Abstract
The cell wall, a major barrier protecting cells from their environment, is an essential compartment of both bacteria and archaea. It protects the organism from internal turgor pressure and gives a defined shape to the cell. The cell wall serves also as an anchoring surface for various proteins and acts as an adhesion platform for bacteriophages. The walls of bacteria and archaea are mostly composed of murein and pseudomurein, respectively. Cell wall binding domains play a crucial role in the non-covalent attachment of proteins to cell walls. Here, we give an overview of the similarities and differences in the biochemical and functional properties of the two major murein and pseudomurein cell wall binding domains, i.e., the Lysin Motif (LysM) domain (Pfam PF01476) and the pseudomurein binding (PMB) domain (Pfam PF09373) of bacteria and archaea, respectively.
Keywords: Murein, Pseudomurein, Motifs and domains
Introduction
Murein and pseudomurein are the major cell wall material of bacteria and some methanogenic archaea, respectively. Murein, also called peptidoglycan, is composed of N-acetylmuramic acid and N-acetyl-d-glucosamine (NAG) linked by β(1→4) glycosidic bonds. Pseudomurein is made up of N-acetyltalosaminuronic acid (NAT) and NAG connected through β(1→3) glycosidic linkages (König and Kandler 1979a, b; König et al. 1983; Leps et al. 1984; Kiener et al. 1987; Luo et al. 2001, 2002; Eichler 2003). Even though cell walls made of either murein or pseudomurein resemble each other in their structural and functional properties, there are some fundamental differences in their biosynthetic pathways and in cell wall chemistry, suggesting that they may not have evolved from a common ancestor but are rather the result of convergent evolution. This hypothesis, proposed by Hartmann et al. (Hartmann and König 1990; Steenbakkers et al. 2006) two decades ago, is supported by recent genome sequencing results and by the phylogenetic distribution of the two types of cell wall-containing organisms. Comparison of the biosynthetic and assembly pathways of murein and pseudomurein did not reveal any similarities of the involved genes until now, indicating the unique nature of the biosynthetic enzymes present in pseudomurein-containing organisms (Steenbakkers et al. 2006).
Moreover, the phylogenetic distribution of murein and pseudomurein-containing organisms is quite contrasting, as murein is highly conserved in eubacteria, while pseudomurein is restricted to a few methanogenic archaeal members (Methanobacteriales and Methanopyrus) (Steenbakkers et al. 2006).
Most bacterial cell wall hydrolases cleave the β(1→4) glycosidic bonds in peptidoglycan, and are ineffective on pseudomurein-containing archaeal cell walls. Surprisingly, no archaeal enzymes are known yet, which cleave the β(1→3) glycosidic bond of pseudomurein. In addition, only two archaeal pseudomurein endoisopeptidases (Pei) have been identified so far that cleave the peptide links that connect adjacent pseudomurein glycan strands; they were shown to cleave the isopeptide bond between the ε-amino group of l-lysine and the carboxyl group of an l-alanine residue in the peptide cross-link between two pseudomurein NAT residues (Kiener et al. 1987; Pfister et al. 1998; Luo et al. 2001, 2002; Steenbakkers et al. 2006; Visweswaran et al. 2010, 2011).
Most murein hydrolases as well as the two pseudomurein endopeptidases are composed of two distinct domains, a catalytic domain and a cell wall binding domain. The catalytic domain possesses a glucosaminidase, muramidase, endopeptidase, or N-acetylmuramyl-l-alanine amidase activity. The cell wall binding domains are crucial for the proper functioning of both murein and pseudomurein hydrolases. Here, we describe two important murein and pseudomurein binding modules, namely the Lysin Motif (LysM) domain and pseudomurein binding (PMB) domain. To date, no reviews have been published on the PMB domains, nor on a comparison of LysM and PMB domains. This review should attract attention of a large group of readers working on microbial cell walls.
LysM and PMB domain-containing cell wall hydrolases
LysM is the most common protein domain of peptidoglycan hydrolases (Ruhland et al. 1993; Bateman and Bycroft 2000; Steen et al. 2005; Arrighi et al. 2006; Andre et al. 2008; Buist et al. 2008). More than 4,000 proteins in prokaryotes and eukaryotes contain the highly conserved LysM domain (Buist et al. 2008), which allows these proteins to bind to the peptidoglycan layer of bacterial cell walls in a non-covalent manner (Steen et al. 2003; Tarahomjoo et al. 2008). LysM was first discovered as a C-terminal motif in the lysozyme of Bacillus subtilis phage ø29. As this motif was originally identified in bacterial lysins, it was termed Lysin Motif (Birkeland 1994; Buist et al. 1995, 2008; Hu et al. 2010). Subsequently, these motifs were shown to be present in various numbers in other murein hydrolases (Table 1, Hu et al. 2010).
