Abstract
Endotoxin tolerance is characterized by the suppression of further TNF release upon recurrent exposure to LPS. This phenomenon is proposed to act as a homeostatic mechanism preventing uncontrolled cytokine release such as that observed in bacterial sepsis. The regulatory mechanisms and inter-individual variation of endotoxin tolerance induction in man remain poorly characterized. Here we describe a genetic association study of variation in endotoxin tolerance amongst healthy individuals. We identify a common promoter haplotype in TNFRSF1B (encoding TNFR2) to be strongly associated with reduced tolerance to LPS (P = 5.82×10−6). This identified haplotype is associated with increased expression of TNFR2 (P = 4.9 ×10−5) and we find basal expression of TNFR2, irrespective of genotype and unlike TNFR1, is associated with secondary TNF release (P <0.0001). Functional studies demonstrate a positive feedback loop via TNFR2 of LPS induced TNF release, confirming this previously unrecognized role for TNFR2 in the modulation of LPS response.
INTRODUCTION
The inflammatory response to LPS, an invariant component of the outer membrane of Gram negative bacteria, is mediated via a TLR dependent transduction cascade (1, 2). This process is tightly regulated and characterized by the coordinate expression and release of multiple cytokines including TNF, IL-6 and IL-1β. Much of the early morbidity associated with Gram negative bacterial infections is attributable to LPS induced TNF release (3). Therefore mechanisms controlling this response in man have vital implications in health and disease. Repeated or chronic exposure to LPS, or indeed other TLR ligands, leads to rapid tolerance to their pro-inflammatory actions both at a cellular level and at the level of the organism. This tolerance to LPS is referred to as endotoxin tolerance and was described over a century ago (4, 5). However, only recently have the underlying molecular mechanisms begun to be elucidated (6). Endotoxin tolerance is now recognized as a multi-factorial process characterized by the induction of negative regulators of TLR signalling that can be proteins such as SHIP (7) and IRAK-M (8) or microRNAs including miR146, miR155 and miR9 (9, 10). Concomitant to this, chronic exposure to LPS leads to chromatin remodeling and the selective silencing of pro-inflammatory genes resulting in a more profound state of tolerance (11).
The multiple conserved biological mechanisms of tolerance across species underline the importance of homeostatic regulation of innate immunity. In animal models, induction of endotoxin tolerance can render lethal doses of LPS sub-lethal, primarily by restricting further LPS induced TNF release. Failure to silence innate immune activity is implicated in many acute and chronic inflammatory states, ranging from sepsis and autoimmune conditions to cancer. Identification of mechanisms regulating innate immune tolerance in man provides targets for therapeutic intervention to silence dysregulated innate immune responses.
Genetic polymorphisms play a significant role in determining inter-individual variation in cytokine responsiveness to LPS (12, 13) but the phenotype of endotoxin tolerance has not been characterized in this respect. Genome-wide association analyses have demonstrated the impact of common genetic variants on multifactorial traits, highlighting the importance of noncoding sequence variants which modulate gene expression (14). For a number of common diseases such as type 1 diabetes and Crohn’s disease, genome-wide association studies have isolated genes whose encoded products had not been previously considered in disease pathophysiology (15). In this study we sought to define the extent of variation in endotoxin tolerance to repeated stimulation with LPS based on TNF release among healthy volunteers. We aimed to identify functional genetic variants associated with tolerance induction and in doing so gain novel insights into the regulation of tolerance induction in man. We show that a common haplotype spanning TNFRSF1B, the gene encoding tumor necrosis factor receptor 2 (denoted by its alternative gene name TNFR2 in the following text) is strongly associated with reduced tolerance to LPS. We proceed to demonstrate that this haplotype is associated with differential nuclear factor binding and is specifically associated with increased basal expression of TNFR2. We subsequently show that LPS induced TNF activity can positively feedback through TNFR2, impacting upon the duration and magnitude of TNF release.
MATERIALS AND METHODS
Ethics Statement
This study was approved by the Oxfordshire Research Ethics Committee (REC reference 06/Q1605/55). All volunteers gave written informed consent.
