Abstract
Many mutations that dramatically extend life span in model organisms come with substantial fitness costs. Although these genetic manipulations provide valuable insight into molecular modulators of life span, it is currently unclear whether life-span extension is unavoidably linked to fitness costs. To examine this relationship, we evolved a genetically heterogeneous population of Caenorhabditis elegans for 47 generations, selecting for early fecundity. We asked whether an increase in early fecundity would necessitate a decrease in longevity or late fecundity (antagonistic pleiotropy). Caenorhabditis elegans experimentally evolved for increased early reproduction and decreased late reproduction but suffered no total fitness or life-span costs. Given that antagonistic pleiotropy among these traits has been previously demonstrated in some cases, we conclude that the genetic constraint is not absolute, that is, it is possible to uncouple longevity from early fecundity using genetic variation segregating within and among natural populations.
Keywords: Aging, Antagonistic pleiotrophy, Trade-off, Life history, Experimental evolution
DURING the past three decades, biologists have identified dozens of single-gene mutations capable of significantly extending life span and delaying age-related decline. Most aging genes have been discovered via mutation or gene silencing in the soil roundworm Caenorhabditis elegans [eg, (1–3)], with significant contributions from the yeast Saccharomyces cerevisiae [eg, (4)], and the fruit fly Drosophila melanogaster [eg, (5)]. Mutations in aging genes are capable of extending the life span of laboratory organisms up to 10 times the wild-type length (6). The qualitative effect of these mutations (life-span extension) is evolutionarily conserved across widely divergent taxa, making research in model organisms relevant to the search for interventions for human aging and age-related disease. Although dramatic life-span extension can be achieved via single mutations in the laboratory, most mutations that extend life span are also involved in key metabolic pathways. These mutations are often lethal in homozygous form, and heterozygous individuals are either sterile or have offspring with delayed or arrested development [worms (2), flies (7), mice (8)].
Trade-offs between longevity and fitness, such as those apparent in aging mutants, are central to general life-history theory and the evolution of aging. They are predicted to arise as a result of the removal of suboptimal fitness trait combinations by the environment (natural selection). For example, assuming that in a given environment an individual can acquire a finite amount of energy and invests that energy into activities like foraging, growth, reproduction, and life span, alleles that increase investment into one life-history trait should decrease the total investment into other traits (constraint via genetic architecture). Likewise, investments into a single trait early in life should reduce available resources for the same trait late in life (9). Trade-offs among traits may also be observed if the physiology of the organism imposes a functional constraint, limiting the suite of possible phenotypes [constraint via physiology (10)]. Alleles responsible for such trade-offs are said to be antagonistically pleiotropic: one locus affects multiple traits (pleiotropy) in opposite directions (antagonism) with respect to age-specific fitness. The Antagonistic Pleiotropy (AP) theory of aging is a specific form of the hypothesis describing a genetic trade-off between longevity and one or more early life traits (11). One hypothesized trade-off that has received much attention in the aging literature is that between fertility and longevity—a hypothesis that is commonly supported by observations in aging mutants, regardless of taxon [worms (2), flies (7), mice (8), but see (12)].
Although life-span extending mutations are generally detrimental to fitness, it is not clear if this apparent trade-off represents an underlying functional constraint. Mutations that extend longevity can vary in the magnitude of fitness cost incurred, may be cost free in some environments, do not negatively affect all fitness traits simultaneously, and can be sex specific in their effects. For example, some studies report that C elegans hermaphrodites carrying mild daf-2 mutations retain wild-type reproductive output when raised at 20°C with unlimited food [(13,14), but see (15)]. In addition, Walker and colleagues (16) showed that long-lived C elegans age-1 (hx546) worms maintain the same appearance, development, locomotion, and reproduction as wild type [20°C, unlimited food (1)]. These findings have been taken to indicate that it is possible to extend the life span of worms without fitness effects [eg, (17)]. This is likely a premature conclusion, given the substantial fitness costs incurred by those same mutants under alternate conditions [co-housed with wild type (14) and food limited (14,16)]. However, these findings do provide evidence for allelic and environmentally dependent variation in the magnitude of the observed life-span fitness trade-off.
Can early life fecundity only be achieved at the expense of life span or vice versa? Laboratory selection experiments are powerful tools for answering questions about evolutionary potential and can be classified into two types: (a) artificial selection in which the experimenter selects upon a particular trait by defining which phenotypes will have the highest fitness and (b) experimental evolution in which the experimenter establishes a set of environmental conditions and allows the population to evolve as they might (18). Experimental evolution studies are uniquely poised to address whether fecundity and life span are constrained via AP. Evolving genetically heterogeneous populations under conditions that favor either early life fitness or life span could facilitate the generation of unique allelic combinations and epistatic interactions that enable the dissociation of apparently constrained relationships between life-history traits. As opposed to using phenotypic correlations [misleading due to shared environmental effects (19)] or calculating genetic correlations [typically having large standard errors, making interpretation difficult (20,21)], experimental evolution studies allow us to observe the change in the selected trait and measure its correlated response to selection in the environment in which the traits evolved, ensuring that we measure the genetic relationships that affect the overall genetic architecture of fitness components.
Here, we experimentally evolved genetically heterogeneous populations of C elegans under conditions favoring early life fitness and asked if early fecundity could evolve independently of longevity. Specifically, for 47 generations, we maintained replicate genetically heterogeneous populations of C elegans in discrete generations, propagating only the offspring produced on the first day of reproductive maturity. Based on both general life-history and AP theories, we predicted that early life fitness components should trade off with late life fitness components. In addition, if changes in life span and reproduction are constrained via AP, then populations evolved under conditions favoring increased early fecundity should have shorter life spans than their ancestral population.