Table 1.
Overview of LysM or PMB motif-containing murein and pseudomurein hydrolases
| Protein | Organism | Number of LysM motifs | Location of LysM motifs | Reference |
| LytE | Bacillus subtilis | 3 | N′ | (Yamamoto et al. 2008) |
| LytF | Bacillus subtilis | 5 | N′ | (Yamamoto et al. 2008) |
| CwIS | Bacillus subtilis | 4 | N′ | (Yamamoto et al. 2008) |
| Sep | Lactobacillus fermentum | 1 | N′ | (Turner et al. 2004) |
| P60 | Listeria monocytogenes | 1 | Middle | (Ruhland et al. 1993) |
| AcmA | Lactococcus lactis | 3 | C′ | (Buist et al. 1995) |
| AcmD | Lactococcus lactis | 3 | C′ | (Huard et al. 2004) |
| AtlA | Enterococcus faecalis | 6 | C′ | (Eckert et al. 2006) |
| AtlB | Enterococcus faecalis | 2 | C′ | (Mesnage et al. 2008) |
| AtlC | Enterococcus faecalis | 2 | C′ | (Mesnage et al. 2008) |
| MurA | Listeria monocytogenes | 4 | C′ | (Carroll et al. 2003) |
| Protein | Organism | Number of PMB motifs | Location of PMB motifs | Reference |
| PeiP | Methanothermobacter marburgensis ΨM2 | 4 | N′ | (Steenbakkers et al. 2006; Visweswaran et al. 2010) |
| PeiW | Methanothermobacter wolfeii ΨM100 | 4 | N′ | (Steenbakkers et al. 2006; Visweswaran et al. 2010, 2011) |
Unlike bacterial cell wall hydrolases, not much is known about the archaeal cell wall hydrolases. No enzymes are known that cleave the glycosidic linkages of pseudomurein, and only two endoisopeptidases have been discovered until now, which cleave the pseudomurein peptide cross-links. These pseudomurein endoisopeptidases (Pei), PeiW and PeiP from the methanogenic archaeal-specific prophages Methanothermobacter wolfeii ΨM100 and M. marburgensis ΨM2, respectively, were shown to act as autolysins of methanogenic archaea (Kiener et al. 1987; Stax et al. 1992; Pfister et al. 1998; Luo et al. 2001, 2002; Steenbakkers et al. 2006; Visweswaran et al. 2010, 2011). PeiW and PeiP have the same molecular architecture: they each contain four pseudomurein cell wall binding (PMB) motifs at their N-terminus fused to a C-terminal cysteine protease domain (Luo et al. 2002; Steenbakkers et al. 2006; Visweswaran et al. 2010, 2011) (Table 1). The PMB motifs in the Pei enzymes are involved in binding of the enzymes to the cell envelope of methanogenic archaea and thereby facilitate cell wall hydrolysis (Steenbakkers et al. 2006; Visweswaran et al. 2010, 2011).
Evolutionary conservation and molecular biology of cell wall binding domains
LysM motifs are present in proteins of both prokaryotes and eukaryotes, but not in those of archaea (Mulder et al. 2006; Buist et al. 2008; Visweswaran et al. 2010). Their amino acid sequences are highly conserved. In contrast, the primary structure of the PMB motif is less conserved, and the distribution of the PMB motif is much narrower, being present only in four methanogenic archaeal genera (Methanobrevibacter, Methanothermobacter, Methanothermus, and Methanosphaera), two prophages (M. wolfeii ΨM100 and M. marburgensis ΨM2) and in five genera of bacteria (Xanthomonas, Novosphingobium, Granulibacter, Erythrobacter, and Roseovarius) (Hunter et al. 2009; http://www.ebi.ac.uk/interpro/IEntry?ac=IPR018975). Interestingly, the first three of these bacterial genera also possess proteins with a LysM domain in addition to proteins carrying a PMB domain.
