Abstract
A mutant allele of the transcription factor gene MYB10 from apple induces anthocyanin production throughout the plant. This gene, including its upstream promoter, gene coding region and terminator sequence, was introduced into apple, strawberry and potato plants to determine whether it could be used as a visible selectable marker for plant transformation as an alternative to chemically selectable markers, such as kanamycin resistance. After transformation, red coloured calli, red shoots and red well-growing plants were scored. Red and green shoots were harvested from apple explants and examined for the presence of the MYB10 gene by PCR analysis. Red shoots of apple explants always contained the MYB10 gene but not all MYB10 containing shoots were red. Strawberry plants transformed with the MYB10 gene showed anthocyanin accumulation in leaves and roots. No visible accumulation of anthocyanin could be observed in potato plants grown in vitro, even the ones carrying the MYB10 gene. However, acid methanol extracts of potato shoots or roots carrying the MYB10 gene contained up to four times higher anthocyanin content than control plants. Therefore anthocyanin production as result of the apple MYB10 gene can be used as a selectable marker for apple, strawberry and potato transformation, replacing kanamycin resistance.
Electronic supplementary material
The online version of this article (doi:10.1007/s11248-011-9490-1) contains supplementary material, which is available to authorized users.
Keywords: Anthocyanin, Alternative selection marker, Malus x domestica, Solanum tuberosum, Fragaria ananassa
Introduction
Plant transformation methods rely on the delivery, integration and expression of defined genes in plant cells, which can be regenerated into plants. Selection is necessary to separate transformed cells from non-transformed ones because the efficiency of stable transformation is low. Selectable marker genes are used to confer positive or negative selection and ensure growth of only genetically modified cells. Selectable marker genes that are widely used encode for resistance against the antibiotics kanamycin and hygromycin and originate from bacteria. In response to the negative image of these markers in green biotechnology, scientists have developed alternative selection strategies to avoid the use of antibiotics, although the reason for this is political rather than scientific (Ramessar et al. 2007). Examples are the use of marker gene-free vectors (de Vetten et al. 2003) or removable marker gene systems like pMF-1 (Schaart et al. 2004). Alternative selection marker genes that result in the production of plant-based products, which are non-toxic and easy to score, have also been considered (Miki and McHugh 2004 ). The red and purple coloured anthocyanins seem to be promising since they are visible by the human eye, do not require addition of substrate, are not essential for development and are non-toxic. Recent reports (Butelli et al. 2009) have even shown that consumption of anthocyanins can be beneficial for health, such as anti-cancer, which could make crops with increased anthocyanin content attractive from a consumer’s point of view.
This paper describes the replacement of resistance to the antibiotic kanamycin with anthocyanin formation as a selectable marker for plant transformation. Previous efforts in wheat and sugarcane with the anthocyanin regulatory genes C1 and B from maize failed due to the inability of the anthocyanin-producing cells to divide (Ludwig et al. 1990, Chawla et al. 1999) or for regenerating shoots to grow beyond a certain developmental stage (Bower et al. 1996. Recently, wheat embryo-specific expression of maize transcription factor genes for anthocyanin was described by Doshi et al. (2007). The PAP1/myb75 transcription factor gene from Arabidopsis was expressed in tobacco cell cultures and resulted in dark red cells (Zhou et al. 2008), and the maize Lc gene expressed in apple (Li et al. 2007) resulted in high anthocyanin accumulation in transgenic plants, showing the feasibility of obtaining plants transformed with these transcription factor genes. However, none of these reports showed the use of anthocyanin formation as selection criterion or the use of the promoter of the transcription factor gene itself to drive expression.
Espley et al. (2007) isolated an apple transcription factor gene, MYB10, responsible for skin colour of apple fruits. A mutant allele was found in the apple cultivar (cv) Red Field that results in purple-stained skin and flesh of fruits. The mutant allele contains an insertion/repeat in the promoter region that causes ectopic expression of anthocyanins (Espley et al. 2009). The apple plants of cv Red Field accumulate anthocyanin in the leaves, stems, flowers and fruits (Espley et al. 2007).
Here we describe the use of the mutant MYB10 allele from cultivar Red Field as a selectable marker gene. The MYB10 gene, under the control of its own promoter and terminator, was transformed into apple, strawberry (another member of the rosaceous family) and potato as model crop species. Regenerated shoots were harvested from explants without any antibiotic selection or on kanamycin selection. For comparison, a GUS reporter gene construct was transformed in parallel.
Results
Accumulation of anthocyanin after transformation of apple with the MYB10 gene
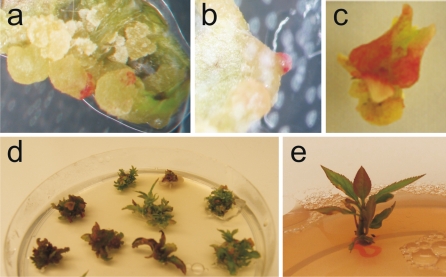
In a pilot experiment apple leaf explants were incubated after transformation on plates without kanamycin and kept in the dark at 24°C for callus induction. Later, shoots regenerated from the calli. Figure 1 shows different stages of regeneration after transformation of apple explants with the MYB10 gene under standard conditions. There was localised accumulation of anthocyanins in some calli (Fig. 1a, b), in shoot-like structures (Fig. 1c), in explants with emerging shoots (Fig. 1d) and in regenerated plantlets (Fig. 1e).
Fig. 1.
