Abstract
Deciphering the biological networks underlying complex phenotypic traits, e.g., human disease is undoubtedly crucial to understand the underlying molecular mechanisms and to develop effective therapeutics. Due to the network complexity and the relatively small number of available experiments, data-driven modeling is a great challenge for deducing the functions of genes/ proteins in the network and in phenotype formation. We propose a novel knowledge-driven systems biology method that utilizes qualitative knowledge to construct a Dynamic Bayesian network (DBN) to represent the biological network underlying a specific phenotype. Edges in this network depict physical interactions between genes and/or proteins. A qualitative knowledge model first translates typical molecular interactions into constraints when resolving the DBN structure and parameters. Therefore, the uncertainty of the network is restricted to a subset of models which are consistent with the qualitative knowledge. All models satisfying the constraints are considered as candidates for the underlying network. These consistent models are used to perform quantitative inference. By in silico inference, we can predict phenotypic traits upon genetic interventions and perturbing in the network. We applied our method to analyze the puzzling mechanism of breast cancer cell proliferation network and we accurately predicted cancer cell growth rate upon manipulating (anti)cancerous marker genes/proteins.
Index Terms: Dynamic Bayesian network, genetic network, phenotype prediction, genetic intervention, systems biology, breast cancer, cell proliferation
1 Introduction
The topology and the dynamic realization of genetic networks often play a dominant role in phenotype formation. In order to understand the cause of a disease and/or develop effective therapeutics, it is important to understand the function and regulation of the underlying biological network. In recent years, studies on this problem have been focused on the data-based (reverse engineering) approaches, i.e., modeling a biological network from the experimental data and prior knowledge by machine learning algorithms, such as learning a genetic regulatory network (GRN) from microarray data using a Bayesian network.
Friedman et al. were the first to use Bayesian networks in identifying a regulatory network structure [17]. In this work, a best Bayesian model structure is learned from gene expression data by maximizing its posterior probability based on the data. The ability of the model to reproduce certain known regulatory interactions is validated against real experiments and the BN model can also predict new regulatory relationships. In general, Bayesian network inference uses two kinds of cost functions, i.e., BIC and BDe score, to learn the BN structure. Following this line, plenty of works have been proposed to learn the genetic regulatory networks and protein-protein interaction networks by analyzing various data resources, such as gene expression data, ChIP-chip data, and protein expression data, etc. [15], [42], [44]. In addition, time-dependent gene activities and their relationships can be inferred from microarray time series data using dynamic Bayesian networks [29], [18]. Other works have recruited data integration schemes to combine different kinds of data together with the prior knowledge into the learning task [22], [36], [50], [51]. Moreover, some works have been proposed to deal with reconstructing the genetic regulatory network with hidden factors and missing observations [2], [14]. All of the above methods and applications inevitably encounter a similar problem, i.e., there are not enough data samples for learning the network structure with given dimensionalities.
In reality, due to the relatively small amount of experimental data available compared to the size of the genetic regulatory network, the learned network often contains a small number of reliable (confident) edges. In addition, the conventional Bayesian network cannot capture cyclic structures in real biological systems, which often results in inaccuracy and/or error. Algorithms of learning cyclic structures from microarray data with dynamic Bayesian network have been proposed [35]. However, these algorithms often need a large amount of data in time series, which is not necessarily available. Moreover, biological networks consist of various interactions, such as protein-protein and protein-DNA interactions. Due to the variation of the techniques used to generate these data, discrepancies between experiments and various types of data often make the data-driven approach difficult.
However, there are plenty of qualitative statements in the literature. For example, TGFβ stimulates tumor invasion and metastasis. This statement indicates a direct functional relationship, stimulation, between a cytokine, TGFβ and phenotypes, tumor invasion and metastasis. Such a qualitative statement lacks quantitative information, e.g., how strongly does TGFβ regulate those phenotypes and its reliability is dependent on the biological experiments that supported it. Nevertheless, the statement is a concrete conclusion supported by various evidences obtained from different experimental measurements including microarray, ELISA, and northern blot experiments. A qualitative statement is often a summary of the most prominent and consistent observations across multiple studies and it should be thusly treated as the most confident information in modeling the underlying biological network. Other links which are less reliable than qualitative statements emphasized in the literatures may be erroneously captured (false positives) in the learned Bayesian model using the data-based reverse engineering approach.
Consequently, it is very important in systems biology to develop methods based on highly confident qualitative statements in the literature (no quantitative experimental data are involved) to establish a genetic network for a specific phenotype (e.g., cancer). In such networks, vertices indicate cellular molecules at multiple levels, such as proteins and RNA molecules. Direct edges from any node(s) to other node(s) in the network represent direct functional regulations from the parental node(s) to the child node(s). Given this genetic network, it is imperative to parameterize its structure. We can thusly use it to interrogate new genetic programs and discover new knowledge about this network and its associated biological phenotypes.
Unfortunately, a major hurdle in developing this knowledge-based approach is the lack of quantitative parameterization information (in qualitative statements) that is crucial for performing quantitative inference. Thus, the problem boils down to constructing parameters from the qualitative statements and encoding this parameter and structure information into a mathematical model for quantitative manipulations. We proposed in this paper, a knowledge-based predictive framework for modeling the recurrent genetic networks based on dynamic Bayesian networks given qualitative knowledge and our model can then perform quantitative inference.
(Dynamic) Bayesian networks (DBNs) are a popular class of graphical probabilistic models which are motivated by Bayes’ theorem [1]. A DBN represents a joint probability distribution over a set of variables. Once known, this joint distribution can be used to calculate the probabilities of any configuration of the variables. In Bayesian probabilistic inference, the conditional probabilities for the values of a set of unconstrained variables are calculated given fixed values of another set of variables, called observations or evidence. Bayesian models have been widely used for efficient probabilistic inference and reasoning [32], [37]. Numerous algorithms for learning the Bayesian network structure and parameters from data have been proposed [23], [24], [16]. However, as we have discussed above, although the maximum a posteriori approximation, i.e., the selection of a single Bayesian network model from the data by learning, is useful for the case of large data sets, independence assumptions among the network variables often make this single model vulnerable to overfitting. In realistic problems, the data basis is often very sparse and hardly sufficient to select one adequate model, i.e., there is considerable model uncertainty. Selecting a single Bayesian model can then lead to strongly biased inference results.
Besides Bayesian networks, other state-of-the-art statistical and deterministic methods have been proposed to infer the genetic regulatory network from the data. These methods can analyze the full range of the behaviors and dynamics of a system under different conditions. (Probabilistic) Boolean networks were initially used to analyze the network stability in the yeast transcriptional regulatory network [27] and to study the dynamics of cell cycle regulation in yeast [33]. Boolean networks can provide important insights in terms of the existence and nature of network steady states and robustness. However, a Boolean network is largely limited by its level of modeling details and computational expense to analyze the dynamics of large networks, as the number of global states is exponential in the number of entities [26]. Petri net is used to analyze the transition sequence of a network from a global state to another. Moreover, Petri net is used to analyze the dynamics of a regulatory network and large-scale metabolic networks [7], [31], [39], [30]. Modulo network module is introduced to infer the regulation logic of gene modules given gene expression data. A regulation logic is represented by a decision tree, in which a path from the root to a leaf is determined by the up or downregulation of regulatory modules, and a leaf determines the expression level of the corresponding genes. Module networks were tested with experimental data and correctly predicted some regulatory modules [43]. Other successful model can predict the genetic regulatory network based on mutual information [34].
As discussed above, a quantitative data set is a sole resource for all these conventional methods. Therefore, these methods’ performance are inevitably limited to the availability and quality of the data. In particular, the performance of these methods will be severely undermined in any of the following cases: 1) the data contain few samples (comparing to number of predictors/features/random variables of the system); 2) the data are contaminated by relatively high-level noise; 3) the data contain no functional measurements. In our method, we try to model the genetic regulatory network structure and parameters and to predict the system behavior based on solely priori qualitative statements. On the contrary, a qualitative knowledge about a physical interaction is usually evaluated by a combination of direct binding and functional regulation experiments. The qualitative knowledge thusly provide a high-confident landscape of the network structure. The major advantage of our proposed method is that we avoid the usage of noisy data yet to construct a confident network structure.
In this paper, we recruit a qualitative knowledge model [5] to map major types of genetic interactions, i.e., 1) transcription factor-DNA regulations and 2) protein-protein interactions, to set a group of constraints over the structure and parameter space of the dynamic Bayesian network. In particular, the qualitative properties of the statements are dealt with by transforming the fuzziness of these statements into a set of prior joint probability distributions over the nodes in the dynamic Bayesian network. The genetic networks are restricted to a subset of models that are consistent with a body of qualitative knowledge. All dynamic Bayesian models satisfying the constraints over the joint probability space are considered as a candidate for the underlying biological network. In this way, we take model uncertainty into account instead of basing our prediction on a single “best” model. With full Bayesian approach, i.e., model averaging, this class of consistent models is used to perform quantitative inference which can be approximated by Monte Carlo methods. This knowledge-based quantitative Bayesian network modeling algorithm preserves the actual network topology derived from knowledge and is able to capture both “correlation” (joint probability) and “causal/influence” (conditional probability) relations in the Bayesian network. When we combine qualitative statements from various studies, statements targeting the same genetic interaction may be inconsistent. In this case, they can be integrated into a unified representation by calculating a priori distribution over the statements [5].