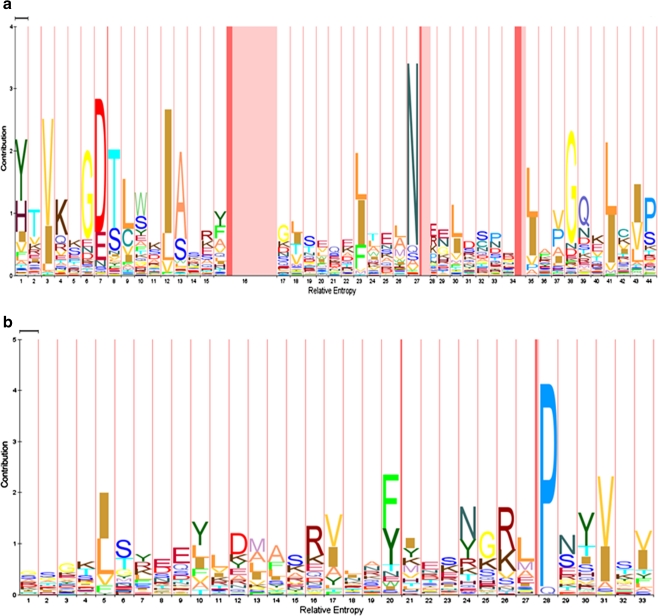
The LysM and PMB motifs share a number of similarities, such as their variable pI values and the variable number and location of the motifs in the proteins that contain them (Buist et al. 2008). An overview of these properties is shown in Table 2. The pI of the LysM motif-containing proteins ranges from 4 to 12 with most ranging between 5 and 10. A similar variability is observed for the PMB domain-containing proteins (pI of 4 to 10). Each LysM or PMB domain can be found at the N- or C -terminus or in the middle of the proteins. The number of motifs varies from 1 to 6 in LysM domains and from 1 to 4 in PMB domains. The LysM motifs in a LysM domain are separated by amino acid linker regions varying in length and composition and mostly consist of serine, threonine, and aspartic acid residues (Buist et al. 2008). In plant receptor-like kinases, a conserved CxC motif is present in the LysM linker regions of which the function is unknown (Madsen et al. 2003; Radutoiu et al. 2003; Arrighi et al. 2006; Mulder et al. 2006; Radutoiu et al. 2007; Buist et al. 2008). Similarly, intervening sequences connecting the adjacent PMB motifs in PMB domains also vary in length and amino acid composition but they share no conserved residues (data not shown). The length of the LysM and PMB motifs differs significantly even though they perform a similar function, i.e., non-covalent cell wall binding. The LysM domain consists of motifs of 44 to 65 amino acid residues, whereas the PMB domain is made up of motifs of 30 to 35 amino acid residues (Fig. 1a, b).
Table 2.
Molecular properties of LysM and PMB motifs
| Characteristics | LysM motif | PMB motif |
|---|---|---|
| Length of motif (amino acids) | 44–65 | 30–35 |
| Number of motifs (range) | 1–6 | 1–4 |
| Location in motif-bearing proteins | Variable (N′/C′/center of the protein) | Variable (N′/C′/center of the protein) |
| Iso-electric points (pI) (range) | 5–10 | 4–10 |
| Nature of conserved amino acid residues | Hydrophobic | Hydrophobic |
| Evolutionary distribution | Wide | Narrow |
| Major binding substrate | Murein (in bacteria) | Pseudomurein (in some methanogenic archaea) |
| Chitin (in fungi and insects) | ||
| Secondary structure | βααβ | Unknown |
| Three-dimensional structure known | Yes (Bateman and Bycroft 2000; Bielnicki et al. 2006) | No |
Fig. 1.
Hidden Markov model (HMM) showing the consensus sequence of the LysM (Pfam database entry PF01476) and the PMB motifs (Pfam database entry PF09373). The HMM logo pictures were generated separately from the Pfam database for a the LysM motifs and b the PMB motifs, using their respective Pfam accession numbers. The X-axis indicates the relative entropy, and the contribution of each amino acid residue is shown on the Y-axis. Numbers on the X-axis indicate the position of the amino acid residue in the total length of the motif. The size of the amino acid residues on Y-axis is directly proportional to their conservation in the consensus sequence
Hidden Markov model (HMM) logos generated separately for the LysM and PMB motifs show some highly conserved amino acid residues (Fig. 1a, b). Especially, the first 16 N-terminal and the last 10 C-terminal residues in the LysM motifs are well-conserved, while the central region (residues 17–34) is much less conserved, except for the residues at positions 23, 27, and 30 (Fig. 1a, Buist et al. 2008). These conserved amino acid residues form the hydrophobic core of the LysM motif located between the anti-parallel β-strands and the first α-helix (Fig. 2). The PMB motif is not very well-conserved except for a very highly conserved proline residue at position 28 and other hydrophobic or aromatic residues at positions 5, 20, and 31 (Fig. 1b). These conserved amino acid residues are all hydrophobic, which suggests that, similar to the LysM motif, they might play a role in stabilizing the hydrophobic core of the PMB motif.
Fig. 2.