Study Volunteers, PBMC purification and LPS tolerance assays
The recruitment and demographics of the healthy volunteer cohort, sample collection, PBMC purification and cell culture conditions have been previously described (13). In summary healthy volunteers without a history of recent viral or bacterial infection were venesected in the morning and PBMCs purified using a Ficoll-paque PLUS (GE Healthcare Charlfont St Giles, UK) gradient within 2 h. Cells were counted with a haemocytometer and resuspended at 2.5×106 cells per ml in RPMI 1640 growth medium (Sigma) supplemented with L-glutamine (2mM), penicillin/streptomycin (100U/ml penicillin, 0.1mg/ml streptomycin) and 10% v/v heat-inactivated Gold standard fetal calf serum, with three biological replicate assays per condition set up for each volunteer. After overnight incubation, cells were washed in fresh media then resuspended in media alone, or with LPS at 2 ng/ml (Sigma L-4391 lot 114K4133), for 6 h prior to separation of cells from supernatant by centrifugation. To assess tolerance, additional naïve or LPS treated cells (2ng/ml for 6 h) were exposed to LPS (20ng/ml) for a further 6 h. Three biological replicate assays per condition were set up for each volunteer and the mean used in subsequent statistical analysis. Supernatants for ELISA were stored at −80°C prior to quantification; cell pellets for RNA extraction were resuspended in RLT buffer (Qiagen, Valencia, CA) supplemented with 2-beta-mercaptoethanol and stored at −80°C. Where indicated, a TNFR2 allosteric modulator antibody (anti-human TNF-R II Clone 80M2 Cat# HM2022 Hycult Biotech, Uden, The Netherlands) or mouse IgG1 isotype control (functional grade) monoclonal antibody (Cat# M075-3M2 Medical & Biological Laboratories, Nagoya, Japan) was added to cultures at a concentration of 2.5μg/ml for the times indicated. For the experiments involving purified monocytes, CD14+ monocytes were positively selected with Miltenyi CD14+ labelled microbeads (catalog number 130-050; Miltenyi Biotec GmbH, Bergisch Gladbach, Germany) according to instructions, with all steps performed at 4°C or on ice. Experiments with monocytes were carried out at the lower concentration of 5×105 cells per ml.
ELISAs
ELISAs were performed on all samples in duplicate as described (13) using TNF, IL-6 and sTNF RII Duoset kits (DY 206, DY210, DY726 R&D Systems, Minneapolis, MN).
RNA extraction, cDNA synthesis and quantitative PCR
Total RNA was prepared using the RNeasy Mini kit (Qiagen) as described with cDNA prepared with SuperScript III (Invitrogen, Paisley, UK) using random hexamers (13). Quantitative PCR was carried out using SYBR Green Supermix (BioRad, Hemel Hempstead UK) on the CFX96 C1000 Thermal Cycler (Bio-Rad). PCR efficiency was determined and melt curve analysis performed for gene specific primer sets (Supplementary Table 1). Relative gene transcript levels were determined by the ΔΔCt method.
Genotyping, haplotype and statistical analysis
The TNFR2 VNTR (Variable Number Tandem Repeat) was genotyped by amplification of a 150bp fragment by PCR (primer sequences shown in Supplementary Table 1) and the number of repeats was directly identified from electrophoresis of a 3% agarose gel. Genotyping of the volunteer cohort using the humanCVD bead array chip (Illumina, San Diego, CA), computation of LD (Linkage Disequilibrium) and haplotypes using HaploView 3.3.2 program (16) and expression quantitative trait mapping using PLINK (17) was performed as previously described (13) with verification using SPSS, R package and SNPTest. Standard QC measures were used with exclusion criteria of maximum per-SNP missing (GENO > 0.1) and MAF < 0.03. The genotyping rate was > 98.8%. For each SNP analysed, the phenotypic mean for the three genotypic states was compared using the Wald test statistic to generate a P value which does not require that the data fits a normal distribution. Covariates age, ethnicity and sex were included in PLINK analysis to further interrogate observed associations. Non-parametric statistics and log transformation were applied where data was not normally distributed, otherwise unpaired t-tests were used for analysis of expression data for specific SNPs, data passing tests of normality. Permutation analysis was performed using a label-swapping procedure and an adaptive algorithm. Analysis was performed with one million permutations using a within-cluster algorithm as described (13). The empirical P value is robust with respect to normality of phenotype and multiple testing issues. All correlations were carried out using linear regression least square fit models with statistical relevance and correlation coefficients described for each association. Where indicated, two tailed t-tests were used.
Nuclear extracts, electrophoretic mobility shift assays (EMSA)
Nuclear extracts were prepared, oligonucleotide probes were radiolabelled with [32P]deoxycytidine 5′-triphosphate (Perkin Elmer, Beaconsfield U.K.) by fill-in of 5′ overhanging ends using Klenow and unlabelled deoxyadenosine 5′-triphosphate, deoxycguanosine 5′-triphosphate, and deoxythymiidine 5′-triphosphate, and EMSA performed as previously described (18, 19). Probes were generated by annealing forward and reverse oligonucleotides. Sequences are shown in the Supplementary Table 1.
RESULTS
Variation in endotoxin tolerance at the level of TNF release in response to LPS
Similar to previous studies (20), we noted that a 6 h pretreatment of cells with LPS established marked tolerance to further LPS induced TNF release. We also observed a degree of inter-individual variation at this time-point, permitting investigation of underlying genetic contributions to this. We therefore based our assay on TNF release in response to LPS stimulation for 6 h, comparing naïve versus pretreated cells. We were interested in the individual silencing of TNF production after LPS treatment and so to normalize for variation in initial response, due to either inter-individual variation in PBMC composition or other intrinsic sensitivities to LPS, values were expressed relative to the naïve response. This approach allowed us to focus upon silencing of TNF responses in a sample most representative to that in circulation. We proceeded to recruit a cohort of 96 healthy volunteers and to define variation in tolerance to LPS based on three replicate stimulation assays for a given volunteer on a particular day, with the mean value used in subsequent analyses.