Anthocyanin accumulation at different stages of apple regeneration. a and b: calli on explants transformed with the MYB10 gene construct 4–8 weeks after transformation. c: shoot-like structure forming on a callus, approximately 12 weeks after transformation. d: regenerated shoots on explants on a Petri dish, 12–16 weeks after transformation and e: a regenerated plantlet on propagation medium, 20 weeks after transformation
Light intensity, non-specifically, influences accumulation of anthocyanin
To determine in which phase of tissue culture (as early as callus or shoot) and under what conditions phenotypic selection of putative transgenic plants was possible, 600 apple explants were transformed with the MYB10 gene construct in the dark. Four weeks after transformation, 563 calli were harvested, of which a low number of nine showed anthocyanin accumulation (Table 1). These nine anthocyanin accumulating calli were placed at standard light condition for regeneration. Two of those nine calli developed a shoot of which one survived that was completely red. In the dark grown calli harvested from explants transformed with the GUS gene construct or the untransformed controls did not show any anthocyanin accumulation (Table 1). So, the MYB10 gene transformation showed infrequently specific accumulation of anthocyanin in, dark initiated and grown, calli.
Table 1.
Occurrence of red calli four weeks after transformation of the apple explants, incubated in the dark
| Gene | Number of explants | Number of calli analyzed | Number of red calli |
|---|---|---|---|
| MYB10 | 600 | 563 | 9 |
| GUS | 600 | 493 | 0 |
| Untransformed control | 180 | 164 | 0 |
In the dark initiated green calli from the transformation experiment mentioned in Table 1 were exposed to different light intensities: low(only reflected; 15 μE), standard (50 μE) and high (100 μE). Twenty-four hours after the calli were exposed to these different light conditions; they were scored for visible anthocyanin accumulation. A callus was considered red if it was completely red or showing red sectors. The results are presented in Table 2, indicating that anthocyanin accumulation in a low frequency of green calli was observed after transformation with the MYB10 construct but also with the GUS construct and even in the untransformed control. Red coloration of calli in the light was more a-specific. The callus material was further incubated in the climate chamber for regeneration of shoots.
Table 2.
The change to red of green apple calli after exposure to different light intensities
| Gene | Light condition | Number of calli tested | Number of red calli after 24 h exposure to light |
|---|---|---|---|
| MYB10 | Low | 97 | 0 |
| Standard | 93 | 12 | |
| High | 97 | 3 | |
| GUS | Low | 78 | 0 |
| Standard | 88 | 4 | |
| High | 96 | 8 | |
| Untransformed control | Low | 30 | 0 |
| Standard | 30 | 2 | |
| High | 30 | 1 |
Red shoots always contain the inserted MYB10 gene and green shoots sometimes
At a later stage after regeneration, shoots were harvested from explants with red and green calli and scored for their phenotypes. PCR analysis of genomic DNA (gDNA) extracted from the red and green shoots showed that red shoots always contained the construct, as represented by a 500-bp PCR product (Online Resource 1). The 400-bp band is representative of the endogenous MYB10 gene. In a pilot experiment 29 shoots were harvested from 50 leaf explants transformed with the pART27-21 construct not subjected to kanamycin selection. Of the harvested shoots, all 29 were positive when tested by PCR for the endogenous 400 bp band. As is shown in Online Resource 1, 20 out of 29 shoots were also PCR positive for the 500 bp band representing the pART27-21 construct. All 9 red colored shoots were PCR positive as expected, whereas 11 out of the 20 green shoots also showed the same specific PCR product. So all the shoots, visibly accumulating anthocyanin, showed the inserted gene but not vice versa, indicating that shoots containing the specific PCR product of the inserted gene, were either red or green. PCR analysis on the VirG gene showed the absence of Agrobacterium contamination in all of the analyzed shoots.
MYB10 induces anthocyanin accumulation in strawberry and potato
To determine the application range of the MYB10 gene as a selectable marker, this gene was also transformed into the plant model species potato and another member of the rosaceous family, strawberry. Shoots regenerated in both cases directly from the explants, without a clear callus phase. Regenerated shoots emerging from strawberry explants transformed with the MYB10 gene were either red or green. Red-leafed plantlets produced red roots (Fig. 2a–d), which did not occur when the GUS gene was used during transformation. PCR analysis of gDNA extracted from red rooted plants showed a direct relation between anthocyanin accumulation and the presence of the inserted MYB10 gene: all anthocyanin accumulating plants were PCR positive for the inserted gene. However, not all regenerated plants that contained the inserted MYB10 gene showed anthocyanin accumulation. PCR analysis on the VirG gene showed the absence of Agrobacterium contamination in all but one of the regenerated shoots. Potato shoots regenerated from stem segments and further propagated in vitro did not show visible accumulation of anthocyanin after transformation with the MYB10 gene, neither in leaves nor in roots (Fig. 2e, f), although the presence of the inserted gene was confirmed by PCR. Absence of a PCR product for the VirG gene in 24 tested regenerated plants indicated this was not due to Agrobacterium contamination. Overnight incubation of leaves or shoots of MYB10-transformed plants in acid methanol showed that they did contain extractable pigments (Fig. 2g). The absorbance of the extracts was measured and the formula A530 − 0.25*A657 was used to roughly estimate the concentration of presumably anthocyanins. Roots and/or shoots of transgenic potato plants contained up to four times more anthocyanin compared to (un)transformed control (Online Resource 2). Statistical analysis (Anova single factor) of the data showed that the group of PCR + plants differed significantly from the group of PCR- plants (and the untransformed control) for anthocyanin content both in the root and shoot.