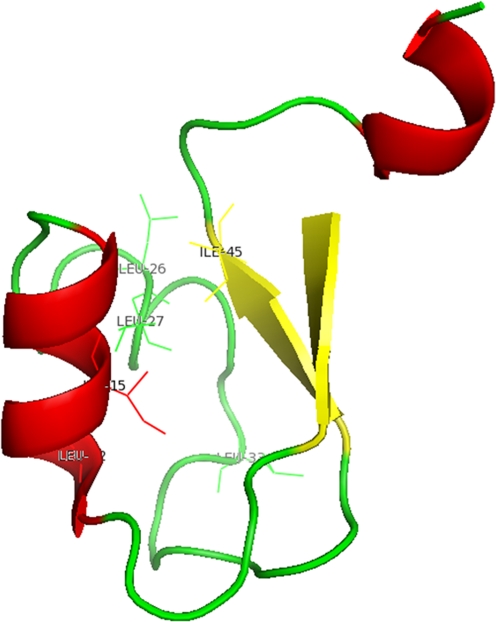
Crystal structure of the LysM motif of YkuD from B. subtilis. The figure was generated using PyMOL (The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC) and PDB entry 1Y7M (Bielnicki et al. 2006). The hydrophobic core formed by the conserved hydrophobic amino acid residues is labeled with side chains. The anti-parallel β-strands and α- helices are colored yellow and red, respectively. The loops are represented in green
Structural aspects of the LysM and PMB domains
The structure of one LysM motif was initially solved by NMR for the Escherichia coli membrane-bound murein lytic transglycosylase-D (MltD; protein data bank (PDB) entry 1EOG; Bateman and Bycroft 2000). It was shown to possess a βααβ secondary structure with the two α-helices packing against one side of the anti-parallel β-strands (Fig. 2) (Bateman and Bycroft 2000). The NMR structure of the human hypothetical protein SB145 (PDB 2DJP (unpublished)) also showed the presence of a LysM motif with the same secondary structure. The X-ray crystal structure of the Bacillus subtilis protein YkuD (Fig. 2) (PDB 1Y7M) (Bielnicki et al. 2006) revealed that this protein also has an N-terminal LysM motif, with a structure that is in agreement with the NMR structures (Bielnicki et al. 2006). The three-dimensional structures of these LysM motifs also suggested the location of a potential ligand-binding site. However, a ligand-bound structure which could explain the substrate binding mode and specificity and show whether any conformational changes occur upon ligand binding is not known yet.
So far, no three-dimensional structures are available of a PMB motif or domain. Circular dichroism (CD) spectra indicate a typical α-helical spectrum for the PMB domain of the M. thermautotrophicus surface (S)-layer protein MTH719, which contains three C-terminal PMB motifs (Visweswaran et al. unpublished results).
Domain stability
The LysM domain from the fern Pteris ryukyuensis chitinase A, which contains two LysM motifs, was thermally stable up to 90°C, whereas a single motif is denatured at a thermal transition value (T m) of 96.5°C (Ohnuma et al. 2008). This high-temperature stability of the LysM domain is due to the presence of four cysteine residues in each motif that form two disulfide bonds (Ohnuma et al. 2008), as shown by differential scanning calorimetry (DSC) experiments: in the presence of reducing agents the T m of the LysM domain with two motifs decreased from 90°C to 77.4°C for β-mercaptoethanol and to 68.4°C for tris(2-carboxyethyl)phosphine (Ohnuma et al. 2008). The thermal stability of the LysM domain was slightly increased upon binding to NAG units (Ohnuma et al. 2008). Thermal denaturation of the LysM domain is irreversible due to aggregation at temperatures above the T m (Ohnuma et al. 2008).
DSC experiments with the PMB domain of the M. thermautotrophicus S-layer protein MTH719 revealed that the domain was stable at temperatures up to 76°C (Visweswaran et al. unpublished results). There is only one cysteine residue in this domain and the observed high T m is thus not the result of stabilization by intramolecular disulfide bonds. The PMB domain also forms aggregates after complete thermal denaturation, a process that is irreversible. As attempts to refold the aggregated domain were not successful, its folding and un-folding kinetics are unknown. Irreversible chemical denaturation of the PMB domain with guanidinium hydrochloride was reached at a concentration of 3.8 M guanidinium hydrochloride (Visweswaran et al. 2011).
Specificity of substrate binding
The LysM domain specifically binds to the NAG moiety of murein in a non-covalent but unknown manner (Ohnuma et al. 2008). LysM domains from plant origin are also capable of binding to chitin, a polymer of NAG (Ohnuma et al. 2008; Petutschnig et al. 2010). A fusion protein generated by combining the LysM domain of the Lactococcus lactis AcmA protein with the PMB domain of the M. thermautotrophicus MTH719 protein bound to murein, pseudomurein, and chitin, and it was shown that the binding of the fusion protein to chitin was caused by the LysM domain (Visweswaran et al., submitted).