In summary, our method demonstrates that we can achieve good predictions on the biological network behaviors given qualitative statements without any quantitative data. In Section 2, we present the quantitative inference methods with a dynamic Bayesian model based on a set of qualitative statements. In Section 3, we apply our framework to model the cell proliferation network in normal and cancerous breast cells and also predict cell growth given regulatory interventions to the network. Conclusions are made in Section 4.
2 Method
In this section, we suggest a way to use qualitative relational statements for inference in the Bayesian framework. We proceed from the general equation for Bayesian inference based on data and knowledge, followed by a detailed recipe to transform knowledge, represented by a set of qualitative statements, into an a priori distribution for models.
2.1 Bayesian Modeling and Inference
A Bayesian model m represents the joint probability distribution of a set of variables X = X1; X2; … ; XD [24]. The model is defined by a graph structure s, which defines the structures of the conditional independence between variables, and a parameter vector θ, the components of which define the entries of the corresponding joint probability tables (CPTs). Hence, a Bayesian network can be written as m = {s, θ}. If we believe that one single model m reflects the true underlying distribution, we can perform inference based on this model. Given some observations or “evidence” E, reflected by fixed measured values of a subset of variables, Xe = E, we wish to query on the distribution of the subset of remaining variables X = Xq. It is provided by their conditional probability given the evidence in light of the model,
| (1) |
which can be efficiently evaluated with known methods [35].
The full Bayesian framework does not attempt to approximate one true underlying distribution. Instead, all available information is used in an optimal way to perform inference, without taking one single model for granted. To formalize this statement for our purposes, let us classify the set of available information into an available set of data, D, and a body of nonnumeric knowledge, Ω. The a posteriori distribution of models m is then given by
| (2) |
The first term in the numerator of (2) is the likelihood of the data given the model, which is not directly affected by nonnumeric knowledge Ω, the second term denotes the model prior, whose task is to reflect the background knowledge. We obtain
| (3) |
where Z is a normalization factor which will be omitted from the equations for simplicity. The first term contains the constraints of the model space by the data, and the second term the constraints imposed by the background knowledge. In the full Bayesian approach, we can perform inference by model averaging. Now, given some observation or evidence E, the (averaged) conditional distribution of the remaining variable X is performed by integrating over the models:
| (4) |
In this paper, we consider the extreme case of no available quantitative data, D = ∅ Even in this case, it is still possible to perform proper Bayesian inference,
| (5) |
Now the inference is based on the general background information contained in Ω alone, and the specific information provided by the measurements E. This is reflected by the fact that inference results are conditioned on both quantities in (5).
In order to determine Pr(m|Ω), we need a formalism to translate a body of qualitative knowledge into an a priori distribution over Bayesian models. For this, we adopt the following notation for a Bayesian model class. A Bayesian model is determined by a graph structure s and by the parameter vector θ needed to specify the conditional probability distributions given that structure. We refer to θ as one specific joint configuration. A Bayesian model class M̃ is then given by 1) a discrete set of model structures S̃ = {s1,s2, …, sK}, and 2) for each structure sk a (eventually continuous) set of CPT configurations Θk. The set of member Bayesian models m ∈ M̃ of that class is then given by m = {(sk, θ)|k ∈ {1,…, K}, θ ∈ Θk}. The model distribution now reads
| (6) |
According to (6), in order to construct a prior distribution of the models, we use each statement to constrain the model space to the subspace which is consistent with that statement. In other words, if a statement describes a relationship between two variables, only structures sk which contain the corresponding edge are assigned a nonzero probability Pr(sk|Ω). Likewise, only the parameter values (joint probability) on that structure, which are consistent with the contents of that statement, are assigned a nonzero probability Pr(θ|sk, Ω). If no further information is available, the distribution is constant in the space of consistent models. Inference is carried out by integrating over the structure space and the structure-dependent parameter space:
| (7) |
As we utilize the nonnumeric knowledge in terms of qualitative statements about a relationship between biological molecules in a cell, we hypothesize that Ω contains a list of qualitative statements on cellular molecular interactions. In this form, the information can be used in a convenient way to determine the model prior, (6): 1) Each entity which is referenced in at least one statement throughout the list is assigned to one variable Xi. 2) Each relationship between a pair of variables constrains the likelihood of an edge between these variables being present. 3) The quality of that statement (e.g., “activates” and “inactivates”) affects the distribution over the local conditional probability distributions (CPDs) entries given the local structures. The joint probability (θ in (6) and (7)) can be decomposed into a product of CPDs of the conditionally independent variables in the network. Thus, in the most general case, the statements can be used to (indirectly) shape the joint distribution over the class of all possible Bayesian models obtained from Ω. In other situation, such as chain graph, the statements can be used to (directly) shape the joint distribution over any subset of variables in the network connected by undirected edges. In addition, these statements can constrain the model parameters with either linear inequality or nonlinear inequalities. In this way, a broad diversity of cellular process can be captured by our approach.
2.2 Dynamic Bayesian Network Inference with Qualitative Knowledge
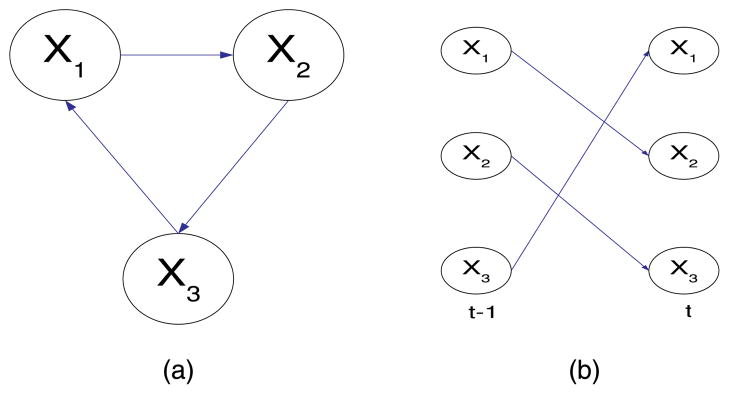
In this section, we give the detail receipt of probabilistic inference in DBN with only qualitative statements. Please note that, from now on, m stands for a Dynamic Bayesian network [35] in stead of a static Bayesian network (BN). A dynamic Bayesian network [11] is a way to extend Bayes nets to model probability distributions over semiinfinite collections of random variables. We only consider discrete-time processes, and we increase the index t by one every time. The variable probability is exclusively dependent on its last step parents state which makes a DBN Markovian process. The conventional Bayesian networks is acyclic graph. Thus, we use DBNs to model the recurrent molecule interactions in cells. An example of DBN is shown in Fig. 1a and it can be unrolled into a series of 2-Time-Slice Bayesian Networks (2TBNs) over time as shown in Fig. 1b. Each 2TBN is a conventional Bayesian model with time-invariant interslice structure s and joint probability distributions θ over the nodes X̄ at time t and (t – 1), i.e., P(X̄t, X̄t–1). If time starts from t = 0, the joint probability of a DBN over time T is
Fig. 1.
Dynamic Bayesian Example. (a) DBN example. (b) 2TBN.
| (8) |
For tth 2TBN, the joint probability of X̄ at time t, i.e., P(X̄t) can be written as
| (9) |
where Xn,t denotes the nth node at time t. The child nodes in the tth 2TBN, Xn,t, are independent given the parents, i.e., the nodes at time (t – 1). The posterior probability distribution of each nodes at time t, i.e., P(Xn,t) in (9) can be calculated by integrating over the parents as in the case of conventional Bayesian network,
| (10) |
where θj denotes the jth entry in the conditional probability table of node Xn given its parents. represent the joint probability of jth configuration of the parents states at time (t – 1). The posterior probability distribution of X̄t can be used as the priori probability for the next time step. Thus, the posterior probability P(Xn,t) can be calculated iteratively over time t = {0, …, T}. More efficient algorithms have also been developed to perform exact probabilistic inference in DBN, such as junction tree, frontier algorithm, and interface algorithm [35]. As demonstrated in the last section, if there is a set of qualitative knowledge retrieved from a publication which defines a class of models, with the structure and its associated parameter space, the inference with full Bayesian approach is calculated by integrating the inference in each model weighted by its posterior probability given the set of hypothesis as in (7). The inference can be written as
| (11) |
Only the structure which is consistent with the hypotheses is assigned with nonzero probability P(sk|Ω). Likewise, only parameter values on that structure, which are consistent with the contents of the hypotheses, are assigned a nonzero probability P(θ|sk, Ω). If no further information is available, the distribution is constant in the space of consistent models. Now, we can perform inference on the marginal probability of Xn at time t in each DBN model mk according to (10)
| (12) |
where θk;j represents the jth entry of the CPT in kth DBN model. E denotes the evidence of the observed nodes and denotes the joint probability distribution of the jth configuration of the parent nodes π(Xn) at time (t – 1) given the observation E. Therefore, the quantitative inference in (11) can be calculated by
| (13) |
The inference in (13) can be calculated for each model mk = (sk, θ) over time T and the predictions are averaged over all models in the model class M̃.