Fig. 2.
Visible anthocyanin accumulation in strawberry and potato plants transformed with the MYB10 gene. Figure 3 a and b show the shoots and roots of strawberries transformed with the MYB10 gene c and d show the shoots and roots of a control line. e and f show a potato line transformed with the MYB10 gene without visible anthocyanin accumulation. g shows the extractable pigments after overnight incubation of roots and leaves of different potato lines transformed with the MYB10 gene construct
Anthocyanin content and composition in lines accumulating visibly the highest anthocyanin
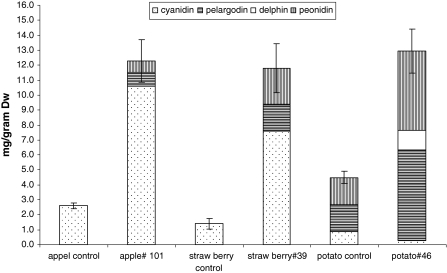
From each species we selected a line that visibly accumulated the highest amount of anthocyanin. These selected lines were PCR+ for the inserted gene construct and were multiplied in vitro. Harvested plant material was analyzed by HPLC to determine total anthocyanin content and anthocyanin composition after acid hydrolysis. The results are shown in Fig. 3. Total anthocyanin content was statistically significantly higher in the MYB10 transformed plants compared to the control lines. P-values using Anova single factor from the analysis tools in Excell software were 0.0137 for apple, 0.0098 for strawberry and 0.022 for potato, respectively.
Fig. 3.
Anthocyanin content expressed as milligram per gram of dry weight (dw) and anthocyanin composition of hydrolysed extracts determined by HPLC of apple, strawberry and potato transgenic line, carrying the MYB10 gene and their (un)transformed control. Anthocyanin chloride standards were used to quantify the anthocyanin aglycones for the hydrolysed samples
In untransformed control apple and strawberry lines, only cyanidin was detected, whereas potato mainly contained pelargonidin and peonidin. The transgenic apple and strawberry lines still accumulated mostly cyanidin-based anthocyanins in addition to detectable levels of pelargonidin and peonidin. Potato plants transformed with the MYB10 gene still accumulated predominantly pelargonidin and peonidin in addition to a considerable amount of the blue delphinidin.
The HPLC analysis was performed with additional MYB10 transformed, anthocyanin accumulating apple, strawberry and potato lines with similar results (data not shown).
Transformation efficiency
Table 3 shows the number of explants, the number of regenerated shoots and PCR positive shoots, and the number of regenerated shoots with a visible phenotype: either GUS blue staining or visible accumulation of anthocyanin. A shoot was considered GUS positive when blue spots or sectors were observed.
Table 3.
Analysis of regenerated shoots from explants transformed with MYB10 or GUS gene construct and selected on kanamycin (50 mg/l) or not
| Species | Gene | Kanamycin added | No. of explants inoculated | No. of shoots tested | No. of shoots with visible phenotype (PCR +) | No. of shoots without visible phenotype (PCR +) | % transformation efficiency for visible phenotype* |
|---|---|---|---|---|---|---|---|
| Apple | MYB10 | No | 150 | 29 | 14a (14) | 15 (6) | 9 |
| Apple | MYB10 | Yes | 95 | 25 | 19a (19) | 6 (3) | 20 |
| Apple | GUS | No | 150 | 53 | 3b | 50 | 2 |
| Apple | GUS | Yes | 95 | 6 | 5b | 1 | 5 |
| Strawberry | MYB10 | No | 50 | 23 | 3 (3) | 20 (4) | 6 |
| Strawberry | MYB10 | Yes | 50 | 31 | 5 (5) | 26 (7) | 10 |
| Strawberry | GUS | No | 50 | 18 | 0 | 18 | 0 |
| Strawberry | GUS | Yes | 50 | 16 | 1 | 15 | 2 |
| Potato | MYB10 | No | 150 | 27 | 10** (10) | 17 (4) | 7 |
| Potato | MYB10 | Yes | 75 | 28 | 13** (13) | 15 (6) | 17 |
| Potato | GUS | No | 150 | 30 | 13 | 17 | 9 |
| Potato | GUS | Yes | 75 | 33 | 18 | 15 | 24 |
These shoots were checked for visible phenotype red anthocyanin accumulation or blue GUS staining, and analysed by PCR for the presence of the MYB10 gene construct (number in brackets)
* Percentage of explants producing a shoot with a visible phenotype
** After incubation in acid methanol
Values with a differ significantly from b with P value < 0.05
nd not determined
It was generally observed in all 3 plant species that the number of PCR positive shoots after MYB10 transformation was higher compared to the number of shoots with a visible phenotype indicating that not all PCR+ shoots showed a visible phenotype. This was not dependent on the presence or absence of kanamycin selection.
Secondly, we observed that transformation efficiencies, expressed as percentage of explants resulting in a shoot with a visible phenotype, were higher when kanamycin was included in the regeneration medium for both the MYB10 and the GUS gene construct in apple, strawberry and potato.