The PMB domain of PeiW specifically binds to the pseudomurein layer in the cell envelopes of methanogenic archaea (Steenbakkers et al. 2006; Visweswaran et al. 2010, 2011). The PMB domain of the (S)-layer protein MTH719 not only binds to the pseudomurein-containing archaeal cell envelope but also to the cell wall components of lysed bacterial cells (Visweswaran et al. 2011). The latter property suggests that the PMB domain recognizes free NAG, the moiety that murein and pseudomurein have in common. However, there is no experimental evidence as yet to underpin this assumption.
An optimal number of motifs are required for proper cell wall-binding of the LysM and PMB domains
AcmA, a well-characterized autolysin from L. lactis, contains three LysM motifs in its C-terminal LysM domain and an N-terminal glucosaminidase domain (Buist et al. 1995; Steen et al. 2005). Motif deletion and addition studies revealed that AcmA requires three LysM motifs for optimal functioning (Steen et al. 2005). AcmA devoid of LysM motifs or with only one motif did not bind to its substrate, whereas constructs with two and four motifs did bind, but to a lesser extent than the native protein with three motifs (Steen et al. 2005).
Similarly, the MTH719 S-layer protein contains a PMB domain with three motifs at its C-terminus. C-terminally GFP-tagged motif deletion constructs with two and one motif failed to bind to pseudomurein-containing archaeal cells, whereas the native GFP-tagged protein with three motifs did bind (Visweswaran et al. 2011). Also in the case of the pseudomurein hydrolases, PeiW and PeiP removal of the PMB domain resulted in loss of binding of the enzymes to M. thermautotrophicus cells (Visweswaran et al. 2010, 2011). In both cases, apparently, the native LysM and the PMB domains are equipped with an optimal number of motifs to perform their cell wall binding function.
Factors affecting the binding of LysM and PMB domains to cell walls
The binding of the LysM (Hu et al. 2010) and PMB (Visweswaran et al. 2011) domains to their respective substrates is mainly dependent on the pH and the salt concentration. The domains are capable of binding to their substrates at a pH close to their pI value. The binding of the LysM domain of the N-acetylmuramidase AcmD from L. lactis takes place at pH 4.0, which is close to its calculated pI of 4.3 (ExPASy) and no binding is observed at other pH conditions (pH 6.0 and 8.0) (Visweswaran et al. unpublished results).
Similarly, the PMB domain of the S-layer protein MTH719 from M. thermautotrophicus shows maximum binding to pseudomurein-containing Methanobacterium sp. cells at pH 9.0, which is close to pI of the PMB domain (pI 10.6) (Visweswaran et al. 2011). Together these results show that positive charges play a key role in favoring the binding of the LysM and PMB domains to their respective substrates (Huard et al. 2004; Visweswaran et al. 2011).
Similarly, pH also plays a crucial role in the optimal functioning of the LysM domain-containing proteins. Huard et al. (2004) have shown that the LysM domain-containing N-acetylmuramidase AcmD is catalytically active only at pH 4.0, which is close to the pI value of the enzyme (4.3). A similar study was performed on the endolysin Ly5C from the Lactobacillus fermentum temperate bacteriophage φPYB5, an enzyme that possesses a C-terminal LysM domain of three motifs and an active site domain at the N-terminus (Hu et al. 2010). The enzyme showed a twofold increase in binding to substrate at pH 11, which is close to the pI value of the enzyme (pI 9.0), compared to lower pHs. These results suggest that pH plays a vital role in binding of LysM domain-containing proteins to their substrates.
The effect of salt concentration on substrate binding has been examined for the LysM domain of Ly5C. NaCl at a concentration of 0.5 M significantly enhanced the binding of Ly5C to host cells by increasing the ionic interactions between them (Hu et al. 2010). Also, the carbon source present in the growth medium influenced the binding of the LysM domain-containing autolysin AcmA from L. lactis. Lactococcal cells grown on galactose bound less AcmA than cells grown on glucose, because of differences in the cell wall composition caused by the growth on galactose (Steen et al. 2008).
Possible perspectives for research and application
The LysM domains of bacterial cell wall hydrolases help in binding of the enzymes to their substrate peptidoglycan and thereby facilitate cell wall hydrolysis. LysM domains of the plant NFR1 and NFR5 receptors are involved in bacterial Nod-factor signal recognition between leguminous plants and their symbiotic hosts (Madsen et al. 2003; Mulder et al. 2006; Radutoiu et al. 2003, 2007). The LysM domain is currently being developed as an anchor to display heterologous proteins and peptides on the surface of lactic acid bacteria, among others for the development of oral vaccines (Bosma et al. 2006; Okano et al. 2008). Similarly, the PMB domain could be used as an anchor for surface display of heterologous proteins and peptides on methanogenic archaea. Since the PMB domains specifically bind to pseudomurein-containing methanogens, these domains could also be used as biomarkers to identify the industrially important methanogens and distinguish them from other organisms.