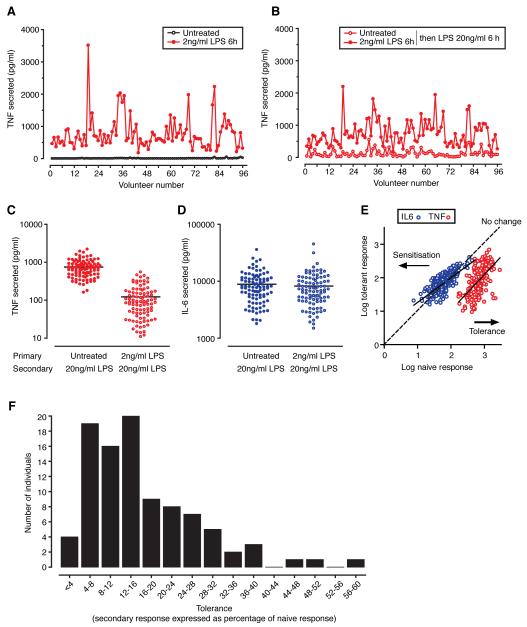
Over a 6 h period, LPS treatment (2ng/ml) resulted in a pronounced release of TNF into cell culture supernatants, with significant inter-individual variability in induced TNF release (geometric mean 664pg/ml, 95% confidence intervals 593-743pg/ml, range 118-3511pg/ml) (Fig. 1). Cells were subsequently washed extensively and re-stimulated with a 10-fold higher LPS concentration (20ng/ml) for a further 6 h. In tandem with this, naïve cells that had been treated identically (washed in same manner), but had been exposed to media alone, were stimulated with LPS at the higher concentration. Cells pre-exposed to LPS showed a pronounced reduction in secondary release of TNF upon re-stimulation and also showed significant variability across the cohort (geometric mean 83pg/ml, 95% CI 69-100pg/ml, range 11-550pg/ml) (Fig. 1).
Figure 1. LPS induced TNF release among 96 healthy volunteers.
A, TNF levels in culture supernatant for untreated and treated PBMCs (2ng/ml LPS for 6 h) or B, comparing response to 20ng/ml LPS for 6 h between naïve cells and cells pretreated with LPS 2ng/ml for 6 h. C, Scatter plot for all volunteers showing TNF secretion and D, IL-6 secretion. E, Comparison of TNF and IL6 naïve vs tolerant response. F, Frequency distribution of tolerance across the cohort.
While most individuals showed a high level of silencing on secondary stimulation (mean 15.8%, SD 10.8, range 1.4-59), several individuals showed reduced induction of tolerance with secondary responses 30-50% that of their primary response (Fig. 1). Interestingly we did not note an association between the magnitude of the primary and secondary TNF response (data not shown). When we analysed the effect of secondary stimulation on IL-6 production at 6 h we did not see tolerance, with most individuals releasing a similar amount or more IL-6 (Fig. 1), confirming continued cell viability and reflecting mechanistically different silencing mechanisms between TNF and other acute phase cytokines at this relatively early time point in endotoxin tolerance.
Common genetic markers associated with reduced endotoxin tolerance
In order to determine genetic modulators of endotoxin tolerance we proceeded to map tolerance as a quantitative trait among the cohort of 96 healthy volunteers. Individuals were genotyped at 45,237 SNPs using the Illumina HumanCVDv1 beadchip which has high density coverage of ~ 2,000 genes implicated in immune and inflammatory responses of specific relevance to vascular pathology, metabolic disorders and inflammatory disease states (21).
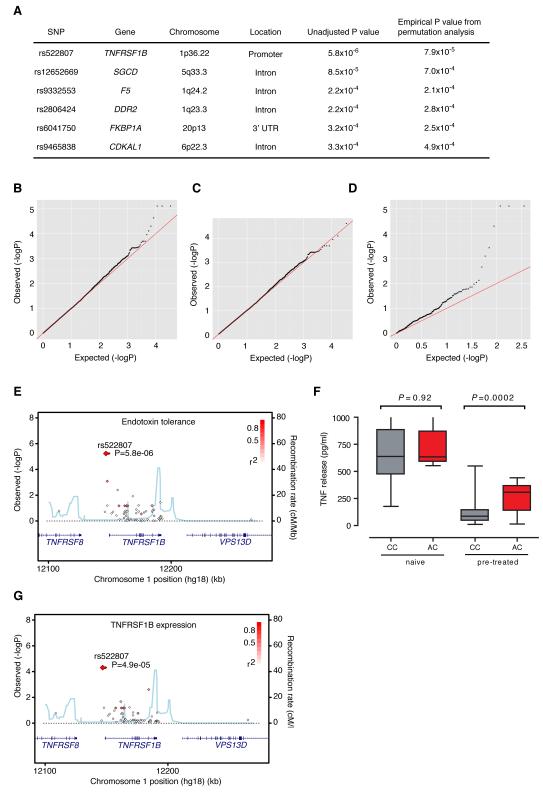
We found the most significant associations were with a cluster of single nucleotide polymorphisms (SNPs) at or near to TNFR2 at chromosome 1p36.22 (Fig. 2). Moreover, analysis of quantile-quantile plots highlighted that SNPs in TNF receptor related genes or TNF Receptor Associated Factor family (TRAF) genes accounted for the majority of the over-dispersion of P values over the expected in terms of statistical association (Fig. 2). Three SNPs in complete linkage disequilibrium (LD) upstream of the TNFR2 gene (rs522807, rs5745938 and rs625847) showed the strongest association with endotoxin tolerance, the mean tolerance was 31.3% (95% CI 17.2-45.5) among individuals possessing a copy of the haplotype defined by rs522807 compared to 14.4% (95% CI 12.5-16.3) among those without (Supplementary Fig. 1). By contrast, we found no association with initial TNF release (Fig. 2). The reduced tolerance associated SNP rs522807 shows marked differences in allele frequency between different populations world wide. The tolerance associated A allele (denoted as the ancestral allele as this is the allele observed in chimp, orangutan and macaque genomes) was present at a frequency of 8% in northern European derived populations, absent in Asian populations but present at a frequency of near 50% in equatorial Africa (Supplementary Fig. 2).