Interestingly, in apple, the transformation efficiency with the MYB10 gene was 9% and with the GUS gene 2% in the regeneration medium without kanamycin and 20 and 5% respectively when kanamycin was included. A similar trend was observed when strawberry explants were transformed with the MYB10 gene or the GUS gene. After transformation with the MYB10 gene, 6% of the explants produced a shoot with visible anthocyanin accumulation. When transforming the GUS construct into explants, no GUS positive shoots were found. In case kanamycin was included in the regeneration medium, a single shoot was found that stained blue for GUS while 10% of the explants produced shoots with visible anthocyanin accumulation. In potato, the transformation efficiencies for the different constructs were of the same order, 7% for the MYB10 gene (determined after overnight acid methanol of leaf material from transformed shoots) compared to 9% for the 35S:: GUS construct (positive staining spots) when kanamycin was omitted. When kanamycin was included the transformation efficiencies were 17% for MYB10 and 24% for GUS.
Detection of chimerism
During the selection for anthocyanin accumulating shoots, those with an overall uniform anthocyanin accumulation were initially chosen for multiple in vitro propagation cycles. Ten apple lines, 12 strawberry lines and 21 potato lines were propagated every 4–6 weeks by cutting the shoot apex and placing that on fresh medium. The plants have been multiplied in vitro for almost 2 years, and after 21 rounds of propagation the apple and strawberry plants still showed uniform red coloration and potato, after extraction in acid methanol, anthocyanin accumulation.
Discussion
MYB10 can be used as a selectable marker gene in apple, strawberry and potato
The results presented in this manuscript show that it is possible to use the MYB10 gene from apple as a selectable marker gene for transformation of apple, strawberry and potato plants using standard transformation protocols. For apple and strawberry anthocyanin accumulation is directly visible in planta, for potato additional extraction in acid methanol is required in order to visualize increased anthocyanin synthesis by red coloration of the extract.
Red shoots, accumulating anthocyanin, always tested positive for the presence of the MYB10 construct. However, not all shoots that are PCR positive for the introduced MYB10 gene showed an anthocyanin accumulating phenotype. This type of variation among individual transgenic plants is often found in plant transformation experiments and can be explained by variation in expression of the introduced MYB10 gene caused by copy number, position of integration or silencing of the transgene (Butaye et al. 2005).
The possibility of chimerism
When the GUS construct was used for transformation, several plants with blue-staining sectors or spots were observed. This patchy GUS staining was observed in regenerating shoots both when kanamycin was omitted and when it was included in the medium (Online Resource 3). This non-uniform GUS staining can be attributed to the staining procedure, gene silencing, or chimerism of the regenerated shoot. Chimeras in apple have been reported in transformants of apple obtained without use of a selectable marker (Malnoy et al. 2010) at a low frequency. In two out of 26 transgenic apple lines that were initially selected on kanamycin but propagated for 4 years in vitro under non selective conditions (Flachowsky et al. 2008) chimerism was detected as well. Similarly, in tobacco transformation under non selective conditions, one third of the recovered shoots were shown to be chimeric (Li et al. 2009) as opposed to 6–7% when kanamycin was used as selection marker. So chimerism, not observed here in our experiments, is expected to occur occasionally. An additional phase after selecting the putative transgenic plants, such as re-regeneration could reduce the problem of chimerism.
In our study, 10 apple, 12 strawberry and 21 potato lines were multiplied for 21 rounds of propagation and the plants still showed uniform red coloration. On the basis of red coloration these are expected to be uniformly transformed, but the ultimate proof will include future experiments where several rounds of regeneration of transformed plants will be analysed for their possible chimerical nature.
Conditions for selecting putative transgenic plants
Our results showed that for apple a possible early phase for selecting putative transformants is callus initiated in the dark. Red calli were observed that produce a red shoot, although this happened at very low frequencies. Upon transformation of potato or strawberry there is a very short callus phase so the shoot stage is the earliest opportunity for testing.
Environmental factors, especially light and temperature are also able to induce the production of anthocyanins (Rabino and Mancinelli 1986). Exposing regenerating plants to high light intensity usually was accompanied by an increased anthocyanin accumulation, even in control plants.
Selection for anthocyanin accumulating shoots in the dark would prevent light induction of endogenous anthocyanins but unfortunately also causes a decrease in shoot outgrowth (Predier and Malavasi 1989). Using our standard transformation conditions we were able to select transgenic plants when we selected for anthocyanin accumulating shoots after regeneration in the light.
Here, in apple and strawberry regenerating shoots with very high anthocyanin accumulation died. Apparently anthocyanin accumulation can be toxic-possibly by influencing the redox state of the cell or by sequestering all available carbon and nitrogen. Similar results were described by Bower et al. 1996, who described constitutive expression of the maize R and C1 gene to be lethal for regenerating sugarcane shoots beyond a certain developmental state. The differences between the experiments are in the promoter that was used to drive expression of the anthocyanin inducing transcription factor genes (35S CaMV versus MYB10), the origin of the coding part of the genes (maize versus apple) and the method to deliver the genes into the genome (particle gun bombardment versus Agrobacterium), and the host plant themselves (sugarcane versus apple/strawberry/potato).
In potato, visible accumulation of anthocyanins in intact plants transformed with the MYB10 gene was observed after overnight incubation of leaf or root material in acid methanol. The total anthocyanin content of the transformed apple, potato and strawberry plants detected by HPLC was similar, around one percent of the total dry weight (Fig. 3) for the plants visually showing the highest anthocyanin accumulation. The structural composition of anthocyanidins from the different sources varied, apple and strawberry mainly contained cyanidin and potato mainly contained the red pelargonidin. In contrast to control plants, MYB10 transformed apple and strawberry accumulated detectable amounts of pelargonidin and peonidin. Potato transformed with the apple MYB10 gene accumulated detectable amounts of delphinidin. The differences in anthocyanin amount and composition were not likely to be the reason for the lack of red colour in the potato plants. Colour is not only determined by the amount and chemical structure of the flavonoids, but also by the presence of other coloured pigments and by the vacuolar pH (Tanaka and Ohmiya 2008). It is possible that the anthocyanins formed in potato were not transported to the vacuole or that the vacuolar pH was too high. The pH of grinded potato leaf material in water was measured with a pH electrode to be 5.75 which is consistently higher compared to the pH 5.45 of grinded strawberry leaf material. Submersion of transgenic potato plants in acidic methanol, but not methanol without acid, immediately turns the material red suggesting that the lack of red colour in intact transgenic potato plants is dependent on a relative high pH.
