Abstract
A multisubunit form of acetyl coenzyme A (CoA) carboxylase (ACCase) from soybean (Glycine max) was characterized. The enzyme catalyzes the formation of malonyl CoA from acetyl CoA, a rate-limiting step in fatty acid biosynthesis. The four known components that constitute plastid ACCase are biotin carboxylase (BC), biotin carboxyl carrier protein (BCCP), and the α- and β-subunits of carboxyltransferase (α- and β-CT). At least three different cDNAs were isolated from germinating soybean seeds that encode BC, two that encode BCCP, and four that encode α-CT. Whereas BC, BCCP, and α-CT are products of nuclear genes, the DNA that encodes soybean β-CT is located in chloroplasts. Translation products from cDNAs for BC, BCCP, and α-CT were imported into isolated pea (Pisum sativum) chloroplasts and became integrated into ACCase. Edman microsequence analysis of the subunits after import permitted the identification of the amino-terminal sequence of the mature protein after removal of the transit sequences. Antibodies specific for each of the chloroplast ACCase subunits were generated against products from the cDNAs expressed in bacteria. The antibodies permitted components of ACCase to be followed during fractionation of the chloroplast stroma. Even in the presence of 0.5 m KCl, a complex that contained BC plus BCCP emerged from Sephacryl 400 with an apparent molecular mass greater than about 800 kD. A second complex, which contained α- and β-CT, was also recovered from the column, and it had an apparent molecular mass of greater than about 600 kD. By mixing the two complexes together at appropriate ratios, ACCase enzymatic activity was restored. Even higher ACCase activities were recovered by mixing complexes from pea and soybean. The results demonstrate that the active form of ACCase can be reassembled and that it could form a high-molecular-mass complex.
The ATP-dependent carboxylation of acetyl CoA to yield malonyl CoA is a primary reaction that occurs during de novo fatty acid biosynthesis. ACCase (EC 6.4.1.2), the enzyme that directs this reaction, carries out a two-step process. The first step in the reaction involves the ATP-dependent carboxylation of a biotin prosthetic group in the enzyme. The second step results in the transfer of the carboxyl from carboxybiotin to acetyl CoA to form malonyl CoA. This is a regulated step in de novo fatty acid biosynthesis (Page et al., 1994) and is subject to feedback inhibition by long-chain fatty acids (Shintani and Ohlrogge, 1995).
Two forms of ACCase have been described in plants. The first is a very large (>200 kD) MF protein similar to the one originally discovered in animals (Gregolin et al., 1966). The second is a MS complex with a structure of yet-unknown configuration. The MF enzyme form, also designated as a homomeric- or eukaryotic-type enzyme in the literature, is found in the cytoplasm of many eukaryotic organisms such as mammals (Lopez-Casillas et al., 1988; Ha et al., 1994), yeast (Al-Feel et al., 1992), algae (Roessler and Ohlrogge, 1993), and plants (Gornicki et al., 1994; Roesler et al., 1994; Anderson et al., 1995; Egli et al., 1995; Yanai et al., 1995). The MS type of ACCase, also known as the heteromeric or prokaryotic form in the literature, is best characterized from bacteria such as Escherichia coli (Li and Cronan, 1992a, 1992b; Waldrop et al., 1994). A MS enzyme has also been described in plant chloroplasts (Kannangara and Stumpf, 1972; Sasaki et al., 1993).
The malonyl CoA generated by the cytoplasmic and chloroplast enzymes are used for different purposes. The malonyl CoA produced in cytosol by the MF form of ACCase is used for fatty acid elongation and the biosynthesis of phytoalexins and flavonoids (Ebel et al., 1984), whereas that produced by the chloroplast enzyme is used for de novo fatty acid biosynthesis. Although chloroplasts from members of the Gramineae harbor a homomeric MF ACCase, chloroplasts from many other higher plants contain MS ACCases (Sasaki et al., 1993, 1995; Alban et al., 1994; Konishi et al., 1996). An exception to this generalization is that chloroplasts from Brassica napus seem to contain both MF and MS ACCases (Elborough et al., 1996; Markham et al., 1997; Schulte et al., 1997). The MS ACCase from pea chloroplasts is organized like the MS form of the enzyme from E. coli. The bacterial enzyme has four subunits: BCCP, BC, and two carboxyl transferases, α- and β-CT (Guchhait et al., 1974; Li and Cronan, 1992b). Whereas the three-dimensional structure of the E. coli BC subunit has been reported (Waldrop et al., 1994), those for the other three components are unknown.
The study of chloroplast ACCase by the traditional methods of protein chemistry has proven difficult because the enzyme is extremely unstable (Alban et al., 1994). An alternative to protein purification involves the isolation of cDNAs that encode each of the chloroplast ACCase components, and their use in producing the various subunits in vitro. In this regard, DNAs that encode the chloroplast ACCase β-CT subunit from pea (Sasaki et al., 1993), BC from tobacco (Shorrosh et al., 1995) and Arabidopsis (Bao et al., 1997; Sun et al., 1997), BCCP from Arabidopsis (Choi et al., 1995) and Brassica spp. (Elborough et al., 1996), and α-CT from pea (Pisum sativum) (Shorrosh et al., 1996) have been reported.
Because ACCase is likely to be a primary control point in the pathway for the biosynthesis of oil storage reserves in soybeans (Glycine max), we have undertaken a study of the MS ACCase from this important oil crop. We isolated cDNA clones that encode each subunit from the MS ACCase in developing soybean seeds. Unlike pea, in which there are single copies of genes that encode each of the four ACCase subunits, evidence is reported that each of the nuclear-encoded subunits of soybean ACCase are encoded by small families of genes. Subunits synthesized at the direction of these soybean cDNAs interact with a MS ACCase when they are imported into isolated pea chloroplasts. By comparing the deduced amino acid sequences of the precursors with the chemically determined sequence of the ACCase subunits after import and assembly in the chloroplast, the N-terminal regions constituting the transit sequences in the precursors could be deduced. Antibodies that specifically recognize each component of the MS ACCase were generated with the cloned cDNAs and used to demonstrate that the soybean chloroplast enzyme separates into two large, high-molecular-mass complexes during purification. Recombining these complexes caused a restoration of ACCase activity. The implications of these observations on the regulation of ACCase are discussed.
MATERIALS AND METHODS
Plant Material
Peas (Pisum sativum var Green Arrow) were grown at 18°C in a growth chamber set on a 12-h light/12-h dark cycle. Plants were watered daily with tap water. Soybeans (Glycine max cv Resnik) were collected in the field at mid-maturation, and after removing the pods, they were immediately frozen in liquid nitrogen. The seeds were transferred to −80°C for storage.
Soybean cDNA Library Construction and Screening
A soybean cDNA library was prepared in λ vector ZAP Express following the protocol from the manufacturer (Stratagene cDNA synthesis kit). Poly(A+) mRNA was isolated from cv Resnik soybean seeds collected on the 3rd d of germination and stored in liquid nitrogen. The library totaled 1.3 × 106 primary clones and was amplified once. The degenerate oligonucleotides for α-CT cDNA amplification were designed after an analysis of conserved segments in α-CT gene sequences from other species. Amplification by PCR was carried out with soybean cDNA and resulted in several fragments to probe the cDNA library. Arabidopsis clone VCVDE11, obtained from the Arabidopsis Biological Resources Center (The Ohio State University, Columbus), and that contained the 3′ portion of a BCCP gene, was used as a probe to screen the library at low stringency. Plaque lifts and hybridizations were carried out with nylon membranes according to the manufacturer's recommendations (Magnalift, MSI, Westborough, MA). Plaques positive after a secondary screening were used for in vivo plasmid excision with ExAssist helper phage and Escherichia coli strain XLORL according to the manufacturer's recommendations (Stratagene). The resulting colonies were randomly screened with PCR and then used for phagemid DNA preparation. Inserts were characterized by restriction analysis and partial readings from T3 and T7 primers. Selected clones were finally sequenced by the shifting-primer method (Sequenase 2.0 kit, United States Biochemical).
Probe Preparation and Labeling
The desired DNA fragment (restriction fragment or PCR product) was purified by gel electrophoresis through low melting point agarose (ultrapure grade; BRL). Fragments were excised from the gel and digested for several hours to overnight with the Gelase enzyme (Epicentre, Madison, WI). Labeling was carried out with the DECAPRIME II kit (Ambion, Austin, TX) in accordance with instructions from the manufacturer. After labeling, the probe was filtered through two layers of nitrocellulose BA85 (0.45 μm, Schleicher & Schuell) in a spin column to reduce background, and then denatured by a 5-min incubation in boiling water.
Nucleic Acid Preparation and DNA-Blot Analysis
Total RNA was obtained from germinating soybean seeds with the TriPure isolation reagent from Boehringer Mannheim. Total RNA was used for mRNA preparation with the PolyATract System kit (Promega). Genomic DNA was isolated from young soybean leaves by phenol/chloroform extraction with a final CsCl purification step (Cho, 1988). Restriction enzyme digestions of DNAs for DNA gel blots were performed overnight with appropriate buffers and conditions. Soybean genomic DNA restriction fragments were separated on 0.5% agarose gels cast with 5× Tris-acetate-EDTA buffer. The same buffer was used for electrophoresis, which was at 1 to 1.5 V/cm for 15 to 25 h at 4°C. A vacuum blotting system (VacuGene XL, Pharmacia) was used to transfer nucleic acids from gels to membranes (GeneScreen Plus, DuPont). Protocols recommended by the manufacturer were used for DNA alkaline vacuum transfer. The same buffers and conditions described for library screening were used for blot hybridization.
Preparation of the Plasmid Constructs for in Vivo and in Vitro Expression
Plasmid vector pET16b (Novagen, Madison, WI) was used for expression in E. coli. The constructs used for in vitro transcription-translation experiments were prepared in the pGEMEX vector (Promega). NdeI sites were created at the 5′ termini of the α-CT1, α-CT2, BCCP precursor, β-CT gene, and BC PCR fragments. The oligonucleotide-directed mutagenesis procedure described by Jung et al. (1992) was used for this purpose, except in the case of the α-CT cDNAs, for which a PCR approach was used. Cloning was carried out at NdeI-XhoI sites of the receptor vectors, except for the BC PCR fragment, which was cloned at the NdeI-BamHI site of pET16b. Triple ligation at an internal BstEII site was required for α-CT cDNAs because of the presence of additional NdeI sites in the gene sequence. The fragments of α-CT and β-CT cDNAs were cloned into pET16b/BamHI and NdeI-BamHI vectors. The constructs obtained were used to transform BL21(DE3) E. coli cells. Growing cells were induced at optical densities of 0.4 to 0.6 by addition of isopropyl-β-thiogalactopyranoside to 1 mm and incubated for 2 to 4 h at 37°C. Expressed proteins were analyzed by direct lysis in gel-loading buffer (Laemmli, 1970) with subsequent SDS-PAGE and Coomassie blue staining. For antibody production, the antigens in protein bodies were isolated from the insoluble fraction of lysed cells, solubilized in 6 m urea or 6 m guanidine-HCl, and purified through the nickel-agarose column according to recommendations of the manufacturer (Novagen). The proteins were purified further by preparative SDS-PAGE, and the bands of interest were excised, ground in PBS, and used to immunize female New Zealand White rabbits at the Antibody Production Core Facility (Purdue University Cancer Center, Lafayette, IN).
