Abstract
Viruses are powerful manipulators of microbial diversity, biogeochemistry, and evolution in the marine environment. Viruses can directly influence the genetic capabilities and the fitness of their hosts through the use of fitness factors and through horizontal gene transfer. However, the impact of viruses on microbial ecology and evolution is often overlooked in studies of the deep subsurface biosphere. Subsurface habitats connected to hydrothermal vent systems are characterized by constant fluid flux, dynamic environmental variability, and high microbial diversity. In such conditions, high adaptability would be an evolutionary asset, and the potential for frequent host–virus interactions would be high, increasing the likelihood that cellular hosts could acquire novel functions. Here, we review evidence supporting this hypothesis, including data indicating that microbial communities in subsurface hydrothermal fluids are exposed to a high rate of viral infection, as well as viral metagenomic data suggesting that the vent viral assemblage is particularly enriched in genes that facilitate horizontal gene transfer and host adaptability. Therefore, viruses are likely to play a crucial role in facilitating adaptability to the extreme conditions of these regions of the deep subsurface biosphere. We also discuss how these results might apply to other regions of the deep subsurface, where the nature of virus–host interactions would be altered, but possibly no less important, compared to more energetic hydrothermal systems.
Keywords: viral ecology, microbial evolution, hydrothermal vents, deep subsurface biosphere
Introduction
Viruses play a crucial role in marine biogeochemical cycles, microbial ecology, and evolution. Several recent reviews (Suttle, 2005; Rohwer and Thurber, 2009; Kristensen et al., 2010) have highlighted our current understanding of the viral impact on the marine environment. Generally, viruses influence the marine environment in three ways: first, through altering biogeochemical cycles by “shunting” the microbial loop through lysis of hosts (Suttle, 2005); second, by modifying the diversity and abundance of their hosts, particularly those that are most abundant, through what is dubbed “kill the winner” (Thingstad and Lignell, 1997); and third, by altering the genetic content of their hosts. Via the latter mechanism, viruses can fundamentally alter the course of evolution in their microbial hosts.
One means by which viruses can manipulate the genetic content of their hosts is by facilitating the process of horizontal gene transfer through transduction. This occurs when, in the process of virion synthesis, host genetic material is also incorporated into the viral genome. The newly synthesized viruses can then transfer the previous host’s genetic material into a new host upon infection. It has been suggested that this could be an important mechanism for horizontal gene transfer in the marine environment: one study estimated that up to 1014 transduction events per year occur in Tampa Bay Estuary alone (Jiang and Paul, 1998). Additionally, viral-like transducing particles known as gene transfer agents (GTAs) are increasingly recognized as an important mechanism for horizontal gene transfer: it has been estimated that GTA transduction rates are over one million times higher than previously reported viral transduction rates in the marine environment (McDaniel et al., 2010). GTAs and their potential impact on the deep subsurface biosphere will be discussed below.
Viruses may be particularly important in facilitating horizontal gene transfer between phylogenetically distinct lineages. Fully sequenced genomes of archaea and bacteria indicate that horizontal gene transfer does occur between lineages that are distantly related, including between domains. Overall, it is thought that interdomain horizontal gene transfer is a relatively common phenomenon, with approximately 3% of genes in bacterial genomes and 4–8% of genes in archaeal genomes involved in transfer events (Yutin and Koonin, 2009). The genome of Methanosarcina mazei, for example, contains many genes of possible bacterial origin including a bacterial chaperonin system (Deppenmeier et al., 2002). Thermophiles appear to have a particularly high percentage of transferred genes (Beiko et al., 2005; Yutin and Koonin, 2009), though it is unclear if this is due to a propensity for acquiring distantly related genes or to the close proximity of archaea and bacteria in high temperature habitats like vents. For example, up to 16.2% of the genes in Aquifex aeolicus had a best hit to archaeal species (Deckert et al., 1998; Yutin and Koonin, 2009), which is much higher than the average for bacteria. The genome of Thermotoga maritima, isolated from geothermally heated sediment, contains genes related to oxygen reduction that are likely to have been transferred through a single transfer event from a member of the Thermococcales (Nelson et al., 1999). Several Thermococcales strains have been found to contain mobile genetic elements or virus-like particles, and thus there is a strong possibility that viruses are responsible for these transfer events (Prieur et al., 2004). Viral transduction and GTAs are attractive as possible mechanisms for transfer events such as these because other mechanisms of genetic transfer do not seem capable of explaining the observed genomic similarities among distantly related species. Conjugation is a specialized process limited to particular lineages, and transformation often transfers only small amounts of genetic material, rather than multi-gene cassettes. While viral host range varies depending on viral type, some, especially GTAs, are thought to be capable of infecting distantly related hosts.
Viruses can manipulate the genetic content of their hosts not only by horizontal gene transfer, but also by expression of viral-encoded genes during the course of infection. The life cycle of lysogenic viruses involves a stage in which the virus integrates its own genome into the host genome, lying latent within the host genome for several generations until it is induced by an environmental stressor or other signal. Many integrated prophage have been found to encode what are termed fitness factors or lysogenic conversion genes: genes expressed by the prophage that can promote host fitness (Paul, 2008). These fitness factors can enhance host survivability in various environmental conditions. The cholera toxin genes in Vibrio cholerae, for example, are expressed by a filamentous bacteriophage that has integrated into the V. cholerae genome (Waldor and Mekalanos, 1996). Studies have shown that prophage genes are upregulated in response to changing environmental conditions (Smoot et al., 2001) or during biofilm formation (Whiteley et al., 2001). Even cryptic prophage, which have been integrated in the host genome for so long that they have decayed and are no longer active as phage, can carry genes that improve survivability during osmotic, oxidative, and acid stresses, and influence biofilm formation (Wang et al., 2010). Viruses infecting marine cyanobacteria, or cyanophage, have been found to carry genes for both photosystems I and II (Mann et al., 1993; Lindell et al., 2004, 2005; Millard et al., 2004; Sullivan et al., 2005; Sharon et al., 2009). These genes are expressed during viral infection, and are thought to enhance phage fitness by supplementing the host’s photosynthetic machinery (Lindell et al., 2005).
While a large amount of work has been dedicated to understanding the viral impact on the marine environment, the implications for the deep subsurface biosphere have been barely explored. Table 1 summarizes current research on viral abundance, production, and diversity in the deep subsurface biosphere, as well as shallow sediments, methane hydrates, and the deep-water column to provide a basis for comparison.
Table 1.
Summary of previous work on viral abundance, activity, and diversity in various environments of the deep subsurface biosphere, deep ocean, and sediments.
| Environment | Work on viruses to date | Reference |
|---|---|---|
| Surface marine sediments | High viral production in benthic ecosystems: may be responsible for up to 80% of cell mortality, thus releasing large amounts of carbon through the “viral shunt.” Viral diversity in sediments is fairly high, and showed a higher incidence of lysogenic than lytic phages | Danovaro et al. (2008), Middelboe et al. (2006), Siem-Jørgensen et al. (2008), Breitbart et al. (2004) |
| Deep sediments | Viral and bacterial abundance and production decrease exponentially with depth in sediments, up to 96 mbsf | Middelboe et al. (2011), Bird et al. (2001), Engelhardt et al. (2011) |
| Mitomycin C experiments revealed that 46% of isolates contained inducible prophage | ||
| Deep basalt | None known | |
| Deep granitic groundwater | Viruses are present and correlated with bacterial abundance (ratio of ∼10:1), similar to many surface environments | Kyle et al. (2008) |
| Diffuse flow hydrothermal fluid | Lysogeny appears to be a dominant lifestyle among vent viruses; viruses in diffuse flow are capable of infecting a wide range of hosts across domains and thermal regimes | Williamson et al. (2008), Anderson et al. (2011) |
| Cold seeps/methane hydrates | Viral activity and abundance vary among seeps, with a virus to prokaryote ratio ranging between relatively low (<0.1) to relatively high (66.36) | Middelboe et al. (2006), Kellogg (2010) |
| Deep-water column | Viral abundance generally tracks bacterial abundance, but the virus:cell ratio at depth varies. In some areas, the ratio increases with depth | Hara et al. (1996), Parada et al. (2007), Steward and Preston (2011) |
| Metagenomic work characterizing viral diversity found most viral sequences had matches to bacteriophages in the Podo-, Sipho-, and Myoviridae, with a few hits to eukaryotic sequences |
As can be seen in Table 1, only a few studies have focused on deep subsurface viruses. Most research to date in the deep ocean has focused on viral production and abundance in the water column and surface sediments. None of these studies, to our knowledge, has focused on the genetic and evolutionary implications of viral infection on the deep subsurface biosphere. Yet the viral impact on the evolution of bacterial and archaeal hosts could have even more profound implications in the deep subsurface than in the upper water column. A primary reason for this is that it is generally accepted that lysogeny is a more important viral lifestyle under suboptimal conditions, when host or nutrient abundance is low (Paul, 2008). This has been demonstrated in laboratory conditions, in which lysogenic viruses have a competitive advantage over lytic viruses in nutrient-limited media (Levin and Lenski, 1983). In natural populations, lysogeny becomes a more common lifestyle during seasons or in regions where host abundance is low (Jiang and Paul, 1998; McDaniel et al., 2002; Williamson et al., 2002; Weinbauer, 2004). Therefore, lysogeny may be a favored lifestyle in the deep subsurface due to difficulties in finding a new host after newly synthesized virions are released. In deep sediments, the main difficulties are likely to be low cell abundance and immobility. In extreme environments, lysogeny may be favored because of reduced viability of viral particles outside the cell. As shown in Table 1, lysogeny appears to be a common lifestyle in sediments and hydrothermal vents. If lysogeny is indeed more common in the deep subsurface biosphere, this would increase the proportion of cells harboring prophage, potentially increasing the number of cells expressing fitness factors encoded by the prophage.
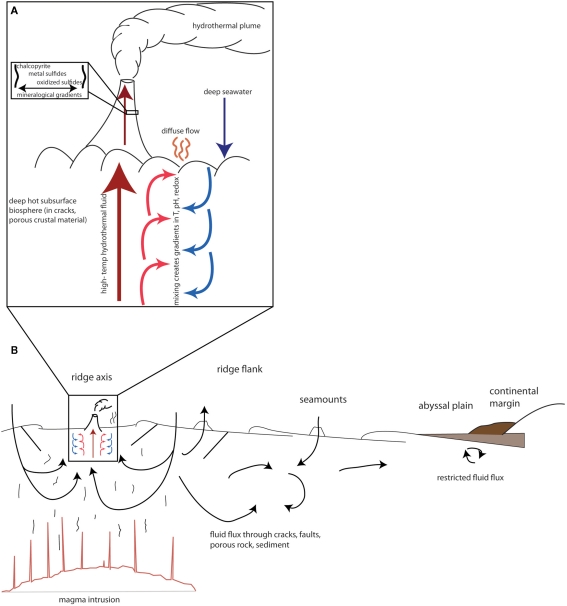
The impact of viruses on the genetic landscape of their hosts may be particularly pronounced in diffuse flow fluids of hydrothermal systems. In these environments, high temperature hydrothermal fluid mixes with seawater both in the subsurface and above the seafloor, resulting in gradients of temperature, pH, and chemical and mineralogical composition (Baross and Hoffman, 1985). A schematic representation of the vent environment, and the accompanying gradients, is shown in Figure 1A. The constant fluid flux through these gradients enables potentially frequent contact between diverse microbial communities.
Figure 1.