METHODS
Strains
To create a genetically heterogeneous population, we used strains AB1, AB3, CB4852, CB4853, CB4855, CB4857, CB4858, N2, PB303, PB306, RC301, PX174, PX178, and PX179 (Caenorhabditis Genetics Center, University of Minnesota, Minneapolis, MN); JU262 and JU345 (received from Marie-Anne Félix, Institut Jacques Monod, Paris, France). Subsequent genomic analysis has revealed that PX174 is identical to RC301 and PX178 is not different from PX179 (E. Anderson, Ph.D., personal communication, 2010). Each strain was inbred by single individual self-fertilization for at least 13 generations to generate isogenic lines and frozen. Escherichia coli strains OP50 and HT115 (DE3; L4440; hereafter HT115) were also obtained from the CGC. Worms were maintained using standard protocols on Nematode Growth Medium-lite (NGM-lite; US Biological; Marblehead, MD), with E coli strain OP50 at 20°C (22,23) during the generation of isogenic lines and the creation of the heterogeneous C elegans population (later).
To generate a heterogeneous population of C elegans, 16 isogenic strains, representing the global diversity of the species, were crossed in a pairwise mating design for eight generations (Supplementary Figure 1). Because outcrossing in C elegans only occurs between males and hermaphrodites and males are present at low frequency in some strains, we first thawed the isogenic strains (earlier) and enriched them for males by mating males that arose spontaneously in each strain with L4 hermaphrodites (fourth larval stage, effectively virgin) of the same strain and then froze the male-enriched populations. The 16 male-enriched isogenic strains were thawed synchronously, and males were mated to L4 hermaphrodites of the same strain for two generations to remove any potential grand-maternal environmental effects. We then mated strain pairs reciprocally (three males to one hermaphrodite) over eight generations to ensure equal contributions of nuclear and mitochondrial DNA from all strains to the resulting heterogeneous strain (Supplementary Figure 1). The resulting heterogeneous strain was grown at large population size and frozen. This strain served as the ancestral population (Generation 0) for experimental evolution.
Experimental Evolution
We evolved replicates of the ancestral population under experimental conditions for 47 generations. These conditions differed from standard in which worms were reared in discrete generations (achieved via hatch-off, see later), and each generation was established exclusively by offspring (embryos) produced by hermaphrodites at age 3 days. Experimental evolution conditions also differed from standard in that worms were reared on NGM-lite supplemented with BactoPeptone (16 g/L), 100 μg/mL ampicillin, and 250 μg/mL Isopropyl β-D-1-thiogalactopyranoside in 100-mm Petri dishes seeded with E coli HT115. Prior to seeding on NGM plates, HT115 was maintained on selective media (Luria Bertani agar with 100 μg/mL ampicillin and 25 μg/mL tetracycline). Single colonies were used to inoculate liquid cultures in selective media (as earlier but without agar), which were grown overnight at 37°C with shaking. These cultures were diluted 1:5 with Luria Bertani plus 100 μg/mL ampicillin and grown at 37°C with shaking an additional 6–8 hours before use. Plates were seeded with 200 μL of the live bacteria culture and will be referred to as ‘EE’ plates. All worms were reared, evolved, and assayed at 20°C.
The ancestral population was thawed on EE plates and allowed to recover from freezing for two generations before being transferred to establish five replicate lines. After approximately one generation, each replicate line was transferred onto five plates per line. The replicate lines (A, B, C, D, and F) were thereafter stage synchronized using a hatch-off procedure [based on Protocol 7, (24)] every generation (approximately 96 hours) for 47 generations. Adults were washed from the plates using S basal and discarded. The remaining eggs were collected in S basal by wiping the agar surface with a glass rod and transferred to a sterile 15-mL plastic culture tube. Eggs from the five plates per line yielded a total of 5 mL of egg/S basal solution when combined in a single 15-mL tube. Residual larval and adult worms were killed, and their corpses dissolved by the addition of 120 μL/mL bleach and 60 μL/mL 4 mM NaOH (5 minutes at room temperature). To remove the bleach and NaOH, the eggs were pelleted by centrifugation (94g for 3–5 minutes) and the supernatant was decanted. The cultures were washed once in S basal, centrifuged, and decanted as earlier. The eggs were then suspended in 5 mL of S basal and incubated on a mechanical rotator for 24 hours. The concentration of live first-larval stage (L1) worms was determined and used to transfer 1,500 worms, by volume, to five replicate plates per line (approximate population size per line per generation = 7,500). Only those embryos produced by adults between approximately 48 and 72 hours post-L1 stage (72–96 hours after the start of the hatch-off) survive the hatch-off treatment and establish the next generation. To maintain the natural ability of C elegans populations to survive freeze–thaw cycles and to facilitate contemporaneous comparison of ancestral and evolved populations, we froze experimentally evolved lines at −80°C (24) after every six generations of selection. Populations remained frozen for at least 3 days before being thawed from the frozen cultures.