Transcription-Translation in Vitro and Import of the Translation Products into Isolated Pea Chloroplasts
Plasmids with DNAs encoding ACCase subunits in pGEMEX-1 were isolated in preparative amounts using the modified alkali lysis/PEG precipitation method described by Beilinson (1992). The purified plasmids were used to generate proteins in a coupled transcription-translation (rabbit reticulocyte) system (Promega) and either [35S]Met or [3H]Leu. Pea chloroplasts were isolated essentially as described by Bruce et al. (1994). Import reactions (usually in a volume of 200 μL) contained 0.25 to 0.3 mg/mL chlorophyll, 3 mm MgATP, and an aliquot of translation reaction diluted by 1× IB buffer (50 mm Hepes-KOH, pH 8.0, and 330 mm sorbitol). It was essential to keep the amount of translation reaction in the total reaction mixture below 5%. The import reaction was carried out for 20 to 30 min at 25°C with occasional mixing. Chloroplasts that survived were reisolated through a 40% Percoll cushion, washed and lysed in 25 mm Hepes, pH 8.0, and kept on ice for 10 min. The fractionation of components from the chloroplasts was performed as described by Bruce et al. (1994), or stromal and membrane fractions were separated by rapid ultracentrifugation at 100,000g for 30 min. Preparative import reactions were scaled up to a volume of 10 to 12 mL. Washed chloroplasts were lysed in 50 mm Hepes, pH 8.0, 1 mm EDTA, 5 mm DDT, 1 mm PMSF, 5 mm ε-aminohexanoic acid, and 1 mm benzamidine-HCl for 10 min on ice with subsequent centrifugation at 10,000g for 10 min. The supernatant was further centrifuged at 85,000g over a layer of 0.6 m Suc. Samples were collected from the top of the tubes. Glycerol was added to a final concentration of 10%, and the samples were concentrated in a Centricon-10 (Amicon, Beverly, MA). All preparations were stored at −80°C until use.
Determination of the Cleavage Sites for α-CT and BCCP Transit Peptides
Chloroplast import reactions were scaled up to obtain preparative amounts of clarified pea stroma that contained soybean α-CT2 and BCCP subunits. The in vitro-translated proteins were labeled with [3H]Leu, and a small amount of [35S]Met-labeled subunit was added to facilitate detection. Pea stromal proteins were separated by preparative SDS-PAGE and transferred to PVDF membranes (Bio-Rad). Radioactively labeled bands were detected by autoradiography. Appropriate portions of the membranes were excised and subjected to 25 cycles of protein sequencing at the Purdue Center for Protein Sequencing (West Lafayette, IN). Cycles from sequencing were collected, spotted on glass-fiber discs, and counted in a liquid-scintillation counter.
Determination of the Cleavage Site for the BC Transit Peptide
Pea stromal extract with CPE activity was prepared basically as described by Abad et al. (1989). Briefly, intact chloroplasts were lysed hypotonically by resuspension in a volume of ice-cold 25 mm Hepes-KOH, pH 8.0, to produce a final chlorophyll concentration between 0.8 and 1 mg/mL. After incubation for 30 to 40 min on ice, the lysed chloroplast suspension was centrifuged at 16,000g for 15 min, and then the supernatant that resulted was centrifuged at 140,000g in an ultracentrifuge. The CPE extract from this procedure was stored on ice until used. A typical cleavage reaction consisted of two parts CPE extract, one part in vitro translation reaction (diluted 1:1 with 2× IB buffer that contained 60 mm of the corresponding cold amino acid), and two parts CPE reaction buffer (100 mm Gly-NaOH, pH 9.0, 220 mm KCl, 6 mm MgCl2). Reactions were incubated at 27°C for 60 to 90 min, and then stopped by adding 0.5 volume of SDS-PAGE 3× loading buffer (2% SDS, 10% glycerol, 100 mm DTT, 60 mm Tris-HCl, pH 6.8, 0.001% bromphenol blue). The samples were boiled for 1 min, separated by SDS-PAGE, transferred to PVDF membranes, and sequenced as described above.
Soybean Extracts
Frozen soybeans were ground to a fine powder in a mortar and suspended in three volumes of extraction buffer. The slurry was stirred for 1 h, filtered through two layers of Miracloth (Calbiochem), and centrifuged for 30 min at 30,000g. The filtrate was brought to 20% saturation with ammonium sulfate, stirred for another hour at 4°C, and centrifuged at 30,000g for 20 min. This supernatant was brought to 60% saturation with ammonium sulfate and stirred for another hour. The precipitate was collected by centrifugation and dissolved in fresh extraction buffer that contained 20 mm Hepes, pH 8.0, 0.5 mm DTT, 1 mm EDTA, 10% glycerol, 1 mm PMSF, 5 mm ε-aminohexanoic acid, and 1 mm benzamidine-HCl (buffer A). That suspension was desalted and concentrated by several repeated rounds of ultrafiltration on 50-kD concentrators (Filtron, Northborough, MA). The resulting protein solution was brought to 40 mg/mL and stored at −80°C.
Protein Gel-Blot Analysis
A minigel system for SDS-PAGE (Bio-Rad) was used with 7.5% acrylamide for α-CT proteins and 9% acrylamide for all other MS ACCase components. After separation the proteins were transferred to PVDF membranes (Bio-Rad) with a Hoefer T70 semidry transfer unit. Membranes were washed with TBS and blocked with a solution containing 0.8% blocking reagent (Boehringer Mannheim) and 0.02% NaN3 in TBST (TBS containing 0.1% [v/v] Tween 20). Primary antibodies against β-CT, BCCP, and BC were used at a 1:2,000 dilution, whereas those against α-CT were used at a 1:10,000 dilution in blocking solution. An anti-rabbit IgG-alkaline phosphatase conjugate (Bio-Rad), used as secondary antibodies, was applied at a 1:3,000 dilution. Biotinylated proteins were detected with a streptavidin-alkaline phosphatase conjugate (Boehringer Mannheim) diluted 1:2,000. Blots were visualized with a 5-bromo-4-chloro-3-indolylphosphate-p-toluidine/nitroblue tetrazolium color reagent kit (Bio-Rad).
Chromatographic Procedures
Pea and soybean protein extracts were separated in 1- × 55-cm glass columns packed with Sephacryl 300 HR or 400 HR (Pharmacia). Columns were eluted with buffer A at 0.05 mL/min. For high-salt chromatography, the buffer was supplemented with 0.5 m KCl. Fractions of 0.75 mL were collected and analyzed. Typically, 2 to 5 mg of total protein was loaded on the column in each run. For anion-exchange chromatography, either a 1- × 23-cm glass column or a small, plastic, 5-mL column was packed with Q-Sepharose (Sigma). Proteins were loaded on the columns in buffer A and washed with 1 to 2 volumes of the same buffer. Columns were eluted with a 0 to 0.5 m gradient of KCl in buffer A. A total of 15 to 20 column volumes were collected in 2-mL fractions at 0.5 mL/min. Finally, the columns were washed with 1 to 2 volumes of 2 m KCl in buffer A.
Affinity Chromatography
Monomeric avidin-agarose resin from Sigma was used to capture biotinylated proteins from seed extracts. A 1-mL column was washed according to the manufacturer's recommendations and equilibrated with buffer containing 20 mm Hepes, pH 8.0, 1 mm EDTA, 0.5 mm DDT, 10% glycerol, 1 mm PMSF, 5 mm ε-aminohexanoic acid, 1 mm benzamidine-HCl, and 0.5 m KCl. A total of 4 mg of soybean seed extract was loaded onto the column and incubated for 16 h at 4°C as described previously (Alban et al., 1994). The column was then washed extensively with the same buffer, and the biotinylated proteins were eluted by applying 1 mg/mL biotin (Sigma). Eluates were concentrated with Microcon-10 units (Amicon) and stored at −80°C for analysis.
Polyclonal antibodies against soybean MS ACCase subunits from whole serum were purified by caproic acid/ammonium sulfate precipitation as described by Harlow and Lane (1988). The antibodies were used to derivatize Affi-Gel-10 activated resin (Bio-Rad) according to the manufacturer's recommendations. Coupling yields ranged from 3 to 5 mg of protein per milliliter of resin. Four small (1-mL) columns were constructed, one with each of the antibody resins. Before loading, each column was washed with PBS, 50 mm Tris-HCl (pH 7.5) containing 0.5 m KCl, 0.1 m Gly, pH 2.5, 50 mm Tris-HCl, pH 8.5, 0.1 m ethanolamine, pH 11.5, and then PBS again. Finally, the columns were equilibrated with PBS containing 1% NP40. A soybean seed extract (10 mg of protein) in equilibration buffer was loaded onto each column and shaken overnight at 4°C. The columns were extensively washed with PBS-NP40, PBS, 50 mm Tris-HCl (pH 7.5) containing 0.5 m KCl, and then eluted with 0.1 m Gly, pH 2.5. The eluates were immediately neutralized with Tris buffer, pH 7.5. Then the columns were washed with Tris-HCl, pH 8.5, and eluted with 0.1 m ethanolamine, pH 11.5. This eluate was neutralized with Tris-HCl, pH 7.5. The eluates were concentrated and desalted by repeated cycles of ultrafiltration with concentrators (Centricon-30, Amicon). Analysis showed that the majority of proteins were eluted with 0.1 m Gly.
Assay for ACCase Activity
The procedure described by Alban et al. (1994) was used for determination of ACCase activity.
Computer Analysis
The Genetics Computer Group (Devereux et al., 1984) software package was used for sequence alignments and calculations of molecular mass and pI. Secondary structure predictions were accomplished with the PHDsec program (Rost and Sander, 1994). The prediction sites where proteins were localized in chloroplasts were determined by the PSORT program (Nakai and Kanehisa, 1992), and the COILs program (Lupas et al., 1991) was used for analysis of putative coiled-coil structures.
RESULTS
Gene Cloning and Characterization
Table I summarizes the clones used for these studies, clones that encode putative BC, BCCP, α-CT, and β-CT subunits of soybean ACCase. Their nucleotide sequences can be found in previous publications by Reverdatto et al. (1995, 1996, 1997). Also provided in Table I is the nomenclature we used to identify each cDNA, together with a summary of various parameters that can be calculated for the protein sequences deduced from them.
Table I.
Features of cDNAs encoding soybean multisubunit ACCases
| Gene and Accession No. | Insert Size | Coding Region | ACCase Subunit | Similarity to E. coli Subunit | Deduced Protein
MMa
|
Calculated pI
|
||
|---|---|---|---|---|---|---|---|---|
| Precursor | Mature | Precursor | Mature | |||||
| bp | % | kD | ||||||
| accD (U26948) | 5600 | 1296 | β-CT | 63 | nab | 48.9 | na | 4.86 |
| accB-1 (U40666) | 1071 | 786 | BCCP | 57 | 27.7 | 22.6 | 9.47 | 6.83 |
| accA-1 (U34392) | 2839 | 2127 | α-CT1 | 67 | 78.5 | 73.7 | 7.64 | 6.5 |
| accA-2 (U40979) | 2607 | 2070 | α-CT2 | 67 | 76.9 | 72.1 | 8.63 | 7.7 |
| accC-1 (partial) (U34393) | 871 | 867 | BC-1 (partial) | 68 | 30.3 (partial) | 5.65 (partial) | ||
| accC-2 (AF007100) | 2030 | 1617 | BC-2 | 70 | 58.9 | 52.3 | 7.22 | 5.82 |
| accC-3 (AF068249) | 2080 | 1617 | BC-3 | 70 | 58.8 | 52.2 | 7.22 | 5.91 |
MM, Molecular mass.
na, Not applicable.