Schematic of fluid flux and gradient formation in (A) hydrothermal systems and (B) various regimes of the deep biosphere. Arrows represent fluid flux. Based on figures in Edwards et al. (2011), Huber et al. (2003), and Baross and Hoffman (1985).
The microorganisms inhabiting diffuse flow fluids are tremendously diverse in terms of taxonomy, metabolism, and thermal regime. Studies of the population structures of archaea and bacteria in diffuse fluids have found thousands of phylotypes within both domains (Huber et al., 2007). The abundance of reduced compounds in vent fluids allows microbial metabolisms to take advantage of a wide range of energy sources, including hydrogen, reduced iron, sulfur, as well as organic compounds. There is, therefore, a wide range of potential hosts for viruses to infect in diffuse flow fluids.
In the extreme and dynamic conditions of hydrothermal vents, genetic exchange could provide a fitness advantage to any organism able to acquire a novel function that is useful in a changing environment. Evidence already exists suggesting that vent habitats are conducive for genetic exchange. As mentioned above, genomic studies have shown that rates of horizontal gene transfer between thermophiles are higher than between other groups, including between domains. This may be due to the predominant physiology of vent organisms: microbial communities often form biofilms on the surfaces of or within vent structures (Jannasch and Wirsen, 1981; Schrenk et al., 2003). Subsurface sediments and crustal habitats also provide abundant surfaces for biofilm formation. The high cell density in a biofilm increases rates of host contact, thus facilitating higher rates of viral infection, and can also foster genetic exchange through transformation. A recent metagenomics study found that transposase sequences in biofilms in the Lost City hydrothermal field are 10 times more abundant than in metagenomes from other environments (Brazelton and Baross, 2009). If this result is characteristic of subsurface biofilms, then genetic exchange in the deep subsurface biosphere is likely to be of great importance.
It has been suggested that viruses may act as a reservoir of genes that can be used as a mechanism for adaptation by their cellular hosts (Goldenfeld and Woese, 2007), effectively expanding the “pan-genome” to include the viral assemblage. Here, we argue that viral-mediated genetic exchange is particularly important as a means to adapt to frequently changing conditions in the diffuse flow vent environment. We will review what is known about viruses in the vent environment, and examine what evidence exists that viruses play a role in modifying the genetic content (and therefore the fitness) of their cellular hosts. We conclude by discussing the larger implications for viruses in the deep subsurface biosphere in general.
Viruses as a “Genetic Repository” in Hydrothermal Vent Systems
Evidence for viral activity in diffuse flow systems
The few studies that have focused on viruses in diffuse flow vent environments have indicated that viruses play an important role in influencing microbial ecology in these ecosystems. One study quantified viral abundance in diffuse flow fluids, finding that on average there were approximately 107 viral-like particles (VLPs) per milliliter of fluid, or about 10 times as many viruses as cells, which is comparable to other marine ecosystems (Ortmann and Suttle, 2005). Another study, focusing on VLP counts at shallow hydrothermal vents, found that viruses were about five times more abundant than cells, though found that VLP counts increased with distance from the vents (Manini et al., 2008). The authors suggested that one possible explanation for this trend was a change in viral lifestyle from lysogenic to lytic, resulting in an increase in the number of apparent viruses. Additionally, diffuse fluids from 9° North, a hydrothermal vent field on the East Pacific Rise, have been found to harbor a higher incidence of inducible prophage than nearby ambient seawater, indicating that lysogeny is a more common lifestyle for vent phage than those in other marine environments (Williamson et al., 2008). A viral metagenome at Hulk vent on the Juan de Fuca Ridge showed that vent viruses are relatively diverse and have the potential to infect hosts from a wide range of taxonomic groups and thermal regimes (Anderson et al., 2011).
Genome sequences of bacteria and archaea may also show evidence of extensive viral activity in diffuse flow hydrothermal vents. Within bacterial and archaeal genomes are regions called clustered regularly interspaced palindromic repeats (CRISPRs), which are thought to facilitate the immune response to viral infection (Barrangou et al., 2007; Brouns et al., 2008; Sorek et al., 2008; van der Oost et al., 2009; Hovarth and Barrangou, 2010; Labrie et al., 2010; Marraffini and Sontheimer, 2010). These loci consist of a series of short repeats, about 20–50 bp long, and are interspersed by a series of spacer regions, each about 25–75 bp in length. These spacer regions are created to match a short sequence on an invading element, such as a virus or a plasmid. The newly synthesized spacers are then inserted between direct repeats on the CRISPR locus (Makarova et al., 2003; Bolotin et al., 2005; Haft et al., 2005; Mojica et al., 2005; Pourcel et al., 2005; Marraffini and Sontheimer, 2008; Hale et al., 2009). If a virus or plasmid invades a cell that possesses a CRISPR spacer matching a sequence on that invader, the CRISPR system will mobilize the immune response. This occurs through the formation of a Cascade complex from nearby Cas (CRISPR-associated) genes in conjunction with small RNAs derived from the CRISPR spacers, which recognize and bind target DNA (Jore et al., 2011). The invading nucleic acid is cleaved as a result (Garneau et al., 2010).
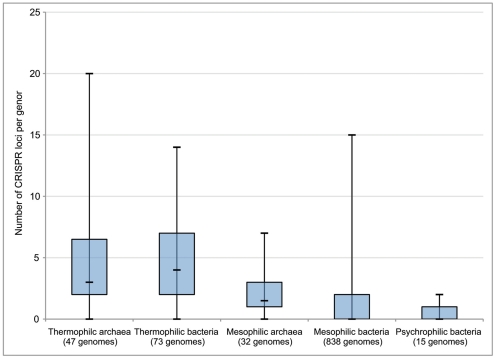
Each of these CRISPR regions therefore acts as a record of previous viral infection, with each spacer thought to represent at least one independent infection event in the history of that strain. Interestingly, it has been observed that thermophilic strains, on average, have a higher number of CRISPR loci in their genomes than mesophiles or psychrophiles (Makarova et al., 2003; Anderson et al., 2011; Figure 2). While there is no definitive explanation for the abundance of CRISPRs in thermophiles, this does indicate that viral infection plays an important role in the evolution and ecology of thermophilic microbial communities. Moreover, the CRISPR immune mechanism itself is unique among viral immunity systems in that it responds in a sequence-specific manner to the invasion of foreign genetic material, rather than through prevention of phage adsorption, blocking phage DNA entry, or random restriction modification. Thus, the abundance of CRISPRs in thermophiles indicates that at least the entry, if not the successful takeover, of foreign genetic material between different hosts is a relatively common phenomenon in high temperature environments. CRISPR loci can also be found in the genomes of bacteria and archaea isolated from other environments in the deep ocean and sediments (Table 2), though there is a clear correlation with temperature. This may serve as evidence that viruses play a particularly important role in the evolution of microbes in high temperature environments. This includes diffuse flow hydrothermal vents but could also include other regions in the subsurface with broad temperature gradients.
Figure 2.
Number of CRISPR loci per genome in thermophilic, mesophilic, and psychrophilic archaea and bacteria. Box boundaries represent first and third quartiles, and markers on lines represent minimum, median, and maximum, respectively. Modified from Anderson et al. (2011).
Table 2.
Habitat, thermal regime, number of CRISPR loci, and number of prophage for archaea and bacteria isolated from the deep-water column, marine sediments, or hydrothermal systems whose genomes have been sequenced.
| Name | No. CRISPR loci | No. prophage | Optimal growth temperature (°C) | Habitat | Reference |
|---|---|---|---|---|---|
| ARCHAEA | |||||
| Aciduliprofundum boonei | 2 | 2 | 70 | Hydrothermal chimney | Reysenbach et al. (2006) |
| Methanocaldococcus jannaschii DSM 2661 | 20 | 0 | 85 | Hydrothermal chimney | Jones et al. (1983) |
| Methanocaldococcus sp. FS406-22 | 23 | 0 | 85 | Hydrothermal fluid | Mehta and Baross (2006) |
| Methanopyrus kandleri strain 116 | 5 | 0 | 122 | Hydrothermal fluid | Takai et al. (2008) |
| Methanothermococcus okinawensis | 7 | 1 | 60–65 | Hydrothermal chimney | Takai et al. (2002) |
| Thermococcus barophilus | 4 | 0 | 85 | Hydrothermal chimney | Marteinsson et al. (1999) |
| Thermococcus gammatolerans | 3 | 3 | 88 | Hydrothermal chimney | Jolivet et al., 2003 |
| BACTERIA | |||||
| Deferribacter desulfuricans | 4 | 1 | 60–65 | Hydrothermal chimney | Takai et al. (2003) |
| Idiomarina loihiensis L2TR | 0 | 3 | 43 | Hydrothermal fluid | Hou et al. (2004) |
| Marinithermus hydrothermalis | 5 | 2 | 67.5 | Hydrothermal chimney | Sako et al. (2003) |
| Nautilia profundicola | 0 | 1 | 41–45 | Polychaete worm | Campbell et al. (2001) |
| Nitratifractor salsuginis | 2 | 3 | 37 | Hydrothermal chimney | Nakagawa et al. (2005) |
| Oceanithermus profundus | 1 | 1 | 60 | Hydrothermal chimney | Miroshnichenko et al. (2003) |
| Oceanobacillus iheyensis HTE831 | 0 | 4 | 30 | Deep sea sediment | Lu et al. (2001) |
| Photobacterium profundum | 2 | 3 | 10 | Deep sea sediment | Nogi et al. (1998) |
| Shewanella piezotolerans WP3 | 1 | 5 | 15–20 | Deep sea sediment | Wang et al. (2008) |
| Shewanella violacea DSS12 | 1 | 1 | 8 | Deep sea sediment | Kato et al. (1995) |
| Sulfurimonas autotrophica | 0 | 1 | 25 | High temperature sediment | Inagaki et al. (2003) |
| Thermosediminibacter oceani DSM 16646 | 6 | 7 | 68 | Deep sea sediment | Lee et al. (2005) |
| Zunongwangia profunda SM-A87 | 2 | 2 | 25–30 | Deep sea sediment | Qin et al. (2010) |
A further piece of evidence for the important role of viruses, one that is not restricted to high temperature organisms, is the presence of prophage in the genomes of sequenced archaea and bacteria. As mentioned above, lysogenic phage can provide supplementary metabolic functions while integrated into the genome as prophage. Table 2 lists the bacteria and archaea from diffuse flow fluids and other deep ocean habitats that have been found with integrated prophage in their genomes. Two of the listed bacterial isolates, one from the deep-water column and one from marine sediments, each possess seven prophage in their genomes. This represents a significant amount of genetic material. Considering the tendency of bacteria to select for faster reproduction rates and thus for smaller genomes (Carlile, 1982), the presence of seven prophage regions on the genome represents a potential fitness cost (through slower replication rates) that must be offset by a fitness benefit, such as the presence of fitness factors expressed by the prophage. Considering the prevalence of prophage in the genomes of isolates from diffuse flow fluids and deep sediments, it would not be surprising if prophage are present in most subsurface organisms.
Evidence for viral-mediated horizontal gene transfer
Having established that viruses are abundant and likely to be active in diffuse flow hydrothermal fluids, we now turn to evidence that viruses mediate the exchange of genes between hosts in these regions. Here, we will do this primarily through analysis of a marine hydrothermal vent viral metagenome, or virome, collected from Hulk vent in the Main Endeavor Field on the Juan de Fuca Ridge, previously described in Anderson et al. (2011). We examine evidence for viral-mediated horizontal gene transfer in the vent environment first by searching the marine vent virome for genes related to lysogeny and gene insertion; and then by using comparative metagenomics to determine which genes are selected to be maintained in the viral gene pool.