Figure 2. Expression quantitative trait mapping of endotoxin tolerance.
A, Top SNP associations with endotoxin tolerance. Quantile-quantile plots considering all genotyped SNPs (B), SNPs excluding those involving all TNF/TRAF genes (C), or including only those SNPs relating to TNF/TRAF genes (D). E, Regional association plot for endotoxin tolerance. F, Allelic association for rs522807 with TNF release following LPS 20 ng/ml for 6h, with and without LPS 2 ng/ml pretreatment for 6h. G, Regional association plot for TNFR2 expression.
rs522807 is associated with increased expression of TNFR2 in the basal state
Given the association of SNPs spanning TNFR2 with endotoxin tolerance, we sought to define if the same SNPs were modulating expression of this gene locally in a cis-acting manner. Using data from real time quantitative RT-PCR we proceeded to carry out an expression quantitative trait analysis for TNFR2. Interestingly, we found that rs522807 marked an eQTL for TNFR2 (P = 4.9 ×10−5). Individuals possessing a copy of the endotoxin associated SNP rs522807 had significantly increased basal expression of TNFR2 (Fig. 2, Supplementary Fig. 1). This indicated that the promoter polymorphisms associated with decreased tolerance enhanced basal expression of the receptor, suggesting the hypothesis that irrespective of genotype, basal expression of TNFR2 positively influences secondary responses by regulating tolerance.
TNFR2 expression correlates with secondary, but not primary, TNF release
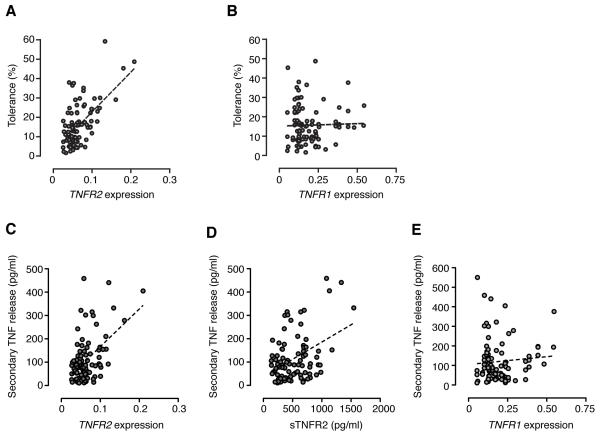
We proceeded to test this hypothesis by investigating whether there was any correlation between the expression of TNFR2 and LPS induced TNF response. We postulated that feedback of LPS induced TNF through basally expressed TNFR2 may influence the secondary response, although we were uncertain whether this would contribute to desensitization and tolerance or have the opposite effect. Using real time quantitative RT-PCR we found there was no association between basal expression of either TNFR1 or TNFR2 and primary release of TNF (data not shown). However, we found a highly significant correlation between the basal expression of TNFR2 mRNA and secondary TNF protein release (P <0.0001) (Fig. 3). LPS stimulation resulted in a robust induction of TNFR2 expression (4.8-fold induction, 95% CI 4.2-5.4), but whilst an association between induced TNFR2 and tolerance remained, this was far more modest (r2 0.09, P 0.002). This implied that the level of basal expression is of greater biological relevance in the determination of later responses. Notably, the association with tolerance was not seen for TNFR1 (Fig 3), suggesting this is a TNFR2 specific phenomenon. Analysis of expression levels of TNF mRNA rather than protein gave similar results (Supplementary Figure 3).
Figure 3. Baseline expression of TNFR2 predicts tolerance to LPS irrespective of genotype.