Fecundity
Reproductive output and timing were assayed in the ancestral and evolved lines (Generations 6, 24, 47) contemporaneously, using populations revived from frozen stocks. Populations were revived and assayed on EE plates. To insure representative sampling of the population, inclusive of variation in rates of development, stage/age-synchronized cohorts were produced as described earlier and reared on plates for 24 hours before randomly selected second- or third-stage larvae (L2/L3: juvenile developmental stages) were moved to 30-mm EE plates, one worm per plate (ancestor N = 80, each evolved line per generation N = 60). Worms were transferred to individual plates prior to sexual maturation. The sex of each worm was identified the following day, and males were discarded before 30 (ancestor N = 40) hermaphrodites were randomly selected for the assay. Assay individuals were assigned random number identifiers and then organized in numerical order (across all lines and generations) to reduce human and experimental bias. Worms were transferred to new 30-mm plates daily for 4 days, and the progeny resulting from each 24-hour period were counted upon maturation to young adult. Lifetime reproductive success (LRS) was calculated as the total of progeny produced during the 5 days assayed. Any hermaphrodites producing 20 or less offspring were considered outliers and were removed from the data set (N = 15). LRS was normally distributed. Outliers were random with respect to line and generation of evolution. The intrinsic rate of increase, r, was calculated from , where lx is age-specific survivorship to day x and mx is the fecundity at day x (25). LRS and r were computed for assayed individuals from each population at each assayed generation (the ancestor only assayed at Generation 0). Results were analyzed using an analysis of variance with number of generations of evolution as the main effect and line as a random nested effect. Least square means contrasts were used to compare reproduction in the ancestor with that of the evolved lines. Pearson’s correlations were used to look for correlations between the first day of reproduction and residual reproduction for both ancestral and evolved populations [JMP 8.0 (26)].
Longevity
In a separate assay, we determined individual life spans for ancestral (Generation 0) and evolved worms (Generation 47 only). The life-span assay was performed in two blocks. This assay was initiated identically to the fecundity assay described earlier except that more worms were collected at the L2/L3 stage to produce sample sizes of 70 for each evolved line and 200 for the ancestor per batch, following the removal of males. The worms were transferred to new 30-mm EE plates daily for 4 days to prevent crowding by offspring and then moved once weekly to new plates with fresh bacterial lawns. The status of each worm (live/dead) was evaluated daily starting 48 hours post-L1. Worms were declared dead when pharyngeal pumping was not observed, and they failed to respond by moving (within 3 seconds) after being prodded with a sterile platinum wire. Worms that were missing for three or more consecutive days and those that died from handling were recorded as right censored on the day they were last seen alive. Fewer than 6% of worms per population were censored (range = 0%–5.71%). We used the Kaplan–Meier method to compute mean life span and standard errors (PROC LIFEREG). We used the Cox proportional hazards method (PROC PHREG) to compare age-specific risk of death (mortality) of evolved lines with the ancestral line, with line and block (line) as main effects, and Breslow’s approximation for handling tied events with censored data (27). We performed Pearson’s correlations to test for an evolutionary genetic correlation between late life survival and early adult fertility [SAS, (28)].
Development
In this experiment, increased early reproduction is favored by selection because only offspring produced on the first day of sexual maturity, age 3 days, survive and reproduce each generation. It is possible that these offspring are produced by hermaphrodites that attain sexual maturity relatively early; thus, increased rates of larval development could evolve as a correlated response to selection under these conditions. To evaluate developmental timing in the ancestor and evolved lines (Generation 47), we repeatedly sampled worms from synchronized populations over the course of development as follows. Stage-synchronized populations of each strain were produced via hatch-off and grown on eight replicate plates per strain. Worms were harvested every 2 hours beginning 34.5–48.5 hours post-L1, fixed in methanol, and stored in S basal at 4°C until they were stained (<2 days) with 333 ng/mL or 666 ng/mL 4′,6′-diamidino-2-phenylindole hydrochloride (DAPI). At least 30 randomly selected worms per strain per time point were photographed on a light microscope at 10× magnification using florescent light and a DAPI filter set. The images were given random numerical identifiers and then sorted by random number to allow blind and unbiased scoring of the images. Worms were scored (0/1) as having discernable spermatids, oocytes, or embryos (Supplementary Figure 2). We compared the mean age at observation of spermatids, oocytes, and embryos for each evolved line with the ancestral using Dunnett’s method. We used an ordinal logistic regression (PROC GENMOD to compute the odds of observing advanced developmental stages (presence of spermatids, oocytes, or embryos) at age 3 days and to compare the odds of each evolved line with that of the ancestral line (proportional odds ratios, evolved:ancestral).
RESULTS
Reproduction
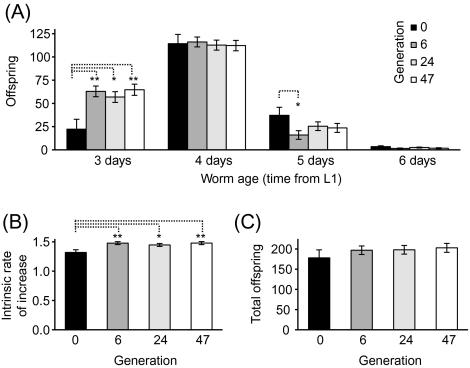
Within only six generations, the number of progeny produced by experimentally evolved worms (average of A, B, C, D, and F) on the first day of sexual maturity, age 3 days, increased to three times that of the ancestral population (F1,16.3 = 11.25, p = .004; Figure 1A). This increase in early reproductive output was maintained through Generations 24 (F1,16.3 = 8.09, p = .012) and 47 (F1,16.3 = 11.98, p = .003) of experimental evolution. The evolved lines decreased reproductive output on Day 5 (significant at Generation 6 [F1,16.5 = 4.68, p = .046]). These changes in reproductive output are reflected by the significant overall increase in population growth rate relative to the ancestor (Whole model: Generation F3,424 = 3.40, p = .043; Figure 1B) in Generations 6 (F1,16.3 = 8.97, p = .008), 24 (F1,16.3 = 5.75, p = .029), and 47 (F1,16.3 = 5.75, p = .028). The average evolved population at Generation 6 increases at a rate 17% faster than the ancestral population. Population growth rate was significantly higher in all lines at all assayed generations with only two exceptions (A-24 and C-24; Table 1). However, worms from the ancestral and evolved populations produced on average the same number of offspring over the course of their reproductive life spans (Whole model: Generations F3,424 = 0.40, p = .752; 6, F1,16.3 = 0.71, p = .410; 24, F1,16.3 = 0.79, p = .396; 47, F1,16.3 = 1.20, p = .288; Figure 1C). LRS differed between the ancestor and evolved lines in only 5 of the 15 comparisons (A-24, D-6, D-47, F-6, and F-47; p < .05, Table 1); in these cases, LRS in the ancestral population was lower than in the evolved lines.