Genes related to lysogeny and gene insertion
As mentioned above, a higher proportion of bacteria and archaea in diffuse flow hydrothermal vents appear to contain inducible prophage within their genomes than bacteria and archaea from other marine environments (Williamson et al., 2008), indicating that lysogeny is an important lifestyle for viruses in diffuse flow fluids. To test whether this finding is supported by publicly available metagenomic data, we searched for genes associated with lysogeny in the marine vent virome from Hulk and compared the number of matches with that of metagenomes from other environments. It is important to note that while most lysogenic phage are integrated into cellular genomes as prophage, the Hulk vent sample would have experienced substantially colder temperatures as well as decreased pressures prior to the filtration steps that removed cells and captured viral-sized particles. This environmental shock may have induced many of the prophage within the microbial community, resulting in their being captured in the viral size fraction. While this has not yet been demonstrated experimentally, it is an important consideration in the sampling of metagenomes from relatively extreme environments.
To determine the overall abundance of genes related to lysogeny, we first created a database of lysogeny-associated proteins with Pfam seed sequences (Finn et al., 2010). These proteins include phage integrases, repressors, and antirepressors expressed during the prophage stage, regulatory proteins that trigger the switch between the lysogenic and lytic stages, and proteins involved in phage integration and excision. We queried this database with the marine vent virome using BLASTX, as well as a set of other viral and cellular metagenomes from the MG-RAST database (Meyer et al., 2008).
Metagenomes with the highest percentages of sequencing reads matching lysogeny domains were cellular rather than viral (Table 3). These hits were most likely matches to prophage incorporated into cellular genomes. Two of the top three metagenomes were sampled from environments that are considered “extreme,” namely, a highly acidic mine drainage and the high-pH, high temperature Lost City hydrothermal field. This supports the hypothesis stated earlier that lysogeny becomes a more common viral lifestyle in extreme environments because viruses have a higher chance of survival as prophage than as virions under harsh conditions. Also in the top three was the whale fall cellular metagenome. While it is unclear exactly why this metagenome had a relatively high percentage of lysogeny-related domains, one possibility is that since whale falls are relatively rare events, the organisms colonizing whale falls must endure extreme periods of relative starvation, followed by periods of plenty. Any viruses infecting these taxa would benefit from a lysogenic lifestyle in order to survive these extreme periods of starvation, during which cells would most likely either be dormant or replicating extremely slowly.
Table 3.
Percent of reads matching a DNA ligase and a transposase domain in 19 sequenced metagenomes.
| Metagenome | Type of metagenome | Percent of reads matching a lysogeny domain (rank) | Percent of reads matching a DNA ligase | MG-RAST ID number | Reference |
|---|---|---|---|---|---|
| Marine vent virome (subset) | Viral | 0.073 (6) | 1.245 | Subset of 4448187.3 | Anderson et al. (2011) |
| Marine vent virome (full) | Viral | 0.067 (7) | 0.594 | 4448187.3 | Anderson et al. (2011) |
| Farm soil | Microbial | 0.192 (4) | 0.131 | 4441091.3 | Tringe et al. (2005) |
| Acid mine drainage | Microbial | 0.468 (1) | 0.109 | 4441137.3, 444138.3 | Tyson et al. (2004) |
| Yellowstone hot springs | Viral | 0.104 (5) | 0.085 | 4443745.3, 4443746.3, 4443747.3, 4443762.3, 4443749.3, 4443750.3 | Bhaya et al. (2007) |
| Whale fall | Microbial | 0.332 (2) | 0.070 | 4441619.3, 4441656.4, 4441620.3 | Tringe et al. (2005) |
| Lost city hydrothermal field | Microbial | 0.291 (3) | 0.069 | 4461585.3 | Brazelton and Baross (2009) |
| Alvinella symbionts | Microbial | 0.044 (9) | 0.068 | 4441102.3 | Grzymski et al. (2008) |
| Peru margin sediments, 32 mbsf | Microbial | 0.020 (11) | 0.047 | 4459940.3 | Biddle et al. (2008) |
| Peru margin sediments, 1 mbsf | Microbial | 0.017 (13) | 0.044 | 4440961.3 | Biddle et al. (2008) |
| Arctic Ocean | Viral | 0.020 (12) | 0.018 | 4441622.3 | Angly et al. (2006) |
| Peru margin sediments, 16 mbsf | Microbial | 0.005 (18) | 0.018 | 4440973.3 | Biddle et al. (2008) |
| Peru margin sediments, 50 mbsf | Microbial | 0.016 (14) | 0.016 | 4459941.3 | Biddle et al. (2008) |
| Fishgut | Microbial | 0.023 (10) | 0.010 | 4441695.3 | Dinsdale et al. (2008) |
| Gulf of Mexico | Viral | 0.008 (15) | 0.006 | 4441625.4 | Angly et al. (2006) |
| Bay of British Columbia | Viral | 0.006 (17) | 0.004 | 4441623.3 | Angly et al. (2006) |
| Microbialites | Viral | 0.063 (8) | 0.003 | 4440320.3, 4440321.3, 4440323.3 | Desnues et al. (2008) |
| Sargasso Sea | Viral | 0.004 (19) | 0.002 | 4441624.3 | Angly et al. (2006) |
| Microbialites | Microbial | 0.008 (16) | 0 | 4440061.3 | Breitbart et al. (2009) |
Metagenomes are ordered according to the percent of reads matching a DNA ligase, with the highest percentage at the top. Column 3 lists the percentage of reads to a lysogeny-related domain, followed by the rank according to the percentage of reads with a hit among other metagenomes. The marine vent virome subset consists of all reads within contigs with a coverage of eight or greater plus all reads within contigs labeled “unknown” (Anderson et al., 2011). Marine vent virome reads were dereplicated prior to analysis. Mbsf, meters below seafloor. Metagenomes were downloaded from the MG-RAST database (www.metagenomics.anl.gov) and compared to a list of lysogeny-related sequences derived from Pfam seed sequences using blastx with a minimum e-value of 10−5.
Fourth on the list was a cellular metagenome sampled from farm soil, in which lysogeny may be the favored lifestyle for viruses as a result of the difficulties in encountering a new host within the sediment matrix, an environment in which mobility is likely to be significantly impaired. However, sequences matching lysogeny domains were not particularly abundant in cellular metagenomes derived from deep Peru Margin sediments, though some depth horizons had a higher abundance than others (Table 3). At this point it is unclear whether this is due to differences in viral lifestyle or simply to a lower abundance of viruses in Peru Margin sediments.
In contrast, the sequences matching lysogeny domains in the Yellowstone hot springs and marine vent viromes (fifth and sixth on the list, respectively) are most likely derived from lysogenic viruses that entered the lytic stage due to an environmental stressor, possibly resulting from the sampling process. Interestingly, these two viromes, both sampled from high temperature environments, had the highest proportion of sequencing reads matching lysogeny domains of all the viromes analyzed here. In contrast, viromes from more temperate environments, such as the Bay of British Columbia or the Sargasso Sea, had a much lower abundance of lysogeny sequences. This parallels the relative abundance of lysogeny-related domains in extreme cellular metagenomes, and again may indicate that natural selection favors lysogenic viruses in extreme environments.
The lysogeny-associated domains chosen for this analysis were selected from genes that are uniquely associated with prophage. However, this would exclude genes that serve other roles in both viruses and cells but may also be crucial in facilitating horizontal gene transfer. This includes genes required for integration of DNA into host or viral genomes, such as DNA ligases. Interestingly, 1.25% of the sequencing reads in the marine vent virome had a match to a DNA ligase, which is almost 10 times higher than any of the 17 other cellular or viral metagenomes analyzed here (Table 3). Moreover, these ligases were especially enriched in the subset of the marine vent virome considered more likely to be “viral” – that is, those reads that were assembled into contigs with an average coverage of at least eight, or within contigs in which a majority of reads were categorized as either “unknown” or “viral” by MG-RAST (Anderson et al., 2011). Therefore, the abundance of ligases appears to be distinctly viral in character.
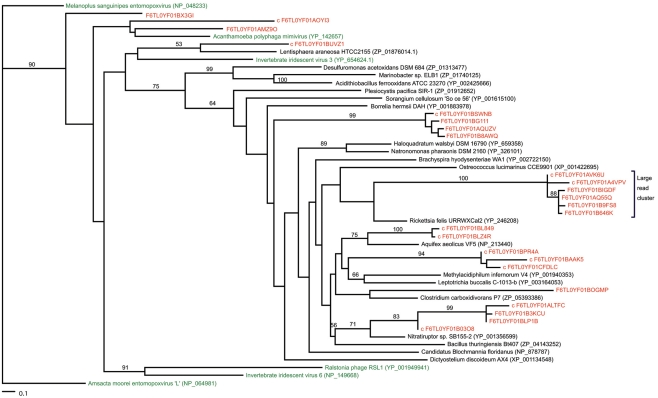
Most of the potential ligase sequences in the marine vent virome had matches to NAD-dependent ligases rather than ATP-dependent ligases. ATP-dependent ligases are fairly widespread among eukaryotes, bacteria, and archaea, while NAD-dependent ligases are generally characteristic of bacteria, though they have been seen in some viruses, eukaryotes, and archaea (Doherty and Suh, 2000; Tomkinson et al., 2006). Furthermore, they appear to be characteristic of marine rather than terrestrial metagenomes, for unknown reasons. To determine whether the ligases in the marine vent virome were most closely related to ligases from a particular group of organisms, we constructed a phylogenetic tree of NAD-dependent DNA ligases including ligase-matching sequences from the marine vent virome, using a reference tree of previously characterized DNA ligases as a constraint (Figure 3). The reference viral ligases do not necessarily group together on this tree, suggesting there is not a viral ligase type that is necessarily distinct from bacterial, archaeal, or eukaryotic ligases. Similarly, the sequences from the marine vent virome are scattered across the tree, with some closely related to ligases found in viruses, and others more closely related to bacterial or archaeal ligases. Yutin and Koonin (2009) have previously highlighted the non-monophyletic nature of viral DNA ligases. This may attest to the fluid nature of viral genomes in that they frequently pick up genes from their hosts and may also be a further indication of the high diversity of the marine vent viral assemblage. While several vent virome sequences were grouped with DNA ligases found in viruses, the majority grouped most closely with a DNA ligase from Rickettsia felis, a Gram-negative bacterium in the Alphaproteobacteria group. R. felis is closely related to SAR11, one of the most common bacterial lineages in marine environments. While SAR11 sequences were not common in the marine vent virome, it is possible that ancestral viruses in the marine vent environment had acquired ligases from SAR11 or similar groups, which then became more abundant in the viral gene pool due to positive selection.
Figure 3.
Maximum-likelihood phylogenetic tree of NAD-dependent DNA ligases with metagenomic reads from the marine vent virome. The “large read cluster” denotes the branch in which the majority of metagenomic reads matching ligases grouped on the tree. Numbers indicate the bootstrap values of internal nodes (where n = 100). Metagenomic reads are colored red. All ligase protein sequences were obtained from NCBI (accession numbers listed). Trees were constructed in RAxML by incorporating metagenomic sequences into a constraint tree of references sequences based on the phylogeny of Yutin and Koonin (2009). Trees imaged with TreeViewX.