To obtain a detailed three-dimensional structure of a complete LysM or PMB domain would be an interesting target for future research as it would help to understand how the domains interact with their substrates. Of course, to have a deep molecular understanding of the non-covalent interactions between the domains and their substrates, or even a crystal structure of the substrate-bound domains, would then be the next challenge.
Acknowledgments
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Andre G, Leenhouts K, Hols P, Dufrêne YF. Detection and localization of single LysM-peptidoglycan interactions. J Bacteriol. 2008;190:7079–7086. doi: 10.1128/JB.00519-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrighi JF, Barre A, Ben Amor B, Bersoult A, Soriano LC, Mirabella R, de Carvalho-Niebel F, Journet EP, Gherardi M, Huguet T, Geurts R, Denarie J, Rouge P, Gough C. The Medicago truncatula lysin motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 2006;142:265–279. doi: 10.1104/pp.106.084657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Bycroft M. The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD) J Mol Biol. 2000;299:1113–1119. doi: 10.1006/jmbi.2000.3778. [DOI] [PubMed] [Google Scholar]
- Bielnicki J, Devedjiev Y, Derewenda U, Dauter Z, Joachimiak A, Derewenda ZS. B. subtilis ykuD protein at 2.0 Å resolution: insights into the structure and function of a novel, ubiquitous family of bacterial enzymes. Proteins. 2006;62:144–151. doi: 10.1002/prot.20702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkeland NK. Cloning, molecular characterization, and expression of the genes encoding the lytic functions of lactococcal bacteriophage ΦLC3: a dual lysis system of modular design. Can J Microbiol. 1994;40:658–665. doi: 10.1139/m94-104. [DOI] [PubMed] [Google Scholar]
- Bosma T, Kanninga R, Neef J, Audouy SAL, van Roosmalen ML, Steen A, Buist G, Kok J, Kuipers OP, Robillard G, Leenhouts K. Novel surface display system for proteins on non-genetically modified Gram-positive bacteria. Appl Environ Microbiol. 2006;72:880–889. doi: 10.1128/AEM.72.1.880-889.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buist G, Kok J, Leenhouts KJ, Dabrowska M, Venema G, Haandrikman AJ. Molecular cloning and nucleotide sequence of the gene encoding the major peptidoglycan hydrolase of Lactococcus lactis, a muramidase needed for cell separation. J Bacteriol. 1995;177:1554–1563. doi: 10.1128/jb.177.6.1554-1563.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buist G, Steen A, Kok J, Kuipers OP. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol Microbiol. 2008;68:838–847. doi: 10.1111/j.1365-2958.2008.06211.x. [DOI] [PubMed] [Google Scholar]
- Carroll SA, Hain T, Technow U, Darji A, Pashalidis P, Joseph SW, Chakraborty T. Identification and characterization of a peptidoglycan hydrolase, MurA, of Listeria monocytogenes, a muramidase needed for cell separation. J Bacteriol. 2003;185:6801–6808. doi: 10.1128/JB.185.23.6801-6808.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert C, Lecerf M, Dubost L, Arthur M, Mesnage S. Functional analysis of AtlA, the major N-acetylglucosaminidase of Enterococcus faecalis. J Bacteriol. 2006;188:8513–8519. doi: 10.1128/JB.01145-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler J. Facing extremes: archaeal surface-layer (glyco) proteins. Microbiol. 2003;149:3347–3351. doi: 10.1099/mic.0.26591-0. [DOI] [PubMed] [Google Scholar]
- Hartmann E, König H. Comparison of the biosynthesis of the methanobacterial pseudomurein and the eubacterial murein. Naturwissenschaften. 1990;77:472–475. doi: 10.1007/BF01135923. [DOI] [PubMed] [Google Scholar]