Figure 1.
Evolution of reproductive patterns in populations evolved under conditions favoring early fecundity. (A) Age-specific offspring production. (B) Intrinsic rate of increase. (C) Total offspring (lifetime reproductive success). All results (mean ± 1 SEM) are based on reproduction by self-fertilized hermaphrodites. Results analyzed via analysis of variance with least squared mean contrasts for comparison of ancestor to the evolved lines. Significances indicated by asterisks: *p ≤ .05, **p ≤ .01.
Table 1.
Life-History Phenotypes of Ancestral and Evolved Populations
| Reproduction |
Life Span |
Development |
|||||||
| Line | Generation | r | LRS | Mean | Hazard Ratio | Time to Spermatids | Time to Oocytes | Time to Embryos | Odds Ratio, Age of 3 Days |
| Ancestor | 0 | 1.32 (0.02) | 177.73 (8.90) | 13.09 (0.18) | NA | 45.90 (0.16) | 47.03 (0.13) | 48.07 (0.16) | NA |
| A | 6 | 1.42 (0.03)** | 177.03 (10.45) | ||||||
| 24 | 1.56 (0.03)*** | 266.34 (10.45)*** | |||||||
| 47 | 1.35 (0.03) | 186.04 (11.74) | 11.98** (0.36) | 1.06*** | 45.44 (0.21) | 46.76 (0.16) | 47.33 (0.16)* | 0.53 (0.23) | |
| B | 6 | 1.46 (0.03)*** | 197.30 (10.83) | ||||||
| 24 | 1.46 (0.03)*** | 181.67 (10.83) | |||||||
| 47 | 1.50 (0.02)*** | 179.43 (10.28) | 14.28** (0.34) | 0.55*** | 45.58 (0.18) | 47.26 (0.14)** | 48.15 (0.15) | 1.86 (0.78) | |
| C | 6 | 1.47 (0.03)*** | 189.85 (11.04) | ||||||
| 24 | 1.34 (0.03) | 163.54 (11.49) | |||||||
| 47 | 1.54 (0.03)*** | 198.33 (10.83) | 13.36 (0.39) | 1.44 | 45.76 (0.16) | 47.02 (0.11) | 47.29 (0.13)** | 0.41 (0.14)* | |
| D | 6 | 1.52 (0.03)*** | 214.96 (10.64)** | ||||||
| 24 | 1.41 (0.03)*** | 193.00 (10.45) | |||||||
| 47 | 1.50 (0.03)*** | 235.57 (10.64)*** | 13.06 (0.38) | 0.90 | 46.01 (0.16) | 47.10 (0.12) | 47.59 (0.11)* | 2.94 (1.37)* | |
| F | 6 | 1.52 (0.03)*** | 205.79 (10.45)* | ||||||
| 24 | 1.47 (0.03)*** | 185.74 (10.83) | |||||||
| 47 | 1.50 (0.03)*** | 215.10 (12.29)* | 13.72 (0.36) | 0.82 | 44.58 (0.18)*** | 46.28 (0.13)** | 47.07 (0.11)*** | 5.97 (3.93)** | |
Note: Summary statistics and comparisons of evolved versus the ancestral populations. r = intrinsic rate of increase. LRS is mean lifetime reproductive success. Mean life span in days was calculated using the Kaplan–Meier method including censored individuals. Hazard ratio is risk of death (life span) relative to the ancestral population. Time to developmental stage was measured in hours post-L1. For all estimates, standard errors are presented in parentheses. All reported significance values are the result of planned contrasts with the ancestor and are indicated via asterisks: *p ≤ .05, ** p ≤ .01, ***p ≤ .0001. Reproduction ancestor N = 40, evolved lines average N = 26. Life span, ancestor N = 393, evolved N = ∼140. Development average N = 46 per line and sample.
Longevity
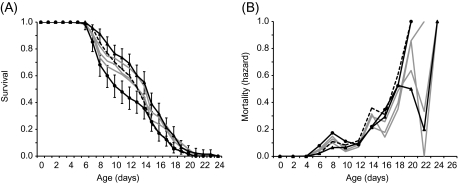
Contrary to expectations from both general life-history and AP theories, we did not observe an overall decrease in mean life span as a correlated response to selection for increased early reproduction (Table 1; Figure 2). Populations A and B evolved significantly different mean life spans relative to the ancestor (Dunnet’s test: Population A is shorter lived, AbsDiff-LSD = 0.28, p = .01; Population B is longer-lived, AbsDiff-LSD = 0.27, p = .01; Table 1; Figure 2A). Age-specific mortality differed significantly among evolved and ancestral lines (χ2 = 27.14, p < .0001; Figure 2B). Mortality was 6% higher than the ancestral population in Population A and 45% lower in Population B (Table 1; Figure 2B). However, no other populations showed life spans that were significantly different from the ancestor. There was no significant correlation between life span and LRS (r2 = .04, p = .22) or early reproduction (adjusted r2 = .001, p = .80).
Figure 2.
(A) Age-specific survival and (B) age-specific hazards of ancestral and evolved populations. Dashed black line indicates ancestor. Gray lines represent evolved populations that do not differ from the ancestor. Solid black lines with circles (Population A) or triangles (Population B) indicate evolved populations that differ significantly from the ancestor (Table 1).