While it is unclear exactly what ecological role these ligases play, their high abundance suggests that they play a uniquely important role in the vent viral assemblage. DNA ligases repair double-stranded breaks in DNA by catalyzing the synthesis of phosphodiester bonds between 5′-phosphoryl and 3′-hydroxyl groups (Lehnman, 1974). DNA ligases are thus important for DNA replication, recombination, and repair across all domains as well as viruses. However, the high abundance of DNA ligases in the vent virome is unusual and may reflect a specific role in the vent ecosystem, possibly in the integration of viral genomes into the host genome through recombination. It is therefore plausible that the abundance of these genes is indicative of prevalent horizontal gene transfer in these regions of the deep subsurface biosphere.
Finally, GTAs may act as an important mechanism for viral-mediated horizontal gene transfer. These phage-like particles were originally discovered in marine Rhodobacteria, but have since been discovered in various other bacterial and archaeal species, particularly in the marine environment (Lang and Beatty, 2000; Matson et al., 2005; Stanton, 2007; Biers et al., 2008, Leung et al., 2010). GTAs randomly incorporate segments of the host genome into a viral capsid, then transfer this to new hosts, including phylogenetically unrelated organisms, without resulting in lysis of the host cell. It has been suggested that GTAs are defective phage (Lang and Beatty, 2000; Matson et al., 2005; Stanton, 2007); if so, it would seem that GTAs have effectively lost their parasitic nature and have instead been usurped by the host for the purposes of gene exchange. While the only sequenced GTA genes are from the Rhodobacter capsulatus (Lang and Beatty, 2000) and Brachyspira hyodysentariae (Matson et al., 2005) GTAs, the marine vent virome included six sequencing reads with matches to known GTA genes. While these are few, the scarcity of sequences from GTAs in public databases to date prohibits a larger-scale search for GTAs in viral metagenomes. Nevertheless, GTAs may play a significant role in transferring genes between hosts in the marine environment and appear to be present in diffuse flow fluids. In the dynamic, extreme conditions of the deep subsurface, selection may be particularly strong for cells that harbor GTAs as a mechanism for obtaining advantageous genes. It has been hypothesized that a large portion of marine viromes may consist of GTAs carrying poorly conserved bacterial genes (Kristensen et al., 2010) and may thus contribute a large portion of the poorly conserved “cloud” of genes in the viral gene pool.
Enrichment for functional genes in the vent virome
This “cloud” of genes, or genetic reservoir, consists of an overlapping pool of genes derived from both viruses and cells. Genes in the viral genetic reservoir are expressed in cellular hosts through either horizontal gene transfer or prophage insertion, and then maintained in the gene pool through positive selection. Thus, the genes maintained in the viral metagenome, in addition to genes for viral synthesis, packaging, and maintenance, are likely to consist of non-housekeeping genes that provide a selective advantage in the event of a shift in environmental conditions. This is expected to be particularly the case in the gradient-dominated, dynamic vent environment.
An initial example of this can be found through analysis of assembled contigs of the marine vent virome from Hulk (assembly and contig analysis described in Anderson et al., 2011). Among the lysogeny domains matching sequences in the marine vent virome was the XerD tyrosine recombinase protein, commonly associated with lysogenic phage and transposases. This gene was assembled onto a contig that also contains the universal stress protein UspA. UspA is known to be phosphorylated in response to stasis stress in Escherichia coli (Nyström and Neidhardt, 1994; Freestone et al., 1997), and thus is induced when growth conditions are not optimal. It would be advantageous to both host and prophage for the prophage-encoded protein to induce a response in the host when growth conditions are poor. As mentioned, prophage have been found to encode genes to enhance host survivability in stressful conditions, and this sequence may be one example of many such instances in the vent ecosystem.
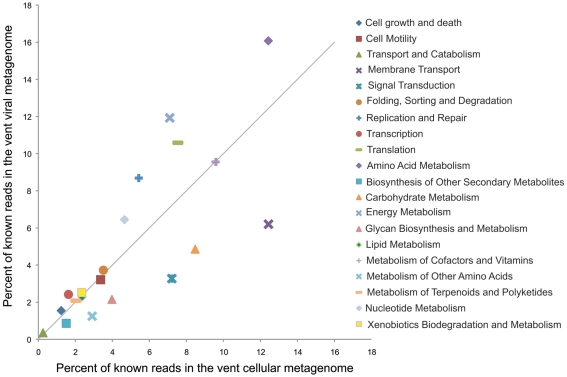
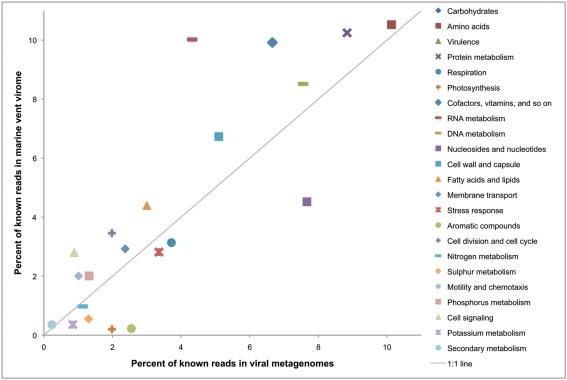
While this is one example of a potential fitness factor encoded by a lysogenic phage, comparative metagenomics can give a more large-scale picture of which functional genes are enriched in the viral fraction. To do this we compared the relative enrichment of several gene categories in the marine vent virome and a cellular metagenome sampled from a sulfide chimney at Mothra hydrothermal field on the Juan de Fuca Ridge (Xie et al., 2011; Figure 4). Each metagenome was analyzed through the MG-RAST pipeline and annotated using the KEGG Orthology database (Meyer et al., 2008). For this analysis, as before, we used only the vent virome subset that we consider to be convincingly viral: that is, sequencing reads assembled into contigs with an average coverage of at least eight or composed of reads identified as “unknown” or “viral” by MG-RAST. The virome was relatively enriched in gene categories such as replication and repair, nucleotide metabolism, and translation, which are important for the synthesis of viral genetic material during the lytic stage. Interestingly, however, the virome was particularly enriched in genes related to energy metabolism, with nearly 12% of all identifiable sequencing reads matching this category in the virome, compared to 7% in the cellular metagenome. Genes in this category include genes used for oxidative phosphorylation, photosynthesis, methane metabolism, carbon fixation, and sulfur and nitrogen metabolism. It seems plausible that these genes are maintained in the viral genetic reservoir through positive selection. For example, if a cell were to be flushed into a region in which the abundance of its conventional electron donor was limited, the acquisition of a gene allowing it to utilize an alternative electron donor, via either transduction or prophage expression, would increase the fitness of that cell. The relative enrichment of genes related to energy metabolism in the virome may be evidence of this type of selection.
Figure 4.
Relative percentages of reads matching gene categories in the marine vent virome (Anderson et al., 2011) and the hydrothermal vent cellular metagenome by Xie et al. (2011). The solid line indicates a 1:1 ratio between the viral and cellular metagenomes. Metagenomes were analyzed with MG-RAST (Meyer et al., 2008), and reads were annotated with the KEGG Orthology database, release 56.
Other sequences in the marine vent virome had matches to metabolic genes that have also been found in viruses from other environments. For example, 11 sequencing reads in the marine vent virome had close matches to PhoH, a phosphate starvation-inducible protein that has previously been found in Prochlorococcus and Synechococcus phage genomes (Sullivan et al., 2009). It has been hypothesized that PhoH, when expressed by phage, could aid the host in phosphorus scavenging during the infection stage. In hydrothermal systems, phosphate is removed from seawater through water–rock reactions in the deep subsurface, thus limiting phosphate availability to native microbial communities (Wheat et al., 1996). The presence of PhoH in the marine vent virome suggests that vent viruses may, in a manner similar to the cyanophage, assist in phosphate acquisition during the course of infection. Similarly, eight reads closely matching transaldolases, also found on cyanophage genomes, were found in the marine vent virome. It is thought that these transaldolases may play a role in metabolizing carbon substrates to assist in energy production and synthesis of important compounds, and thus may be yet another example of fitness factors expressed by phage (Sullivan et al., 2005).
This positive selection for genes that can increase host fitness would suggest that the genes found in the marine vent virome can give an indication of the unique environmental conditions from which it was sampled, such as limitations or extremes in factors such as nutrients, energy availability, temperature, or pH. For example, 301 sequencing reads in the marine vent virome were annotated by MG-RAST as uptake [NiFe]-hydrogenases, enzymes involved in the oxidation of hydrogen to produce energy (Vignais and Billoud, 2007). Hydrogen oxidation is known to be an important metabolic strategy in hydrothermal vent systems, where it provides a greater energy return than the oxidation of methane or sulfur (Amend and Shock, 2001), which are also common reduced compounds in vent environments. The [NiFe]-hydrogenases encoded in the marine vent virome could enable utilization of an important alternative energy source for viral hosts if transferred into their genome via transduction, or if expressed by prophage as a fitness factor, which could bolster host metabolism during viral infection.
To present a larger-scale illustration of how the genetic profile of a virome might act as a signature of the environment from which it was sampled, we compared the relative enrichment of functional gene categories in the marine vent virome with a set of 42 other viromes, initially analyzed by Dinsdale et al., 2008; Figure 5). As in the previous analysis, only the convincingly viral subset of the marine vent virome from Hulk was used. In this analysis, genes were annotated with the SEED subsystems in MG-RAST to make a more direct comparison with the study by Dinsdale et al. (2008). The results show that the marine vent virome is particularly enriched in genes related to regulation and cell signaling (enriched by 300%) and RNA metabolism (enriched by 230%). The genes assigned to the “cell signaling” category include those related to biofilm formation and quorum sensing, regulation of virulence, and sensing environmental stimuli. For example, 118 sequencing reads in the marine vent virome have close matches to genes in the “regulation of virulence” category. Most of these matched the BarA–UvrY two-component system, a system regulating virulence in pathogenic E. coli (Herren et al., 2006). The BarA–UvrY system functions by sensing changes in environmental conditions to induce a metabolic switch, a function that would be useful in the rapidly changing conditions of the vent environment.
Figure 5.
Relative percentages of reads matching gene categories in the marine vent virome and a set of 42 viral metagenomes isolated from other environments (listed in Dinsdale et al., 2008). The solid line indicates a 1:1 ratio between the vent metagenome and the other viral metagenomes. Metagenomes were analyzed with MG-RAST (Meyer et al., 2008) and annotated using the SEED subsystems database.
We expect that the pool of genes undergoing positive selection for retention in the virus genetic reservoir would represent those genes that may be occasionally necessary, though not strictly required, in the environment from which it was sampled. Thus, we expect accessory genes such as those related to secondary metabolisms to be selected for maintenance in the virome, rather than housekeeping genes such as ribosomal proteins. In addition to representing the “cloud” of poorly conserved genes from both cellular and viral pangenomes (Kristensen et al., 2010), we expect positive selection to customize this “genetic reservoir” to match the needs of the hosts drawing from it.
Extrapolating the Viral Impact from Hydrothermal Vents to Other Regimes of the Deep Subsurface Biosphere
We have suggested here that viruses are likely to play an important role in modifying the genetic content of their hosts in diffuse flow fluids of hydrothermal systems. We now turn to a discussion of whether the characteristics of viruses in diffuse fluids is likely to be the case for other regimes of the deep subsurface biosphere. The vent ecosystem has several unique attributes that distinguish it from other deep subsurface habitats, so extrapolation from the vent subsurface to the rest of the deep subsurface biosphere must be done cautiously. The two most distinctive attributes of the vent system are the extreme gradients in temperature, pH, redox state, chemical composition, and mineralogy, and the constant fluid flux between microenvironments. On the ridge axis, hot hydrothermal fluid mixing with cool, oxygenated seawater facilitates interaction among diverse cellular communities and their accompanying viral assemblages. For this reason, we have argued that this environment may be conducive to gene flow from one microenvironment to the next via viral transfer. However, the impact of viruses is likely to extend to other provinces of the deep subsurface biosphere, which we detail below.
