Abstract
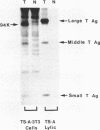
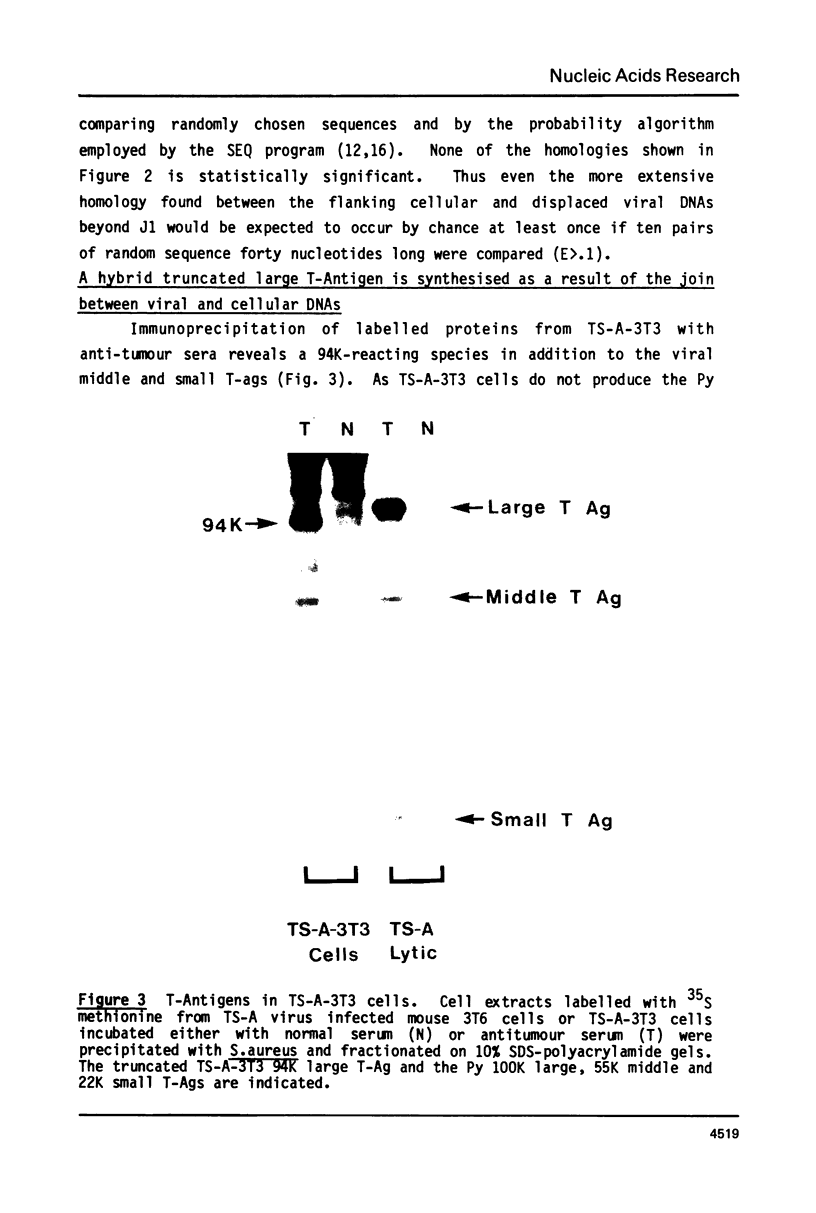
The DNA sequences at and around the junctions between viral and cellular DNA in the polyoma virus transformed mouse cell line, TS-A-3T3, have been determined. No common sequence specificity or structural features at the joins have been observed. The sequence indicates that the 94K truncated large T antigen found in TS-A-3T3 cells is a hybrid protein in which the carboxy-terminal 19 amino acids are encoded by adjacent host sequences. Moreover, the three early region transcripts initiated in viral sequences are also hybrid in nature and appear to utilize a host polyadenylation signal associated with the hexanucleotide, AATAAA, found 100 bp beyond a viral-host join.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benoist C., O'Hare K., Breathnach R., Chambon P. The ovalbumin gene-sequence of putative control regions. Nucleic Acids Res. 1980 Jan 11;8(1):127–142. doi: 10.1093/nar/8.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchan M., Stringer J., Mitchison T., Sambrook J. Integration and excision of SV40 DNA from the chromosome of a transformed cell. Cell. 1980 May;20(1):143–152. doi: 10.1016/0092-8674(80)90242-1. [DOI] [PubMed] [Google Scholar]
- Brutlag D. L., Clayton J., Friedland P., Kedes L. H. SEQ: a nucleotide sequence analysis and recombination system. Nucleic Acids Res. 1982 Jan 11;10(1):279–294. doi: 10.1093/nar/10.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calos M. P., Miller J. H. Transposable elements. Cell. 1980 Jul;20(3):579–595. doi: 10.1016/0092-8674(80)90305-0. [DOI] [PubMed] [Google Scholar]
- Dailey L., Colantuoni V., Fenton R. G., La Bella F., Zouzias D., Gattoni S., Basilico C. The evolution of polyoma-transformed rat cell lines during propagation in vitro. Virology. 1982 Jan 15;116(1):207–220. doi: 10.1016/0042-6822(82)90414-7. [DOI] [PubMed] [Google Scholar]
- Fenton R. G., Basilico C. Viral gene expression in polyoma virus-transformed rat cells and their cured revertants. J Virol. 1981 Oct;40(1):150–163. doi: 10.1128/jvi.40.1.150-163.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin B. E., Fried M., Cowie A. Polyoma DNA: a physical map. Proc Natl Acad Sci U S A. 1974 May;71(5):2077–2081. doi: 10.1073/pnas.71.5.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassell J. A., Topp W. C., Rifkin D. B., Moreau P. E. Transformation of rat embryo fibroblasts by cloned polyoma virus DNA fragments containing only part of the early region. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3978–3982. doi: 10.1073/pnas.77.7.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israel M. A., Vanderryn D. F., Meltzer M. L., Martin M. A. Characterization of polyoma viral DNA sequences in polyoma-induced hamster tumor cell lines. J Biol Chem. 1980 Apr 25;255(8):3798–3805. [PubMed] [Google Scholar]