2.3 Qualitative Knowledge Model
2.3.1 Constraints over Joint and Conditional Probability Space
In this paper, we recruit the qualitative knowledge model in [6] to translate the “causal”(causal-like) qualitative statements into constraints over conditional probability distribution. In addition, we define here a novel form of inequality to translate the “correlate”(correlate-like) qualitative statements into constraints over joint probability distribution. The joint probability distribution of a DBN and BN (θ in (6) and (7)) can be decomposed into a product of local conditional probabilities of each child node given its parental nodes in the network, i.e., Pr(Xn, π(Xn)) = Pr(Xn|π(Xn))Pr(π(Xn)). The conditional probabilities θk;j in (12) and (13) denotes the jth CPT entry in kth dynamic Bayesian model. (Here, we assume the Dynamic Bayesian network parameters are time-invariant). In general case, it is equivalent to use either joint or local CPDs to parameterize a BN and DBN’s structure since these two distributions can be calculated from each other. However, it is still worthy to distinguish them in a modeling task depending on the specific information that qualitative statements provide. In case the qualitative statements explicitly specify a direct functional regulations between molecules, it is desired to translate these “causal” (or causal-like) relationships into constraints on the conditional probability distributions. In case the qualitative statements indicates two or more molecules’ level correlates to each other, it is more proper to transform this “correlate”(or correlate-like) relationship to constraints on the joint probability distributions instead of conditional probability distribution. In case of two or more molecules correlate to each other, the edges among these molecules are undirected resulting in a mixture of directed and undirected graph if “causal” relationships present among other variables in the network.
2.3.2 Correlate Influence
Definition 1
If a child node B has a neighbor node(s) A and these nodes impose isolated bidirectional influences from one to the other, then qualitative influence between this node and its neighbor node(s) is referred to as Correlate influence. Correlate influence can be further classified into positive correlation and negative correlation.
Definition 2
If presence/absence of one node A or B renders the presence/absence of neighbor node(s) B or A more likely than only one of them is present and the other is absent, then these nodes is said to have a positive correlate influence on each other. This can be represented by the inequality
| (14) |
Definition 3
If presence/absence of one node A or B renders the presence/absence of neighbor node(s) B or A less likely than only one of them is present and the other is absent, then these nodes is said to have a negative correlate influence on each other. This can be represented by the inequality
| (15) |
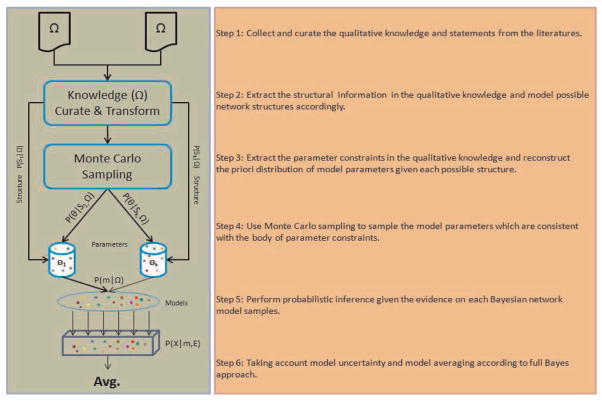
In summary, we demonstrate our algorithm of utilizing solely qualitative knowledge to generate quantitative probabilistic inference with Bayesian networks in Fig. 2.
Fig. 2.
Demonstration and pipeline of our algorithm.
2.3.3 Toy Examples
In the following paragraphs, we will demonstrate how to make use of the qualitative knowledge model to construct the parameter distributions and network structures from qualitative statements. In the first example, if two entities A and B are positively correlated to each other, inequality constraints in (14) can be imposed over the joint probability distribution of A and B. Monte Carlo sampling can be used to draw the valid joint probability samples which are consistent with the constraints as shown in Fig. 3a. In the second example, entity A activates B (single positive influence in [5], P(B|A) ≥ P(B|Ā)), then the valid conditional probability samples based on these “causal” statements is shown in Fig. 3b.
Fig. 3.
An example of applying the inequality constraints and Monte Carlo method to draw samples in joint probability space and conditional probability space. (a) Samples in Joint Probability Space. (b) Samples in Conditional Probability Space.
3 Experiments
We apply our method to investigate the underpinnings of the mammary epithelial carcinoma cell proliferation and to predict the breast cancerous cell growth upon genetic interventions.
As the first step of our experiment, we model the core genetic network of breast cancer cell proliferation program by collecting qualitative knowledge on the direct regulations. In our model, each node represents either a gene or a protein and each edge describes either a physical transcriptional binding or a protein-protein interaction. In addition, an artificial node is added to denote the phenotype, i.e., cell proliferation. Several cell cycle regulating proteins in the signaling pathway(s) are determined to be the direct cause of cell proliferation in breast cancer. Therefore, we could link these proteins to this phenotype.
Second, we transform this network into a mathematically manipulatable representation with a directed cyclic structure. For this purpose, we utilize dynamic Bayesian model which encodes these physical interactions into a (cyclic) directed graph with the CPD.
Last, we perform two sets of in silico intervention simulation in various human breast cancer as well as normal mammalian cell lines. Specifically, we both knockdown and overexpress two cell cycle regulators: cyclinD1 and TGFβ. Upon interference, our method (in Section 2) quantitatively predicts the level of cell proliferation in these breast cancer cell lines. In the first prediction, we evaluate the cell growth rate in MCF-7 breast cancer cell in the case that cyclinD1 is interfered. In the second simulation, we predict the cell proliferation rate in three distinct human mammary cell lines: 1) human normal mammary cell line MCF-10A; 2) human breast cancer cell line: MDA-MB-231; and 3) RAS/ErbB2-transfected breast carcinoma cell line: MCF-10A(Ras/ErbB2).
3.1 Mammalian Cell Proliferation Network
Deregulated cell cycle activity is one of the main cause for cancer development, such as breast carcinomas. The cell cycle is controlled by a set of regulators which forms a complex system including the cyclins, cyclin-dependent kinases (CDKs), and CDK inhibitors. The progression through the cell cycle requires sequential activation and inactivation of these modulators. The complex signaling networks and pathways mediated by cytokines and hormones can influence cell proliferation in positive or negative ways and activates a cascade of intracellular biochemical events and is ultimately responsible for the biological phenotypic observations.
Among these pathways, signaling from transforming growth factor β (TGFβ) plays key roles in regulation of a wide variety of biological end points from early embryonic patterning to the control of cell differentiation and growth in adult cells. TGFβ can activate antiproliferation gene responses in G1 phase and impede the completion of the ongoing cell cycle. TGFβ responses in human epithelial cell lines from skin, lung, and mammary gland originals have revealed a shared proliferation program. This program consists of cyclins and cyclin-dependent kinases. Meanwhile, the cell proliferation is realized by the sequential activation of CDK inhibitors and repression of the growth promoting transcription factor c-MYC.
Besides the cell growth regulating signals mediated by cytokine TGFβ, Ras genes are the most common targets for mutations in human breast cancer. Ras protein is activated in response to a very wide spectrum of extracellular stimuli, such as cytokines, growth factors, hormones, neurotransmitters, and extracellular matrix components. These factor stimulation leads to a rapid and transient increase of active Ras-GTP [20]. Ras signal transduction involves the passage of information along a chain of proteins which exert control over a host of signal transduction pathways. The best characterized Ras signal transduction pathway is Ras-MAPK cascade. Ras activates the Raf-1 kinase, which, in turn, activates the MEK-ERK kinase cascade. ERK phosphorylates transcription factors in cytosol and nucleus which leads to activation of the genes involved in cell proliferation.