Transformation frequencies for MYB10 and GUS gene constructs
Analysis of the results of transformation frequencies (Table 3) showed that the percentage of transformed plants with the MYB10 gene was higher relatively to the ones transformed with the GUS construct. According to Yao et al. (1995), the transformation efficiency of apple cultivar Royal Gala was described to be to 2.8%, which is comparable to the transformation frequency found using the GUS gene (2–5%). When using the MYB10 construct, the transformation frequency was to 9–20%, suggesting that the presence of the MYB10 gene promotes regeneration. The same was found in strawberry transformation experiments; higher transformation frequency for the MYB10 gene, though in potato the transformation frequency when using the MYB10 gene stayed similar to that of the GUS gene. Logistic regression analysis was performed for experiments with binominal output (success/no success) where a shoot with a phenotype, either GUS positive staining or visible anthocyanin accumulation was defined as success. Possible explaining factors considered were: kanamycin_added, species (apple, strawberry, potato) and gene (MYB10, GUS). The P values were 0.06 for kanamycin added, 0.418 for species and 0.314 for gene type, respectively. From this we conclude that the differences observed in transformation frequencies were not statistically significant. More experiments will have to be carried out to determine whether the transformation efficiency increases when the MYB10 gene is used as a selectable marker or not. Speculation about a possible explanation for increased transformation frequency when using the MYB10 gene is, that the additional anthocyanins help to protect the regenerating cells against oxidative stress by acting as antioxidants (Terahara et al. 2001). Although the precise mechanism is not fully understood, antioxidants in tissue culture improve plant transformation (Dan 2008) probably by protecting tissue against reactive oxygen species (ROS), which is the normal initial plant response after pathogen attack.
Another explanation for the differences between regeneration frequencies may lie in the structure of the vectors used, pART27 versus pK2GW7. To prove that anthocyanin accumulation is a result of expression of the MYB10 gene and not that the binary vector structure is primarily responsible for a higher transformation frequency, experiments will have to include the same binary vector for transformation.
MYB10 gene as a cisgenic or intragenic selectable marker
For apple, the MYB10 gene can even be used as what is referred to as a cisgenic selectable marker. Cisgenesis is a new emerging technology based on the use of natural genes that originate from the target plant itself (Schouten et al. 2006) or from crossable species that could be introgressed through sexual crossing. Similarly, intragenesis is a method based on using all native plant derived functional DNA from the same species or crossable wild species for transformation vectors (Rommens 2004). Both, cisgenesis and intragenesis, combine the benefits of genetic modification while at the same time avoiding many of the societal issues about the expression of foreign genes and proteins in food crops. The application of cisgenic and intragenic technology requires that crop-specific selectable marker genes must be identified. Anthocyanin regulatory and biosynthetic genes seem to be ideal because the biosynthesis pathway is conserved among plant species and a large number of genes involved in the pathway have been identified. Natural mutants with colour phenotypes and altered anthocyanins have been identified in many species, such as grape, apple, pear, potato, maize and petunia (Winkel-Shirley 2001 and references therein), and would be likely candidates to be used as selectable marker genes. Recently, the sweet potato IbMYB1 gene was evaluated for being used as a intragenic marker for sweet potato transformation (Kim et al. 2010), showing the potential application of visible marker genes in transformation of different plant species.
Experimental procedures
Plasmids used for transformation
Two binary plasmids were used for transformation experiments. The T-DNA region of plasmid pART27-21 carries the apple MYB10 genomic region from cultivar Red Field, including 1.8 kb upstream of the translational start, the coding region and 2 kb after the translational stop codon in plasmid pART27 (Gleave 1992) as well as the NPTII gene (Fraley et al. 1983) The plasmid pK2GW7.0 (Karimi et al. 2002) carries the GUS gene (Joersbo and Okkels 1996) under the control of the 35S Cauliflower Mosaic Virus promoter (Odell et al. 1985) and the NPTII gene for resistance against kanamycin.
Plant material, transformation procedure and selection of transformed plants
Plant material of apple (cv Gala), strawberry (cv Calypso) and potato (cv Desiree) were obtained from established tissue cultures in the lab. For transformation purposes, these were micro-propagated on medium including macro- and micronutrients, vitamins (Murashige and Skoog 1962) and 30 g/l sucrose unless stated otherwise. All plant material was placed in a climate chamber with a day/night cycle of 16/8 h and set at 24°C at 15 μE/m2/s unless stated otherwise.
Agrobacterium tumefaciens strain AGL-0 (Lazo et al. 1991), with either binary plasmid pART27-21 or pK2GW7.0, was used for all transformation experiments.