- Hu S, Kong J, Kong W, Guo T, Ji M. Characterization of a novel LysM domain from Lactobacillus fermentum bacteriophage endolysin and its use as an anchor to display heterologous proteins on the surfaces of lactic acid bacteria. Appl Environ Microbiol. 2010;76:2410–2418. doi: 10.1128/AEM.01752-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huard C, Miranda G, Redko Y, Wessner F, Foster SJ, Chapot-Chartier MP. Analysis of the peptidoglycan hydrolase complement of Lactococcus lactis: identification of a third N-acetylglucosaminidase, AcmC. Appl Environ Microbiol. 2004;70:3493–3499. doi: 10.1128/AEM.70.6.3493-3499.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, Finn RD, Gough J, Haft D, Hulo N, Kahn D, Kelly E, Laugraud A, Letunic I, Lonsdale D, Lopez R, Madera M, Maslen J, McAnulla C, McDowall J, Mistry J, Mitchell A, Mulder N, Natale D, Orengo C, Quinn AF, Selengut JD, Sigrist CJA, Thimma M, Thomas PD, Valentin F, Wilson D, Wu CH, Yeats C. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiener A, König H, Winter J, Leisinger T. Purification and use of Methanobacterium wolfeii pseudomurein endopeptidase for lysis of Methanobacterium thermoautotrophicum. J Bacteriol. 1987;169:1010–1016. doi: 10.1128/jb.169.3.1010-1016.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König H, Kandler O. The amino acid sequence of the peptide moiety of the pseudomurein from Methanobacterium thermoautotrophicum. Arch Microbiol. 1979;121:271–275. doi: 10.1007/BF00425067. [DOI] [PubMed] [Google Scholar]
- König H, Kandler O. N-Acetyltalosaminuronic acid a constituent of the pseudomurein of the genus Methanobacterium. Arch Microbiol. 1979;123:295–299. doi: 10.1007/BF00406664. [DOI] [Google Scholar]
- König H, Kandler O, Jensen M, Rietschel ET. The primary structure of the glycan moiety of pseudomurein from Methanobacterium thermoautotrophicum. Hoppe Seylers Z Physiol Chem. 1983;364:627–636. doi: 10.1515/bchm2.1983.364.1.627. [DOI] [PubMed] [Google Scholar]
- Leps B, Labischinski H, Barnickel G, Bradaczek H, Giesbrecht P. A new proposal for the primary and secondary structure of the glycan moiety of pseudomurein. Conformational energy calculations on the glycan strands with talosaminuronic acid in 1 C conformation and comparison with murein. Eur J Biochem. 1984;144:279–286. doi: 10.1111/j.1432-1033.1984.tb08461.x. [DOI] [PubMed] [Google Scholar]
- Luo Y, Pfister P, Leisinger T, Wasserfallen A. The genome of archaeal prophage ΨM100 encodes the lytic enzyme responsible for autolysis of Methanothermobacter wolfeii. J Bacteriol. 2001;183:5788–5792. doi: 10.1128/JB.183.19.5788-5792.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Pfister P, Leisinger T, Wasserfallen A. Pseudomurein endoisopeptidases PeiW and PeiP, two moderately related members of a novel family of proteases produced in Methanothermobacter strains. FEMS Microbiol Lett. 2002;208:47–51. doi: 10.1111/j.1574-6968.2002.tb11059.x. [DOI] [PubMed] [Google Scholar]
- Madsen EB, Madsen LH, Radutoiu S, Olbryt M, Rakwalska M, Szczyglowski K, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J. A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature. 2003;425:637–640. doi: 10.1038/nature02045. [DOI] [PubMed] [Google Scholar]
- Mesnage S, Chau F, Dubost L, Arthur M. Role of N-acetylglucosaminidase and N-acetylmuramidase activities in Enterococcus faecalis peptidoglycan metabolism. J Biol Chem. 2008;283:19845–19853. doi: 10.1074/jbc.M802323200. [DOI] [PubMed] [Google Scholar]
- Mulder L, Lefebvre B, Cullimore J, Imberty A. LysM domains of Medicago truncatula NFP protein involved in Nod factor perception. Glycosylation state, molecular modeling and docking of chitooligosaccharides and Nod factors. Glycobiology. 2006;16:801–809. doi: 10.1093/glycob/cwl006. [DOI] [PubMed] [Google Scholar]
- Ohnuma T, Onaga S, Murata K, Taira T, Katoh E. LysM domains from Pteris ryukyuensis chitinase-A: a stability study and characterization of chitin-binding site. J Biol Chem. 2008;283:5178–5187. doi: 10.1074/jbc.M707156200. [DOI] [PubMed] [Google Scholar]