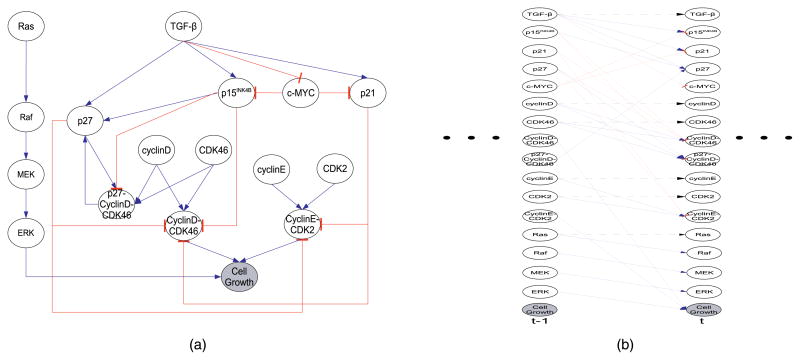
In this study, we include the compact representation of TGFβ and Ras signaling pathways in the cell proliferation network (Fig. 4). The signal transduction along these two pathways are extracted from a collection of qualitative knowledge in the publications (see Appendix). TGFβ represses the growth promoter c-MYC and activates the cell growth inhibitor p15 of the INK4-family and p21, p27 of the Cip1/Waf1/Kip1-2-family. Ras influences the transcription of cyclin and CDK genes through the Raf/MEK/ERK cascade. It is also known that many intermediate factors in the cytosol, e.g., smad proteins and other coexpressors, e.g., E2F4/5, p38, p53, p107, p300, ID1, ID2, and Miz-1 coregulates the cell proliferation [48] which can be integrated into our later studies consistently [4]. The loopy circuit in the network serves to providing tight and robust control to this program.
Fig. 4.
Mammary Cell Proliferation Network. Blue arrow means activation and red arrow means inhibition. Cytokine TGFβ inhibits cell growth promoter, c-MYC. In additional, c-MYC promotes cell proliferation by repressing several cell growth suppressor proteins, p15, p21. TGFβ elevates activity of three cyclin-dependent kinase’s inhibitor: p15, p21 and p27. p15, p21 and p27 inhibit the complex formation between cyclinD and CDK4,6 and p27, p21 further prevent cyclinE-CDK2’s activation. TGFβ elevates expression of CDK4/6-specific inhibitor p15. p27 binds to CDK4,6 to form a complex, meanwhile, p27 is released from this protein complex under the presence of p15. p15 indirectly stimulates the surge of p27. CyclinD1 and CDKs 4,6 form a complex which drives the cell proliferation in combination with complex formed by cyclinE and CDK2. Besides TGFβ pathway, hyperactive Ras signaling regulates cell developments and promotes cell growth. (a) Dynamic Bayesian Network. (b) 2-Time-Slice Bayesian Network.
3.2 DBN Model of Cell Proliferation Network
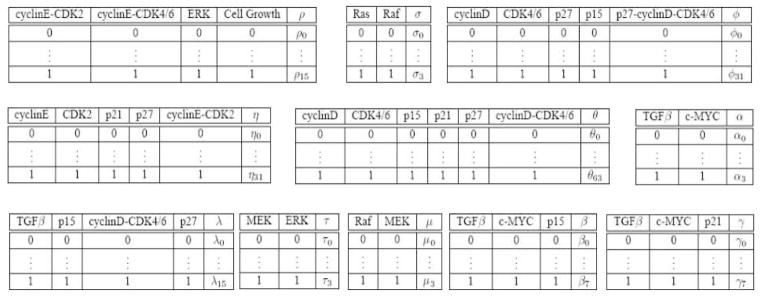
The cell proliferation network of mammalian epithelial cell can be modeled by a DBN as shown in Fig. 4a. The DBN can be unrolled over the time into a series of 2TBNs as shown in Fig. 4b. The parameters are described by the joint probability distribution (Fig. 5). In [6], a qualitative knowledge model is proposed to define a set of inequality constraints over the conditional probability space. In this study, we project this set of conditional-space constraints onto the joint probability space where we transform the conditional probability into a set of joint probabilities.
Fig. 5.
The parameters in the dynamic Bayesian network of mammary cell proliferation program are denoted by joint probability tables of child nodes and their parents. The joint condition probability is listed in the rightmost column of each table. The secondary rightmost column denotes the child node and its left columns indicates the parent nodes.
The conditional probability distribution (a0; a1) transformed from α can be modeled by Single Negative Influence, i.e., a0 ≥ a1, where and Similarly, the conditional probability distribution transformed from σ can be modeled by by Single Positive Influence and the conditional probability distribution (b,r) transformed from (β, γ) can be described by Mixed Joint Influence, i.e.,
| (16) |
where the condition probability entries in (16) can be described by joint probability table. For example, . The conditional probability distribution transformed from (ρ) can be modeled by Plain Synergy with Positive Individual Influence, i.e.,
| (17) |
where p0 is the probability of breast cancer cell growth given its parents being underexpressed, thus, . λ can be defined similarly. The parameters (θ, Ø, η) can be defined by a set of constraints hierarchically. First, the conditional probabilities transformed from these parameters can be modeled by Mixed Joint Influence since there are multiple input signals to activate and to repress the complexes CyclinD-CDK46, CyclinE-CDK2, and p27-CyclinD-CDK46 from their parents. Therefore, the parameters can be classified according to the number of repressors being overexpressed. Second, these probabilities in each class can be further defined by Plain Synergy with Positive Individual Influence based on the number of parental activators being overexpressed. For example, the conditional probability (g) based on the parameter 3 can be first classified into four classes of parameters based on the configuration of p21 and p27, i.e., G0 = {g0, g4, g8, g12}, G1;1 = {g1, g5, g9, g13}, G1;2 = {g2, g6, g10, g14}, and G2 = {g3, g7, g11, g15}. If we assume the inhibitive effects of p21 and p27 on CyclinD-CDK46 are symmetric (In general, it is possible to model the unsymmetrical effects given specific knowledge), we could merge the parameters of G1;1 and G1;2 into one class, G1. With the same configuration of the parents, the parameters across the classes can be constrained as
| (18) |
Second, within each class, the parameters can be further classified by the number of activators being overexpressed as
| (19) |
where the condition probability is defined by the network parameters η. Similarly, Ø and θ can be modeled as (18) and (19).
Besides this basic knowledge, further constraints can be added to regulate the sensitivity and specificity in parameter space. In Fig. 4a, p21 and p15’s activities are completely blocked by c-MYC (see the Appendix). We can confine those corresponding conditional parameters close to zero, i.e., and .
The inference on cell growth is computed for each possible model with parameters Π = {α, σ, γ, ρ, ø, β, η, θ, λ, τ, μ} by (13). However, since the parameter space is rather high-dimensional, we can use Monte Carlo method to approximate the integration. For each parameter in Π, its distribution in the parameter space is defined by a set of constraints as in (16) to (19). By using Monte Carlo Accept-Reject method, we simulate K = 500,000 CPT samples and together with the structure s in Fig. 4a define a consistent model class, M̃ = {mk(s, Πk)|k = 1, …, Kg.
3.3 Interfering Prediction on Breast Cancer Cell Proliferation
After we build up the DBN model of cell proliferation network in mammalian epithelial cells, we can interrogate this network with quantitative in silico interventional simulation and gain insight on the functional mechanism of cell proliferation program in breast cancer cells.
3.3.1 CyclinD Interference in MCF-7 Cell
In [13], the relationship of cyclinD1 and breast cancer cell progression is investigated. Transcription factors regulating the cyclinD1 gene(CCND1) are silenced by transfecting the MCF-7 breast cancer cells with RNA interfering (RNAi) vectors. The RNAi against two transcription factors, FoxA1 and NFIC, significantly increases and reduces the mRNA and protein level of cyclinD1 in the cell nucleus. In addition, the effects of estrogen (estradiol) on CCND1’s expression level is explored in combination with the RNAi against CCND1’s transcription factors. In the experiment, real-time PCR assays determined the mRNA level of CCND1 in MCF-7 under each case of the RNAi interference with or without estradiol. The cell proliferation efficiency is evaluated as percentage change of cell number relative to the control. The control is indicated by RNA interference against luciferase gene in MCF-7 cell.
Therefore, in the first prediction, we like to predict the breast cancer lineage MCF-7’s cell proliferation efficiency in each case of RNAi interference to CCND1’s transcription factors. To this end, we first determine the expression levels of several key cell progression regulators, i.e., TGFβ, Ras protein, cyclinD1, cyclinE, and CDKs in MCF-7 cells. It is known that TGFβ is a cell growth inhibitor. Ras, cyclinD,E, and CDKs are cell growth promoters. For example, a breast cell line bearing high levels of cyclinD,E will show proactive cell proliferation than a cell with low levels of these regulators. These regulators do not have any parent node(s) in the cell proliferation network (Fig. 4a), therefore, their levels eventually determine the cell state. To define the cell state, we collect the level of these regulators in MCF-7 cells from a set of publications (please see legend of Table 3). In Table 3, the expression level of each regulators is indicated by a set of six symbols. We transform this discrete set of symbols into probability value by equally dividing [0, 1] into six intervals. We set the initial probability of these regulators in MCF-7 cell according to the transformed probability value which is indicated by the value in bracket in Table 3. In [13], cyclinD1 is interfered to five different levels by RNAi silencing experiments. The ratio between cyclinD1’s level and the control level is evaluated. We transfer these ratio values into probability and then we clamp the level of cyclinD1 to these probabilities as evidence (E) in our interfering prediction (13). The ratio is transformed to the fold change which is calculated as F = log2 R and this log-value indicates the expression level of cyclinD1 in each case. The probability of cyclinD1 equals to 1 if the fold change is greater than 2 and 0 if the fold change is less than −1. Given these initial settings, we can predict the probability of cell proliferation in each interference experiment by (13). The experimental cell proliferation efficiency is measured as the relative cell number changes to the control. Thus, we calculate the predicted cell growth efficiency by referring the change in cell proliferation probability to the probability in the control case. The probability change is assessed between the interfered case and the control case. The ratio, fold change, initial probability of cyclinD1, cell proliferation probability, and cell proliferation efficiency are summarized in Table 2.