Crustal environments: unsedimented ridge flanks, crustal outcrops, seamounts, arc systems
This fluid flux from one environment to the next is characteristic of many regimes in the deep subsurface biosphere. As sediments covering the seafloor tend to be relatively impermeable, fluid flux depends on outcropping of igneous crust (Edwards et al., 2011). Fluids flux most vigorously at mid-ocean ridges at the hydrothermal systems discussed above, and degree of fluid flux decreases with distance from the ridge axis, as shown in Figure 1B. However, these environments are not completely stagnant. On ridge flanks and recharge zones, which can extend hundreds of kilometers from the ridge axis, oxygenated seawater flows into the ocean floor, with residence times of days to years (Johnson et al., 2010). Some of this fluid emerges in discharge zones in the form of hydrothermal vents on ridge axes, while some fluids may circulate more locally. Off-axis, fluid flows through seamounts; and farther afield, fluids may circulate in a more restricted manner (Edwards et al., 2005). Overall, however, the volume of fluid flux is large, as at least 60% of the oceanic crust is hydrologically active, and the fluid-crust reservoir in which these processes might be expected to occur is approximately 10 times the size of the sedimentary reservoir (Edwards et al., 2011). Fluid-flux-dominated environments, therefore, constitute the majority of the deep subsurface. This fluid flux facilitates transfer of chemicals throughout the crust, thus exposing microbial communities to varying conditions and also transporting microorganisms from one region to the next. The regions of the oceanic crust dominated by fluid flux are therefore analogous to the hydrothermal systems described above, though characterized by fewer extremes in temperature, pH, and redox conditions.
By means of this flux, viruses or their accompanying hosts are transported between biomes, and can facilitate horizontal gene transfer between organisms native to drastically different environments. Viral metagenomics has suggested that while local diversity of viral assemblages is high, global diversity may be low, suggesting that viruses frequently migrate between biomes (Breitbart and Rohwer, 2005). One study found that viruses from sediments, soils, and lakes were able to propagate in marine waters, suggesting that viruses have a broad enough host range that they can successfully move between biomes (Sano et al., 2004). Fluid flux between the seafloor–ocean interface, within sediments, and within cracks in the Earth’s crust may act as a conduit for viruses or the microbes that bear them, thus potentially sharing genes among habitats. Moreover, if biofilms are hotspots for horizontal gene transfer and viral infection (as discussed above), we may expect that regions of the subsurface characterized by biofilm formation, which could include any habitat with surfaces available for attachment, will be exposed to higher rates of gene transfer than in environments with lower cell density.
Sedimented environments: abyssal plains, continental margins
As shown in Figure 1B, some provinces of the deep subsurface biosphere, particularly those characterized by deep sedimentation, experience more restricted fluid flux and therefore have potentially limited contact between hosts of different environment niches. Yet even in these regions, viruses may alter host fitness through lysogeny. As stated above, lysogeny appears to become a more predominant lifestyle in regions with suboptimal conditions, such as low nutrient abundance, low host replication rates, or low host contact rates. Some sedimented regions of the deep subsurface are characterized by particularly low cell abundance, such as in sediments within oligotrophic gyres (D’Hondt et al., 2009), or are exposed to extremely limited organic matter, nutrient, or free energy availability (Schrenk et al., 2010). Moreover, the lack of mobility in the sediment matrix may encourage a lysogenic lifestyle as well, as viruses may have difficulty in contacting a new host within a sediment matrix, especially in regions with low cell abundance. Therefore, the lysogenic lifestyle is likely to be much more common among viruses in these regions, and the archaeal and bacterial inhabitants of these regions have an even higher likelihood of expressing fitness factors encoded by prophage. Initial studies of prophage in the deep subsurface biosphere seem to support this case (Engelhardt et al., 2011). These prophage may aid in host survivability. For example, in deeply buried marine sediments with limited organic carbon or other nutrients, viruses may carry genes to aid in scavenging these compounds or in providing secondary metabolisms to take advantage of alternative energy or nutrient sources. However, more work needs to be done on sequencing viral or cellular isolates from these regions to gain further support for this hypothesis.
One final viral influence that is likely to impact all provinces of the deep subsurface biosphere is the input of viruses from surface waters. Marine sediments receive large inputs of allochthonous material daily, much of it bearing particle-associated microbes that sink through the water column. If these microbes carry prophage or lytic viruses in the process of replicating, these viruses could potentially encounter a different host in its new sediment-bound habitat. These viruses, delivered from the upper water column, could then deliver genes from more pelagic habitats to the deep subsurface. One might even expect induction of prophage to occur more frequently in sinking microbes as they are exposed to increased pressures, which has been found to induce prophage in E. coli (Aertsen et al., 2004). In this sense, viruses may serve as a highway for gene exchange between the surface marine realm and the deep subsurface biosphere.
Conclusion
The presence of lysogeny-related genes as well as potentially advantageous cellular functional genes in the virome of a marine, diffuse flow hydrothermal vent bolster the hypothesis that viruses modify the genetic landscape of the hydrothermal habitat, both through expression of prophage genes during lysogenic infection and through the process of transduction. Indeed, this habitat – as well as several others in the deep subsurface biosphere – is uniquely poised for fostering an almost symbiotic relationship between viruses and their hosts. As mentioned above, deep sea microbes have higher numbers of integrated prophage and transposable elements in their genomes (Campanaro et al., 2005; Vezzi et al., 2005; Ivars-Martinez et al., 2008), and lysogeny appears to be favored over lysis in the extreme regions of the marine realm (Paul, 2008). In regions of the deep subsurface biosphere with low host contact rates, low nutrient flux, or rapidly changing conditions, expression by latent prophage or viral transduction may play a crucial role in bolstering host fitness.
More work certainly remains to be done to determine the scope of the viral impact on the deep subsurface biosphere. Sequencing the genomes of archaeal and bacterial isolates from various regions of the deep subsurface can indicate what prophage elements are present. Viral metagenomics can indicate what types of genes are harbored by viruses, and therefore can indicate which genes may either be transferred into host genomes or expressed during the prophage stage. Thus far, the evidence suggests that viruses play a crucial role in enhancing the fitness of their hosts by modifying their genetic content. They may help their hosts adapt to the unique challenges of the various habitats of the deep subsurface biosphere, acting as a genetic repository for new, adaptive functions. Viruses are emerging as a profound evolutionary force whose impact we have yet to fully assess, particularly in the realms of the deep.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Jon Kaye at the Gordon and Betty Moore Foundation for help with virome sequencing. Sequencing of virome sample G2810 was funded by the Gordon and Betty Moore Foundation through a grant to the Broad Institute. Rika E. Anderson was funded by an NSF Graduate Research Fellowship through NSF grant number DGE-0718124, an NSF IGERT grant to the University of Washington Astrobiology Program, and the ARCS Foundation. William J. Brazelton was funded by a NASA Astrobiology Institute Postdoctoral Fellowship. Research funding was provided through a NASA Astrobiology Institute grant through Cooperative Agreement NNA04CC09A to the Geophysical Laboratory at the Carnegie Institution for Science.
References
- Aertsen A., Van Houdt R., Vanoirbeek K., Michiels C. W. (2004). An SOS response induced by high pressure in Escherichia coli. J. Bacteriol. 186, 6133–6141 10.1128/JB.186.18.6133-6141.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amend J. P., Shock E. L. (2001). Energetics of overall metabolic reactions of thermophilic and hyperthermophilic archaea and bacteria. FEMS Microbiol. Rev. 25, 175–243 10.1111/j.1574-6976.2001.tb00576.x [DOI] [PubMed] [Google Scholar]
- Anderson R. E., Brazelton W. J., Baross J. A. (2011). Using CRISPRs as a metagenomic tool to identify microbial hosts of a diffuse flow hydrothermal vent viral assemblage. FEMS Microbiol. Ecol. 77, 120–133 10.1111/j.1574-6941.2011.01090.x [DOI] [PubMed] [Google Scholar]
- Angly F. E., Felts B., Breitbart M., Salamon P., Edwards R. A., Carlson C., Chan A. M., Haynes M., Kelley S., Liu H., Mahaffy J. M., Mueller J. E., Nulton J., Olson R., Parsons R., Rayhawk S., Suttle C. A., Rohwer F. (2006). The marine viromes of four oceanic regions. PLoS Biol. 4, e368. 10.1371/journal.pbio.0040368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baross J. A., Hoffman S. E. (1985). Submarine hydrothermal vents and associated gradient environments as sites for the origin and evolution of life. Orig. Life Evol. Biosph. 15, 327–345 10.1007/BF01808177 [DOI] [Google Scholar]
- Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D. A., Horvath P. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 10.1126/science.1138140 [DOI] [PubMed] [Google Scholar]
- Beiko R. G., Harlow T. J., Ragan M. A. (2005). Highways of gene sharing in prokaryotes. Proc. Natl. Acad. Sci. U.S.A. 102, 14332–14337 10.1073/pnas.0504068102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaya D., Grossman A. R., Steunou A.-S., Khuri N., Cohan F. M., Hamamura N., Melendrez M. C., Bateson M. M., Ward D. M., Heidelberg J. F. (2007). Population level functional diversity in a microbial community revealed by comparative genomic and metagenomic analyses. ISME J. 1, 703–713 10.1038/ismej.2007.46 [DOI] [PubMed] [Google Scholar]