Development
We tracked developmental timing (Supplementary Figure 2) in ancestral and evolved (Generation 47) hermaphrodites to determine if selection for increased early reproduction produced an increased rate of development as a correlated response. Mean developmental timing with 95% confidence intervals is shown in Figure 3. For most of the evolved populations, the age at which sperm or oocytes were observed did not significantly differ from the ancestral population (Table 1). However, four of the five evolved populations had a significantly shorter time to observation of embryos. Population F showed a particularly remarkable compression of developmental timing: Spermatids, oocytes, and embryos were all observed earlier than in the ancestor (spermatids 41%, χ2 = 11.03, p = .0009; oocytes 55%, χ2 = 10.23, p = .001; embryos 230%, χ2 = 38.08, p < .0001). Population D did not develop sperm or eggs earlier but did produce embryos significantly earlier (117%, χ2 = 17.16, p < .0001) than the ancestral population (Table 1).
Figure 3.
Mean developmental timing of ancestral and evolved populations. Proportion mature refers to the proportion of individuals examined in which spermatids, oocytes, or embryos were observed. Dashed gray lines indicate ancestor, solid black lines indicate evolved populations. Triangles = spermatids; circles = oocytes; squares = embryos.
DISCUSSION
The existence of trade-offs between longevity and early fecundity has been proposed as a mechanism for the origin and maintenance of aging in populations (11) and as a possible example of a fundamental constraint on life-history evolution (29,30). We used an experimental evolution paradigm to test predictions of life-history evolution in general and specifically for the presence of an antagonistically pleiotropic relationship (trade-off) between early reproduction and longevity. Our heterogeneous populations of C elegans tripled early reproduction in response to direct selection on that trait (Figure 1A; Table 1). This dramatic increase in early reproduction was accompanied by decreased residual reproduction, resulting in no change in total reproductive output. This suggests a trade-off between early and residual reproduction in the evolved lines and provides empirical support for evolutionary theory on life-history trade-offs. However, the evolved populations did not show a significant overall decrease in life span (Figure 2), failing to provide support for an AP relationship between early fecundity and life span. The lack of a trade-off between longevity and reproduction in our evolved populations seems to contradict existing empirical data from both mutant studies and previous experimental evolution studies, as most have found the relationship (31–33) [but see (34)].
Early Versus Late Reproduction
Assuming a fixed energy budget, life-history theory predicts that increased early resource expenditure, early reproduction in this case, will be accommodated at a cost, for example, reduced late reproduction (9,19,30). Our data are consistent with this prediction. Within six generations of experimental evolution, hermaphrodites increased reproduction at age 3 days to three times that of ancestral hermaphrodites (Figure 1A) and significantly increased population growth rate (Figure 1B), which takes into account both the timing and number of offspring produced. Increased early reproduction was accompanied by decreased late reproduction and occurred without a significant change in LRS (Figure 1C), indicating a clear trade-off between early and late reproductive investment.
The decrease in reproductive output after the age of 3 days could be explained as the result of the accumulation of age-specific mutations. Under our experimental conditions, any allele with age-specific effects later than the age at which adults were sacrificed (4 days) was freed from selection pressure. Although it is possible that mutations specific to ages older than 3 days arose and increased in frequency, empirical evidence from mutation accumulation studies indicates that it is unlikely that this occurred to an appreciable degree: Estimates of the effect sizes of single mutations on fitness traits in C elegans indicate that 47 generations of mutation accumulation cannot account for the magnitude of the observed decrease in late reproductive output unless populations sizes are restricted to one individual per generation (25). Given our large estimates of effective population size (4,500–7,500,Supplementary Table 1), we would expect it to take at least 18,000 generations (4Ne) for neutral mutations to arise and be fixed by genetic drift (35). As such, we can confidently conclude that the observed trade-off between early and late reproduction is a true example of AP and not mutation accumulation due to relaxed selection after age 4 days. The plateau in the response to selection after six generations strongly suggests that the evolutionary response is generated by variation segregating in the ancestral population that is rapidly exhausted after the initial response to selection. We see no evidence for additional novel mutations contributing to the response to selection in later generations.
How can a change in reproductive timing of this magnitude be achieved? It is possible that hermaphrodites that attain sexual maturity relatively quickly produce more early offspring and that these two traits coevolve. Evolved hermaphrodites generally do begin producing offspring earlier than ancestral hermaphrodites (Figure 3; Table 1); however, the average difference in timing is less than 2 hours. Given that the common laboratory strain N2 produces between three and nine eggs per hour at 20°C (36,37), the increased rate of development we observe could account for at most 6–18 of the additional 40 offspring per hermaphrodite in the evolved lines. Thus, evolved hermaphrodites appear to both begin reproducing earlier and produce offspring faster than their ancestors. It is noteworthy that the compression of developmental timing in evolved populations is generally seen only in the time to embryo production and is not observed in the time to spermatid or oocyte production (but see Population F), which delimits the duration of spermatid production in this experiment. Because total spermatid production increases with time and C elegans hermaphrodites are sperm limited, decreases in duration of spermatid production would likely reduce self-fecundity in C elegans (38). These results suggest that the evolution of developmental timing may be constrained but that latency to embryo production or the rate of embryo production can otherwise evolve independently.