TABLE 3.
Protein Expression Levels of the Key Cell Cycle Regulators in Mammalian Normal Epithelial and Cancerous Cells
| Cell line | TGFβ | c-MYC | Ras(Ras-GTP) | cyclinD1 | cyclinE | CDK2 | CDK4 | CDK6 |
|---|---|---|---|---|---|---|---|---|
| MCF-7 | *4 | * | + | +++ | ++ | +++ | ++ | +/− |
| MDA-MB-231 | * | +++++5 | ++ | ++ | ++ | ++ | ++ | ++ |
| MCF-10A | * | * | + | −2 | −2 | −2 | −2 | −2 |
| MCA-10A(Ras/ErbB2) | * | +++++5 | +++++1 | +++++3 | +++++3 | +++++3 | +++++3 | +++++3 |
Relative intensities are scored as following. −: not detected (0); +/− : weak expression (0.16); +: low expression (0.33); ++: moderate expression (0.5); +++: strong expression (0.67); ++++: very strong expression (0.83); +++++: highly overexpressed (1.0); The protein expression level of cyclinD1, cyclinE, CDK2/4/6 in MCF-7, and MDA-MB-231 cell lines are based on the data in [49]. The expression level of the active form of Ras protein, i.e., Ras-GTP, in MCF-7, MCF-10A, MDA-MB-231 are based on the data in [12]. 1: Ras and ErbB2 protein’s expression is transfected to be highly overexpressed in MCF-10A(Ras/ErbB2) cells [8]; 2: The expression level of cyclinD and cyclinE in MCF-10A are undetectable [52], [28]. 3: In MCF-10A(Ras/ErbB2), the overexpression of cyclinD is strongly induced by ErbB2 and Ras significantly stimulates the cyclinE/CDK2’s activity [3], [25]. 4: TGFβ’s level is assigned arbitrarily to low expression and strong expression in MCF-7 cell due to the lack of knowledge in the literature. 5: c-MYC is overexpressed in MDA-MB-231 and MCF-10A(Ras/ErbB2) cells due to the lost repression by TGFβ [8]. *: TGFβ’s level is interfered in MCF-10A, MCF-10A(Ras/ErbB2), and MDA-MB-231 cells [8]. c-MYC’s level in MCF-7 and MCF-10A is determined by TGFβ. The expression levels are normalized across studies.
TABLE 2.
The Ratio (R), other symbols denote the same as Table 1
| Control | I | II | III | IV | V | |
|---|---|---|---|---|---|---|
| R | 1 | 0.85 | 1.25 | 1.65 | 2.2 | 3.15 |
| F | 0 | −0.23 | 0.32 | 0.72 | 1.14 | 1.66 |
| PC | 0.33 | 0.26 | 0.44 | 0.57 | 0.71 | 0.89 |
| CG* | 0.09 | 0.08 | 0.10 | 0.11 | 0.13 | 0.14 |
| Eff* | 0 | −10.51% | 11.11% | 24.43% | 38.12% | 55.1% |
| Exp | 0 | −14.4% | 4.4% | 14.4% | 55% | 67.8% |
: Number is calculated in case that Pr(TGFβ) = 0:8.
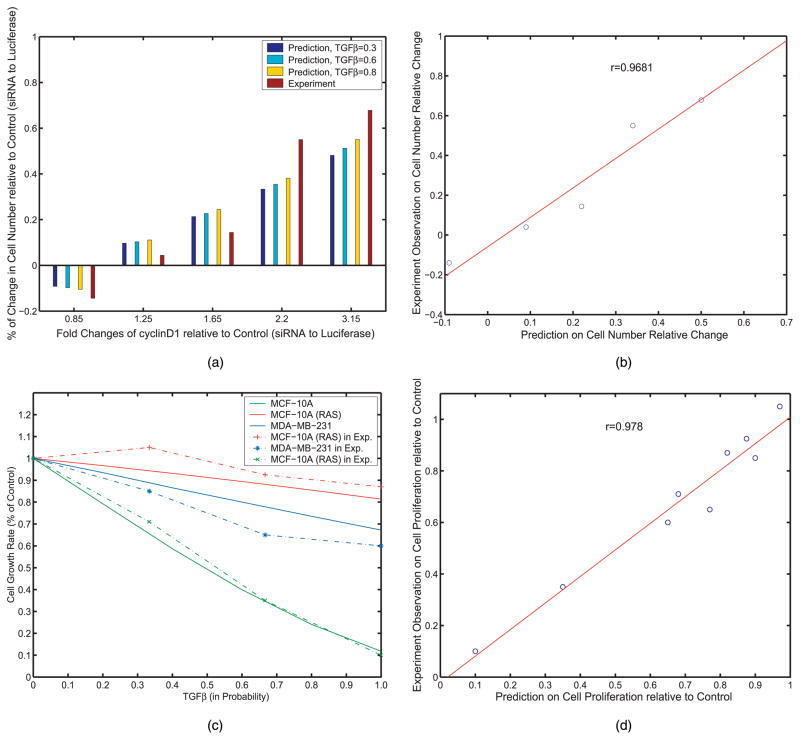
The prediction and experiment observations in each interference study is shown in Fig. 6a. Since the TGFβ level is unknown in MCF-7, we simulated three cases where the TGFβ level varies from low (0.3) to high (0.8). The correlation between prediction and experimental observations is shown in Fig. 6b. The Pearson correlation coefficient equals to 0.9681. We can see that the relative cell proliferation efficiency in case of cyclinD1 overexpression and knockdown is relatively independent on the level of TGFβ, i.e., TGFβ in the MCF-7 cell state, exhibits weak inhibitions on the cell growth. CyclinD1’s changes dominate the TGFβ’s changes. This observation may be partially due to that cyclinD1 is a cell growth promoter in nucleus and is located at the very downstream of signaling pathways.
Fig. 6.
Prediction on Cell Proliferation Efficiency with Interfered TGFβ and cyclinD1 in Breast Cancer. (a) Prediction on Cell Proliferation in MCF-7 cell. (b) Correlation between predictions and experiments. (c) Prediction on Cell Proliferation in three breast normal and cancer cell. (d) Correlation between predictions and experiments.
3.3.2 TGFβ Interference in Other Breast Normal and Carcinoma Cell Lines
In [8], it has been identified that the loss of TGFβ growth inhibition often occurs without a loss of receptors and/or smad cofactors in breast cancer. Instead, the repression of a key cell growth promotion component c-MYC is selectively lost. In breast cancer cell line, MDA-MB-231 and MCF-10A(Ras/ErbB2), this repressive response of c-MYC to the cytokine TGFβ is lost [8]. However, the normal breast cell line, MCF-10A reserves this repression. In addition, in MCF-10A(Ras/ErbB2) cell, Ras protein is overexpressed. In the experiment, the concentration of TGFβ is manipulated to different levels from 0 to 100 (μm) in these cell cultures and cell proliferation efficiency is measured.
In the second prediction, we predict the inhibition of cell growth by TGFβ in these three breast cancer cell lines. Similar to the first experiment, we first determine the probability of the key cell progression regulators in MCF-10A, MCF-10A(Ras/ErbB2), and MDA-MB-231 cells according to Table 3. The level of these regulators defines the distinct cell context of these three cell lines. MCF-10A is a normal human mammary epithelial cell line. MDA-MB-231 and MCF-10A(Ras/ErbB2) are the human immortal cell lines showing aggressive cell progression. We set the initial probability of these regulators in these cell lines according to the transformed probability value which is indicated by the value in bracket in Table 3.
The level of TGFβ is similarly discretized into probability with log-transformation as cyclinD1. In addition, the TGFβ’s repression on c-MYC is lost in MCF-10A(Ras/ErbB2) and MDA-MB-231 cell lines. Thus, the level of c-MYC in these two cell lines are set to 1 according to the experiment [8]. The experimental cell proliferation efficiency is measured as the ratio between cell number given specific TGFβ’s concentration and the control. The control case of each cell line is indicated by TGFβ knockout, i.e., TGFβ = 0 [8]. Thus, we calculate the predicted cell growth efficiency by referring the cell proliferation probability to the probability in the control case. The concentration, fold change, initial probability of TGFβ, predicted cell proliferation probability, and cell proliferation efficiency are summarized in Table 1.