Scatter plots of tolerance assayed at the level of TNF protein secretion showing correlation with TNFR2 mRNA expression (r2 0.36, P <0.0001) (A) ,and with TNFR1 mRNA expression (r2 <0.01, P ns) (B). Correlation of secondary TNF protein with TNFR2 mRNA expression (r2 0.88, P <0.0001) (C), sTNFR2 protein (r2 0.18, P <0.0001) (D), and TNFR1 mRNA expression (r2 0.02, P ns) (E). All transcript abundance expressed relative to ACTB.
Fine mapping association at TNFRSF1B and analysis of transcription factor binding
In order to fine map the causative functional genetic variant we re-sequenced a 3.5kb region of the TNFR2 promoter that included rs522807 (chr1:12,146,659-12,149,575). In addition to the three single nucleotide substitutions identified from the array (rs522807, rs5745938, rs625847) a further single nucleotide substitution (rs520916) and insertion (rs35752907) were found to be in complete LD with the haplotype defined by rs522807. Additionally we noted a previously described Variable Number Tandem Repeat (VNTR) comprising a 15bp insertion within the promoter that exists in two allelic forms (either one or two repeats) (22) that has been associated with both infective and autoimmune disease processes (23, 24), although as yet it is unclear whether it influences expression of TNFR2. To answer this question we genotyped this VNTR and found a weak association between possession of a single copy of the VNTR and TNFR2 expression, but this effect was lost when the analysis was conditioned on rs522807.
In order to determine if any of the SNPs within the endotoxin tolerance associated haplotype modulated allele-specific protein-DNA interactions, electrophoretic mobility shift assays (EMSAs) were performed. Radiolabelled oligonucleotides corresponding to each SNP allele were incubated with nuclear extracts from the human leukemic monocyte lymphoma cell line U937. We observed that rs522807 showed markedly different binding profile between alleles using nuclear extracts from U937 cells that was specific on competition EMSA (Fig. 4). In silico analysis shows that the associated A allele of rs522807 introduces a cyclic AMP response element (CRE) binding site and competition experiments demonstrated that the upper allele-specific complex I is specifically competed by an oligonucleotide containing a CRE binding site, but the identity of the allele-specific CRE binding protein recruited at rs522807 remains to be defined.
Figure 4. Allele-specific protein-DNA interactions.
A, EMSA using radiolabelled probes corresponding to the two alleles of rs522807 show allele-specific differences in binding when incubated with nuclear extracts for U937 cells, either unstimulated (○) or after induction with LPS for 6 h (●). B, The specificity of these interactions was confirmed on competition with 10- or 100-fold molar excess of unlabelled probe corresponding to each of the two alleles, a CRE binding site or an unrelated sequence (corresponding to a YY1 binding site).
As this study neared completion, data from the 1000 Genomes Project (www.1000genomes.org) became available. This shows that the haplotype tagged by rs522807 extends for 15.9kb upstream of TNFR2 with 17 SNPs in complete LD (Supplementary Table 2) which will require further characterization to define the causative regulatory variant(s).
Allosteric modulation demonstrates a role for TNFR2 in augmenting LPS responsiveness
Our data suggests that the expression of TNFR2 plays a direct role in modulating tolerance to LPS induced TNF release. However, the overriding effect of increased basal expression of TNFR2 was unclear. TNFR2 is cleaved at the cell surface by ADAM 17 (ADAM metallopeptidase domain 17), a disintegrin and metalloproteinase also referred to as TACE (TNF-alpha converting enzyme) to form soluble TNFR2 that can antagonize the activity of TNF at its receptors with important functional consequences (25, 26). It was therefore possible that the predominant effect of over-expression of TNFR2 would be to antagonize the action of TNF. To dissect out the potential effect of TNFR2 activity in the development of endotoxin tolerance we took advantage of an allosteric modulating monoclonal antibody raised against this receptor (anti-human TNF-R II Clone 80M2). Whilst 80M2 has no independent agonist activity, it markedly increases the affinity for TNF to TNFR2 and thus confers enhanced TNFR2 signaling in the presence of TNF only (27).
Prior to LPS application cells were treated with 80M2 or isotype control IgG1 for one h before extensive washing to remove any unbound antibody. Cells were then treated with LPS for up to 10 h, with supernatants and RNA harvested in the absence of LPS and at 2, 6 and 10 h. Pretreatment with antibodies did not elicit TNF release in the absence of LPS. When LPS was applied, 80M2 had no effect on early induced TNF release at 2 h, but at later time points cells pretreated with 80M2 showed significantly enhanced LPS induced TNF release (Fig. 5). Furthermore, expression of TNF, which was maximal at 2 h, was not significantly different from IgG1. However at 6 h there was delayed normalization in TNF expression (Fig. 5).
Figure 5. Allosteric modulator of TNFR2 enhances late LPS induced TNF release.
PBMCs were pretreated for 1 h with allosteric modulating antibody or isotype control (2mg/ml) prior to extensive washing and LPS treatment (2ng/ml). A, TNF protein (n=7) B, TNF transcript (relative to baseline with IgG control) (n=5). CD14+ monocytes were pretreated for 1 h with allosteric modulating antibody or isotype control (2mg/ml) prior to extensive washing and LPS treatment (2ng/ml). TNF protein released was assayed. Following stimulation with LPS for 6 h (2ng/ml) (C), or following pretreatment with LPS (2ng/ml) for 6 h (D), then restimulation with LPS (20ng/ml) for 3 h. *** P <0.01, ** P = 0.01, * P < 0.05.