For apple, leaf material of cultivar Gala was used for transformation, according to the method described by Yao et al. (1995). Leaf explants were cocultivated with the bacteria for 3 days and then placed on callus induction medium SIM (including MS salts with vitamins and 3% w/v sorbitol) and incubated in the dark at 24°C, either with or without kanamycin (50 mg/l). After 4 weeks, calli were harvested, put on fresh medium and placed in the light (day/night cycle of 16 h light, 8 h dark). Emerging shoots were placed on SEM medium (including MS salts with vitamins, 2% sucrose, 1% galactose and BAP/GA3) with or without kanamycin. Shoots with several leaves were placed on SPM [including MS salts and vitamins, 3% sucrose and 96 mg/l FeEDDHA (ferric ethylenediaminedi(hydroxyphenylacetate))].
Strawberry cultivar Calypso leaf explants were cocultivated for 3 days with A. tumefaciens carrying either the pART27-21 or pK2GW7.0 plasmids, according to the method described by Schaart et al. (2002) with or without kanamycin (50 mg/l). Shoots regenerated from the explants without a visible callus phase and were harvested 12–16 weeks after start of the experiment.
Potato stem segments of cultivar Desiree were cocultivated for 3 days with A. tumefaciens carrying the pART27-21 or pK2GW7.0 plasmids, according to the method described by Visser (1991). Regenerated shoots were harvested from stem segments, with minimal callus formation, after 4–10 weeks and cultured on MS medium including 3% sucrose, with or without kanamycin (50 mg/l).
Histochemical assay and microscopic analysis
Plant tissues were analysed for ß-glucuronidase activity according to the method described by Jefferson (1987), using a modified buffer containing 100 mM sodium phosphate buffer, pH 7.5, 10 mM EDTA, 0.1% Triton X-100, 1 mM K3[Fe(CN)6/K4[Fe(CN)6.3H2O]] and 2 mM X-gluc (5-bromo-4-chloro-3-indolyl-β-D-glucuronic acid). Tissues were incubated overnight at 37°C, and chlorophyll was extracted for 2 h in 80% ethanol. Blue staining was detected by eye and with the help of a microscope.
PCR detection of MYB10, GUS and VirG
Genomic DNA was isolated from regenerated shoots with a CTAB-based extraction buffer according to the protocol described by Rogers and Bendich (1988).
PCR primers, designed to amplify a 500 base pair region of the MYB10 promoter containing the R6 repeat and a 400 base pair region of the endogenous promoter, have been described by Espley et al. (2009). PCR products were analysed by agarose gel electrophoresis according to standard molecular biology protocols (Sambrook et al. 1989). Primers used to amplify a 819 bp region of the VirG gene to detect the presence of Agrobacterium were forward primer (5′–3′) CCGCGGTCAGCCGCAATTCT, reverse primer (5′–3′) CCTGCACGTCCGCGTCAAAGAAATA.
Extraction and determination of anthocyanins
Total anthocyanin content was determined spectrophotometrically according to the method described by Fuleki and Francis (1968). At least 100 mg of leaf or root material was cut from plants grown in vitro and incubated overnight at 4°C in a solution of 1% (w/v) hydrochloric acid in methanol. The supernatant was collected after centrifugation and transferred to a fresh tube. Absorbance was measured at 530 nm and 657 nm (to correct for chlorophyll) and expressed in absorbance units as anthocyanin content per gram fresh weight.
For determination of the total amount and the composition of the anthocyanins, 1–10 g of plants grown in vitro was freeze-dried and ground in liquid nitrogen. Anthocyanin was extracted in 2 ml of 1% (w/v) acetic acid in methanol with 0.1% butylated hydroxyanisol (BHA). After centrifuging for 5 min, the supernatant was diluted 1:1 with milliQ water and a 100 μl aliquot was separated by reverse phase HPLC using a C18 Thermo column with a gradient of 1% H3PO4:H3PO4 in formic acid/TFA in acetonitrile. For hydrolysis, 2 volumes of 2 N HCl were added, and the sample was heated to 99°C for 2 h. Extraction was carried out twice. Samples were run on the column twice. The anthocyanidin standards (cyanidin, pelargonidin, peonidin and delphinidin chloride) were obtained from Sigma–Aldrich Chemie B.V. (Zwijndrecht, the Netherlands).
pH measurement of leaf homogenates
One hundred milligram of leaf material was grinded in liquid nitrogen. Six millilitre of distilled water was added and the pH was measured immediately with a pH electrode (pH meter CG840, Schott).