The β-CT DNA was obtained as a part of 5.6-kb fragment of soybean ctDNA, whereas several copies each of the BC, BCCP, and α-CT genes were obtained by screening of a cDNA library prepared from seeds 3 d after germination. A cDNA encoding a partial BC subunit was obtained from soybean cDNA by PCR with an oligonucleotide primer to a consensus sequence from a conserved region of BCs described previously. Two additional clones that contained the full-length coding sequences for BC were later obtained after PCR on soybean cDNA using oligonucleotide primers based on the nucleotide sequence of the partial BC clone.
Data obtained from PCR amplification experiments revealed that the accA genes probably contain introns (data not shown). To demonstrate this, PCR amplifications performed with degenerate oligonucleotides resulted in a product approximately 1 kb larger when genomic DNA served as a template, compared with when cDNA was used. A partial nucleotide-sequence analysis of the genomic fragment indicated that the position of the insertion was after the Val-107 codon in the α-CT sequence ATT GAT GTA-(intron)-CAG/A AAG ATG. This DNA sequence is conserved among all of the α-CT genes that have been described.
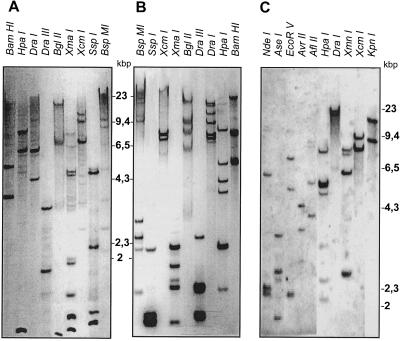
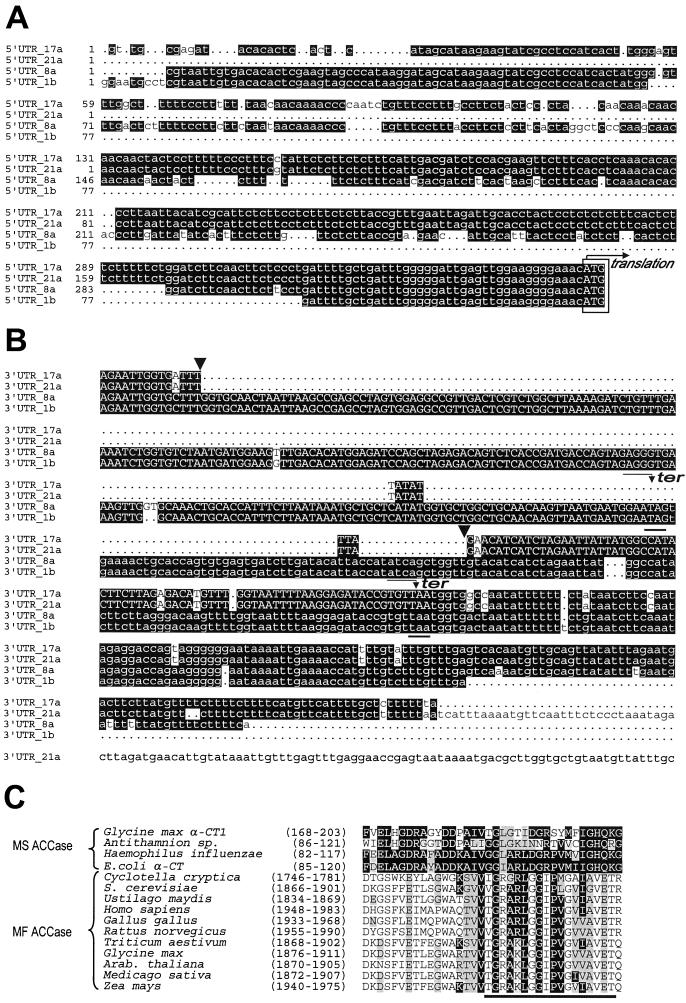
Figure 1 shows the results of a DNA-blot analysis of soybean genomic DNA when probed with accA (α-CT), accB (BCCP), or accC (BC) cDNA. The complex hybridization patterns obtained suggest that in soybean there are several genes that encode each subunit. Consistent with this conclusion, four clones with different nucleotide sequences that encoded α-CT subunits were isolated from the cDNA library. Although the inserts in these clones all had very similar coding regions, they differed from one another in both their 3′- and 5′-UTRs. As is apparent from the data in Figure 2, the α-CT cDNAs can be divided into two groups based on their nucleotide sequence homology. Clones 1b and 8a make up one group, and clones 17a and 21a make up the second. Members of the two groups are easily distinguished by their 5′-UTRs, where considerably more sequence heterogeneity is evident among members of the accA-1 group of clones than among those in the accA-2 group (Fig. 2A). The clones with the longest nucleotide sequences in each group were submitted to GenBank; these correspond to accession numbers U34392 and U40979, respectively.
Figure 1.
DNA gel-blot analysis of soybean genomic DNA. Blots were probed with coding parts of soybean BCCP (A), α-CT (B), and BC (C) cDNAs at high stringency (65°C; see Methods for details). DNA was digested overnight; the restriction enzymes are indicated above each lane. The complexity of the restriction patterns suggests multiple gene copies for each of these ACCase subunits. Positions of λ/HindIII DNA size markers are indicated in thousands of bp.
Figure 2.
Alignment of the 5′-UTR (A) and 3′-UTR (B) regions of soybean accA cDNAs. The ATG start codons are boxed and the terminator codons are underlined. Note that the deletion in clones 17a and 21a (marked with arrowheads) result in a frame shift in the coding region (shown by uppercase letters) such that the C terminus of the protein encoded by these clones is different from the one encoded in clones 1b and 8a. C, Alignment of a segment of ACCase α-CT subunits from different organisms. The region considered to be involved in acetyl CoA binding (Li and Cronan, 1992b) is underlined. All sequences are aligned to the E. coli protein.
Figure 2B shows that there is a 270-bp deletion in the 3′-UTR of accA-2 cDNA compared with accA-1. The deletion includes the end of the accA-1 coding region and the beginning of the 3′-UTR. The deletion in accA-2 results in proteins with different C-terminal ends compared with accA-1. It causes the last 28 codons of the accA-2 cDNA to be included in the 3′-UTR instead of lying at the C terminus of the encoded protein, as in accA-1. As indicated in Table I, the open reading frames in the sequenced inserts translate into α-CT proteins. Those have a high proportion of positively (>17%) and negatively charged (>14%) amino acid residues, which account for roughly one-third of the residues in the entire deduced primary structure of the subunit. There are also a considerable number of hydrophobic amino acids (>25%), almost half of which are Leu (>11%). Secondary structure analysis predicted that the α-CTs contained 58.5% α-helices, 5.8% extended β-sheets, and 35.7% loops.
The cDNA clones that encode complete BC subunits translated into precursors with about 540 amino acids and molecular masses around 60 kD. About one-fourth of the amino acids in the deduced BC proteins are charged, one-half positive and the rest negative. Secondary structure predictions for BC-2 indicate that it is a protein with 34.4% α-helix, 17.6% β-sheet secondary structure, and the remainder loops. It is interesting that the deduced protein sequence from the partial accC-1 cDNA clone was different from that encoded by accC-2 and accC-3. Like the α-CT cDNA described above, the two full-length BC cDNAs (accC-2 and accC-3) are quite homologous (approximately 97% identical) but diverge from each other in both 5′- and 3′-UTRs.
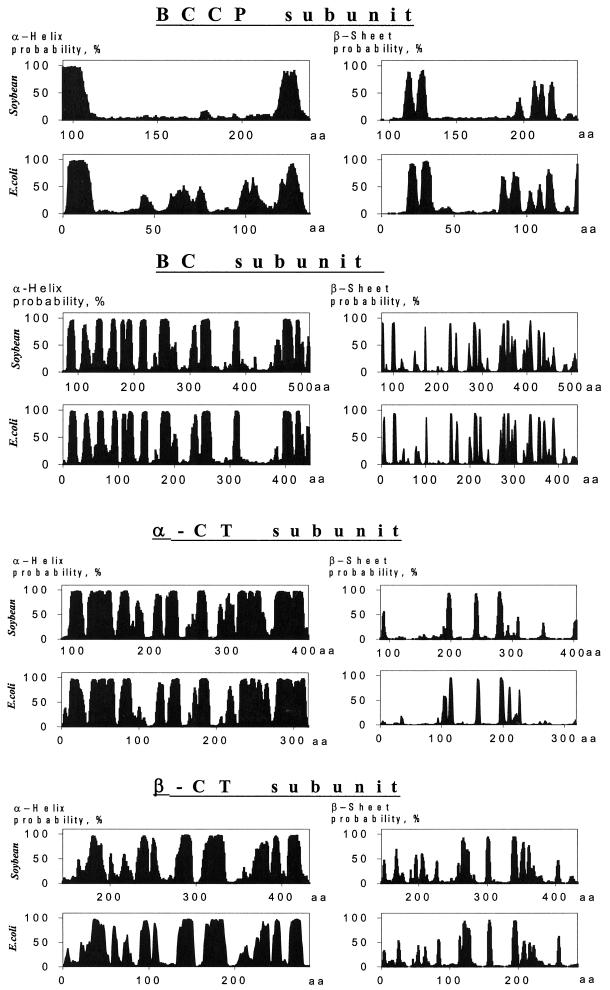
As anticipated, the primary structure of proteins that were deduced from the putative soybean MS ACCase cDNAs exhibited considerable homology to the corresponding ACCase subunits from E. coli (Table I) and from other higher plants (Sasaki et al., 1993; Choi et al., 1995; Shorrosh et al., 1995, 1996; Elborough et al., 1996). Even more striking, however, were the homologies among the predicted secondary structures for the E. coli and soybean ACCase subunits (Fig. 3).
Figure 3.
Alignment of predicted secondary structures for subunits from soybean chloroplast and E. coli ACCases. Plots represent potential α-helix rods and β-sheets. The homologous regions of soybean and E. coli proteins are indicated.
Certain structural elements shared by the two types of ACCases can be found in the deduced sequences of the soybean proteins. For example, the biotinylation motif (C/G/M)-I-(V/I/L)-G-A-M-K-(M/L)-(M/E)-(N/I), which is conserved among all of the BCCPs characterized to date, is present at amino acids 239 to 248 in the soybean BCCP primary structure. The Lys at amino acid 245 is the probable biotin attachment site (Samols et al., 1988). Alignment of the regions proposed to be involved in acetyl CoA binding in α-CTs from different species (Li and Cronan, 1992b) is presented in Figure 2C. That the primary structure is conserved is evident even between distant members of this family. The alignments exhibit more homology between E. coli protein and α-CT subunits from MS enzymes than with the equivalent domains of the MF ACCases.
The α-CT subunit from soybean is about 300 amino acids longer than the corresponding subunit from E. coli. As can be seen in Figure 3, amino acids from residues 100 to 400 of the soybean subunit correspond in predicted secondary structure to those of the E. coli subunit; the remaining 300 C-terminal amino acids of the soybean subunit are unique. A computer-assisted structure prediction indicates that the 300 amino acids at the C terminus of α-CT might assume a coiled-coil structural conformation.