- Okano K, Zhang Q, Kimura S, Narita J, Tanaka T, Fukuda H, Kondo A. system using tandem repeats of the cA peptidoglycan-binding domain from Lactococcus lactis for display of both N- and C-terminal fusions on cell surfaces of lactic acid bacteria. Appl Environ Microbiol. 2008;74:1117–1123. doi: 10.1128/AEM.02012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petutschnig EK, Jones AME, Serazetdinova L, Lipka U, Lipka V. The lysin motif receptor-like kinase (LysM-RLK) CERK1 is a major chitin-binding protein in Arabidopsis thaliana and subject to chitin-induced phosphorylation. J Biol Chem. 2010;285:28902–28911. doi: 10.1074/jbc.M110.116657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister P, Wasserfallen A, Stettler R, Leisinger T. Molecular analysis of Methanobacterium phage psiM2. Mol Microbiol. 1998;30:233–244. doi: 10.1046/j.1365-2958.1998.01073.x. [DOI] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Grønlund M, Sato S, Nakamura Y, Tabata S, Sandal N, Stougaard J. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature. 2003;425:585–592. doi: 10.1038/nature02039. [DOI] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Jurkiewicz A, Fukai E, Quistgaard EMH, Albrektsen AS, James EK, Thirup S, Stougaard J. LysM domains mediate lipochitin-oligosaccharide recognition and Nfr genes extend the symbiotic host range. EMBO J. 2007;26:3923–3935. doi: 10.1038/sj.emboj.7601826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhland GJ, Hellwig M, Wanner G, Fiedler F. Cell-surface location of Listeria-specific protein p60—Detection of Listeria cells by indirect immunofluorescence. J Gen Microbiol. 1993;139:609–616. doi: 10.1099/00221287-139-3-609. [DOI] [PubMed] [Google Scholar]
- Stax D, Hermann R, Falchetto R, Leisinger T. The lytic enzyme in bacteriophage ΨM1-induced lysates of Methanobacterium thermoautotrophicum Marburg. FEMS Microbiol Lett. 1992;100:433–438. [Google Scholar]
- Steen A, Buist G, Leenhouts KJ, El Khattabi M, Grijpstra F, Zomer AL, Venema G, Kuipers OP, Kok J. Cell wall attachment of a widely distributed peptidoglycan binding domain is hindered by cell wall constituents. J Biol Chem. 2003;278:23874–23881. doi: 10.1074/jbc.M211055200. [DOI] [PubMed] [Google Scholar]
- Steen A, Buist G, Horsburgh GJ, Venema G, Kuipers OP, Foster SJ, Kok J. AcmA of Lactococcus lactis is an N-acetylglucosaminidase with an optimal number of LysM domains for proper functioning. FEBS J. 2005;272:2854–2868. doi: 10.1111/j.1742-4658.2005.04706.x. [DOI] [PubMed] [Google Scholar]
- Steen A, Buist G, Kramer NE, Jalving R, Benus GFJD, Venema G, Kuipers OP, Kok J. Reduced lysis upon growth of Lactococcus lactis on galactose is a consequence of decreased binding of the autolysin AcmA. Appl Environ Microbiol. 2008;74:4671–4679. doi: 10.1128/AEM.00103-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenbakkers PJM, Geerts WJ, Ayman-Oz NA, Keltjens JT. Identification of pseudomurein cell wall binding domains. Mol Microbiol. 2006;62:1618–1630. doi: 10.1111/j.1365-2958.2006.05483.x. [DOI] [PubMed] [Google Scholar]
- Tarahomjoo S, Katakura Y, Shioya S. Expression of C-terminal repeat region of peptidoglycan hydrolase of Lactococcus lactis IL1403 in methylotrophic yeast Pichia pastoris. J Biosci Bioeng. 2008;105:134–139. doi: 10.1263/jbb.105.134. [DOI] [PubMed] [Google Scholar]
- Turner MS, Hafner LM, Walsh T, Giffard PM. Identification and characterization of the novel LysM domain-containing surface protein Sep from Lactobacillus fermentum BR11 and its use as a peptide fusion partner in Lactobacillus and Lactococcus. Appl Environ Microbiol. 2004;70:3673–3680. doi: 10.1128/AEM.70.6.3673-3680.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visweswaran GRR, Dijkstra BW, Kok J (2010) Two major archaeal pseudomurein endoisopeptidases: PeiW and PeiP. Archaea. doi:10.1155/2010/480492 [DOI] [PMC free article] [PubMed]
- Visweswaran GRR, Dijkstra BW, Kok J. A minimum of three motifs is essential for optimal binding of pseudomurein cell wall-binding domain of Methanothermobacter thermautotrophicus. PLoS One. 2011;6(6):e21582. doi: 10.1371/journal.pone.0021582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Miyake Y, Hisaoka M, Kurosawa SI, Sekiguchi J. The major and minor wall teichoic acids prevent the sidewall localization of vegetative DL-endopeptidase LytF in Bacillus subtilis. Mol Microbiol. 2008;70:297–310. doi: 10.1111/j.1365-2958.2008.06397.x. [DOI] [PubMed] [Google Scholar]