Statistical analysis
Statistical analysis was performed with Excell software version 2003.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Green or red phenotypes of regenerated apple shoots, 18 weeks after transformation and presence or absence of the MYB10 transgene (500 bp) and endogenous gene (400 bp) in regenerated apple shoots detected by PCR (PDF 46 kb)
Anthocyanin content in Absorbance Units of potato plants not amplifying the inserted gene (PCR- group) including untransformed control plants and PCR + plants that amplify the MYB10 gene construct. PCR- and PCR + group differ significantly for anthocyanin content. Anova single factor analysis showed P values of P = 0.018 for root and P = 2.54E-05 for shoot respectively (PDF 10 kb)
Histochemical GUS assay on regenerated shoots from explants transformed with the 35S::GUS gene construct (PDF 6072 kb)
Acknowledgments
We would like to thank Dr. Geert Jan de Klerk and Dr. Frans Krens from Wageningen UR Plant Breeding for stimulating discussions, Christel Denneboom, Aranka van de Burgh and Luuk Groenestein for help with the analysis of the transgenic plants and tissue culture work.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Bower R, Elliott AR, Potier BAM, Birch RG. High-efficiency, micro projectile-mediated co transformation of sugarcane, using visible or selectable markers. Mol Breeding. 1996;2:239–249. doi: 10.1007/BF00564201. [DOI] [Google Scholar]
- Butaye KMJ, Cammue BPA, Delauré SL, De Bolle MFC. Approaches to minimize variation of transgene expression in plants. Mol Breeding. 2005;16:79–91. doi: 10.1007/s11032-005-4929-9. [DOI] [Google Scholar]
- Butelli E, Titta L, Giorgo M, Mock HP, Matros A, Peterek S, Schijlen EGWM, Hall RD, Bovy AG, Luo J, Martin C. Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nature Biotech. 2009;26:1301–1308. doi: 10.1038/nbt.1506. [DOI] [PubMed] [Google Scholar]
- Chawla HS, Cass LA, Simmons JA. Developmental and environmental regulation of anthocyanin pigmentation in wheat tissues transformed with anthocyanin regulatory genes. In Vitro Cell. Dev Biol Plant. 1999;35:403–408. doi: 10.1007/s11627-999-0055-4. [DOI] [Google Scholar]
- Dan Y. Biological functions of antioxidants in plant transformation. In Vitro Cell Dev Biol Plant. 2008;44:149–161. doi: 10.1007/s11627-008-9110-9. [DOI] [Google Scholar]
- de Vetten N, Wolters AM, Raemakers K, van der Meer I, Stege R, Heeres E, Heeres P, Visser R. A transformation method for obtaining marker-free plants of a cross-pollinating and vegetatively propagated crop. Nature Biotech. 2003;21:439–442. doi: 10.1038/nbt801. [DOI] [PubMed] [Google Scholar]
- Doshi KM, Eudes F, LaRoche A, Gaudet D. Transient embryo-expression of anthocyanin in wheat. In Vitro Cell Dev Biol Plant. 2007;42:432–438. [Google Scholar]
- Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MYB10. Plant J. 2007;49:414–427. doi: 10.1111/j.1365-313X.2006.02964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espley RV, Brendolise C, Chagne D, Kutty-Amma S, Green S, Volz R, Putterill J, Schouten HJ, Gardiner SE, Hellens R, Allan AC. Multiple repeats of a promoter segment causes transcription factor auto regulation in red apples. Plant Cell. 2009;21:168–183. doi: 10.1105/tpc.108.059329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flachowsky H, Riedel M, Reim S, Hanke MV (2008) Evaluation of the uniformity and stability of T-DNA integration and gene expression in transgenic apple plants. Electronic Journal of Biotechnology 11 (1)1–15. DOI 10.2225/vol11-issue 1-fulltext-10
- Fraley RT, Rogers SG, Horsch RB, Sanders PR, Flick JS, Adams SP, Bittner ML, Brand LA, Fink CL, Fry JS, Galuppi GR, Goldberg SB, Hoffmann NL, Woo SC. Expression of bacterial genes in plant cells. Proc Natl Acad Sci USA. 1983;80:4803–4807. doi: 10.1073/pnas.80.15.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuleki T, Francis FJ. Quantitative methods for anthocyanins. 1. Extraction and determination of total anthocyanin in cranberries. J of Food Sci. 1968;33:72–77. doi: 10.1111/j.1365-2621.1968.tb00887.x. [DOI] [Google Scholar]
- Gleave AP. A versatile binary vector system with a T-DNA organisational structure conductive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol. 1992;20:1203–1207. doi: 10.1007/BF00028910. [DOI] [PubMed] [Google Scholar]
- Jefferson RA. Assaying chimeric genes in plants: the gene fusion system. Plant Mol Biol Rep. 1987;5:387–405. doi: 10.1007/BF02667740. [DOI] [Google Scholar]
- Joersbo M, Okkels FT. A novel principle for selection of transgenic plant cells: positive selection. Plant Cell Rep. 1996;16:219–221. doi: 10.1007/BF01890871. [DOI] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A. Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7:193–195. doi: 10.1016/S1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- Kim CY, Ahn YO, Kim SH, Kim YH, Lee HS, Catanach AS, Jacobs JME, Conner AJ, Kwak SS (2010) The sweet potato IbMYB1 gene as a potential visible marker for sweet potato intragenic vector system. Phys Plant 2010 doi:10.1111/j.1399-3054.2010.01.01365.x [DOI] [PubMed]
- Lazo G, Stein PA, Ludwig RA. A DNA Transformation-competent Arabidopsis genomic library in Agrobacterium. Bio/technology. 1991;9:963–967. doi: 10.1038/nbt1091-963. [DOI] [PubMed] [Google Scholar]
- Li H, Flachowsky H, Fischer TC, Hanke MV, Forkmann G, Treutter D, Schwab W, Hoffmann T, Szankowski I. Maize Lc transcription factor enhances biosynthesis of anthocyanins distinct proanthocyanins and phenylpropanoids in apple (Malus domestica Borkh.) Planta. 2007;226:1243–1254. doi: 10.1007/s00425-007-0573-4. [DOI] [PubMed] [Google Scholar]