Expression of Putative Soybean MS ACCase cDNAs in a Cell-Free System
The deduced precursors from the soybean cDNAs were examined for transit sequences that would guide cytoplasm-produced proteins into chloroplasts. The accA and accB cDNA translation products have N-terminal sequences that are rich in the hydroxylated amino acids Ser and Thr, contain multiple small hydrophobic residues such as Ala and Val, and have positively charged amino acids such as Arg and Lys. These features are characteristic of many transit sequences. The transit peptides also have a characteristic Met-Ala sequence at their N termini. Analysis of the protein-sorting signals performed by the PSORT program indicated a high probability for chloroplast localization of the proteins, with a predicted likelihood of 0.934 for stromal localization in the case of α-CT, and 0.61 for BCCP.
We investigated the possibility that the proteins encoded by the various cDNAs could interact with MS ACCase from pea. Accordingly, the accA, accB, and accC cDNA inserts were introduced into pGEMEX-1 under the control of T7 bacteriophage promoters (Table II). The subunits encoded by these chimeric genes were synthesized in vitro with the TNT system (Promega). Data reported earlier (Reverdatto et al., 1997) demonstrated that the α-CT1, α-CT2, and BCCP precursors could successfully be imported into isolated pea chloroplasts. This was indicated by the observation that translation products became protected from protease (thermolysin) digestion, and that there was an apparent shift in electrophoretic mobility. It is interesting that the distribution of imported proteins among chloroplast compartments seemed to be dependent on the fractionation scheme. Upon application of the Suc step-gradient protocol for chloroplast fractionation described by Bruce et al. (1994), BCCP was found mainly in stroma. In these experiments the α-CT subunits distributed almost equally between stromal and chloroplast membrane fractions (thylakoid plus envelope membranes). No labeled product was found in the thylakoid lumen fraction in either case. On the other hand, when a rapid fractionation scheme was used to separate components of lysed chloroplasts into stromal and membrane fractions, almost 100% of the labeled, mature subunits were in the stromal fraction. We believe the inconsistency between the two results to be attributable to an instability of the complex upon chloroplast fractionation, and that there are differences in the hydrophobicity of subunit subcomplexes derived from ACCase. The dissociation of ACCase into subcomplexes will be described in more detail below.
Table II.
Summary of the E. coli expression plasmids encoding ACCase proteins
| Name | Vector | Insert Size | Subunit | Expected Protein Size | Size of Expressed Protein (SDS-PAGE) |
|---|---|---|---|---|---|
| bp | kD | ||||
| pA1-1x | pET-16b | 2487 | α-CT-1 precursor, 709 aaa | 80.9 | No expression |
| pA1-2x | pGEMEX-1 | 2487 | α-CT-1 precursor, 709 aa | 78.5 | No expression |
| pA2-1x | pET-16b | 2247 | α-CT-2 precursor, 690 aa | 79.3 | No expression |
| pA2-2x | pGEMEX-1 | 2247 | α-CT-2 precursor, 690 aa | 76.9 | No expression |
| pB-1x | pET-16b | 1068 | BCCP precursor, 262 aa | 30 | ∼40 |
| pB-2x | pGEMEX-1 | 1068 | BCCP precursor, 262 aa | 27.6 | ∼38 |
| pC-1x | pET-16b | 850 (PCR fragment) | BC (fragment), 273 aa | 32.4 | ∼36 |
| pC-2x | pGEMEX-1 | 1940 | BC-2 precursor, 539 aa | 58.9 | 53 |
| pD-1x | pET-16b | 1350 | β-CT, 432 aa | 51.3 | ∼50 |
| pA1-3x | pET-16b | 680 | α-CT-1 fragment, aa 454-681, total 228 aa | 27.4 | ∼30 |
| pA1-4x | pET-16b | 969 | α-CT1, fragment, aa (158–453) + (682–709), total 324 aa | 38.3 | ∼40 |
| pD-3x | pET-16b | 660 | β-CT fragment, aa 1-223 | 27.2 | ∼36 |
aa, Amino acid(s).
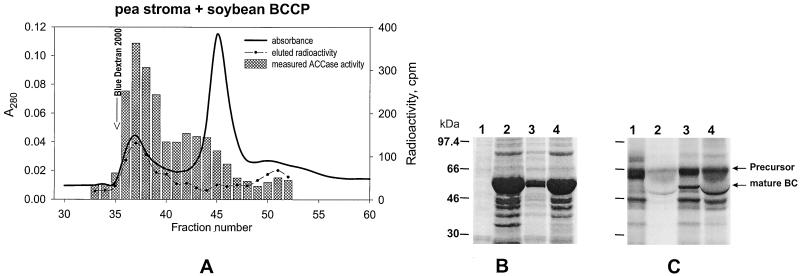
To test whether radioactive soybean ACCase subunits that were synthesized in vitro could be incorporated into functional pea ACCase complexes after import, clarified pea stromal preparations containing imported soybean proteins were resolved by gel-filtration chromatography using Sephacryl S300. The data in Figure 4A show that radioactively labeled soybean BCCP subunit coeluted from the columns together with ACCase activity in the high-molecular-mass region. Similar results were obtained for the soybean α-CT subunit (see Reverdatto et al., 1997). We concluded, therefore, that the soybean accA and accB cDNAs encode members of a soybean ACCase complex, and that they are incorporated into the ACCase complex of pea chloroplasts. The latter result implies that pea ACCase is structurally and functionally similar to soybean ACCase.
Figure 4.
A, Coelution of pea ACCase and [3H]BCCP. [3H]BCCP precursors were synthesized in vitro and imported into isolated pea chloroplasts. Plastid ACCase activity in the stromal fractions was determined after chromatography with a Sephacryl S300 column. Identical results obtained with α-CT precursors produced from accA were used in the same type of experiment (Reverdatto et al., 1997). B and C, Processing of the in vitro-synthesized soybean BC precursor by pea chloroplasts. Lanes 1, Translation with [35S]Met. Lanes 2, Intact pea chloroplasts incubated with the BC precursor and reisolated through a Percoll cushion (total chloroplast protein). Lanes 3, BC precursor incubated with pea stromal preparation that has CPE activity. Lanes 4, Same as for lanes 3, but 5× concentrated (by ultrafiltration through Microcon-30) pea stroma. Note that lanes 2 and 4 contain sizable amounts of ribulose bisphosphate carboxylase, which causes the band attributable to mature BC to be distorted, whereas in lanes 3 there is no distortion. B, Coomassie blue stain for total protein. C, Autoradiogram of the gel from B. These data show that the soybean BC precursors processed after import into chloroplasts, and by cleavage with CPE, are of the same apparent size.
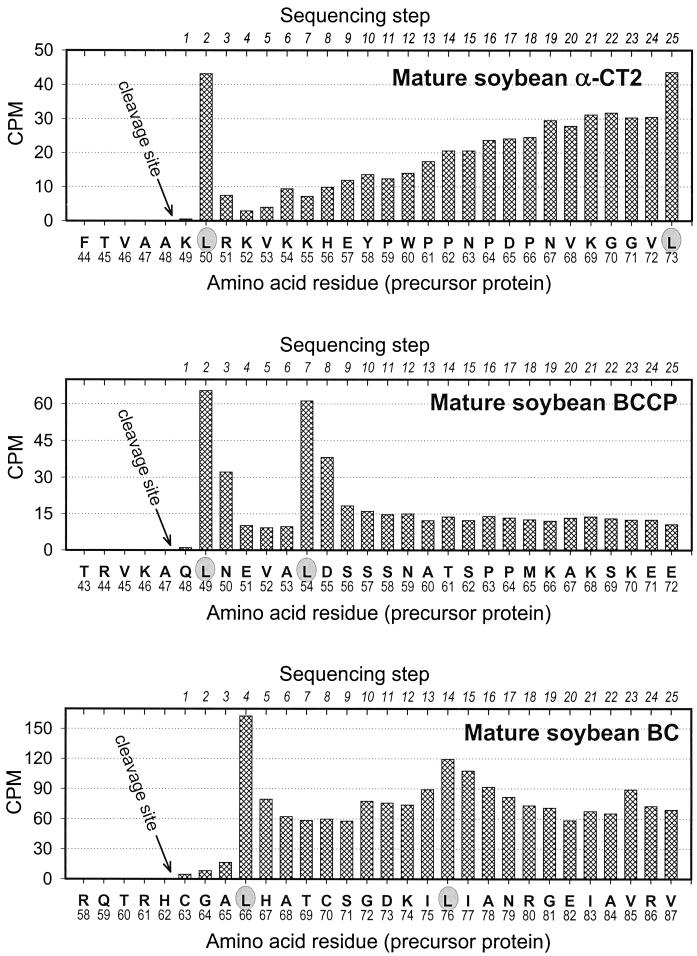
A determination of the amino acid sequence of the labeled mature soybean α-CT and BCCP subunits isolated from pea stroma after import permitted a determination of the position where cleavage of the transit peptide took place. The data in Figure 5 show the amount of radioactivity recovered in each cycle of Edman degradation as the imported soybean subunits were sequenced. In both α-CT and BCCP, the positions where the precursors were cleaved were remarkably close to the location predicted on the basis of β-sheet/α-helix transitions. The position determined for α-CT2 was: Thr-45–Val-46–Ala-47–Ala-48 ↓ Lys-49–Leu-50–Arg-51–Lys-52–Val-53; the position determined for BCCP was: Arg-44–Val-45–Lys-46–Ala-47 ↓ Gln-48–Leu-49–Asn-50–Glu-51–Val-52.
Figure 5.
Microsequence analysis of the N termini of soybean α-CT2, BCCP, and BC after import into isolated pea chloroplasts. Proteins synthesized in vitro as precursors were labeled with [3H]Leu. The lower horizontal axis in each plot records the deduced amino acid sequences of the corresponding precursor, and the upper horizontal axis in each plot records the sequencer cycle. Vertical bars indicate the [3H]Leu recovered from each sequencer cycle.
The transit peptides of both subunits were about the same size, and the amino acid sequences surrounding the cleavage sites (identical amino residues are underlined) were similar. We speculate that α-CT1 protein has the same cleavage-site position as α-CT2 because of the similarity of the two proteins in this region (see Fig. 5 for sequencing data).
A different approach was necessary to determine where the transit peptide of the BC was cleaved. Because the recovery of mature protein from isolated pea chloroplasts was extremely low after the import of labeled precursor into chloroplasts, we treated the BC precursor with the CPE from pea (Robinson and Ellis, 1984; Abad et al., 1989, 1991; Musgrove et al., 1989). Figure 4, B and C, shows the results from such an experiment. As is evident from Figure 4C, the BC precursor was the predominant product from in vitro translation but was accompanied by less prevalent products of an unknown origin. When these translation products were imported into chloroplast and the chloroplast was washed extensively, only intact BC and the cleavage products were present in chloroplast preparations (Fig. 4C, lane 2). When freshly prepared pea stromal extract was used to process the radioactive products from translation, a product of the same size as the soybean BC precursor was recovered in the translation mixture (Fig. 4C, lanes 3 and 4). By using CPE, enough mature BC protein was recovered after isolation by SDS-PAGE so that the cleavage-site position could be identified by N-terminal protein sequence analysis (see Fig. 5): Gln-59–Thr-60–Arg-61–His-62 ↓ Cys-63–Gly-64–Ala-65–Leu-66–His-67.