- Ito Y. Polyoma virus-specific 55K protein isolated from plasma membrane of productively infected cells is virus-coded and important for cell transformation. Virology. 1979 Oct 15;98(1):261–266. doi: 10.1016/0042-6822(79)90545-2. [DOI] [PubMed] [Google Scholar]
- Ito Y., Spurr N. Polyoma virus T antigens expressed in transformed cells: significance of middle T antigen in transformation. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):149–157. doi: 10.1101/sqb.1980.044.01.017. [DOI] [PubMed] [Google Scholar]
- Kamen R., Favaloro J., Parker J., Treisman R., Lania L., Fried M., Mellor A. Comparison of polyoma virus transcription in productively infected mouse cells and transformed rodent cell lines. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):63–75. doi: 10.1101/sqb.1980.044.01.009. [DOI] [PubMed] [Google Scholar]
- Korn L. J., Queen C. L., Wegman M. N. Computer analysis of nucleic acid regulatory sequences. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4401–4405. doi: 10.1073/pnas.74.10.4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lania L., Gandini-Attardi D., Griffiths M., Cooke B., De Cicco D., Fried M. The polyoma virus 100K large T-antigen is not required for the maintenance of transformation. Virology. 1980 Feb;101(1):217–232. doi: 10.1016/0042-6822(80)90497-3. [DOI] [PubMed] [Google Scholar]
- Lania L., Hayday A., Bjursell G., Gandini-Attardi D., Fried M. Organization and expression of integrated polyoma virus DNA sequences in transformed rodent cells. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):597–603. doi: 10.1101/sqb.1980.044.01.062. [DOI] [PubMed] [Google Scholar]
- Lania L., Hayday A., Fried M. Loss of functional large T-antigen and free viral genomes from cells transformed in vitro by polyoma virus after passage in vivo as tumor cells. J Virol. 1981 Aug;39(2):422–431. doi: 10.1128/jvi.39.2.422-431.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. Sequence at the 3' end of globin mRNA shows homology with immunoglobulin light chain mRNA. Nature. 1974 Nov 29;252(5482):359–362. doi: 10.1038/252359a0. [DOI] [PubMed] [Google Scholar]
- Soeda E., Arrand J. R., Smolar N., Walsh J. E., Griffin B. E. Coding potential and regulatory signals of the polyoma virus genome. Nature. 1980 Jan 31;283(5746):445–453. doi: 10.1038/283445a0. [DOI] [PubMed] [Google Scholar]
- Stringer J. R. Integrated simian virus 40 DNA: nucleotide sequences at cell-virus recombinant junctions. J Virol. 1981 May;38(2):671–679. doi: 10.1128/jvi.38.2.671-679.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver R. F., Weissmann C. Mapping of RNA by a modification of the Berk-Sharp procedure: the 5' termini of 15 S beta-globin mRNA precursor and mature 10 s beta-globin mRNA have identical map coordinates. Nucleic Acids Res. 1979 Nov 10;7(5):1175–1193. doi: 10.1093/nar/7.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]