- Li B, Xie C, Qiu H. Production of selectable-marker-free transgenic tobacco plants using a non-selection approach: chimerism or escape, transgene inheritance, and efficiency. Plant Cell Rep. 2009;28:373–386. doi: 10.1007/s00299-008-0640-8. [DOI] [PubMed] [Google Scholar]
- Ludwig SR, Bowen B, Beach L, Wessler SR. A regulatory gene as a novel visible marker for maize transformation. Science. 1990;247:449–450. doi: 10.1126/science.247.4941.449. [DOI] [PubMed] [Google Scholar]
- Malnoy M, Boresjza-Wysocka EE, Norelli JL, Flaismann MA, Gidoni D, Aldwinckle HS. Genetic transformation of apple (Malus x domestica) without use of a selectable marker. Tree Gen Gen. 2010;6:423–433. doi: 10.1007/s11295-009-0260-7. [DOI] [Google Scholar]
- Miki B, McHugh S. Selectable marker genes in transgenic plants:applications, alternatives and biosafety. J of Biotech. 2004;107:193–232. doi: 10.1016/j.jbiotec.2003.10.011. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- Odell JT, Nagy F, Chua NH. Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature. 1985;313:810–812. doi: 10.1038/313810a0. [DOI] [PubMed] [Google Scholar]
- Predier S, Malavasi FFF. High frequency shoot regeneration from leaves of apple rootstock M26 (Malus pumila Mill) Plant Cell Tiss Org Cult. 1989;17:133–142. doi: 10.1007/BF00046858. [DOI] [Google Scholar]
- Rabino I, Mancinelli AL. Light, temperature, and anthocyanin production. Plant Physiol. 1986;81:922–924. doi: 10.1104/pp.81.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramessar K, Peremarti A, Gomez-Galera S, Naqvi S, Moralejo M, Munoz P, Capell T, Christou P. Biosafety and risk assessment framework for selectable marker genes in transgenic crop plants: a case of the science not supporting the politics. Transgenic Res. 2007;16:261–280. doi: 10.1007/s11248-007-9083-1. [DOI] [PubMed] [Google Scholar]
- Rogers SO, Bendich AJ. Extraction of DNA from plant tissues. In Plant Molecular Biology Manual A6 (eds. Gelvin, S.B. & Schilperoort R.A.Sec A6, 1–10. Dordrecht: Kluwer Academic Publishers; 1988. [Google Scholar]
- Rommens CM. All-native DNA transformation:a new approach to plant genetic engineering. Trends Plant Sci. 2004;9:457–464. doi: 10.1016/j.tplants.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: a laboratory Manual. 2. Cold Spring Harbour: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schaart JG, Salentijn EMJ, Krens FA. Tissue-specific expression of a β-glucuronidase reporter gene in transgenic strawberry (Fragaria×ananassa) plants. Plant Cell Rep. 2002;21:313–319. doi: 10.1007/s00299-002-0514-4. [DOI] [Google Scholar]
- Schaart JG, Krens FA, Pelgrom KTB, Mendes O, Rouwendaal GJA. Effective production of marker-free transgenic strawberry plants using inducible site-specific recombination and a bifunctional selectable marker gene. Plant Biotech J. 2004;2(3):233–240. doi: 10.1111/j.1467-7652.2004.00067.x. [DOI] [PubMed] [Google Scholar]
- Schouten HJ, Krens FA, Jacobsen E. Do cisgenic plants warrant less stringent oversight. Nature Biotech. 2006;24(7):753. doi: 10.1038/nbt0706-753. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Ohmiya A. Seeing is believing: engineering anthocyanin and carotenoid biosynthetic pathways. Current Opin Biotech. 2008;19:190–197. doi: 10.1016/j.copbio.2008.02.015. [DOI] [PubMed] [Google Scholar]
- Terahara N, Callebout A, Ohba R, Nagata T, Ohnishi-Kameyama M, Suzuki M. Acetylated anthocyanin 3-sophoroside-5-glucosides from Ajuga reptans flowers and the corresponding cell cultures. Phytochemistry. 2001;58:493–500. doi: 10.1016/S0031-9422(01)00172-8. [DOI] [PubMed] [Google Scholar]
- Visser RGF (1991) Regeneration and transformation of potato by Agrobacteriumtumefaciens. In Plant Tissue Culture Manual (ed. Lindsey, K.) Sec. B5, 1–9 (Kluwer Academic Publishers, Dordrecht, Boston, London. 1991
- Winkel-Shirley B. It takes a garden. How work on diverse plant species has contributed to an understanding of flavonoid metabolism. Plant Physiol. 2001;127:1389–1404. doi: 10.1104/pp.010675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao JL, Cohen D, Atkinson R, Richardson K, Morris B. Regeneration of transgenic plants from the commercial apple cultivar Royal Gala. Plant Cell Rep. 1995;14:407–412. doi: 10.1007/BF00234044. [DOI] [PubMed] [Google Scholar]
- Zhou LL, Zeng HN, Shi MZ, Xie DY. Development of tobacco callus cultures over expressing Arabidopsis PAP/MYB75 transcription factor and characterization of anthocyanin biosynthesis. Planta. 2008;229:37–51. doi: 10.1007/s00425-008-0809-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Green or red phenotypes of regenerated apple shoots, 18 weeks after transformation and presence or absence of the MYB10 transgene (500 bp) and endogenous gene (400 bp) in regenerated apple shoots detected by PCR (PDF 46 kb)
Anthocyanin content in Absorbance Units of potato plants not amplifying the inserted gene (PCR- group) including untransformed control plants and PCR + plants that amplify the MYB10 gene construct. PCR- and PCR + group differ significantly for anthocyanin content. Anova single factor analysis showed P values of P = 0.018 for root and P = 2.54E-05 for shoot respectively (PDF 10 kb)
Histochemical GUS assay on regenerated shoots from explants transformed with the 35S::GUS gene construct (PDF 6072 kb)