TABLE 1.
The Concentration (C), Fold Change (F), Probability of TGFβ(PT), Probability of Cell Growth (CG), Predicted Cell Growth Efficiency (Eff), and Experimental Cell Growth Efficiency (Exp) is Listed
| Control | I | II | III | |
|---|---|---|---|---|
| C | 0 | 1 | 10 | 100 |
| F | -Inf | 0 | 1 | 2 |
| PT | 0 | 0.33 | 0.66 | 1 |
| CG* | [0.29,0.85,0.52] | [0.22,0.82,0.49] | [0.11,0.76,0.42] | [0.03,0.69,0.35] |
| Eff* | [1,1,1] | [79.26%,96.7%,93.48%] | [39.92%,89.43%,80.03%] | [11.81%,81.38%,67.22%] |
| Exp* | [1,1,1] | [71%,100.5%,85%] | [35%,92.5%,65%] | [10%,87%,60%] |
: Number is listed in MCF-10A, MCF-10A(Ras/ErbB2), MDA-MB-231 from left to right.
The prediction on the cell growth efficiency in each cell line is depicted by solid line in Fig. 6c. For comparison, the experimental observation is adopted from [8] and shown as dashed line in Fig. 6c. The correlation between simulation results and experimental observation is shown in Fig. 6d and the pearson correlation equals to 0.978. We observe that the cell proliferation ratio in the two breast cancer cell lines, MCF-10A(Ras/ErbB2) and MDA-MB-231 is much higher than that of the human normal mammary epithelial cell line MCF-10A at any TGFβ’s level. This may be due to the difference in cell state determined by the key cell cycle regulators. In addition, depending on different cell state, TGFβ inhibits the cell proliferation distinctly. In the normal epithelial cell line MCF-10A, TGFβ exhibits prominent inhibition effect on cell proliferation while in the two breast carcinoma cell lines TGFβ exerts relative faint inhibitions on cell growth.
4 Conclusion
Several state-of-the-art statistical and deterministic methods have been used to infer the genetic regulatory network from the data. These methods include but are not limited to (Probabilistic) Boolean networks, Petri nets, Modulo networks and mutual-information-based model, and Bayesian networks. Besides their mathematical differences, they encounter a common problem: learning performance and generalization accuracy of these methods are subject to the availability and the quality of the observation data. The performance of these methods will be severely undermined (with slight variance) in any of the following cases: 1) the data contain few samples (comparing to number of predictors/ features/random variables of the system); 2) the data are contaminated by a relatively high-level noise; 3) the data contain no functional measurements.
Comparatively, we propose in this paper, a knowledge-driven systems biology approach using dynamic Bayesian network based solely on knowledge. In this approach, the model structure and parameters are exclusively determined by qualitative knowledge. Given a collection of physical interactions between genes and proteins without parameter details, we can construct not only the structure of the genetic regulatory network, but also automatically recover the parameter distribution to parameterize the network. Consequently, quantitative generalization and prediction can be generated by a model averaging scheme upon genetic interventions. In this study, the prediction of breast cancer phenotypes closely matches the experimental observations.
There are several significant advantages in our proposed knowledge-based framework over the state-of-the-art data-based methods. First, knowledge describing the same biological interactions is often redundant across literatures and knowledge contains qualitative information other than quantitative measurements. These properties of the knowledge help rule out incorrect and noisy information. Second, knowledge can usually identify direct binding and functional regulation through in vitro and in vivo small-scale biochemistry experiments, such as chromo immunoprecipitation, vector reporter, etc. However, the large-scale high-throughput data either cannot provide information on in-/ direct interactions, e.g., mRNA microarray data, or cannot provide functional regulatory information, such as ChIP-chip or ChIP-seq data. Not to mention that in any data format, the information is buried under considerable noise. Third, the constructed network complexity and system dimensionality is solely subject to the amount of knowledge available. The direct benefit is that we need not be concerned about the insufficient statistics in any given set of knowledge since the system dimensionality is limited by the information in the knowledge set. Thus, we do not have the “dimension curse” and “overfitting” problem. Finally, our method has a good scaling ability. By compiling pieces of knowledge together, we can eventually construct a network with an infinite number of variables. Because each piece of knowledge is describing a local structure of a whole network, i.e., substructure of the network, we can obtain the complete network by assembling these substructures together. In addition, the parameters of the whole network are decomposable into a product of local parameters, namely, parameters can be determined locally at each nodes given their parents. No heuristic search for model structures is invoked and the problem remains solvable in linear time.
Like any method, there are inherent limitations with our proposed knowledge-based method. These limitations largely stem from the nature of qualitative knowledge. First, knowledge can be inconsistent, i.e., there can be contradicting information regarding to the same genetic regulation(s). In this case, the entropy of the qualitative knowledge and the dynamic Bayesian model uncertainty is increased. We sort to model the distribution of the inconsistent information and to integrate the knowledge entropy into our proposed method. Following this line, a solution is proposed in [5]. Second, knowledge is usually incomplete. Though it is important to interrogate the system behavior by assembling existing knowledge into a whole network, the existing knowledge does not disclose any new information regarding to the interactions. Knowledge is useful to produce a high-confident network, instead, data-based methods do a good job at discovering new links between molecules but the outcome is noisy and edges are low confident (as discussed in the last paragraph). Therefore, it is natural to combine the two strengths into one framework to recover new links with high confident as well as to generate accurate predictions on the system behavior. Third, we can improve our modeling complexity to cope with more forms of knowledge. Currently, we use basic forms of linear inequality constraints to model “activating” and “repressing” genetic regulations. We can further expand our inequality to incorporate ratios, difference, and boundary features of the parameters. We can even employ nonlinear inequality to model the relations between the parameters. This new set of features is very useful in modeling qualitative knowledge such as a transcription factor A increases the expression level of gene B several times.
One possible reason for the achieved prediction power by our method is that the complex of genetic regulatory network structure with feed-forward and feed-backward regulations ensured the parametric robustness, i.e., the regulatory network behavior becomes robust against parameter configurations under certain structure complexes. It seems that nature tends to preserve certain redundant functional feed-forward and feed-backward links in the biological network so that its functions become robust over insignificant disruptions to the network kinetics. We observed feedback links and feed-forward links in our breast cancer cell growth network, in addition, this hypothesis has been well observed and confirmed in other studies in various organisms [33], [19]. On the other hand, for a simpler network, the network behavior is more sensitive to parameter configurations of the network. Our method may become less predictive than the complex network.
In summary, we believe that our method provides a promising pathway for understanding the underpinnings of a genetic regulatory network. With this method, it is possible to integrate currently available knowledge on genetic regulations and signaling pathways in a disease to a large network and to decipher the biological mechanisms by producing quantitative predictions on interventions based on the network. Thus, it may be especially interesting to drug target discovery research.
Acknowledgments
This work is partially supported by NIH (R01GM072856 to W.W.).
Biographies
 Rui Chang received the PhD degree in computer science and informatics from the Technical University of Munich, Germany, in 2008. He is currently a postdoctoral scholar at the University of California, San Diego. His research focuses on developing and applying statistical algorithms to human genomics. From 2005 to 2007, he worked as a PhD research scientist at Siemens Corporate Technology in Munich, Germany. During this time, he worked on developing novel statistical models and on modeling signaling pathways in breast cancer. He has several approved patents in Germany and USA. He currently works on the development and application of several novel statistical methods in modeling the human embryonic stem cell. He has first-authored several peer-reviewed journal publications and proceeding publications. He has been invited to author a bookchapter and give presentations at several international conferences. He is a member of the IEEE.
Rui Chang received the PhD degree in computer science and informatics from the Technical University of Munich, Germany, in 2008. He is currently a postdoctoral scholar at the University of California, San Diego. His research focuses on developing and applying statistical algorithms to human genomics. From 2005 to 2007, he worked as a PhD research scientist at Siemens Corporate Technology in Munich, Germany. During this time, he worked on developing novel statistical models and on modeling signaling pathways in breast cancer. He has several approved patents in Germany and USA. He currently works on the development and application of several novel statistical methods in modeling the human embryonic stem cell. He has first-authored several peer-reviewed journal publications and proceeding publications. He has been invited to author a bookchapter and give presentations at several international conferences. He is a member of the IEEE.
 Robert Shoemaker received the BS degree in biochemistry and the BA degree in German literature from the University California at San Diego, and the PhD degree from the University of California, San Diego. His research interest includes the human epigenome with an emphasis on DNA methylation. Currently, he is exploring DNA methylations influence on cell line phenotypes.