In addition to modulating the primary response to LPS, we were keen to investigate whether 80M2 could modify secondary responses to LPS, and hence tolerance. Use of 80M2 in crude PBMC populations had variable effects, with cells from some individuals showing pronounced alteration in tolerance, whilst others were not affected (data not shown). We reasoned that this may be secondary to varying cellular composition of PBMC fractions. 80M2 has been shown to have activity at specific T-cell populations (28) and it was reasoned that activity of the antibody at non-monocyte populations may confound effects on tolerance. To avoid this possibility we purified CD14+ monocytes from the PBMC fraction of unrelated healthy volunteers, thus focusing on the cells mainly responsible for LPS induced cytokine release. Similar to the PBMC fraction, we found pretreatment with 80M2 had no independent agonist activity (data not shown), but greatly enhanced LPS induced TNF release (2363 ng vs. 1010 ng, n=4, p<0.05) (Fig. 5). After 6 h of 2ng/ml LPS, monocytes were washed and restimulated for a further 3 h with 20ng/ml LPS before measuring secondary responses. We found that this pretreatment significantly enhanced secondary TNF release in response to LPS, demonstrating activity through TNFR2 can modulate tolerance (Fig. 5). These results supported the hypothesis generated from the genetic screen whereby LPS induced TNF acts to further amplify late release of TNF in a TNFR2 specific manner. This data shows that manipulation of basally expressed TNFR2 can dictate the both the magnitude and durability of LPS induced TNF release.
DISCUSSION
Whilst the phenomenon of endotoxin tolerance has been described for over a century, the biological mechanisms underlying this critical immunological process are only now beginning to be elucidated. These include the concomitant upregulation of negative regulators of TLR signaling, induction of micro-RNAs which specifically target products of the NF-κB pathway and the selective silencing of multiple pro-inflammatory genes. Most data emanates from mouse models and there is a degree of discrepancy between genes implicated in the induction of endotoxin tolerance in animal studies and in man such that only IRAK-M has consistently been demonstrated to play a role in both mice and men (8, 20). The significance of a more comprehensive understanding of the basis of endotoxin tolerance are underlined by the clear role for this phenomenon in the pathogenesis of diseases such as sepsis. The tolerant state is associated with increased susceptibility to subsequent infections and indeed may reflect an underlying pro-inflammatory but paradoxically immunosupressed state (20). Nonetheless, the highly conserved nature of tolerance and the multiplicity of mechanisms whereby it can be achieved indicate it plays a key role in the regulation of the innate immune response. Indeed it is postulated that endotoxin tolerance has evolved to act as a protective mechanism to prevent the ‘cytokine storm’ associated with onset of sepsis and strongly associated with shock (6).
In this study we were interested in addressing the degree to which individuals tolerised to LPS over a short period, thus reflecting silencing of TNF release. We specifically chose a 6 h pretreatment duration because in a clinical setting the onset of sepsis can often be abrupt, with patients deteriorating rapidly over such a time period. Whilst studies of tolerance over 24 h show universal silencing of most inflammatory cytokines (11), we find there is a large degree of inter-individual variability in the silencing of TNF over 6 h and therefore this timepoint is more applicable to an association study. It is of note that in our study we found little silencing of the IL-6 response, illustrating both the differential regulation of these genes and also the continued viability of cells.
Our interest in identifying genetic markers of endotoxin tolerance has been driven by the observations that responses to innate immune stimuli show a large degree of heritability (12, 29). We were curious to investigate whether there may be genetic variants associated with differential silencing of TNF release. Our initial finding that the most significantly associated SNP from a panel of over 48000 SNPs was located in the promoter region for TNFR2, a receptor for the measured parameter (TNF release), was of high biological plausibility. We proceeded to demonstrate that this SNP, rs522807, marks a haplotype associated with increased basal expression of TNFR2 in our dataset and directly modulated allele-specific recruitment of a CRE binding protein. The observation that the basal expression of TNFR2 relates to secondary TNF release was replicated across the cohort, independently of genotype, with a highly significant correlation between expression and secondary response. By contrast there was no association with the expression of TNFR1, suggesting a specific role for TNFR2 in the regulation of LPS induced TNF release. This may reflect the differential signaling pathways of TNFR1 versus TNFR2, with TNFR2 specifically activated by membrane bound TNF and showing relative insensitivity to free TNF (27). It is plausible that the early expression of membrane bound TNF in response to LPS and subsequent signaling via TNFR2 has later effects on tolerance. This would also explain why the basal expression levels on TNFR2 seem to be of greatest importance in influencing subsequent later responses, with individuals expressing higher levels having consequentially reduced tolerance. We substantiated this finding by using monoclonal antibodies specific for TNFR2 that enhance the affinity of the receptor for TNF (27). Pretreatment of cells with antibody has no stimulatory effect in its own right, demonstrating that TNFR2 activity only acts to amplify other responses in this experimental model. Moreover, at 2 h cells pretreated with antibody show similar LPS induced TNF release to those pretreated with isotype control. However by 6 h, a time when endotoxin tolerance has developed (20), there is a highly significant difference between isotype control treated cells and those treated with TNFR2 specific antibodies, indicating positive feedback of LPS induced TNF release, acting through the receptor to enhance further TNF production. Using a purified monocyte subset we were able to separately demonstrate that activity involving TNFR2 could modulate tolerance, with monocytes pretreated showing a significant increase in secondary TNF release compared to control cells.
