs of the immune system. Annals of the New York Academy of Sciences. 2010;1183:183–194. doi: 10.1111/j.1749-6632.2009.05121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong MD, Simmons CP, Thanh TT, Hien VM, Smith GJD, Chau TNB, Hoang DM, Van Vinh Chau N, Khanh TH, Dong VC, Qui PT, Van Cam B, Ha DQ, Guan Y, Peiris JSM, Chinh NT, Hien TT, Farrar J. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med. 2006;12:1203–1207. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Shi C, Manduchi E, Civelek M, Davies PF. MicroRNA-10a regulation of proinflammatory phenotype in athero-susceptible endothelium in vivo and in vitro. Proceedings of the National Academy of Sciences. 2010;107:13450–13455. doi: 10.1073/pnas.1002120107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavett SH, O’Hearn DJ, Li X, Huang SK, Finkelman FD, Wills-Karp M. Interleukin 12 inhibits antigen-induced airway hyperresponsiveness, inflammation, and Th2 cytokine expression in mice. J Exp Med. 1995;182:1527–1536. doi: 10.1084/jem.182.5.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hien TT, Liem NT, Dung NT, San LT, Mai PP, Chau NvV, Suu PT, Dong VC, Mai LTQ, Thi NT, Khoa DB, Phat LP, Truong NT, Long HT, Tung CV, Giang LT, Tho ND, Nga LH, Tien NTK, San LH, Tuan LV, Dolecek C, Thanh TT, de Jong M, Schultsz C, Cheng P, Lim W, Horby P, Farrar J. Avian Influenza A (H5N1) in 10 Patients in Vietnam. New England Journal of Medicine. 2004;350:1179–1188. doi: 10.1056/NEJMoa040419. [DOI] [PubMed] [Google Scholar]
- Hu G, Zhou R, Liu J, Gong AY, Eischeid AN, Dittman JW, Chen XM. MicroRNA-98 and let-7 Confer Cholangiocyte Expression of Cytokine-Inducible Src Homology 2-Containing Protein in Response to Microbial Challenge. The Journal of Immunology. 2009;183:1617–1624. doi: 10.4049/jimmunol.0804362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu SJ, Ren G, Liu JL, Zhao ZA, Yu YS, Su RW, Ma XH, Ni H, Lei W, Yang ZM. MicroRNA Expression and Regulation in Mouse Uterus during Embryo Implantation. Journal of Biological Chemistry. 2008;283:23473–23484. doi: 10.1074/jbc.M800406200. [DOI] [PubMed] [Google Scholar]
- Iliopoulos D, Hirsch HA, Struhl K. An Epigenetic Switch Involving NF-[kappa]B, Lin28, Let-7 MicroRNA, and IL6 Links Inflammation to Cell Transformation. Cell. 2009;139:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, Brummelkamp TR, Fleming MD, Camargo FD. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- Karupiah G, Chen JH, Mahalingam S, Nathan CF, MacMicking JD. Rapid interferon gamma-dependent clearance of influenza A virus and protection from consolidating pneumonitis in nitric oxide synthase 2-deficient mice. J Exp Med. 1998;188:1541–1546. doi: 10.1084/jem.188.8.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelliher MA, Grimm S, Ishida Y, Kuo F, Stanger BZ, Leder P. The Death Domain Kinase RIP Mediates the TNF-Induced NF-[kappa]B Signal. Immunity. 1998;8:297–303. doi: 10.1016/s1074-7613(00)80535-x. [DOI] [PubMed] [Google Scholar]
- Koziczak-Holbro M, Joyce C, Glück A, Kinzel B, Müller M, Tschopp C, Mathison JC, Davis CN, Gram H. IRAK-4 Kinase Activity Is Required for Interleukin-1 (IL-1) Receptor- and Toll-like Receptor 7-mediated Signaling and Gene Expression. Journal of Biological Chemistry. 2007;282:13552–13560. doi: 10.1074/jbc.M700548200. [DOI] [PubMed] [Google Scholar]
- Li T, Morgan MJ, Choksi S, Zhang Y, Kim YS, Liu Z-g. MicroRNAs modulate the noncanonical transcription factor NF-[kappa]B pathway by regulating expression of the kinase IKK[alpha] during macrophage differentiation. Nat Immunol. 2010a;11:799–805. doi: 10.1038/ni.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Chan EY, Li J, Ni C, Peng X, Rosenzweig E, Tumpey TM, Katze MG. MicroRNA Expression and Virulence in Pandemic Influenza Virus-Infected Mice. J Virol. 2010b;84:3023–3032. doi: 10.1128/JVI.02203-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu TX, Munitz A, Rothenberg ME. MicroRNA-21 Is Up-Regulated in Allergic Airway Inflammation and Regulates IL-12p35 Expression. The Journal of Immunology. 