Robert Shoemaker received the BS degree in biochemistry and the BA degree in German literature from the University California at San Diego, and the PhD degree from the University of California, San Diego. His research interest includes the human epigenome with an emphasis on DNA methylation. Currently, he is exploring DNA methylations influence on cell line phenotypes.
 Wei Wang is an associate professor in the Department of Chemistry and Biochemistry at the University of California, San Diego (UCSD). His research is focused on understanding the principles governing the topology and dynamics of biological networks. He is particularly interested in how biological networks regulate phenotype formation in response to environmental or developmental signals from both genetic and epigenetic perspectives. His interdisciplinary research integrates statistics, machine learning, chemistry, and physics to uncover biological mechanisms.
Wei Wang is an associate professor in the Department of Chemistry and Biochemistry at the University of California, San Diego (UCSD). His research is focused on understanding the principles governing the topology and dynamics of biological networks. He is particularly interested in how biological networks regulate phenotype formation in response to environmental or developmental signals from both genetic and epigenetic perspectives. His interdisciplinary research integrates statistics, machine learning, chemistry, and physics to uncover biological mechanisms.
Appendix
A set of qualitative statements, Ω̄ = {Ωi|i = 1 … 9}, with regarding to this network can be extracted from a group of publications [48], [21], [40], [10], [38], [9], [46], [45], [41] where Ω1: CDK2, CDK4, and CDK6-drive progression through the G1 phase of the cell cycle. In G1, CDK4/6 activation requires association with D-type cyclins whereas cyclin E binding activates CDK2. [48]; Ω2: The cyclin-dependent kinase inhibitor p15, is induced by treatment with TGFβ, suggesting p15 may act as an effector of TGFβ-mediated cell cycle arrest. [21]; Ω3: TGFβ elevates expression of CDK4/6-specific inhibitor p15 and induces the release of p27 from CDK4 and CDK6 complex and this release coincides with the increased binding of p27 from CDK4 to CDK2 in vivo, suggesting that the the release of CDK4-bound p27 in TGFβ treated cells is caused by the surge in p15 levels. [40]; Ω4: TGFβ can induce the cyclin-dependent kinase inhibitor p21 through a p53-independent pathway. [10]; Ω5: TGFβ can induce the cyclin-dependent kinase inhibitor p27 which associates with cyclinE-CDK2 complex in vivo and prevents their activation. [38], [41]; Ω6: A complex containing Smad3, E2F4/5, DP1, and p107, in response to TGFβ, associates with Smad4 and recognize a composite Smad-E2F site on c-MYC for repression. [9]; Ω7: TGFβ signalling prevents recruitment of c-MYC to the p15 transcription initiator by Miz-1. Two separate TGFβ-dependent inputs keep tight control over p15 activation: Smad-mediated transcription and relief of repression by c-MYC. [46]; Ω8: Transcription factor c-MYC is directly recruited to the p21 promoter by the DNA-binding protein Miz-1. This interaction blocks p21 induction by p53 and other activators. [45]; Ω9: TGFβ activates Ras and ErbB2 which induces formation of proliferative structures in noninvasive early stage mammary epithelial lesions [47].
Footnotes
For information on obtaining reprints of this article, please send e-mail to: tcbb@computer.org, and reference IEEECS Log Number TCBB-2010-02-0047.
Contributor Information
Rui Chang, Email: chang.rui@hotmail.com.
Robert Shoemaker, Email: rfshoema@ucsd.edu.
Wei Wang, Email: wei-wang@ucsd.edu.
References
- 1.Bayes T. An Essay Towards Solving a Problem in the Doctrine of Chances. Philosophical Trans Royal Soc of London. 1763;53:370–418. [Google Scholar]
- 2.Beal MJ, Falciani F, Ghahramani Z, Rangel C, Wild DL. A Bayesian Approach to Reconstructing Genetic Regulatory Networks with Hidden Factors. Bioinformatics. 2005;21:349–356. doi: 10.1093/bioinformatics/bti014. [DOI] [PubMed] [Google Scholar]
- 3.Caldon CE, Daly RJ, Sutherland RL, Musgrove EA. Cell Cycle Control in Breast Cancer Cells. J Cellular Biochemistry. 2006;97(2):261–274. doi: 10.1002/jcb.20690. [DOI] [PubMed] [Google Scholar]
- 4.Chang R. Consistent Modeling, Integration and Simulation of Molecular Interaction Networks in Space-Time Dimension. Proc. Seventh IEEE Int’l Conf. Bioinformatics and Bioengineering (BIBE ’07); 2007. pp. 1254–1259. [Google Scholar]
- 5.Chang R, Brauer W, Stetter M. Modeling Semantics of Inconsistent Qualitative Knowledge for Quantitative Bayesian Network Inference. Neural Networks. 2008;21(2–3):182–192. doi: 10.1016/j.neunet.2007.12.042. [DOI] [PubMed] [Google Scholar]
- 6.Chang R, Stetter M, Brauer W. Quantitative Inference by Qualitative Semantic Knowledge Mining with Bayesian Model Averaging. IEEE Trans Knowledge and Data Eng. 2008 Dec;20(12):1587–1600. [Google Scholar]
- 7.Chaouiya C, Remy E, Ruet P, Thieffry D. Petri Net Modelling of Biological Regulatory Networks. Proc. 25th Int’l Conf. Applications and Theory of Petri Nets; 2004. [Google Scholar]
- 8.Chen C-R, Kang Y, Massagué J. Defective Repression of c-MYC in Breast Cancer Cells: A Loss at the Core of the Transforming Growth Factor β Growth Arrest Program. Proc Nat’l Academy of Sciences USA. 2001;98(3):992–999. doi: 10.1073/pnas.98.3.992. http://www.pnas.org/content/98/3/992.abstract. [DOI] [PMC free article] [PubMed]
- 9.Chen CR, Kang Y, Siegel PM, Massague J. E2F4/5 and p107 as Smad Cofactors Linking the TGF-beta Receptor to c-myc Repression. Cell. 2002;110:19–32. doi: 10.1016/s0092-8674(02)00801-2. [DOI] [PubMed] [Google Scholar]
- 10.Datto MB, Li Y, Panus J, Howe DJ, Xiong Y, Wang XF. Transforming Growth Factor Beta Induces the Cyclin-Dependent Kinase Inhibitor p21 through a p53-Independent Mechanism. Proc Nat’l Academy of Science USA. 1995;92:5545–5549. doi: 10.1073/pnas.92.12.5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dean T, Kanazawa K. A Model for Reasoning about Persistence and Causation. Computational Intelligence. 1989;5:142–150. [Google Scholar]
- 12.Eckert LB, Repasky GA, Ulku AS, McFall A, Zhou H, Sartor CI, Der CJ. Involvement of Ras Activation in Human Breast Cancer Cell Signaling, Invasion, and Anoikis. Cancer Research. 2004;64:4585–4592. doi: 10.1158/0008-5472.CAN-04-0396. [DOI] [PubMed] [Google Scholar]
- 13.Eeckhoute J, Carroll JS, Geistlinger TR, Torres-Arzayus MI, Brown M. A Cell-Type-Specific Transcriptional Network Required for Estrogen Regulation of Cyclin D1 and Cell Cycle Progression in Breast Cancer. Genes & Development. 2006 Sept;20(18):2513–2526. doi: 10.1101/gad.1446006. http://dx.doi.org/10.1101/gad.1446006. [DOI] [PMC free article] [PubMed]
- 14.Friedman N. Learning Belief Networks in the Presence of Missing Values and Hidden Variables. Proc. 14th Int’l Conf. Machine Learning (ICML ’97); 1997. pp. 125–133. [Google Scholar]
- 15.Friedman N. Inferring Cellular Networks Using Probabilistic Graphical Models. Science. 2004;303:799–805. doi: 10.1126/science.1094068. [DOI] [PubMed] [Google Scholar]
- 16.Friedman N, Goldszmidt M. Learning in Graphical Models. MIT Press; 1999. Learning Bayesian Networks with Local Structure. [Google Scholar]
- 17.Friedman N, Linial M, Nachman I, Peer D. Using Bayesian Networks to Analyze Expression Data. J Computational Biology. 2000;7:601–620. doi: 10.1089/106652700750050961. [DOI] [PubMed] [Google Scholar]