Our data has highlighted the extent of individual variation in the endotoxin tolerance phenotype among healthy individuals and has implications for the tailored use of therapeutic interventions to modulate the inflammatory response. We have shown how expression quantitative trait mapping in primary human tissue can be used to identify functionally relevant polymorphisms for complex immunological responses. This non-biased approach allows the elucidation of pathway genes whose role may have been previously unsuspected, in this case the TNFR2 gene in endotoxin tolerance. This result has significant physiological implications for our understanding of the biological mechanisms regulating endotoxin tolerance and highlights the value of large scale genotyping in the identification of functional variation in innate immune responses in man.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank all the volunteers who participated in this study and members of the Knight lab for helpful discussion and suggestions.
This work was supported by the Wellcome Trust (Grants 074318 [to J.C.K.], 088891 [to B.P.F.] and 075491/Z/04 [to core facilities Wellcome Trust Centre for Human Genetics].
Abbreviations used in this paper
- EMSA
electrophoretic mobility shift assay
- GWA
genome-wide association
- SNPs
single nucleotide polymorphisms
- TNFR2
TNF Receptor 2
- TRAF
TNF Receptor Associated Factor
- VNTR
Variable Number Tandem Repeat
Footnotes
The online version of this paper contains supplemental material.
REFERENCES
- 1.Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 3.Beutler B, Grau GE. Tumor necrosis factor in the pathogenesis of infectious diseases. Crit Care Med. 1993;21:S423–435. [PubMed] [Google Scholar]
- 4.Van Epps HL. Ignoring endotoxin. J Exp Med. 2006;203:1137. doi: 10.1084/jem.2035fta. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beeson PB. Tolerance to Bacterial Pyrogens : I. Factors Influencing Its Development. J Exp Med. 1947;86:29–38. doi: 10.1084/jem.86.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biswas SK, Lopez-Collazo E. Endotoxin tolerance: new mechanisms, molecules and clinical significance. Trends Immunol. 2009;30:475–487. doi: 10.1016/j.it.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 7.Sly LM, Rauh MJ, Kalesnikoff J, Song CH, Krystal G. LPS-induced upregulation of SHIP is essential for endotoxin tolerance. Immunity. 2004;21:227–239. doi: 10.1016/j.immuni.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 8.van ’t Veer C, van den Pangaart PS, van Zoelen MA, de Kruif M, Birjmohun RS, Stroes ES, de Vos AF, van der Poll T. Induction of IRAK-M is associated with lipopolysaccharide tolerance in a human endotoxemia model. J Immunol. 2007;179:7110–7120. doi: 10.4049/jimmunol.179.10.7110. [DOI] [PubMed] [Google Scholar]
- 9.Baltimore D, Boldin MP, O’Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nat Immunol. 2008;9:839–845. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- 10.Bazzoni F, Rossato M, Fabbri M, Gaudiosi D, Mirolo M, Mori L, Tamassia N, Mantovani A, Cassatella MA, Locati M. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc Natl Acad Sci U S A. 2009;106:5282–5287. doi: 10.1073/pnas.0810909106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foster SL, Hargreaves DC, Medzhitov R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 2007;447:972–978. doi: 10.1038/nature05836. [DOI] [PubMed] [Google Scholar]
- 12.de Craen AJ, Posthuma D, Remarque EJ, van den Biggelaar AH, Westendorp RG, Boomsma DI. Heritability estimates of innate immunity: an extended twin study. Genes Immun. 2005;6:167–170. doi: 10.1038/sj.gene.6364162. [DOI] [PubMed] [Google Scholar]
- 13.Fairfax BP, Vannberg FO, Radhakrishnan J, Hakonarson H, Keating BJ, Hill AV, Knight JC. An integrated expression phenotype mapping approach defines common variants in LEP, ALOX15 and CAPNS1 associated with induction of IL-6. Hum Mol Genet. 2010;19:720–730. doi: 10.1093/hmg/ddp530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cookson W, Liang L, Abecasis G, Moffatt M, Lathrop M. Mapping complex disease traits with global gene expression. Nat Rev Genet. 2009;10:184–194. doi: 10.1038/nrg2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lettre G, Rioux JD. Autoimmune diseases: insights from genome-wide association studies. Hum Mol Genet. 2008;17:R116–121. doi: 10.1093/hmg/ddn246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 17.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schreiber E, Matthias P, Muller MM, Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Udalova IA, Knight JC, Vidal V, Nedospasov SA, Kwiatkowski D. Complex NF-kappaB interactions at the distal tumor necrosis factor promoter region in human monocytes. J Biol Chem. 1998;273:21178–21186. doi: 10.1074/jbc.273.33.21178. [DOI] [PubMed] [Google Scholar]