These results show that the transit peptide on the BC precursors (62 amino acid residues) is slightly larger than the transit peptides in precursors from the two other ACCase subunits. In addition, there is no apparent homology among the sequences immediately upstream or downstream of the cleavage site in BC compared with either α-CT or BCCP. Finally, the alignments of the N-terminal sequences corresponding to the transit peptide of BC from soybean, Arabidopsis (Bao et al., 1997; Sun et al., 1997), and tobacco (Shorrosh et al., 1995) demonstrate that they are not well conserved.
Expression of ACCase cDNAs in E. coli and Antibody Preparation
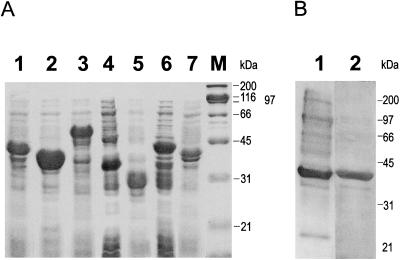
The cDNAs encoding soybean chloroplast ACCase subunits (accA-1 and -2, accB, accD, and part of accC) were introduced into the prokaryotic expression vector pET16b (Novagen). Translation products from these vector constructions that were accumulated by the bacteria were used to generate antibodies against individual components of the complex. Each of the products carried a cleavable His-tag extension on its N terminus to facilitate purification. Unfortunately, as indicated in Table II, the full-size α-CT genes were not expressed. All attempts to obtain an expression product with the portion of the gene encoding amino acids 155 to 450 failed. This is the part of the molecule that is homologous with the E. coli α-CT subunit. To solve the problem, a chimeric construction (pA1-4x) was generated that was composed of the two pieces of the accA-1 gene that did not correspond to the E. coli coding region (i.e. DNA encoding amino acids 158–453 and 682–709 plus the soybean α-CT gene 3′-UTR). A second clone (pA1-4X) that encoded plant-specific protein sequence was also generated. These clones resulted in the synthesis of polypeptides of the expected sizes by the bacteria and were used for antibody production (Fig. 6A). Proteins for each of the other ACCase subunits were successfully produced in a similar manner, and the translation products that were accumulated had the expected sizes.
Figure 6.
A, Expression of soybean chloroplast ACCase subunits in E. coli. Total cell lysates were separated on a 9% SDS-PAGE gel. Gels were stained with Coomassie blue and photographed. Lane 1, Construct pB-1x; lane 2, pC-1x; lane 3, pD-1x; lane 4, pD-3x; lane 5, pA1-3x; lane 6, pA1-4x; lane 7, pB-2x. (See Table II for overview of expression constructs.) B, Refolding of E. coli-expressed BCCP subunit precursor. Soluble protein was run on SDS-PAGE and then probed with streptavidin (lane 1) or stained with Coomassie blue for total protein (lane 2). A substantial proportion of the bacterially expressed BCCP became soluble after refolding. In addition, the BCCP synthesized in E. coli is biotinylated.
Each of the expressed proteins accumulated in protein inclusion bodies. All of the ACCase subunit proteins except for β-CT were soluble in 6 m urea after their purification, and it took 6 m guanidine chloride to solubilize β-CT and its fragment. The solubilized proteins were purified by nickel-agarose column chromatography and used to immunize rabbits. It is interesting that the purified recombinant soybean BCCP protein was biotinylated when isolated from E. coli (Fig. 6B), an indication that this modification took place before accumulation of proteins in the inclusion bodies.
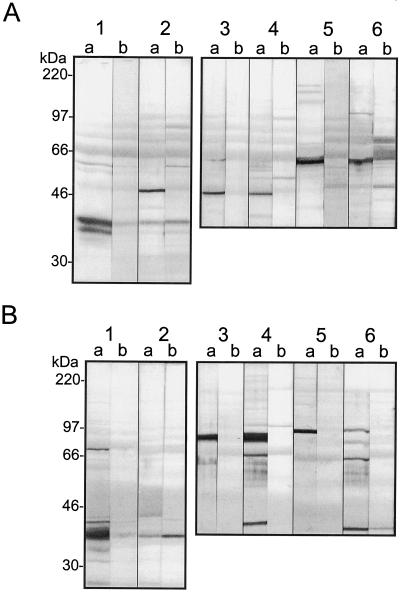
The polyclonal antibodies obtained from rabbit serum varied considerably in their affinity and specificity for the various subunits. Because the proteins used for antibody production were from E. coli, other bacterial proteins inevitably contaminated them and caused a certain degree of cross-reactivity. This resulted in the appearance of the background bands, but the signal ratio of specific binding over the background remained quite high. All of the antibodies recognized proteins of the expected sizes in protein blots, not only when total soybean seed extracts were used, but also in stroma prepared from pea chloroplasts (Fig. 7). Unexpectedly, antibodies raised against the conserved part of the α-CT subunit (construct pA1-4x) had lower specificity (Fig. 7) and were therefore abandoned. Antibody preparations selected for additional purification were pB-1x, which recognized BCCP; pC-1x, which bound to BC; pA1-3x, which recognized α-CT; and pD-3x, which detected β-CT. The antibodies were purified by a combination of caproic acid/ammonium sulfate precipitation (Harlow and Lane, 1988) or with the DEAE Affi-Gel resin (Bio-Rad).
Figure 7.
SDS-PAGE gel blots probed with antibodies against soybean MS ACCase subunits. Soybean total seed (A) and pea leaf chloroplast stroma (B) extracts were probed. Lanes 1, Anti-BCCP; lanes 2, anti-BC; lanes 3, anti-β-CT (anti-pD-1x); lanes 4, anti-β-CT (anti-pD-3x); lanes 5, anti-α-CT (anti-pA1-3x); and lanes 6, anti-α-CT (anti-pA1-4x). (See Table II for overview of expression constructs.) Membranes were treated with both immune (lanes a) and preimmune (lanes b) sera. Whole membranes were cut into strips and each was incubated with a different antibody. Primary antibodies were used at 1:10,000 dilutions.
The Purification of ACCase Components by Affinity-Antibody Columns
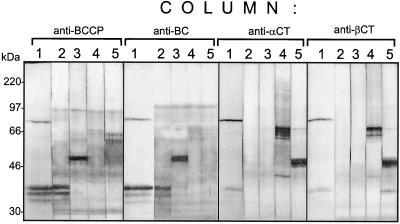
To purify components from the stromal ACCase, affinity-purification columns were prepared by coupling the purified antibodies to Affi-Gel resins (Bio-Rad). Seed extracts were passed through these columns, and any proteins retained by the column were eluted with an acidic Gly solution (pH 2.5). Protein blots were prepared after electrophoresis of eluates from the columns, and these were probed with each of the four antibodies and with streptavidin. Figure 8 reveals that proteins recognized by the anti-BCCP antibody (and also streptavidin) are detected in both the anti-BCCP and anti-BC column eluates. The same eluates were also positive for BC. In the same manner, eluates from the anti-α-CT and anti-β-CT columns each contained both kinds of subunits. The results demonstrate that ACCase components separate but tend to form the stable complexes containing either BC/BCCP or α-CT/ β-CT. Such complexes are stable even in the presence of the 1% nonionic detergent NP40 used in these studies.
Figure 8.
Analysis of column fractions from total soybean seed extracts after chromatography through antibody-affinity columns. Lanes 1, Protein-gel blot probed with streptavidin-alkaline phosphatase conjugate; lanes 2, anti-BCCP probe; lanes 3, anti-BC probe; lanes 4, anti-α-CT probe; lanes 5, anti-β-CT probe. Gly eluates from each column (indicated above the lanes) were separated on 9% SDS-PAGE and transferred to PVDF membranes. Each membrane was subsequently cut into strips and probed with each ACCase antibody.
A similar behavior has been reported for pea ACCase (Sasaki et al., 1993; Alban et al., 1994; Shorrosh et al., 1995, 1996; Roesler et al., 1996). Apparently, a minimal amount of intact MS ACCase complex survived the dissociation conditions used for protein isolation, because traces of α-CT subunit were captured by anti-BCCP and anti-BC columns, and the anti-α-CT and anti-β-CT column eluates contained traces of BCCP (Fig. 8). It is interesting that up to three bands can be seen in those lanes probed with streptavidin or anti-BCCP antibodies. These lie between the positions of the 30- and 46-kD proteins. This observation could mean that at least three BCCP-like proteins are contained in the extracts. The lanes probed with streptavidin also contained a protein of approximately 85 kD. That signal is probably attributable to 3-methylcrotonyl carboxylase, and is recognized by the antibodies because of structural homology between components of ACCase and 3-methylcrotonyl carboxylase (Song et al., 1994). When soybean seed extracts were separated by chromatography with monomeric avidin-agarose (Sigma), a BC/BCCP complex was also captured by the column (data not shown).
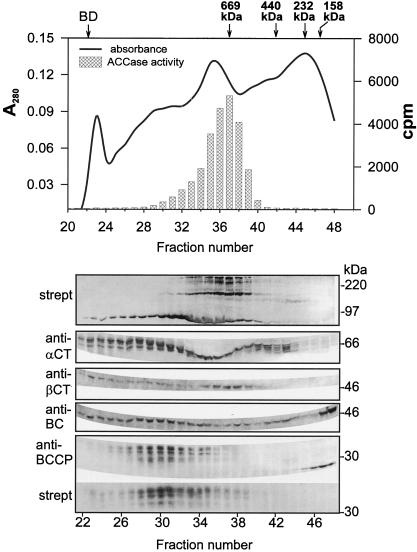
Gel-Filtration Chromatography of ACCase with Sephacryl S400
The distinctive behavior of ACCase during gel-permeation chromatography is well documented for pea (Sasaki et al., 1993; Alban et al., 1994; Roesler et al., 1996; Shorrosh et al., 1996). In these reports the MS ACCase complexes elute from Sephacryl S300 immediately after the void volume and have an estimated molecular mass of 600 to 700 kD. Sephacryl S400 was used for our experiments with the soybean ACCase because this chromatography medium provided better resolution in high-molecular-mass regions than did S300. The chromatographic elution profile and analysis of fractions from such an experiment are presented in Figure 9. Protein-blot analysis performed with either antibodies against each subunit or streptavidin showed the majority of the four ACCase components to be located in fractions that eluted before the 669-kD marker (thyroglobulin). It is interesting that the strong signals against both BCCP and β-CT were confined to a relatively small number of fractions, whereas signals for the BC and α-CT subunits seemed to be present in many fractions. Those fractions that contained BCCP also had strong signals with BC antibodies, which by analogy with the pea enzyme probably contain BC/BCCP complexes. The point at which most of the BC/BCCP pair eluted seemed to be shifted slightly toward higher apparent molecular mass, compared with those fractions that contained strong signals against both α-CT and β-CT. We believe that the latter contains both subunits as part of α-CT/β-CT subcomplexes.
Figure 9.