Longevity Versus Reproduction
Early reproduction increased 300% in our evolved populations, indicating the presence of extensive segregating genetic variation for this trait in the ancestor. The lack of consistent directional change in longevity demonstrates that in our populations, longevity and early reproduction are not antagonistically pleiotropic on average [see also (34)]. The real question then becomes whether or not this observation is relevant to understanding the genetic basis of aging or whether it is a possible artifact of the experimental design. For example, many previous attempts at artificial selection on life span have resulted in inconsistent or nonsignificant responses among replicate populations. This inconsistency is likely due to problems with small population sizes, namely genetic drift and inbreeding depression [cf (31)]. Artificial generation of inbreeding depression can either induce or obscure naturally segregating pleiotropic effects on reproduction and longevity. One might expect this problem to be magnified in C elegans due to its androdioecious mating system (composed of hermaphrodites and males) and predominant hermaphroditic self-fertilization. However, inbreeding depression is not generally observed in this species due to the effective purging of deleterious mutations and genotypes from C elegans populations (39–42). Additionally, unlike some isogenic C elegans strains, which maintain very few males (0.1%), males were present at high frequency in the ancestral population and remained at high frequency (nearly 30%) throughout experimental evolution (43). Because outcrossing can only be achieved by mating between males and hermaphrodites and outcrossing frequency is positively related to male frequency (44), it is likely that outcrossing was relatively frequent in our populations, keeping levels of inbreeding low.
Alternatively, it is possible that derivation of the genetically heterogeneous ancestral population from crosses between 16 strains of C elegans from around the world resulted in outbreeding depression, which could also induce an abnormal genetic relationship between reproduction and longevity. Under this scenario, both reproductive fitness and longevity could be positively correlated due to purging of initial outbreeding depression and adaptation to laboratory conditions. Although C elegans has relatively low genetic diversity overall, the existing variation is sufficient to generate notable phenotypic variation among strains [eg, thermal preference (45), male frequency (43)], and hybrids among strains have been shown to suffer from outbreeding depression in some cases (39). If outbreeding depression exists in an ancestral population and is purged during evolution, we should observe a significant increase in fitness from the ancestor to the evolved population(s). The intrinsic rate of increase is often considered the most appropriate metric of fitness in age-structured populations undergoing population growth (19). Unfortunately, because r increases with greater early reproduction and early reproduction was the target of our selective regime, this metric of fitness is completely confounded with response to selection in our experiment (ie, r was expected to increase no matter what). As such, we cannot use measures of r to unequivocally detect outbreeding depression and subsequent purging. However, other evidence suggests that a significant level of outbreeding depression is unlikely in our populations. Specifically, we would not expect outbreeding depression to affect only one component of fitness (offspring production at age 3 days) to the exclusion of all others, and yet neither LRS nor life span is significantly lower overall for the ancestor in comparison with the evolved lines. Thus, it is likely that our results reflect an evolutionary response to the experimental evolution regime applied.
Thus, our results do appear to reflect a true lack of significant genetic coupling between reproduction and longevity in the ancestral mixed population. This result is contrary to empirical evidence from artificial selection and experimental evolution experiments in other taxa. In arguably the best-known foray into the experimental evolution of life span, Rose and Charlesworth (31) found that lines selected for late reproduction showed an increase in life span and a correlated decrease in early reproduction. They concluded that the simultaneous increase in life span and decrease in early reproduction are evidences that alleles for these traits are antagonistically pleiotropic (31,46). Similar results have been reported in other studies in D melanogaster (32,33,47) and the melon fly Bactrocera cucurbitae (48). The likely explanation for the discrepancy between these studies and the current results is that our ancestral population had yet to achieve an evolutionary equilibrium that would allow for a trade-off between reproduction and longevity to evolve.
Trade-offs are expected to emerge when mutations that are either universally advantageous or universally deleterious are either fixed or eliminated from the population, leaving only alleles with conflicting patterns of pleiotropic effects to segregate in populations (49). In other words, we expect AP patterns to emerge after selection has acted upon the segregating variation within a population and the population has reached mutation selection balance. Our ancestral population contained alleles from 16 different populations and was frozen soon after creation; it is unlikely that our ancestral population was at equilibrium. We have shown that, although AP between early reproduction and longevity may eventually emerge from the milieu of segregating genetic variation at evolutionary equilibrium, the majority of alleles responsible for segregating variation in early reproduction exhibit no functional trade-off with longevity; as such, the evolution of these traits need not be functionally constrained.
Variation in Antagonistic Pleiotropy—An Emerging View
The trade-off between longevity and early fecundity is a commonly observed phenomenon both in mutation and population genetic studies and has been proposed as a possible example of a fundamental evolutionary constraint on life histories (29,30). We used an experimental evolution paradigm to test for the presence of this genetic relationship in a genetically diverse population of C elegans. We tested fecundity and longevity under the same conditions in which the worms were evolved, thereby avoiding interactions among genes and environment. In addition, our populations had large effective population sizes, eliminating the potential for confounding effects from inbreeding and mutation accumulation. We conclude that our populations show an AP relationship between early and late reproduction and not between life span and early fecundity, illustrating that it is possible to uncouple longevity from early fecundity using segregating genetic variation from natural isolates.
Our data, in combination with existing comparative evidence on the genetic architecture of life span and reproduction, suggest that while AP can dictate relationships among fitness components, it does not always represent a strict and unbreakable evolutionary constraint. It is becoming evident that genetically based trade-offs among life-history traits can differ among taxa [(33,50,51), results herein], among populations (31,34), within populations over time (31,52), among fitness components [(13,53), results herein], and across environments (14,16). Whether or not a particular hypothesized trade-off will actually have an evolutionary consequence is therefore best addressed in the context of an evolutionary experiment.
FUNDING
Funding support from the National Science Foundation (DEB-0236180 and DEB-0641066); the National Institutes of Health (P01-AG022500); Ruth L. Kirschstein NRSA Postdoctoral Fellowships (HD055057 to J.L.A. and AG032900 to R.M.R.); and The Ellison Medical Research Foundation.