- 18.Geier F, Timmer J, Fleck1 C. Reconstructing Gene-Regulatory Networks from Time Series, Knock-Out Data, and Prior Knowledge. BMC Systems Biology. 2007;1:article no. 11. doi: 10.1186/1752-0509-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonze D, Goldbeter A. Circadian Rhythms and Molecular Noise. Chaos: An Interdisciplinary J Nonlinear Science. 2006;16(2):026110. doi: 10.1063/1.2211767. [DOI] [PubMed] [Google Scholar]
- 20.Gutkind JS. Cancer Drug Discovery and Development. Vol. 5. Humana Press; 2000. Signaling Networks and Cell Cycle Control: The Molecular Basis of Cancer and Other Diseases. [Google Scholar]
- 21.Hannon GJ, Beach D. pl5INK4B is a Potential Effector of TGF-[Beta]-Induced Cell Cycle Arrest. Nature. 1994;371:257–261. doi: 10.1038/371257a0. [DOI] [PubMed] [Google Scholar]
- 22.Hartemink AJ, Gifford DK, Jaakkola T, Young RA. Combining Location and Expression Data for Principled Discovery of Genetic Regulatory Network Models. Proc Pacific Symp Biocomputing. 2002:437–449. [PubMed] [Google Scholar]
- 23.Heckerman D. “Learning Bayesian Networks: The Combination of Knowledge and Statistical Data,” technical report. KDD Workshop; 1994. [Google Scholar]
- 24.Heckerman D. “A Tutorial on Learning with Bayesian Networks,” technical report. Microsoft Research; 1996. [Google Scholar]
- 25.Hu W, Bellone CJ, Baldassare JJ. RhoA Stimulates p27Kip Degradation through Its Regulation of Cyclin E/CDK2 Activity. J Biological Chemistry. 1999;274:3396–3401. doi: 10.1074/jbc.274.6.3396. [DOI] [PubMed] [Google Scholar]
- 26.Karlebach G, Shamir R. Modelling and Analysis of Gene Regulatory Networks. Nature Rev Molecular Cell Biology. 2008;9:770–780. doi: 10.1038/nrm2503. [DOI] [PubMed] [Google Scholar]
- 27.Kauffman S, Peterson C, Samuelsson B, Troein C. Random Boolean Network Models and the Yeast Transcriptional Network. Proc Nat’l Academy of Sciences USA. 2003;100:14796–14799. doi: 10.1073/pnas.2036429100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keyomarsi K, Pardee AB. Redundant Cyclin Overexpres-sion and Gene Amplification in Breast Cancer Cells. Proc Nat’l Academy of Sciences USA. 1993;90:1112–1116. doi: 10.1073/pnas.90.3.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim SY, Imoto S, Miyano S. Inferring Gene Networks from Time Series Microarray Data Using Dynamic Bayesian Networks. Briefings in Bioinformatics. 2003;4:228–235. doi: 10.1093/bib/4.3.228. [DOI] [PubMed] [Google Scholar]
- 30.Koch I, Schueler M, Heiner M. STEPP—Search Tool for Exploration of Petri Net Paths: A New Tool for Petri Net-Based Path Analysis in Biochemical Networks. In Silico Biology. 2005;5:129–137. [PubMed] [Google Scholar]
- 31.Kuffner R, Zimmer R, Lengauer T. Pathway Analysis in Metabolic Databases via Differential Metabolic Display (DMD) Bioinformatics. 2000;16:825–836. doi: 10.1093/bioinformatics/16.9.825. [DOI] [PubMed] [Google Scholar]
- 32.Lauritzen S, Spiegelhalter D. Local Computations with Probabilities on Graphical Structures and Their Application to Expert Systems. J Royal Statistical Soc Series B. 1988;50:157–224. [Google Scholar]
- 33.Li F, Long T, Lu Y, Ouyang Q, Tang C. The Yeast Cell-Cycle Network is Robustly Designed. Proc Nat’l Academy of Sciences USA. 2004;101:4781–4786. doi: 10.1073/pnas.0305937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Margolin AA, Nemenman I, Basso K, Wiggins C, Stolovitzky G, Favera RD, Califano A. ARACNE: An Algorithm for the Reconstruction of Gene Regulatory Networks in a Mammalian Cellular Context. BMC Bioinformatics. 2006;7(Suppl 1):S7. doi: 10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy K. PhD dissertation. Univ. of California; Berkeley: 2002. Dynamic Bayesian Networks: Representation, Inference and Learning. [Google Scholar]
- 36.Pan Y, Durfee T, Bockhorst J, Craven M. Connecting Quantitative Regulatory-Network Models to the Genome. Bioinformatics. 2007;23:367–376. doi: 10.1093/bioinformatics/btm228. [DOI] [PubMed] [Google Scholar]
- 37.Pearl J. Probabilistic Reasoning in Intelligent Systems: Networks of Plausible Inference. Morgan Kaufmann Publishers; 1988. [Google Scholar]
- 38.Polyak K, Lee M-H, Erdjument-Bromage H, Koff A, Roberts JM, Tempst P, Massague J. Cell. 1994 doi: 10.1016/0092-8674(94)90572-x. [DOI] [PubMed] [Google Scholar]
- 39.Reddy VN, Liebman MN, Mavrovouniotis ML. Qualitative Analysis of Biochemical Reaction Systems. Computers in Biology and Medicine. 1996;26:9–24. doi: 10.1016/0010-4825(95)00042-9. [DOI] [PubMed] [Google Scholar]
- 40.Reynisdottir I, Polyak K, Iavarone A, Massague J. Kip/Cip and Ink4 Cdk Inhibitors Cooperate to Induce Cell Cycle Arrest in Response to TGF-Beta. Genes and Development. 1995;9:1831–1845. doi: 10.1101/gad.9.15.1831. [DOI] [PubMed] [Google Scholar]
- 41.Robson CN, Gnanapragasam V, Byrne RL, Collins AT. Transforming Growth Factor-Beta1 Up-Regulates p15, p21 and p27 and Blocks Cell Cycling in G1 in Human Prostate Epithelium. J Endocrinology. 1999;160:257–266. doi: 10.1677/joe.0.1600257. [DOI] [PubMed] [Google Scholar]
- 42.Sachs K, Perez O, Peer D, Lauffenburger DA, Nolan GP. Causal Protein-Signaling Networks Derived from Multipara-meter Single-Cell Data. Science. 2005;308:523–529. doi: 10.1126/science.1105809. [DOI] [PubMed] [Google Scholar]
- 43.Segal E, Shapira M, Pe’er RAD, Botstein D, Koller D, Friedman N. Module Networks: Identifying Regulatory Modules and Their Condition-Specific Regulators from Gene Expression Data. Nature Genetics. 2003;34:166–176. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- 44.Segal E, Wang H, Koller D. Discovering Molecular Pathways from Protein Interaction and Gene Expression Data. Bioinformatics. 2003;19:264–272. doi: 10.1093/bioinformatics/btg1037. [DOI] [PubMed] [Google Scholar]
- 45.Seoane J, Le HV, Massague J. Myc Suppression of the p21Cip1 Cdk Inhibitor Influences the Outcome of the p53 Response to DNA Damage. Nature. 2002;419:729–734. doi: 10.1038/nature01119. [DOI] [PubMed] [Google Scholar]
- 46.Seoane J, Pouponnot C, Staller P, Schader M, Eilers M, Massague J. TGF-Beta Influences Myc, Miz-1 and Smad to Control the CDK Inhibitor p15INK4b. Nature Cell Biology. 2001;3:400–408. doi: 10.1038/35070086. [DOI] [PubMed] [Google Scholar]
- 47.Seton-Rogers SE, Lu Y, Hines LM, Koundinya M, LaBaer J, Muthuswamy SK, Brugge JS. Cooperation of the ErbB2 Receptor and Transforming Growth Factor Beta in Induction of Migration and Invasion in Mammary Epithelial Cells. Proc Nat’l Academy of Sciences USA. 2003;101:1257–1262. doi: 10.1073/pnas.0308090100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Siegel PM, Massagué J. Cytostatic and Apoptotic Actions of TGF-Beta in Homeostasis and Cancer. Nature Rev Cancer. 2003;3(11):807–821. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- 49.Sweeney KJ, Swarbrick A, Sutherland RL, Musgrove EA. Lack of Relationship between CDK Activity and G1 Cyclin Expression in Breast Cancer Cells. Oncogene. 1998;16:2865–2878. doi: 10.1038/sj.onc.1201814. [DOI] [PubMed] [Google Scholar]
- 50.Troyanskaya OG, Dolinski K, Owen AB, Altman RB, Botstein D. A Bayesian Framework for Combining Heterogeneous Data Sources for Gene Function Prediction (in Saccharomyces cerevisiae) Proc Nat’l Academy of Sciences USA. 2003;100:8348–8353. doi: 10.1073/pnas.0832373100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Werhli AV, Husmeier D. Gene Regulatory Network Reconstruction by Bayesian Integration of Prior Knowledge and/or Different Experimental Conditions. J Bioinformatics and Computational Biology. 2008;6:543–572. doi: 10.1142/s0219720008003539. [DOI] [PubMed] [Google Scholar]
- 52.Zhou Q, Stetler-Stevenson M, Steeg PS. Inhibition of Cyclin D Expression in Human Breast Carcinoma Cells by Retinoids In Vitro. Oncogene. 1997;15:107–115. doi: 10.1038/sj.onc.1201142. [DOI] [PubMed] [Google Scholar]