- 20.del Fresno C, Garcia-Rio F, Gomez-Pina V, Soares-Schanoski A, Fernandez-Ruiz I, Jurado T, Kajiji T, Shu C, Marin E, Gutierrez del Arroyo A, Prados C, Arnalich F, Fuentes-Prior P, Biswas SK, Lopez-Collazo E. Potent phagocytic activity with impaired antigen presentation identifying lipopolysaccharidetolerant human monocytes: demonstration in isolated monocytes from cystic fibrosis patients. J Immunol. 2009;182:6494–6507. doi: 10.4049/jimmunol.0803350. [DOI] [PubMed] [Google Scholar]
- 21.Keating BJ, Tischfield S, Murray SS, Bhangale T, Price TS, Glessner JT, Galver L, Barrett JC, Grant SF, Farlow DN, Chandrupatla HR, Hansen M, Ajmal S, Papanicolaou GJ, Guo Y, Li M, Derohannessian S, de Bakker PI, Bailey SD, Montpetit A, Edmondson AC, Taylor K, Gai X, Wang SS, Fornage M, Shaikh T, Groop L, Boehnke M, Hall AS, Hattersley AT, Frackelton E, Patterson N, Chiang CW, Kim CE, Fabsitz RR, Ouwehand W, Price AL, Munroe P, Caulfield M, Drake T, Boerwinkle E, Reich D, Whitehead AS, Cappola TP, Samani NJ, Lusis AJ, Schadt E, Wilson JG, Koenig W, McCarthy MI, Kathiresan S, Gabriel SB, Hakonarson H, Anand SS, Reilly M, Engert JC, Nickerson DA, Rader DJ, Hirschhorn JN, Fitzgerald GA. Concept, design and implementation of a cardiovascular gene-centric 50 k SNP array for large-scale genomic association studies. PLoS ONE. 2008;3:e3583. doi: 10.1371/journal.pone.0003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keen L, Wood N, Olomolaiye O, Bidwell J. A bi-allelic VNTR in the human TNFR2 (p75) gene promoter. Genes Immun. 1999;1:164–165. doi: 10.1038/sj.gene.6363644. [DOI] [PubMed] [Google Scholar]
- 23.Lee YH, Rho YH, Choi SJ, Ji JD, Song GG. The biallelic variable number of tandem repeats of the tumor necrosis factor receptor 2 promoter in systemic lupus erythematosus. Rheumatol Int. 2003;23:108–111. doi: 10.1007/s00296-002-0248-1. [DOI] [PubMed] [Google Scholar]
- 24.Sainz J, Perez E, Hassan L, Moratalla A, Romero A, Collado MD, Jurado M. Variable number of tandem repeats of TNF receptor type 2 promoter as genetic biomarker of susceptibility to develop invasive pulmonary aspergillosis. Hum Immunol. 2007;68:41–50. doi: 10.1016/j.humimm.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 25.Bell JH, Herrera AH, Li Y, Walcheck B. Role of ADAM17 in the ectodomain shedding of TNF-alpha and its receptors by neutrophils and macrophages. J Leukoc Biol. 2007;82:173–176. doi: 10.1189/jlb.0307193. [DOI] [PubMed] [Google Scholar]
- 26.Horiuchi K, Kimura T, Miyamoto T, Takaishi H, Okada Y, Toyama Y, Blobel CP. Cutting edge: TNF-alpha-converting enzyme (TACE/ADAM17) inactivation in mouse myeloid cells prevents lethality from endotoxin shock. J Immunol. 2007;179:2686–2689. doi: 10.4049/jimmunol.179.5.2686. [DOI] [PubMed] [Google Scholar]
- 27.Grell M, Douni E, Wajant H, Lohden M, Clauss M, Maxeiner B, Georgopoulos S, Lesslauer W, Kollias G, Pfizenmaier K, Scheurich P. The transmembrane form of tumor necrosis factor is the prime activating ligand of the 80 kDa tumor necrosis factor receptor. Cell. 1995;83:793–802. doi: 10.1016/0092-8674(95)90192-2. [DOI] [PubMed] [Google Scholar]
- 28.Ban L, Zhang J, Wang L, Kuhtreiber W, Burger D, Faustman DL. Selective death of autoreactive T cells in human diabetes by TNF or TNF receptor 2 agonism. Proc Natl Acad Sci U S A. 2008;105:13644–13649. doi: 10.1073/pnas.0803429105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Westendorp RG, Langermans JA, Huizinga TW, Elouali AH, Verweij CL, Boomsma DI, Vandenbroucke JP. Genetic influence on cytokine production and fatal meningococcal disease. Lancet. 1997;349:170–173. doi: 10.1016/s0140-6736(96)06413-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.