SUPPLEMENTARY MATERIAL
Supplementary material can be found at: http://biomedgerontology.oxfordjournals.org/
Acknowledgments
We thank Tina Tague, Lori Albergotti, Sarah Martha, Aki Ohdera, and the rest of Team Worm for substantial laboratory assistance; Danielle Hamill, Tim Schedl, and Paul Fox for helpful suggestions regarding DAPI staining in C elegans; and Bill Cresko and Brendan Bohannon for use of reagents and equipment. We would also like to thank the members of the Bill Cresko, Joe Thornton, and Henrique Teotónio laboratories for fruitful discussions about this work. We are grateful to two anonymous reviewers for their helpful suggestions and insights. Strains used in the work were provided by the Caenorhabditis Genetics Center (funded by the National Institutes of Health National Center for Research Resources) and generously donated by Marie-Anne Félix (Institut Jacques Monod, Paris, France).
References
- 1.Friedman DB, Johnson TE. A mutation in the age-1 gene in Caenorhabditis elegans lengthens life and reduces hermaphrodite fertility. Genetics. 1988;118:75–86. doi: 10.1093/genetics/118.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 3.Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: an HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- 4.Kennedy BK, Austriaco NR, Zhang JS, Guarente L. Mutation in the silencing gene sir4 can delay aging in Saccharomyces cerevisiae. Cell. 1995;80:485–496. doi: 10.1016/0092-8674(95)90499-9. [DOI] [PubMed] [Google Scholar]
- 5.Clancy DJ, Gems D, Harshman LG, et al. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science. 2001;292:104–106. doi: 10.1126/science.1057991. [DOI] [PubMed] [Google Scholar]
- 6.Ayyadevara S, Alla R, Thaden JJ, Shmookler Reis RJ. Remarkable longevity and stress resistance of nematode PI3K-null mutants. Aging Cell. 2008;7:13–22. doi: 10.1111/j.1474-9726.2007.00348.x. [DOI] [PubMed] [Google Scholar]
- 7.Tatar M, Kopelman A, Epstein D, Tu MP, Yin CM, Garofalo RS. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science. 2001;292:107–110. doi: 10.1126/science.1057987. [DOI] [PubMed] [Google Scholar]
- 8.Holzenberger M, Dupont J, Ducos B, et al. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature. 2003;421:182–187. doi: 10.1038/nature01298. [DOI] [PubMed] [Google Scholar]
- 9.Williams GC. Natural selection, the costs of reproduction, and a refinement of Lack's Principle. Am Nat. 1966;100:687–690. [Google Scholar]
- 10.Arnold SJ. Constraints on phenotypic evolution. Am Nat. 1992;140(suppl 1):S85–S107. doi: 10.1086/285398. [DOI] [PubMed] [Google Scholar]
- 11.Williams GC. Pleiotropy, natural selection, and the evolution of senescence. Evolution. 1957;11:398–411. [Google Scholar]
- 12.Luo S, Shaw WM, Ashraf J, Murphy CT. TGF-beta sma/mab signaling mutations uncouple reproductive aging from somatic aging. PLoS Genet. 2009;5:e1000789. doi: 10.1371/journal.pgen.1000789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gems D, Sutton AJ, Sundermeyer ML, et al. Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction and longevity in Caenorhabditis elegans. Genetics. 1998;150:129–155. doi: 10.1093/genetics/150.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jenkins NL, McColl G, Lithgow GJ. Fitness cost of extended lifespan in Caenorhabditis elegans. Proc R Soc Lond B Biol Sci. 2004;271:2523–2526. doi: 10.1098/rspb.2004.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen JJ, Senturk D, Wang JL, et al. A demographic analysis of the fitness cost of extended longevity in Caenorhabditis elegans. J Gerontol A Biol Sci Med Sci. 2007;62:126–135. doi: 10.1093/gerona/62.2.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walker DW, McColl G, Jenkins NL, Harris J, Lithgow GJ. Natural selection—evolution of lifespan in C. elegans. Nature. 2000;405:296–297. doi: 10.1038/35012693. [DOI] [PubMed] [Google Scholar]
- 17.Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- 18.Garland T, Rose MR. Experimental Evolution: Concepts, Methods, and Applications of Selection Experiments. Berkeley, CA: University of California Press; 2009. [Google Scholar]
- 19.Roff DA. The Evolution of Life Histories: Theory and Analysis. New York: Chapman & Hall; 1992. [Google Scholar]
- 20.Gromko MH. Unpredictability of correlated response to selection: pleiotropy and sampling interact. Evolution. 1995;49:685–693. doi: 10.1111/j.1558-5646.1995.tb02305.x. [DOI] [PubMed] [Google Scholar]
- 21.Lynch M, Walsh B. Genetics and Analysis of Quantitative Traits. Sunderland, MA: Sinauer Associates; 1998. [Google Scholar]
- 22.Brenner S. Genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sulston JE, Hodgkin J. Methods. In: Wood WB, editor. The Nematode Caenorhabditis elegans. Cold Spring Harbor, New York: Cold Spring Harbor Laboratories; 1988. pp. 110–156. [Google Scholar]
- 24.Stiernagle T. Maintenance of C. elegans. The practical approach series. In: Hope IA, editor. C. elegans: A Practical Approach. Oxford, UK: Oxford University Press; 1999. pp. 51–67. [Google Scholar]
- 25.Vassilieva LL, Lynch M. The rate of spontaneous mutation for life-history traits in Caenorhabditis elegans. Genetics. 1999;151:119–129. doi: 10.1093/genetics/151.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.SAS, Inc. I Jmp® 8. Version 8. Carey, NC: SAS Institute; 1989. [Google Scholar]