- Biddle J. F., Fitz-Gibbon S., Schuster S. C., Brenchley J. E., House C. H. (2008). Metagenomic signatures of the Peru Margin subseafloor biosphere show a genetically distinct environment. Proc. Natl. Acad. Sci. U.S.A. 105, 10583–10588 10.1073/pnas.0709942105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biers E. J., Wang K., Pennington C., Belas R., Chen F., Moran M. A. (2008). Occurrence and expression of gene transfer agent (GTA) genes in marine bacterioplankton. Appl. Environ. Microbiol. 74, 2933–2939 10.1128/AEM.02129-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird D. F., Juniper S. K., Ricciardi-Regault M., Martineu P., Prairie Y. T., Calvert S. E. (2001). Subsurface viruses and bacteria in Holocene/Late Pleistocene sediments of Saanich Inlet, BC: ODP Holes 1033B and 1034B, Leg 169S. Mar. Geol. 174, 227–239 10.1016/S0025-3227(00)00152-3 [DOI] [Google Scholar]
- Bolotin A., Quinquis B., Sorokin A., Ehrlich S. D. (2005). Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151, 2551–2561 10.1099/mic.0.28048-0 [DOI] [PubMed] [Google Scholar]
- Bose M., Barber R. (2006). Prophage finder: a prophage loci prediction tool for prokaryotic genome sequences. In Silico Biol. 6, 0020. [PubMed] [Google Scholar]
- Brazelton W. J., Baross J. A. (2009). Abundant transposases encoded by the metagenome of a hydrothermal chimney biofilm. ISME J. 3, 1420–1424 10.1038/ismej.2009.79 [DOI] [PubMed] [Google Scholar]
- Breitbart M., Felts B., Kelley S., Mahaffy J. M., Nulton J., Salamon P., Rohwer F. (2004). Diversity and population structure of a near-shore marine-sediment viral community. Proc. Biol. Sci. 271, 565–574 10.1098/rspb.2003.2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart M., Hoare A., Nitti A., Siefert J., Haynes M., Dinsdale E., Edwards R., Souza V., Rohwer F., Hollander D. (2009). Metagenomic and stable isotopic analyses of modern freshwater microbialites in Cuatro Cienegas, Mexico. Environ. Microbiol. 11, 16–34 10.1111/j.1462-2920.2008.01725.x [DOI] [PubMed] [Google Scholar]
- Breitbart M., Rohwer F. (2005). Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 13, 278–284 10.1016/j.tim.2005.04.003 [DOI] [PubMed] [Google Scholar]
- Brouns S. J. J., Jore M. M., Lundgren M., Westra E. R., Slijkhuis R. J. H., Snijders A. P. L., Dickman M. J., Makarova K. S., Koonin E. V., Van der Oost J. (2008). Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321, 960–964 10.1126/science.1159689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanaro S., Vezzi A., Vitulo N., Lauro F., D’Angelo M., Simonato F., Cestaro A., Malacrida G., Bertoloni G., Valle G., Bartlett D. H. (2005). Laterally transferred elements and high pressure adaptation in Photobacterium profundum strains. BMC Genomics 6, 122. 10.1186/1471-2164-6-122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell B. J., Jeanthon C., Kostka J. E., Luther G. W., III, Cary S. C. (2001). Growth and phylogenetic properties of novel bacteria belonging to the epsilon subdivision of the proteobacteria enriched from Alvinella pompejana and deep-sea hydrothermal vents. Appl. Environ. Microbiol. 67, 4566–4572 10.1128/AEM.67.1.110-117.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile M. (1982). Prokaryotes and eukaryotes: strategies and successes. Trends Biochem. Sci. 7, 128–130 10.1016/0968-0004(82)90011-1 [DOI] [Google Scholar]
- Danovaro R., Dell’Anno A., Corinaldesi C., Magagnini M., Noble R., Tamburini C., Weinbauer M. (2008). Major viral impact on the functioning of benthic deep-sea ecosystems. Nature 454, 1084–1087 10.1038/nature07268 [DOI] [PubMed] [Google Scholar]
- Deckert G., Warren P. V., Gaasterland T., Young W. G., Lenox A. L., Graham D. E., Overbeek R., Snead M. A., Keller M., Aujay M., Huber R., Feldman R. A., Short J. M., Olsen G. J., Swanson R. V. (1998). The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature 392, 353–358 10.1038/32831 [DOI] [PubMed] [Google Scholar]
- Deppenmeier U., Johann A., Hartsch T., Merkl R., Schmitz R. A., Martinez-Arias R., Henne A., Wiezer A., Bäumer S., Jacobi C., Brügemann H., Lienard T., Christmann A., Bömeke M., Steckel S., Bhattacharyya A., Lykidis A., Overbeek R., Klenk H. P., Gunsalus R. P., Fritz H. J., Gottschalk G. (2002). The genome of Methanosarcina mazei: evidence for lateral gene transfer between bacteria and archaea. J. Mol. Microbiol. Biotechnol. 4, 453–461 [PubMed] [Google Scholar]
- Desnues C., Rodriguez-Brito B., Rayhawk S., Kelley S., Tran T., Haynes M., Liu H., Furlan M., Wegley L., Chau B., Ruan Y., Hall D., Angly F. E., Edwards R. A., Li L., Thurber R. V., Reid R. P., Siefert J., Souza V., Valentine D., Swan B. K., Breitbart M., Rohwer F. (2008). Biodiversity and biogeography of phages in modern stromatolites and thrombolites. Nature 452, 340–344 10.1038/nature06735 [DOI] [PubMed] [Google Scholar]
- D’Hondt S., Spivack A. J., Pockalny R., Ferdelman T. G., Fischer J. P., Kallmeyer J., Abrams L. J., Smith D. C., Graham D., Hasiuk F., Schrum H., Stancin A. M. (2009). Subseafloor sedimentary life in the South Pacific Gyre. Proc. Natl. Acad. Sci. U.S.A. 106, 11651–11656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinsdale E. A., Edwards R. A., Hall D., Angly F., Breitbart M., Brulc J. M., Furlan M., Desnues C., Haynes M., Li L., McDaniel L., Moran M. A., Nelson K. E., Nilsson C., Olson R., Paul J., Brito B. R., Ruan Y., Swan B. K., Stevens R., Valentine D. L., Thurber R. V., Wegley L., White B. A., Rohwer F. (2008). Functional metagenomic profiling of nine biomes. Nature 452, 629–632 10.1038/nature06810 [DOI] [PubMed] [Google Scholar]
- Doherty A. J., Suh S. W. (2000). Structural and mechanistic conservation in DNA ligases. Nucleic Acids Res. 28, 4051–4058 10.1093/nar/28.21.4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K. J., Bach W., McCollom T. M. (2005). Geomicrobiology in oceanography: microbe-mineral interactions at and below the seafloor. Trends Microbiol. 13, 449–456 10.1016/j.tim.2005.07.005 [DOI] [PubMed] [Google Scholar]
- Edwards K. J., Wheat C. G., Sylvan J. B. (2011). Under the sea: microbial life in volcanic oceanic crust. Nat. Rev. Microbiol. 9, 703–712 10.1038/nrmicro2647 [DOI] [PubMed] [Google Scholar]
- Engelhardt T., Sahlberg M., Cypionka C., Engelen B. (2011). Induction of prophages from deep-subseafloor bacteria. Environ. Microbiol. Rep. 3, 459–465 10.1111/j.1758-2229.2010.00232.x [DOI] [PubMed] [Google Scholar]
- Finn R. D., Mistry J., Tate J., Coggill P., Heger A., Pollington J. E., Gavin O. L., Gunasekaran P., Ceric G., Forslund K., Holm L., Sonnhammer E. L., Eddy S. R., Bateman A. (2010). The Pfam protein families database. Nucleic Acids Res. 38, D211–D222 10.1093/nar/gkp985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freestone P., Nyström T., Trinei M., Norris V. (1997). The universal stress protein, UspA, of Escherichia coli is phosphorylated in response to stasis. J. Mol. Biol. 274, 318–324 10.1006/jmbi.1997.1397 [DOI] [PubMed] [Google Scholar]
- Garneau J. E., Dupuis M. È., Villion M., Romero D. A., Barrangou R., Boyaval P., Fremaux C., Horvath P., Magadán A. H., Moineau S. (2010). The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67–71 10.1038/nature09523 [DOI] [PubMed] [Google Scholar]
- Goldenfeld N., Woese C. (2007). Biology’s next revolution. Nature 445, 369. 10.1038/445369a [DOI] [PubMed] [Google Scholar]
- Grissa I., Vergnaud G., Pourcel C. (2007a). CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35, W52–W57 10.1093/nar/gkl1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grissa I., Vergnaud G., Pourcel C. (2007b). The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics 8: 172. 10.1186/1471-2105-8-172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzymski J. J., Murray A. E., Campbell B. J., Kaplarevic M., Gao G. R., Lee C., Daniel R., Ghadiri A., Feldman R. A., Cary S. C. (2008). Metagenome analysis of an extreme microbial symbiosis reveals eurythermal adaptation and metabolic flexibility. Proc. Natl. Acad. Sci. U.S.A. 105, 17516–17521 10.1073/pnas.0802782105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft D. H., Selengut J., Mongodin E. F., Nelson K. E. (2005). A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 1, e60. 10.1371/journal.pcbi.0010060.eor [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale C. R., Zhao P., Olson S., Duff M. O., Graveley B. R., Wells L., Terns R. M., Terns M. P. (2009). RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 139, 945–956 10.1016/j.cell.2009.07.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara S., Koike I., Terauchi K., Kamiya H., Tanoue E. (1996). Abundance of viruses in deep oceanic waters. Mar. Ecol. Prog. Ser. 145, 269–277 10.3354/meps145269 [DOI] [Google Scholar]
- Herren C. D., Mitra A., Palaniyandi S. K., Coleman A., Elankumaran S., Mukhopadhyay S. (2006). The BarA-UvrY two-component system regulates virulence in avian pathogenic Escherichia coli O78:K80:H9. Infect. Immun. 74, 4900–4909 10.1128/IAI.00412-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou S., Saw J. H., Lee K. S., Freitas T. A., Belisle C., Kawarabayasi Y., Donachie S. P., Pikina A., Galperin M. Y., Koonin E. V., Makarova K. S., Omelchenko M. V., Sorokin A., Wolf Y. I., Li Q. X., Keum Y. S., Campbell S., Denery J., Aizawa S.-I., Shibata S., Malahoff A., Alam M. (2004). Genome sequence of the deep-sea gammaproteobacterium Idiomarina loihiensis reveals amino acid fermentation as a source of carbon and energy. Proc. Natl. Acad. Sci. U.S.A. 101, 18036–18041 10.1073/pnas.0407638102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovarth P., Barrangou R. (2010). CRISPR/Cas, the immune system of bacteria and archaea. Science 327, 167–170 10.1126/science.1179555 [DOI] [PubMed] [Google Scholar]
- Huber J. A., Butterfield D. A., Baross J. A. (2003). Bacterial diversity in a subseafloor habitat following a deep-sea volcanic eruption. FEMS Microbiol. Ecol. 43, 393–309 10.1111/j.1574-6941.2003.tb01080.x [DOI] [PubMed] [Google Scholar]
- Huber J. A., Mark Welch D. B., Morrison H. G., Huse S. M., Neal P. R., Butterfield D. A., Sogin M. L. (2007). Microbial population structures in the deep marine biosphere. Science 318, 97. 10.1126/science.1146689 [DOI] [PubMed] [Google Scholar]
- Inagaki F., Takai K., Kobayashi H., Nealson K. H., Horikoshi K. (2003). Sulfurimonas autotrophica gen. nov., sp. nov., a novel sulfur-oxidizing ε-proteobacterium isolated from hydrothermal sediments in the Mid-Okinawa trough. Int. J. Syst. Evol. Microbiol. 53, 1801–1805 10.1099/ijs.0.02682-0 [DOI] [PubMed] [Google Scholar]