Separation of soybean seed protein extract by gel filtration on Sephacryl S400. The column was eluted with buffer A (see Methods) at 0.05 mL/min. Fractions (0.75 mL) were collected, and a high-molecular-mass gel-filtration calibration kit (Pharmacia) was used to calibrate the column. Eluted protein fractions were separated on SDS-PAGE, blotted to nylon membranes, and analyzed with different antibodies as indicated. strept, Streptavidin-alkaline phosphatase conjugate. The results from the protein-gel blot are aligned with the chromatographic profile. Approximate positions of electrophoretic molecular-mass markers are shown at the right of the protein-blot panels. The position of Blue Dextran 2000 (BD) and other markers used to calibrate the column are shown above the elution profile. The markers were thryoglobulin (667 kD), ferritin (440 kD), catalase (232 kD), and aldolase (158 kD).
A similar shift in the elution of the two subunit pairs was reported by Sasaki et al. (1993) and Shorrosh et al. (1996), who attributed this effect to a probable partial dissociation of the enzyme complex during chromatography. The shift seems more apparent on the larger-pore-sized Sephacryl S400 than on S300, perhaps because of an increased resolution of high-molecular-mass complexes on S400. We observed that most of the measurable ACCase activity coincided with the elution of MF ACCase (determined by streptavidin binding to approximately 220-kD proteins in protein blots). The elution of subunits of the MS ACCase overlapped the elution position of the large MF ACCase and appeared at a position where high-molecular-mass proteins would elute. This result was not expected in view of the report that MS ACCase accounted for 80% of total ACCase activity in pea leaves (Alban et al., 1994). It is possible that the ratio of MF to MS forms of the enzymes is different in leaves and seeds. Alternatively, the apparent instability of the MS ACCase from soybean may be greater than in the pea system because of our use of Sephacryl S400 to increase the resolution of partially dissociated enzyme complexes.
The protein gel blots shown in Figure 9 indicate that there are several molecular species each of α-CT and BCCP after separation of the soybean seed extracts by gel-permeation chromatography. The pattern of bands attributable to the BC and β-CT subunits detected by their respective antibodies were less complicated than those of α-CT and BCCP. What seems to be a degraded form of BCCP is found in the fractions with molecular mass < 200 kD relative to the markers. This band is recognized by anti-BCCP antibodies but not by streptavidin, a result indicative of nonbiotinylated BCCP. These fractions emerged from the column soon after the aldolase marker (e.g. 158 kD) but before the inner volume of the column (20 kD). The same fractions from the column also contained proteins that reacted with anti-BC antibodies, although the most intense BC signals seemed to be shifted to slightly lower molecular mass than those fractions with the most intense signals for BCCP.
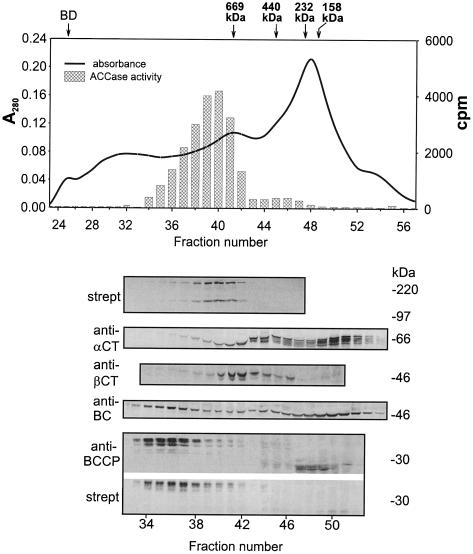
Chromatographic Behavior of the ACCase System during Gel Filtration on Sephacryl S400 in High Salt (0.5 m KCl)
There have been some reports (Alban et al., 1994; Shorrosh et al., 1996) that indicate that high ionic strength causes the MS ACCase from pea to dissociate into the BC/BCCP and α-CT/β-CT subcomplexes with a loss of enzyme activity. To test the effect of high ionic strength on the soybean enzyme, seed extracts were resolved by chromatography on Sephacryl S400 using a buffer that contained 0.5 m KCl. After the proteins in fractions from the column were separated by SDS-PAGE, protein blots were prepared and probed either with antibodies against individual ACCase subunits or with streptavidin. We interpret the data shown in Figure 10 as indicating that soybean ACCase dissociated into two complexes in 0.5 m KCl. The BC/BCCP pair eluted from the column first, and in a region where proteins with an unexpectedly high molecular mass would be expected to emerge (>800 kD). These fractions from the column were characterized by strong signals in protein-gel blots with both the BC and BCCP antibodies. It is interesting that strong signals for BCCP in the protein-gel blots were confined to a fairly narrow range of fractions, whereas signals to anti-BC were observed in nearly every fraction. In the area of the elution profile from the column where ≤200-kD proteins would emerge, a number of strong signals for BCCP and BC subunits were detected immunologically. As in the case of chromatography in low-salt conditions, those BCCP-positive proteins did not react with streptavidin, were apparently not biotinylated, and were probably degraded. These subunit complexes, which presumably contained BC and degraded BCCP, did not form high-molecular-mass assemblies like BC/BCCP with the intact biotin carrier proteins. The α-CT/β-CT subunits eluted from the column after the BC/BCCP pair and seemed to overlap the fractions where the cytoplasmic MF ACCase eluted from the gel-permeation column. Strong signals for β-CT in the protein gel blots were confined to only a few fractions (i.e. 40–44), whereas strong signals against α-CT were much more widely distributed, especially in fractions that would contain lower-molecular-mass proteins. Thus, biotinylated BCCP and β-CT seemed to be found only in fractions where they were associated with either BC or α-CT, respectively.
Figure 10.
The same experiment as in Figure 9, except that buffer A contained 0.5 m KCl. Each fraction was concentrated and desalted with Microcon-30 concentrators. Other conditions are as in Figure 9. Note that the BC/BCCP and α-CT/β-CT complexes were resolved from one another and from the MF ACCase more completely at 0.5 m KCl than they were at lower salt, and that the salt did not cause disassembly of the two complexes.
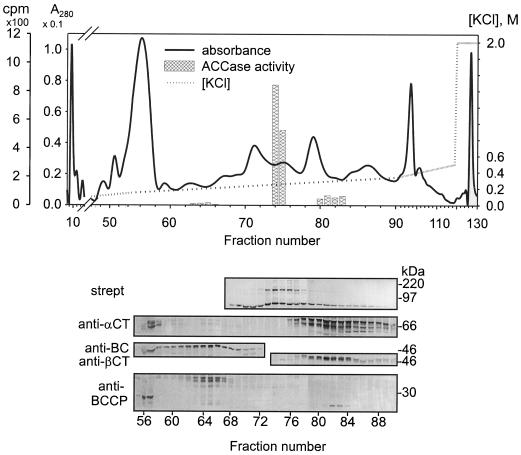
Purification of ACCase Components by Anion-Exchange Chromatography on Q-Sepharose
A shallow salt gradient was reported to separate the two forms of maize ACCase on a Q-Sepharose column (Herbert et al., 1996). Figure 11 shows the results of a similar experiment carried out with an extract from soybeans. As indicated by these data, the various components of ACCase were well resolved by this approach. When antibodies were used to probe the fractions eluted from the column, the BC/BCCP subunit pair emerged from the column first, followed by the MF ACCase, and finally by the α-CT/β-CT subunit pair. It is interesting that the strong anion-exchange matrix apparently dissociated the MS ACCase complex, even in low-salt conditions, because the elution of the BC/BCCP pair at approximately 0.2 m KCl was not accompanied by detectable amounts of other ACCase subunits. The broad elution range for the α-CT/β-CT pair may be attributable to several different α-CT subunits in the extract, each with a different apparent pI (see Table I). The small amounts of α-CT subunits that emerged from the column at the low-salt concentration were probably attributable to nonpaired proteins. The pI values for α-CT (6.5–7.7) might lead one to expect that they would be released from the column at low-salt concentrations, a consideration that suggests that the α-CT/β-CT pair has a substantially higher net pI. Note also that a small proportion of total BCCP eluted in fractions 56 and 57, well before the bulk of these proteins were released from the column. This was attributable to the nonbiotinylated BCCP described above, which remains associated with BC. It is unclear whether a special function can be ascribed to this form of BCCP or whether it represents a portion of the protein modified during purification (e.g. proteolytically).
Figure 11.
Anion-exchange chromatography (Q-Sepharose column) of a soybean seed extract. After loading, the column was washed with buffer A (see Methods) at 0.5 mL/min, and then a KCl gradient (0–0.5 m) was applied. Final wash was with 2 m KCl. Fractions of 2 mL were collected, desalted, and concentrated with Microcon-10 concentrators. Proteins were separated on 9% SDS-PAGE, transferred to PVDF membranes, and probed with antibodies as indicated. strept, Streptavidin-alkaline phosphatase conjugate. The central part of the chromatographic profile is expanded to align with the lanes on the protein-blot panels. The positions of electrophoretic protein molecular-mass markers are shown at the right side of the panels for the protein blots.
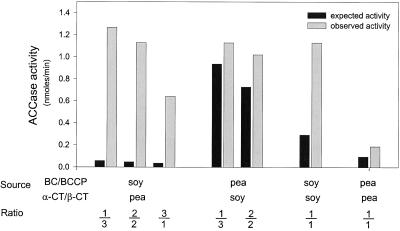
The availability of BC/BCCP and α-CT/β-CT fractions that were essentially free of the MF form of ACCase facilitated reconstitution of the complexes in vitro. By mixing aliquots (see Fig. 11) from fraction 65 (BC/BCCP) with an aliquot of fraction 81 (α-CT/β-CT), a sample was obtained in which 4-fold stimulation of ACCase activity was achieved compared with the sum of the individual fraction activities (Fig. 12). The combination of the BC/BCCP and α-CT/β-CT components of MS ACCase from pea chloroplast stroma (see Shorrosh et al., 1996) was carried out in a similar manner, and resulted in only a 2-fold increase in activity. This is less than was reported by Shorrosh et al. (1996), and could be attributable to a lower concentration of enzyme used in our experiments. As indicated by the data in Figure 12, an attempt was also made to construct hybrid ACCase complexes to test for interspecies compatibility. Mixing of the BC/BCCP and α-CT/β-CT pairs from both plants at different ratios resulted in surprisingly large increases in ACCase activity for some of the combinations. These results support our conclusion that the proteins synthesized at the direction of the cDNAs we isolated are indeed components of the MS ACCase from soybean.
Figure 12.
Reconstitution of ACCase activity. The BC/BCCP and α-CT/β-CT components of soybean and pea ACCase from chloroplasts were each purified by ion-exchange chromatography as illustrated in Figure 11. Aliquots from fractions that contained either the BC/BCCP or α-CT/β-CT complexes were assayed for ACCase activity both individually and when combined with one another in various ratios (v/v). The expected activity refers to the sum of residual ACCase activity from each fraction from the column. Observed activity refers to that actually recovered after combining fractions at various ratios. In each case combining the BC/BCCP and α-CT/β-CT complexes stimulated ACCase activity. Remarkably higher activities than expected were observed when the soybean BC/BCCP and pea α-CT/β-CT complexes were combined.