2009;182:4994–5002. doi: 10.4049/jimmunol.0803560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maines TR, Lu XH, Erb SM, Edwards L, Guarner J, Greer PW, Nguyen DC, Szretter KJ, Chen LM, Thawatsupha P, Chittaganpitch M, Waicharoen S, Nguyen DT, Nguyen T, Nguyen HHT, Kim JH, Hoang LT, Kang C, Phuong LS, Lim W, Zaki S, Donis RO, Cox NJ, Katz JM, Tumpey TM. Avian Influenza (H5N1) Viruses Isolated from Humans in Asia in 2004 Exhibit Increased Virulence in Mammals. J Virol. 2005;79:11788–11800. doi: 10.1128/JVI.79.18.11788-11800.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J, Qiao X, Luo H, Wu J. Transgenic drak2 overexpression in mice leads to increased T cell apoptosis and compromised memory T cell development. J Biol Chem. 2006;281:12587–12595. doi: 10.1074/jbc.M600497200. [DOI] [PubMed] [Google Scholar]
- O’Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10:111–122. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- Olaru AV, Ghiaur G, Yamanaka S, Luvsanjav D, An F, Popescu I, Alexandrescu S, Allen S, Pawlik TM, Torbenson M, Georgiades C, Roberts LR, Gores GJ, Ferguson-Smith A, Almeida MI, Calin GA, Mezey E, Selaru FM. A microRNA downregulated in human cholangiocarcinoma controls cell cycle through multiple targets involved in the G1/S checkpoint. Hepatology. 2011 doi: 10.1002/hep.24591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Lee JH, Ha M, Nam JW, Kim VN. miR-29 miRNAs activate p53 by targeting p85[alpha] and CDC42. Nat Struct Mol Biol. 2009;16:23–29. doi: 10.1038/nsmb.1533. [DOI] [PubMed] [Google Scholar]
- Ramachandran S, Liu P, Young AN, Yin-Goen Q, Lim SD, Laycock N, Amin MB, Carney JK, Marshall FF, Petros JA, Moreno CS. Loss of HOXC6 expression induces apoptosis in prostate cancer cells. Oncogene. 2005;24:188–198. doi: 10.1038/sj.onc.1207906. [DOI] [PubMed] [Google Scholar]
- Recchiuti A, Krishnamoorthy S, Fredman G, Chiang N, Serhan CN. MicroRNAs in resolution of acute inflammation: identification of novel resolvin D1-miRNA circuits. The FASEB Journal. doi: 10.1096/fj.10-169599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szretter KJ, Gangappa S, Lu X, Smith C, Shieh WJ, Zaki SR, Sambhara S, Tumpey TM, Katz JM. Role of Host Cytokine Responses in the Pathogenesis of Avian H5N1 Influenza Viruses in Mice. J Virol. 2007;81:2736–2744. doi: 10.1128/JVI.02336-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumpey TM, García-Sastre A, Taubenberger JK, Palese P, Swayne DE, Basler CF. Pathogenicity and immunogenicity of influenza viruses with genes from the 1918 pandemic virus. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:3166–3171. doi: 10.1073/pnas.0308391100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumpey TM, Garcia-Sastre A, Taubenberger JK, Palese P, Swayne DE, Pantin-Jackwood MJ, Schultz-Cherry S, Solorzano A, Van Rooijen N, Katz JM, Basler CF. Pathogenicity of Influenza Viruses with Genes from the 1918 Pandemic Virus: Functional Roles of Alveolar Macrophages and Neutrophils in Limiting Virus Replication and Mortality in Mice. J Virol. 2005;79:14933–14944. doi: 10.1128/JVI.79.23.14933-14944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Bloom O, Zhang M, Vishnubhakat JM, Ombrellino M, Che J, Frazier A, Yang H, Ivanova S, Borovikova L, Manogue KR, Faist E, Abraham E, Andersson J, Andersson U, Molina PE, Abumrad NN, Sama A, Tracey KJ. HMG-1 as a Late Mediator of Endotoxin Lethality in Mice. Science. 1999;285:248–251. doi: 10.1126/science.285.5425.248. [DOI] [PubMed] [Google Scholar]
- Yuen KY, Chan PKS, Peiris M, Tsang DNC, Que TL, Shortridge KF, Cheung PT, To WK, Ho ETF, Sung R, Cheng AFB. Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. The Lancet. 1998;351:467–471. doi: 10.1016/s0140-6736(98)01182-9. [DOI] [PubMed] [Google Scholar]
- Zhou R, Hu G, Liu J, Gong AY, Drescher KM, Chen XM. NF-kappaB p65-dependent transactivation of miRNA genes following Cryptosporidium parvum infection stimulates epithelial cell immune responses. PLoS Pathog. 2009;5:e1000681. doi: 10.1371/journal.ppat.1000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.