- Ivars-Martinez E., Martin-Cuadrado A.-B., D’Auria G., Mira A., Ferriera S., Johnson J., Friedman R., Rodriguez-Valera F. (2008). Comparative genomics of two ecotypes of the marine planktonic copiotroph Alteromonas macleodii suggests alternative lifestyles associated with different kinds of particulate organic matter. ISME J. 2, 1194–1212 10.1038/ismej.2008.74 [DOI] [PubMed] [Google Scholar]
- Jannasch H. W., Wirsen C. O. (1981). Morphological survey of microbial mats near deep-sea thermal vents. Appl. Environ. Microbiol. 41, 528–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S. C., Paul J. H. (1998). Gene transfer by transduction in the marine environment. Appl. Environ. Microbiol. 64, 2780–2787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson H. P., Tivey M. A., Bjorklund T. A., Salmi M. S. (2010). Hydrothermal circulation within the Endeavour segment, Juan de Fuca Ridge. Geochem. Geophys. Geosyst. 11, Q05002. 10.1029/2009GC002957 [DOI] [Google Scholar]
- Jolivet E., L’Haridon S., Corre E., Forterre P., Prieur D. (2003). Thermococcus gammatolerans sp. nov., a hyperthermophilic archaeon from a deep-sea hydrothermal vent that resists ionizing radiation. Int. J. Syst. Evol. Microbiol. 53, 847–851 10.1099/ijs.0.02503-0 [DOI] [PubMed] [Google Scholar]
- Jones W. J., Leigh J. A., Mayer F., Woese C. R., Wolfe R. S. (1983). Methanococcus jannaschii sp. nov., an extremely thermophilic methanogen from a submarine hydrothermal vent. Arch. Microbiol. 136, 254–261 10.1007/BF00425213 [DOI] [Google Scholar]
- Jore M. M., Lundgren M., van Duijn E., Bultema J. B., Westra E. R., Waghmare S. P., Wiedenheft B., Pul Ü., Wurm R., Wagner R., Beijer M. R., Barendregt A., Zhou K., Snijders A. P., Dickman M. J., Doudna J. A., Boekema E. J., Heck A. J., van der Oost J., Brouns S. J. (2011). Structural basis for CRISPR RNA-guided DNA recognition by cascade. Nat. Struct. Mol. Biol. 18, 529–536 10.1038/nsmb.2019 [DOI] [PubMed] [Google Scholar]
- Kato C., Sato T., Horikoshi K. (1995). Isolation and properties of barophilic and barotolerant bacteria from deep-sea mud samples. Biodivers. Conserv. 4, 1–9 10.1007/BF00115311 [DOI] [Google Scholar]
- Kellogg C. A. (2010). Enumeration of viruses and prokaryotes in deep-sea sediments and cold seeps of the Gulf of Mexico. Deep Sea Res. Part 2 Top. Stud. Oceanogr. 57, 2002–2007 10.1016/j.dsr2.2010.05.006 [DOI] [Google Scholar]
- Kristensen D. M., Mushegian A. R., Dolja V. V., Koonin E. V. (2010). New dimensions of the virus world discovered through metagenomics. Trends Microbiol. 18, 11–19 10.1016/j.tim.2009.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyle J. E., Eydal H. S. C., Ferris F. G., Pedersen K. (2008). Viruses in granitic groundwater from 69 to 450 m depth of the Äspö hard rock laboratory, Sweden. ISME J. 2, 571–574 10.1038/ismej.2008.18 [DOI] [PubMed] [Google Scholar]
- Labrie S. J., Samson J. E., Moineau S. (2010). Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8, 317–327 10.1038/nrmicro2315 [DOI] [PubMed] [Google Scholar]
- Lang A. S., Beatty J. T. (2000). Genetic analysis of a bacterial genetic exchange element: the gene transfer agent of Rhodobacter capsulatus. Proc. Natl. Acad. Sci. U.S.A. 97, 859–864 10.1073/pnas.97.2.859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.-J., Wagner I. D., Brice M. E., Kevbrin V. V., Mills G. L., Romanek C. S., Wiegel J. (2005). Thermosediminibacter oceani gen. nov., sp. nov. and Thermosediminibacter litoriperuensis sp. nov., new anaerobic thermophilic bacteria isolated from Peru Margin. Extremophiles 9, 375–383 10.1007/s00792-005-0453-4 [DOI] [PubMed] [Google Scholar]
- Lehnman I. R. (1974). DNA ligase: structure, mechanism, and function. Science 186, 790–797 10.1126/science.186.4166.790 [DOI] [PubMed] [Google Scholar]
- Leung M. M., Florizone S. M., Taylor T. A., Lang A. S., Beatty J. T. (2010). The gene transfer agent of Rhodobacter capsulatus. Adv. Exp. Med. Biol. 675, 253–264 10.1007/978-1-4419-1528-3_14 [DOI] [PubMed] [Google Scholar]
- Levin B. R., Lenski R. E. (1983). “Coevolution in bacteria and their viruses and plasmids,” in Coevolution, eds Futuyma D. J., Slatkin M. (Sunderland, MA: Sinauer Associates; ), 99–127 [Google Scholar]
- Lindell D., Jaffe J. D., Johnson Z. I., Church G. M., Chisholm S. W. (2005). Photosynthesis genes in marine viruses yield proteins during host infection. Nature 438, 86–89 10.1038/nature04111 [DOI] [PubMed] [Google Scholar]
- Lindell D., Sullivan M. B., Johnson Z. I., Tolonen A. C., Rohwer F., Chisholm S. W. (2004). Transfer of photosynthesis genes to and from Prochlorococcus viruses. Proc. Natl. Acad. Sci. U.S.A. 101, 11013–11018 10.1073/pnas.0401526101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Nogi Y., Takami H. (2001). Oceanobacillus iheyensis gen. nov., sp. nov., a deep-sea extremely halotolerant and alkaliphilic species isolated from a depth of 1050 m on the Iheya Ridge. FEMS Microbiol. Lett. 205, 291–297 10.1111/j.1574-6968.2001.tb10963.x [DOI] [PubMed] [Google Scholar]
- Makarova K. S., Wolf Y. I., Koonin E. V. (2003). Potential genomic determinants of hyperthermophily. Trends Genet. 19, 172–176 10.1016/S0168-9525(03)00047-7 [DOI] [PubMed] [Google Scholar]
- Manini E., Luna G. M., Corinaldesi C., Zeppilli D., Bortoluzzi G., Caramanna G., Raffa F., Danovaro R. (2008). Prokaryote diversity and virus abundance in shallow hydrothermal vents of the Mediterranean Sea (Panarea Island) and the Pacific Ocean (North Sulawesi-Indonesia). Microb. Ecol. 55, 626–639 10.1007/s00248-007-9306-2 [DOI] [PubMed] [Google Scholar]
- Mann N. H., Cook A., Millard A., Bailey S., Clokie M. (1993). Bacterial photosynthesis genes in a virus. Environ. Microbiol. 59, 3736–3743 [Google Scholar]
- Marraffini L. A., Sontheimer E. J. (2008). CRISPR interference limits horizontal gene transfer in Staphylococci by targeting DNA. Science 322, 1843–1845 10.1126/science.1165771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini L. A., Sontheimer E. J. (2010). Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature 463, 568–571 10.1038/nature08703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marteinsson V. T., Birrien J. L., Reysenbach A. L., Vernet M., Marie D., Gambacorta A., Messner P., Sleytr U. B., Prieur D. (1999). Thermococcus barophilus sp. nov., a new barophilic and hyperthermophilic archaeon isolated under high hydrostatic pressure from a deep-sea hydrothermal vent. Int. J. Syst. Bacteriol. 49, 351–359 10.1099/00207713-49-2-351 [DOI] [PubMed] [Google Scholar]
- Matson E. G., Thompson M. G., Humphrey S. B., Zuerner R. L., Stanton T. B. (2005). Identification of genes of VSH-1, a prophage-like gene transfer agent of Brachyspira hyodysenteriae. J. Bacteriol. 187, 5885–5892 10.1128/JB.187.17.5885-5892.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel L., Houchin L. A., Williamson S. J., Paul J. H. (2002). Plankton blooms: lysogeny in marine Synechococcus. Nature 415, 496. 10.1038/415496a [DOI] [PubMed] [Google Scholar]
- McDaniel L. D., Young E., Delaney J., Ruhnau F., Ritchie K. B., Paul J. H. (2010). High frequency of horizontal gene transfer in the oceans. Science 330, 50. 10.1126/science.1192243 [DOI] [PubMed] [Google Scholar]
- Mehta M. P., Baross J. A. (2006). Nitrogen fixation at 92°C by a hydrothermal vent archaeon. Science 314, 1783–1786 10.1126/science.1134772 [DOI] [PubMed] [Google Scholar]
- Meyer F., Paarmann D., D’Souza M., Olson R., Glass E., Kubal M., Paczian T., Rodriguez A., Stevens R., Wilke A., Wilkening J., Edwards R. A. (2008). The metagenomics RAST server – a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9, 386. 10.1186/1471-2105-9-386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middelboe M., Glud R. N., Filippini M. (2011). Viral abundance and activity in the deep sub-seafloor biosphere. Aquat. Microb. Ecol. 63, 1–8 10.3354/ame01485 [DOI] [Google Scholar]
- Middelboe M., Glud R. N., Wenzhöfer F., Oguri K., Kitazato H. (2006). Spatial distribution and activity of viruses in the deep-sea sediments of Sagami Bay, Japan. Deep Sea Res. Part 1 Oceanogr. Res. Pap. 53, 1–13 10.1016/j.dsr.2005.09.008 [DOI] [Google Scholar]
- Millard A., Clokie M. R. J., Shub D. A., Mann N. H. (2004). Genetic organization of the psbAD region in phages infecting marine Synechococcus strains. Proc. Natl. Acad. Sci. U.S.A. 101, 11007–11012 10.1073/pnas.0401478101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miroshnichenko M. L., L’haridon S., Jeanthon C., Antipov A. N., Kostrikina N. A., Tindall B. J., Schumann P., Spring S., Stackebrandt E., Bonch-Osmolovskaya E. A. (2003). Oceanithermus profundus gen. nov., sp. nov., a thermophilic, microaerophilic, facultatively chemolithoheterotrophic bacterium from a deep-sea hydrothermal vent. Int. J. Syst. Evol. Microbiol. 53, 747–752 10.1099/ijs.0.02367-0 [DOI] [PubMed] [Google Scholar]
- Mojica F. J. M., Díez-Villaseñor C., García-Martínez J., Soria E. (2005). Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 60, 174–182 10.1007/s00239-004-0046-3 [DOI] [PubMed] [Google Scholar]
- Nakagawa S., Takai K., Inagaki F., Horikoshi K., Sako Y. (2005). Nitratiruptor tergarcus gen. nov., sp. nov. and Nitratifractor salsuginis gen. nov., sp. nov., nitrate-reducing chemolithoautotrophs of the Epsilonproteobacteria isolated from a deep-sea hydrothermal system in the Mid-Okinawa trough. Int. J. Syst. Evol. Microbiol. 55, 925–933 10.1099/ijs.0.63351-0 [DOI] [PubMed] [Google Scholar]
- Nelson K. E., Clayton R. A., Gill S. R., Gwinn M. L., Dodson R. J., Haft D. H., Hickey E. K., Peterson J. D., Nelson W. C., Ketchum K. A., McDonald L., Utterback T. R., Malek J. A., Linher K. D., Garrett M. M., Stewart A. M., Cotton M. D., Pratt M. S., Phillips C. A., Richardson D., Heidelberg J., Sutton G. G., Fleischmann R. D., Eisen J. A., White O., Salzberg S. L., Smith H. O., Venter J. C., Fraser C. M. (1999). Evidence for lateral gene transfer between archaea and bacteria from genome sequence of Thermotoga maritima. Nature 399, 323–329 10.1038/20601 [DOI] [PubMed] [Google Scholar]
- Nogi Y., Masui N., Kato C. (1998). Photobacterium profundum sp. nov., a new, moderately barophilic bacterial species isolated from a deep-sea sediment. Extremophiles 2, 1–8 10.1007/s007920050036 [DOI] [PubMed] [Google Scholar]
- Nyström T., Neidhardt F. C. (1994). Expression and role of the universal stress protein, UspA, of Escherichia coli during growth arrest. Mol. Microbiol. 11, 537–544 10.1111/j.1365-2958.1994.tb00334.x [DOI] [PubMed] [Google Scholar]