DISCUSSION
We isolated cDNA clones that encode BC, BCCP, α-CT, or β-CT, the four known components of MS ACCase in legume chloroplasts. Several lines of evidence support this conclusion. First, each of the proteins encoded by the cDNAs share structural similarities with subunits of bacterial MS ACCase. Not only is there extensive homology between portions of their primary structures, but extended regions of their predicted secondary structures are also similar. Second, the subunits of soybean ACCase are homologous with the corresponding proteins from MS ACCases described from other higher plants. Nonetheless, significant differences in structure exist between certain deduced soybean proteins and their higher-plant counterparts. For example, β-CT from soybean lacked a 96-amino acid insert in the interior of the molecule found in the pea protein. The insert in pea contained a number of repetitive elements and probably arose by duplication (Sasaki et al., 1993). In the case of the α-CT, the deduced soybean proteins are almost 200 amino acids shorter than the corresponding pea subunit. When compared, the soybean α-CT protein lacks five regions in its primary structure ranging in size from 2 to just over 100 amino acids. The largest deletion contains four of five of the 26-amino acid repeats found in the pea subunit (Shorrosh et al., 1996). The third piece of evidence supporting the conclusion that these cDNAs encode subunits of soybean MS ACCase is that the domain where biotin is bound to BCCP is present, and a putative acetyl CoA binding site is conserved in the deduced α-CT subunit (Fig. 2C). Finally, after expression of the cDNAs in vitro, the translation products are imported into isolated pea chloroplasts, processed to remove a transit sequence, and then incorporated into high-molecular-mass complexes, which coelute with MS ACCase enzymatic activity. Together, this evidence strongly supports the conclusion that the cDNAs we described encode subunits of the chloroplast MS ACCase, an enzyme considered to be involved in the regulation of the fatty acid biosynthesis pathway in this oil seed.
Unlike pea, in which only a single gene has been identified that encodes each subunit for the plastid ACCase, there seem to be small families of genes that encode the BC, BCCP, and α-CT subunits in soybean. Several observations are consistent with this conclusion. First, not only do multiple restriction fragments hybridize to probes for each of these cDNAs in DNA blots of genomic DNA, but so far, two different BCCP, three different BC, and four different α-CT cDNA clones have been recovered that vary in their nucleotide sequences. Second, antibodies generated against BCCP and α-CT recognize several different soybean proteins in protein-gel blots prepared from seed extracts from soybeans. The multiple proteins that are recognized may correspond to products from the multiple genes. The presence of multiple genes that encode MS ACCase subunits in soybean may resemble the situation reported to occur in Brassica napus, in which several BCCPs have been reported (Elborough et al., 1996). The reason for the existence of multiple genes for BC, BCCP, and α-CT in the soybean genome is not known. One possibility is that they reflect the allotetraploid nature of soybean, but the number of genes involved suggests that the answer may be more complex.
We generated antibodies that recognize BC, BCCP, α-CT, and β-CT. These antibodies were used to identify members of the MS ACCase complex isolated from extracts of developing soybeans. As with the pea enzyme (Alban et al., 1994; Shorrosh et al., 1995, 1996; Roesler et al., 1996), soybean ACCase appears to resolve into two stable complexes during purification with red 120-agarose (data not shown), affinity chromatography with monovalent avidin (data not shown), anion-exchange chromatography on Q-Sepharose, and chromatography with antibody-affinity columns. The two subcomplexes, which consist of either the BC/BCCP or the α-CT/β-CT subunit pair, were remarkably stable and tended to form large aggregates. Even in the presence of 0.5 m KCl, the BC/BCCP and α-CT/β-CT subcomplexes eluted from Sephacryl S400 gel-permeation columns with retention times characteristic of proteins with molecular masses that are greater than 800 and 600 kD, respectively. For complexes of this size, there would be at least 10 BC/BCCP pairs, each with an aggregate size of about 75 kD, and at least 5 α-CT/β-CT pairs, each with a combined size of approximately 120 kD. An active complex could be at least partially reconstituted by combining fractions containing each of the subunit pairs. We do not know if these large, aggregated subcomplexes function in vivo or are artifacts from purification.
The MF ACCase seemed to emerge from the Sephacryl S400 column between the BC/BCCP and α-CT/β-CT subcomplexes at low ionic strength (Fig. 9), and was clearly resolved and between them after elution of the column with 0.5 m KCl (Fig. 10). In both cases the MF enzyme emerged with an apparent molecular mass greater than that of the thyroglobulin standard (669 kD). Although the MF ACCase has been reported to be a homodimer of 450 to 550 kD (Everson et al., 1994), there are a number of reports in the literature that indicate that the enzyme emerges with retention times characteristic of proteins with a higher apparent molecular mass. For example, the parsley and wheat MF ACCase had molecular masses estimated to be around 840 and 700 kD, respectively, when purified through Sepharose 6B (Egin-Bühler et al., 1980; Egin-Bühler and Ebel, 1983). It is interesting that the same authors reported an apparent molecular mass of 420 kD after purification of the parsley enzyme through Sephacryl S300. Italian ryegrass is reported to have an apparent molecular mass of about 840 kD when purified by Superose 6 (Everson et al., 1997). Thus, both the MF and MS forms of plant ACCase seem prone to aggregation under the conditions typically used for their purification. The meaning of the tendency of these proteins to aggregate is not clear. The aggregation certainly could be an artifact that arises during extraction and purification. However, the occurrence of aggregation effects is of interest, considering that the animal ACCase undergoes a citrate-induced activation that is accompanied by its assembly into polymers of several million kilodaltons (Gregolin et al., 1966). Although the plant enzyme does not respond to citrate, it seems premature to disregard the possibility that the aggregation effects of the plant enzymes could be related to a physiological function.
Our data indicated that the majority of activity associated with extracts from developing soybeans was attributable to MF ACCase and not to the MS form of the enzyme. The BC/BCCP and α-CT/β-CT subcomplexes, therefore, are labile and they separate from one another easily. Although activity could be restored by combining fractions identified immunologically to contain the two subcomplexes from soybean, experiments in which appropriate soybean and pea subcomplexes were mixed suggested that considerably higher activities were possible. A more-than 20-fold stimulation was obtained by mixing the BC/BCCP subcomplex from soybean with the α-CT/β-CT complex from pea (Fig. 12). Mixed complexes made in the reciprocal manner were not nearly as active. The reason for this difference is unclear. Nonetheless, it is possible to conclude from these data that the ACCase complex can be reassembled from its component subcomplexes, and that the pea and soybean components are to some extent interchangeable. Although many explanations could account for the stimulation, these results could imply that more active MS ACCases could be engineered by biotechnological approaches.
Another interesting observation that emerged during purification of ACCase subunits was that neither BCCP nor β-CT appeared in fractions from the columns unless their putative counterparts from the subcomplexes were present in the same fractions. This observation could mean that they were stable only when associated with their respective partners in the subcomplexes. BC and α-CT, on the other hand, were found in fractions that contained little or no BCCP or β-CT. These phenomena can be observed in the data shown in Figures 9, 10, and 11. The same phenomenon held for fractions in which BCCP was recognized by anti-BCCP antibodies but not by streptavidin, an observation that indicated that these subunits apparently were devoid of biotin. Some of the fractions that contained nonbiotinylated BCCP subunits apparently also contained BC, because they reacted with anti-BC antibodies. Although fractions that contained these molecules emerged immediately before fractions derived from the inner volume of the gel-permeation columns, it is possible that a complex between the degraded BCCP and a BC molecule existed. This notion was supported by the observation that both BC and the degraded BCCP emerged together among the first fractions from the anion-exchange column (Fig. 11) and were not separated by this treatment. We have no explanation for the lack of free BCCP and β-CT in the column fractions other than they may be rapidly degraded when separated from their partners in the subcomplexes.
There is uncertainty about the localization of the MS ACCase complex within chloroplasts. Kannangara and Stumpf (1972) and Sasaki et al. (1993) both suggested that one or more of the ACCase components were associated with thylakoid membranes. Shorrosh et al. (1996) demonstrated that the carboxyltransferase subunits in the total pea leaf extracts tended to accumulate in the insoluble fraction, but the BC subunit remained in solution. Nonetheless, the same authors used stromal fractions of isolated pea chloroplasts for ACCase isolation, as did Alban et al. (1994, 1995). Our own results (Reverdatto et al., 1997) demonstrated that imported ACCase subunits tend to remain soluble after chloroplast lysis if organelle fractionation is done rapidly. These seemingly contradictory results could be explained if the ACCase components redistribute among organellar fractions after dissociation of the complex according to the hydrophobic nature of individual subunits or subcomplexes. Rapid fractionation results in purification of the ACCase components before the more hydrophobic subunits have an opportunity to become associated with membranes.
Abbreviations:
- ACCase
acetyl CoA carboxylase
- BC
biotin carboxylase
- BCCP
biotin carboxyl carrier protein
- CPE
chloroplast processing enzyme
- α- and β-CT
the α- and β-subunits of carboxyltransferase, respectively
- MF
multifunctional
- MS
multisubunit
- UTR
untranslated region
Footnotes
This research was supported in part by American Soybean Association grant no. SPR-2305 to N.C.N.