- 27.Breslow N. Covariance analysis of censored survival data. Biometrics. 1974;30:89–99. [PubMed] [Google Scholar]
- 28.SAS, II. Sas 9.1.3. Carey, NC: SAS Institute; 2002. [Google Scholar]
- 29.Parker GA, Maynard Smith J. Optimality theory in evolutionary biology. Nature. 1990;348:27–33. [Google Scholar]
- 30.Stearns SC. The Evolution of Life Histories. Oxford, UK: Oxford University Press; 1992. [Google Scholar]
- 31.Rose MR, Charlesworth B. Genetics of life-history in Drosophila melanogaster. 2. Exploratory selection experiments. Genetics. 1981;97:187–196. doi: 10.1093/genetics/97.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luckinbill LS, Arking R, Clare MJ, Cirocco WC, Buck SA. Selection for delayed senescence in Drosophila melanogaster. Evolution. 1984;38:996–1003. doi: 10.1111/j.1558-5646.1984.tb00369.x. [DOI] [PubMed] [Google Scholar]
- 33.Zwaan B, Bijlsma R, Hoekstra RF. Direct selection on life span in Drosophila melanogaster. Evolution. 1995;49:649–659. doi: 10.1111/j.1558-5646.1995.tb02301.x. [DOI] [PubMed] [Google Scholar]
- 34.Partridge L, Fowler K. Direct and correlated responses to selection on age at reproduction in Drosophila melanogaster. Evolution. 1992;46:76–91. doi: 10.1111/j.1558-5646.1992.tb01986.x. [DOI] [PubMed] [Google Scholar]
- 35.Hartl DL, Clark AG. Principles of Population Genetics. 3rd ed. Sunderland, MA: Sinauer; 1997. [Google Scholar]
- 36.Ward S, Carrel JS. Fertilization and sperm competition in the nematode Caenorhabditis elegans. Dev Biol. 1979;73:304–321. doi: 10.1016/0012-1606(79)90069-1. [DOI] [PubMed] [Google Scholar]
- 37.Byerly L, Cassandra RC, Russell RL. Life-cycle of nematode Caenorhabditis elegans. 1. Wild-type growth and reproduction. Dev Biol. 1976;51:23–33. doi: 10.1016/0012-1606(76)90119-6. [DOI] [PubMed] [Google Scholar]
- 38.Cutter AD. Sperm-limited fecundity in nematodes: how many sperm are enough? Evolution. 2004;58:651–655. [PubMed] [Google Scholar]
- 39.Dolgin ES, Charlesworth B, Baird SE, Cutter AD. Inbreeding and outbreeding depression in Caenorhabditis nematodes. Evolution. 2007;61:1339–1352. doi: 10.1111/j.1558-5646.2007.00118.x. [DOI] [PubMed] [Google Scholar]
- 40.Johnson TE, Hutchinson EW. Absence of strong heterosis for life-span and other life-history traits in Caenorhabditis elegans. Genetics. 1993;134:465–474. doi: 10.1093/genetics/134.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson TE, Wood WB. Genetic analysis of life-span in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1982;79:6603–6607. doi: 10.1073/pnas.79.21.6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chasnov JR, Chow KL. Why are there males in the hermaphroditic species Caenorhabditis elegans? Genetics. 2002;160:983–994. doi: 10.1093/genetics/160.3.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anderson JL, Morran LT, Phillips PC. Outcrossing and the maintenance of males within C. elegans populations. J Hered. 2010;101(suppl 1):S62–S74. doi: 10.1093/jhered/esq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morran LT, Cappy BJ, Anderson JL, Phillips PC. Sexual partners for the stressed: facultative outcrossing in the self-fertilizing nematode Caenorhabditis elegans. Evolution. 2009;63:1473–1482. doi: 10.1111/j.1558-5646.2009.00652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anderson JL, Albergotti L, Proulx S, Peden C, Huey RB, Phillips PC. Thermal preference of Caenorhabditis elegans: a null model and empirical tests. J Exp Biol. 2007;210:3107–3116. doi: 10.1242/jeb.007351. [DOI] [PubMed] [Google Scholar]
- 46.Rose MR. Laboratory evolution of postponed senescence in Drosophila melanogaster. Evolution. 1984;38:1004–1010. doi: 10.1111/j.1558-5646.1984.tb00370.x. [DOI] [PubMed] [Google Scholar]
- 47.Gasser M, Kaiser M, Berrigan D, Stearns SC. Life-history correlates of evolution under high and low adult mortality. Evolution. 2000;54:1260–1272. doi: 10.1111/j.0014-3820.2000.tb00559.x. [DOI] [PubMed] [Google Scholar]
- 48.Miyatake T. Genetic trade-off between early fecundity and longevity in Bactrocera cucurbitae (Diptera: Tephritidae) Heredity. 1997;78:93–100. doi: 10.1038/sj.hdy.6880900. [DOI] [PubMed] [Google Scholar]
- 49.Rose MR. Evolutionary Biology of Aging. New York: Oxford University Press; 1991. [Google Scholar]
- 50.Sokal RR. Senescence and genetic load: evidence from Tribolium. Science. 1970;167:1733–1734. doi: 10.1126/science.167.3926.1733. [DOI] [PubMed] [Google Scholar]
- 51.Tatar M, Carey JR, Vaupel JW. Long-term cost of reproduction with and without accelerated senescence in Callosobruchus maculatus analysis of age-specific mortality. Evolution. 1993;47:1302–1312. doi: 10.1111/j.1558-5646.1993.tb02156.x. [DOI] [PubMed] [Google Scholar]
- 52.Leroi AM, Chippindale AK, Rose MR. Long-term laboratory evolution of a genetic life-history trade-off in Drosophila melanogaster. 1. The role of genotype-by-environment interaction. Evolution. 1994;48:1244–1257. doi: 10.1111/j.1558-5646.1994.tb05309.x. [DOI] [PubMed] [Google Scholar]
- 53.Estes S, Ajie BC, Lynch M, Phillips PC. Spontaneous mutational correlations for life-history, morphological and behavioral characters in Caenorhabditis elegans. Genetics. 2005;170:645–653. doi: 10.1534/genetics.104.040022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.