- Orcutt B. N., Sylvan J. B., Knab N. J., Edwards K. J. (2011). Microbial ecology of the dark ocean above, at, and below the seafloor. Microbiol. Mol. Biol. Rev. 75, 361–422 10.1128/MMBR.00039-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortmann A. C., Suttle C. A. (2005). High abundances of viruses in a deep-sea hydrothermal vent system indicates viral mediated microbial mortality. Deep Sea Res. Part 2 Top. Stud. Oceanogr. 52, 1515–1527 10.1016/j.dsr.2005.04.002 [DOI] [Google Scholar]
- Parada V., Sintes E., Van Aken H. M., Weinbauer M. G., Herndl G. J. (2007). Viral abundance, decay, and diversity in the meso-and bathypelagic waters of the North Atlantic. Appl. Environ. Microbiol. 73, 4429–4438 10.1128/AEM.00029-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul J. H. (2008). Prophages in marine bacteria: dangerous molecular time bombs or the key to survival in the seas? ISME J. 2, 579–589 10.1038/ismej.2008.35 [DOI] [PubMed] [Google Scholar]
- Pourcel C., Salvignol G., Vergnaud G. (2005). CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 151, 653–663 10.1099/mic.0.27437-0 [DOI] [PubMed] [Google Scholar]
- Prieur D., Erauso G., Geslin C., Lucas S., Gaillard M., Bidault A., Mattenet A. -C., Rouault K., Flament D., Forterre P., Le Romancer M. (2004). Genetic elements of Thermococcales. Biochem. Soc. T. 32, 184–187 10.1042/BST0320184 [DOI] [PubMed] [Google Scholar]
- Qin Q.-L., Zhang X.-Y., Wang X.-M., Liu G.-M., Chen X.-L., Xie B.-B., Dang H.-Y., Zhou B.-C., Yu J., Zhang Y.-Z. (2010). The complete genome of Zunongwangia profunda SM-A87 reveals its adaptation to the deep-sea environment and ecological role in sedimentary organic nitrogen degradation. BMC Genomics 11, 247. 10.1186/1471-2164-11-247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reysenbach A. L., Liu Y., Banta A., Beveridge T. J., Kirshtein J. D., Schouten S., Tivey M. K., Von Damm K. L., Voytek M. A. (2006). A ubiquitous thermoacidophilic archaeon from deep-sea hydrothermal vents. Nature 442, 444–447 10.1038/nature04921 [DOI] [PubMed] [Google Scholar]
- Rohwer F., Thurber R. V. (2009). Viruses manipulate the marine environment. Nature 458, 207–212 10.1038/nature08060 [DOI] [PubMed] [Google Scholar]
- Sako Y., Nakagawa S., Takai K., Horikoshi K. (2003). Marinithermus hydrothermalis gen. nov., sp. nov., a strictly aerobic, thermophilic bacterium from a deep-sea hydrothermal vent chimney. Int. J. Syst. Evol. Microbiol. 53, 59–65 10.1099/ijs.0.02364-0 [DOI] [PubMed] [Google Scholar]
- Sano E., Carlson S., Wegley L., Rohwer F. (2004). Movement of viruses between biomes. Appl. Environ. Microbiol. 70, 5842–5846 10.1128/AEM.70.10.5842-5846.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrenk M. O., Huber J. A., Edwards K. J. (2010). Microbial provinces in the subseafloor. Ann. Rev. Mar. Sci. 2, 279–304 10.1146/annurev-marine-120308-081000 [DOI] [PubMed] [Google Scholar]
- Schrenk M. O., Kelley D. S., Delaney J. R., Baross J. A. (2003). Incidence and diversity of microorganisms within the walls of an active deep-sea sulfide chimney. Appl. Environ. Microbiol. 69, 3580–3592 10.1128/AEM.69.6.3580-3592.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon I., Alperovitch A., Rohwer F., Haynes M., Glaser F., Atamna-Ismaeel N., Pinter R. Y., Partensky F., Koonin E. V., Wolf Y. I., Nelson N., Béjà O. (2009). Photosystem I gene cassettes are present in marine virus genomes. Nature 461, 258–262 10.1038/nature08284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siem-Jørgensen M., Glud R. N., Middelboe M. (2008). Viral dynamics in a coastal sediment: seasonal pattern, controlling factors and relations to the pelagic–benthic coupling. Mar. Biol. Res. 4, 165–179 10.1080/17451000801888718 [DOI] [Google Scholar]
- Smoot L. M., Smoot J. C., Graham M. R., Somerville G. A., Sturdevant D. E., Migliaccio C. A. L., Sylva G. L., Musser J. M. (2001). Global differential gene expression in response to growth temperature alteration in group A Streptococcus. Proc. Natl. Acad. Sci. U.S.A. 98, 10416–10421 10.1073/pnas.191267598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R., Kunin V., Hugenholtz P. (2008). CRISPR – a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat. Rev. Microbiol. 6, 181–186 10.1038/nrmicro1793 [DOI] [PubMed] [Google Scholar]
- Stanton T. B. (2007). Prophage-like gene transfer agents–Novel mechanisms of gene exchange for Methanococcus, Desulfovibrio, Brachyspira, and Rhodobacter species. Anaerobe 13, 43–49 10.1016/j.anaerobe.2007.03.004 [DOI] [PubMed] [Google Scholar]
- Steward G. F., Preston C. M. (2011). Analysis of a viral metagenomic library from 200 m depth in Monterey Bay, California constructed by direct shotgun cloning. Virol. J. 8, 287. 10.1186/1743-422X-8-287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M. B., Coleman M. L., Weigele P., Rohwer F., Chisholm S. W. (2005). Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol. 3, e144. 10.1371/journal.pbio.0030144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M. B., Krastins B., Hughes J. L., Kelly L., Chase M., Sarracino D., Chisholm S. W. (2009). The genome and structural proteome of an ocean siphovirus: a new window into the cyanobacterial “mobilome.” Environ. Microbiol. 11, 2935–2951 10.1111/j.1462-2920.2009.02081.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttle C. A. (2005). Viruses in the sea. Nature 437, 356–361 10.1038/nature04160 [DOI] [PubMed] [Google Scholar]
- Takai K., Inoue A., Horikoshi K. (2002). Methanothermococcus okinawensis sp. nov., a thermophilic, methane-producing archaeon isolated from a Western Pacific deep-sea hydrothermal vent system. Int. J. Syst. Evol. Microbiol. 52, 1089–1095 10.1099/ijs.0.02106-0 [DOI] [PubMed] [Google Scholar]
- Takai K., Kobayashi H., Nealson K. H., Horikoshi K. (2003). Deferribacter desulfuricans sp. nov., a novel sulfur-, nitrate-and arsenate-reducing thermophile isolated from a deep-sea hydrothermal vent. Int. J. Syst. Evol. Microbiol. 53, 839–846 10.1099/ijs.0.02773-0 [DOI] [PubMed] [Google Scholar]
- Takai K., Nakamura K., Toki T., Tsunogai U., Miyazaki M., Miyazaki J., Hirayama H., Nakagawa S., Nunoura T., Horikoshi K. (2008). Cell proliferation at 122°C and isotopically heavy CH4 production by a hyperthermophilic methanogen under high-pressure cultivation. Proc. Natl. Acad. Sci. U.S.A. 105, 10949–10954 10.1073/pnas.0712334105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thingstad T., Lignell R. (1997). Theoretical models for the control of bacterial growth rate, abundance, diversity and carbon demand. Aquat. Microb. Ecol. 13, 19–27 10.3354/ame013019 [DOI] [Google Scholar]
- Tomkinson A. E., Vijayakumar S., Pascal J. M., Ellenberger T. (2006). DNA ligases: structure, reaction mechanism, and function. Chem. Rev. 106, 687–699 10.1021/cr040498d [DOI] [PubMed] [Google Scholar]
- Tringe S. G., von Mering C., Kobayashi A., Salamov A. A., Chen K., Chang H. W., Podar M., Short J. M., Mathur E. J., Detter J. C., Bork P., Hugenholtz P., Rubin E. M. (2005). Comparative metagenomics of microbial communities. Science 22, 554–557 10.1126/science.1107851 [DOI] [PubMed] [Google Scholar]
- Tyson G. W., Chapmon J., Hugenholtz P., Allen E. E., Ram R. J., Richardson P. M., Solovyev V. V., Rubin E. M., Rokhsar D. S., Banfield J. F. (2004). Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428, 37–43 10.1038/nature02340 [DOI] [PubMed] [Google Scholar]
- van der Oost J., Jore M. M., Westra E. R., Lundgren M., Brouns S. J. J. (2009). CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem. Sci. 34, 401–407 10.1016/j.tibs.2009.05.002 [DOI] [PubMed] [Google Scholar]
- Vezzi A., Campanaro S., D’Angelo M., Simonato F., Vitulo N., Lauro F. M., Cestaro A., Malacrida G., Simionati B., Cannata N., Romualdi C., Bartlett D. H., Valle G. (2005). Life at depth: Photobacterium profundum genome sequence and expression analysis. Science 307, 1459–1461 10.1126/science.1103341 [DOI] [PubMed] [Google Scholar]
- Vignais P. M., Billoud B. (2007). Occurrence, classification, and biological function of hydrogenases: an overview. Chem. Rev. 107, 4206–4272 10.1021/cr050196r [DOI] [PubMed] [Google Scholar]
- Waldor M. K., Mekalanos J. J. (1996). Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272, 1910–1914 10.1126/science.272.5270.1910 [DOI] [PubMed] [Google Scholar]
- Wang F., Wang J., Jian H., Zhang B., Li S., Wang F., Zeng X., Gao L., Bartlett D. H., Yu J., Hu S., Xiao X. (2008). Environmental adaptation: genomic analysis of the piezotolerant and psychrotolerant deep-sea iron reducing bacterium Shewanella piezotolerans WP3. PLoS ONE 3, e1937. 10.1371/journal.pone.0004027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Kim Y., Ma Q., Hong S. H., Pokusaeva K., Sturino J. M., Wood T. K. (2010). Cryptic prophages help bacteria cope with adverse environments. Nat. Commun. 1, 147. 10.1038/ncomms1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinbauer M. G. (2004). Ecology of prokaryotic viruses. FEMS Microbiol. Rev. 28, 127–181 10.1016/j.femsre.2003.08.001 [DOI] [PubMed] [Google Scholar]
- Wheat G. C., Feely R. A., Mottl M. J. (1996). Phosphate removal by oceanic hydrothermal processes: an update of the phosphorus budget in the oceans. Geochim. Cosmochim. Acta 60, 3593–3608 10.1016/0016-7037(96)00189-5 [DOI] [Google Scholar]
- Whiteley M., Bangera M. G., Bumgarner R. E., Parsek M. R., Teitzel G. M., Lory S., Greenberg E. P. (2001). Gene expression in Pseudomonas aeruginosa biofilms. Nature 413, 860–864 10.1038/35101627 [DOI] [PubMed] [Google Scholar]
- Williamson S. J., Cary S. C., Williamson K. E., Helton R. R., Bench S. R., Winget D., Wommack K. E. (2008). Lysogenic virus–host interactions predominate at deep-sea diffuse-flow hydrothermal vents. ISME J. 2, 1112–1121 10.1038/ismej.2008.73 [DOI] [PubMed] [Google Scholar]
- Williamson S. J., Houchin L. A., McDaniel L., Paul J. H. (2002). Seasonal variation in lysogeny as depicted by prophage induction in Tampa Bay, Florida. Appl. Environ. Microbiol. 68, 4307–4314 10.1128/AEM.68.9.4307-4314.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W., Wang F., Guo L., Chen Z., Sievert S. M., Meng J., Huang G., Li Y., Yan Q., Wu S., Wang X., Chen S., He G., Xiao X., Xu A. (2011). Comparative metagenomics of microbial communities inhabiting deep-sea hydrothermal vent chimneys with contrasting chemistries. ISME J. 5, 414–426 10.1038/ismej.2010.144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N., Koonin E. V. (2009). Evolution of DNA ligases of nucleo-cytoplasmic large DNA viruses of eukaryotes: a case of hidden complexity. Biol. Direct 4, 51. 10.1186/1745-6150-4-51 [DOI] [PMC free article] [PubMed] [Google Scholar]