LITERATURE CITED
- Abad MS, Clark SE, Lamppa GK. Properties of a chloroplast enzyme that cleaves the chlorophyll a/b binding protein precursor. Optimization of an organelle-free reaction. Plant Physiol. 1989;90:117–124. doi: 10.1104/pp.90.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abad MS, Oblong JE, Lamppa GK. Soluble chloroplast enzyme cleaves preLHCP made in Escherichia colito a mature form lacking a basic N-terminal domain. Plant Physiol. 1991;96:1220–1227. doi: 10.1104/pp.96.4.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alban C, Baldet P, Douce R. Localization and characterization of two structurally different forms of acetyl-CoA carboxylase in young pea leaves, of which one is sensitive to aryloxyphenoxypropionate herbicides. Biochem J. 1994;300:557–565. doi: 10.1042/bj3000557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alban C, Jullien J, Job D, Douce R. Isolation and characterization of biotin carboxylase from pea chloroplasts. Plant Physiol. 1995;109:927–935. doi: 10.1104/pp.109.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Feel W, Chirala SS, Wakil SJ. Cloning of the yeast FAS3 gene and primary structure of yeast acetyl-CoA carboxylase. Proc Natl Acad Sci USA. 1992;89:4534–4538. doi: 10.1073/pnas.89.10.4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JV, Lutz SM, Gengenbach BG, Gronwald JW. Genomic sequence for a nuclear gene encoding acetyl-coenzyme A carboxylase (accession no. L42814) in soybean (PGR 95-055) Plant Physiol. 1995;109:338. [Google Scholar]
- Bao X, Shorrosh BS, Ohlrogge JB. Isolation and characterization of an Arabidopsisbiotin carboxylase gene and its promoter. Plant Mol Biol. 1997;35:539–550. doi: 10.1023/a:1005881006620. [DOI] [PubMed] [Google Scholar]
- Beilinson VA (1992) Cloning, mutagenesis and expression of the genes for herbicide-binding protein from barley photosystem II and zein from maize. PhD thesis. Shemyakin Institute of Bioorganic Chemistry, Russian Academy of Sciences, Moscow
- Bruce BD, Perry S, Froehlich J, Keegstra K. In vitro import of proteins into chloroplasts. In: Gelvin SE, Shilperoort RA, editors. Plant Molecular Biology Manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. J1:1–15. [Google Scholar]
- Cho T-J (1988) Soybean storage protein genes: gene structure and organization. PhD thesis. Purdue University, West Lafayette, IN
- Choi J-K, Yu F, Wurtele ES, Nikolau BJ. Molecular cloning and characterization of the cDNA coding for the biotin-containing subunit of the chloroplastic acetyl-coenzyme A carboxylase. Plant Physiol. 1995;109:619–625. doi: 10.1104/pp.109.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J, Heaberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebel J, Schmidt WE, Loyal R. Phytoalexin synthesis in soybean cells: elicitor induction of phenylalanine ammonia-lyase and chalcone synthase mRNAs and correlation with phytoalexin accumulation. Arch Biochem Biophys. 1984;232:240–248. doi: 10.1016/0003-9861(84)90540-x. [DOI] [PubMed] [Google Scholar]
- Egin-Bühler B, Ebel J. Improved purification and further characterization of acetyl-CoA carboxylase from cultured cells of parsley (Petroselinum hortense) Eur J Biochem. 1983;133:335–339. doi: 10.1111/j.1432-1033.1983.tb07467.x. [DOI] [PubMed] [Google Scholar]
- Egin-Bühler B, Loyal R, Ebel J. Comparison of acetyl-CoA carboxylases from parsley cell cultures and wheat germ. Arch Biochem Biophys. 1980;203:90–100. doi: 10.1016/0003-9861(80)90156-3. [DOI] [PubMed] [Google Scholar]
- Egli MA, Lutz SM, Somers DA, Gengenbach BG. A maize acetyl-coenzyme A carboxylase cDNA sequence. Plant Physiol. 1995;108:1299–1300. doi: 10.1104/pp.108.3.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elborough KM, Winz R, Deka RK, Markham JE, White JA, Rawsthorne S, Slabas AR. Biotin carboxyl carrier protein and carboxyltransferase subunits of the multi-subunit form of acetyl-CoA carboxylase from Brassica napus: cloning and analysis of expression during oilseed rape embryogenesis. Biochem J. 1996;315:103–112. doi: 10.1042/bj3150103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everson KJ, Gronwald JW, Wyse DL. Plant Physiol. 1994;105:671–680. doi: 10.1104/pp.105.2.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everson KJ, Gronwald JW, Wyse DL. Isoforms of acetyl-coenzyme A carboxylase in Lolium multiflorum. Plant Physiol Biochem. 1997;35:265–272. doi: 10.1104/pp.105.2.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gornicki P, Podkowinski J, Scappino L, DiMaio J, Ward E, Haselkorn R. Wheat acetyl-coenzyme A carboxylase: cDNA and protein structure. Proc Natl Acad Sci USA. 1994;91:6860–6864. doi: 10.1073/pnas.91.15.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregolin C, Ryder E, Kleinschmidt AK, Warner RC, Lane MD. Molecular characterization of liver acetyl CoA carboxylase. Proc Natl Acad Sci USA. 1966;56:148–155. doi: 10.1073/pnas.56.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guchhait RB, Polakis SE, Dimroth P, Stoll E, Moss J, Lane MD. Acetyl coenzyme A carboxylase system of Escherichia coli: purification and properties of the biotin carboxylase, carboxyl transferase, and carboxyl carrier protein components. J Biol Chem. 1974;249:6633–6645. [PubMed] [Google Scholar]
- Ha J, Daniel S, Kong IS, Park CK, Tae HJ, Kim KH. Cloning of human acetyl-CoA carboxylase cDNA. Eur J Biochem. 1994;219:297–306. doi: 10.1111/j.1432-1033.1994.tb19941.x. [DOI] [PubMed] [Google Scholar]
- Harlow ED, Lane DP. Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Herbert D, Price LJ, Alban C, Dehaye L, Job D, Cole DJ, Pallett KE, Harwood JL. Kinetic studies on two isoforms of acetyl-CoA carboxylase from maize leaves. Biochem J. 1996;318:997–1006. doi: 10.1042/bj3180997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung R, Scott MP, Oliveira LO, Nielsen NC. A simple and efficient method for the oligodeoxyribonucleotide-directed mutagenesis of double-stranded plasmid DNA. Gene. 1992;121:17–24. doi: 10.1016/0378-1119(92)90157-k. [DOI] [PubMed] [Google Scholar]
- Kannangara CG, Stumpf PK. Fat metabolism in higher plants: a procaryotic type acetyl CoA carboxylase in spinach chloroplasts. Arch Biochem Biophys. 1972;152:83–91. doi: 10.1016/0003-9861(72)90196-8. [DOI] [PubMed] [Google Scholar]
- Konishi T, Shinohara K, Yamada K, Sasaki Y. Acetyl-CoA carboxylase in higher plants: most plants other than Gramineae have both the prokaryotic and the eukaryotic forms of this enzyme. Plant Cell Physiol. 1996;37:117–122. doi: 10.1093/oxfordjournals.pcp.a028920. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Li S-J, Cronan JE. The gene encoding the biotin carboxylase subunit of Escherichia coliacetyl-CoA carboxylase. J Biol Chem. 1992a;267:855–863. [PubMed] [Google Scholar]
- Li S-J, Cronan JE. The genes encoding the two carboxyltransferase subunits of Escherichia coliacetyl-CoA carboxylase. J Biol Chem. 1992b;267:16841–16847. [PubMed] [Google Scholar]
- Lopez-Casillas F, Bai D-H, Luo X-N, Kong I-S, Hermodson MA, Kim K-H. Structure of the coding sequence and primary amino acid sequence of acetyl-coenzyme A carboxylase. Proc Natl Acad Sci USA. 1988;85:5784–5788. doi: 10.1073/pnas.85.16.5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled-coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Markham JE, Elborough KM, Winz R, Evans M, Slabas AR. Acetyl-CoA carboxylase from Brassica napus. In: Williams JP, Khan MU, Lem NW, editors. Physiology, Biochemistry and Molecular Biology of Plant Lipids. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1996. pp. 11–13. [Google Scholar]
- Musgrove JE, Elderfield PD, Robinson C. Endopeptidases in the stroma and thylakoids of pea chloroplasts. Plant Physiol. 1989;90:1616–1621. doi: 10.1104/pp.90.4.1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page RA, Okada S, Harwood JL. Acetyl-CoA carboxylase exerts strong flux control over lipid synthesis in plants. Biochim Biophys Acta. 1994;1210:369–372. doi: 10.1016/0005-2760(94)90242-9. [DOI] [PubMed] [Google Scholar]
- Reverdatto SV, Beilinson VA, Nielsen NC. The rps16, accD, psaI, ORF 203, ORF 151, ORF 103, ORF 229 and petA gene cluster in the chloroplast genome of soybean (accession no. U26948) (PGR 95-051) Plant Physiol. 1995;109:338. [Google Scholar]
- Reverdatto SV, Beilinson VA, Nielsen NC. Characterization of a cDNA clone encoding a BCCP subunit of acetyl-CoA carboxylase from soybean (accession no. U40666) (PGR 96-040) Plant Physiol. 1996;111:652. [Google Scholar]
- Reverdatto SV, Beilinson VA, Nielsen NC. Soybean chloroplast acetyl-CoA carboxylase. In: Williams JP, Khan MU, Lem NW, editors. Physiology, Biochemistry and Molecular Biology of Plant Lipids. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1997. pp. 29–31. [Google Scholar]
- Robinson C, Ellis RJ. Transport of proteins into chloroplasts: partial purification of a chloroplast protease involved in the processing of imported precursor polypeptides. Eur J Biochem. 1984;142:337–342. doi: 10.1111/j.1432-1033.1984.tb08291.x. [DOI] [PubMed] [Google Scholar]
- Roesler KR, Savage LJ, Shintani DK, Shorrosh BS, Ohlrogge JB. Co-purification, co-immunoprecipitation, and coordinate expression of acetyl-coenzyme A carboxylase activity, biotin carboxylase, and biotin carboxyl carrier protein of higher plants. Planta. 1996;198:517–525. doi: 10.1007/BF00262637. [DOI] [PubMed] [Google Scholar]
- Roesler KR, Shorrosh BS, Ohlrogge JB. Structure and expression of an Arabidopsisacetyl-coenzyme A carboxylase gene. Plant Physiol. 1994;105:611–617. doi: 10.1104/pp.105.2.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler PG, Ohlrogge JB. Cloning and characterization of the gene that encodes acetyl-coenzyme A carboxylase in the algae Cyclotella cryptica. J Biol Chem. 1993;268:19254–19259. [PubMed] [Google Scholar]
- Rost B, Sander C. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins. 1994;19:55–72. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- Samols D, Thornton CG, Murtif VL, Kumar GK, Haase FC, Wood HG. Evolutionary conservation among biotin enzymes. J Biol Chem. 1988;263:6461–6464. [PubMed] [Google Scholar]
- Sasaki Y, Hakamada K, Suama Y, Nagano Y, Furusawa I, Matsuno R. Chloroplast-encoded protein as a subunit of acetyl-CoA carboxylase in pea plant. J Biol Chem. 1993;268:25118–25123. [PubMed] [Google Scholar]
- Sasaki Y, Konishi T, Nagano Y. The compartmentation of acetyl-coenzyme A carboxylase in plants. Plant Physiol. 1995;108:445–449. doi: 10.1104/pp.108.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani DK, Ohlrogge JB. Feedback inhibition of fatty acid synthesis in tobacco suspension cells. Plant. 1995;7:577–587. [Google Scholar]
- Shorrosh BS, Roesler KR, Shintani D, van de Loo FJ, Ohlrogge JB. Structural analysis, plastid localization, and expression of the biotin carboxylase subunit of acetyl-coenzyme A carboxylase from tobacco. Plant Physiol. 1995;108:805–812. doi: 10.1104/pp.108.2.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorrosh BS, Savage LJ, Soll J, Ohlrogge JB. The pea chloroplast membrane-associated protein, IEP96, is a subunit of acetyl-CoA carboxylase. Plant J. 1996;10:261–268. doi: 10.1046/j.1365-313x.1996.10020261.x. [DOI] [PubMed] [Google Scholar]
- Shulte W, Töpfer R, Stracke R, Schell J, Martini N. Multifunctional acetyl-CoA carboxylase from Brassica napusis encoded by a multi-gene family: indication for plastidic localization of at least one isoform. Proc Natl Acad Sci USA. 1997;94:3465–3470. doi: 10.1073/pnas.94.7.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Wurtele ES, Nikolau BJ. Molecular cloning and characterization of the cDNA coding for the biotin-containing subunit of 3-methylcrotonyl-CoA carboxylase: identification of the biotin carboxylase and biotin-carrier domains. Proc Natl Acad Sci USA. 1994;91:5779–5783. doi: 10.1073/pnas.91.13.5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Ke J, Johnson JL, Nikolau BJ, Wurtele ES. Biochemical and molecular biological characterization of CAC2, the Arabidopsis thalianagene coding for the biotin carboxylase subunit of the plastidic acetyl-coenzyme A carboxylase. Plant Physiol. 1997;115:1371–1383. doi: 10.1104/pp.115.4.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldrop LG, Rayment I, Holden HM. Three-dimensional structure of the biotin carboxylase subunit of acetyl-CoA carboxylase. Biochemistry. 1994;33:10249–10256. doi: 10.1021/bi00200a004. [DOI] [PubMed] [Google Scholar]
- Yanai Y, Kawasaki T, Shimada H, Wurtele ES, Nikolau BJ, Ichikawa N. Genomic organization of 251 kDa acetyl-CoA carboxylase genes in Arabidopsis: tandem gene duplication has made two differentially expressed isozymes. Plant Cell Physiol. 1995;36:779–787. doi: 10.1093/oxfordjournals.pcp.a078822. [DOI] [PubMed] [Google Scholar]