Abstract
We have previously reported that HIV-1 preferentially infects Foxp3+ Treg cells in vitro and in vivo, and Foxp3 enhances the HIV-1 LTR expression through epigenetic mechanisms in T cells. We report here that histone deacetylase inhibitor (HDACi) failed to further enhance HIV gene expression in FoxP3+ T cells. We discovered that Foxp3 inhibited cellular HDAC activity in T cells, and mutations in the forkhead domain that ablate Foxp3 function also abolished its ability to inhibit HDAC. When co-expressed, Foxp3 specifically inhibited the deacetylase activity of HDAC1. We further showed that Foxp3 was associated with HDAC1, and mutations in the forkhead domain that ablate Foxp3 function in Treg cells also inhibited Foxp3 association with and inhibition of HDAC1. Finally, Foxp3 failed to enhance HIV-1 gene expression in human T cells expressing HDAC1-specific shRNA. We conclude that Foxp3 modulates gene expression in human T cells at least partly by inhibiting HDAC1 activity.
Keywords: Foxp3, HDAC, HIV-1, LTR, Treg
Introduction
Regulatory T cells (Treg) control autoimmunity induced by neonatal thymectomy (Sakaguchi et al., 1995). The forkhead winged-helix family member FoxP3 is identified as the essential transcription factor for Treg development and function (Fontenot et al., 2003; Khattri et al., 2003). In response to T cell stimulation, Foxp3+CD4+ Treg cells fail to produce IL2 or proliferate in vitro. Recent studies have revealed multiple mechanisms of Foxp3-mediated repression of the IL-2 gene (Ziegler, 2006). FoxP3 is reported to interact with the T cell-specific transcription factor NFAT to displace the NFAT/AP-1 complex at the IL-2 enhancer (Wu et al., 2006). A Foxp3/Runx complex binding at a distal IL2 promoter region also contributes to Foxp3-mediated IL-2 repression (Ono et al., 2007).
Although described as a transcriptional repressor of effector cytokines in T cells, Foxp3 also enhances expression of genes associated with Treg functions (Fontenot et al., 2005; Holmes et al., 2007). Interestingly, Treg cells support higher levels of HIV-1 or FIV infection than conventional CD4+ T cells in vitro (Joshi et al., 2005; Oswald-Richter et al., 2004). We have recently reported that in the humanized mouse model, CD4+FoxP3+ T cells in lymphoid organs are also preferentially infected by HIV-1 in vivo (Jiang et al., 2008). We have also reported that HIV gene expression is enhanced in human CD4+ T cells ectopically expressing FoxP3 (Holmes et al., 2007). This enhancement requires NF-κB sites in the LTR, associated with increased histone 3 acetylation at the LTR (Holmes et al., 2007).
HDAC inhibitors such as TSA repress the IL-2 promoter but enhance HIV LTR (Laughlin et al., 1993; Moreira et al., 2003), similarly as Foxp3 (Holmes et al., 2007). In this study we report that TSA failed to further enhance HIV gene expression in Foxp3+ T cells. We showed that Foxp3 specifically inhibited the activity of HDAC1. In addition, Foxp3 failed to enhance HIV-1 gene expression in human T cells expressing HDAC1-specific shRNA.
Results
Foxp3 modulates HIV-1 gene expression via HDAC-related mechanisms
To determine the role of HDACs in FoxP3-mediated gene regulation, Jurkat T cells transduced with control or Foxp3 vector (>95% efficiency) were infected with VSV-g pseudotyped HIV-1 NL4-luciferase reporter virus. Treatment of these cells at 24 hours post infection with the HDAC inhibitor TSA resulted in a 5-6 fold increase in HIV-1 gene expression in unstimulated or CD3/CD28 stimulated T cells (Fig. 1A/1B). As we have previously reported (Holmes et al., 2007), Foxp3 expression also enhanced LTR activity. However, addition of TSA to FoxP3-expressing human T cells did not further enhance LTR activity (Fig. 1A/B), suggesting an overlapping role of HDAC inhibition and FoxP3 in activation of the HIV LTR promoter. We next determined the repression of IL-2 by TSA and FoxP3. Jurkat cells were transduced with vector or FoxP3 retrovirus and stimulated with PMA and Ionomycin for 16 hours in the presence or absence of TSA 50 or 200 nM). High dose TSA inhibited IL-2 expression about 4 fold, to similar levels seen with Foxp3 expression. In the presence of TSA, FoxP3 marginally repressed IL-2 expression (Fig. 1C). To confirm this finding to the IL-2 promoter, Jurkat T cells were transfected with IL-2 luciferase plasmid in the presence or absence of TSA. FoxP3 inhibited the IL-2 promoter by 2-3 fold, and TSA similarly inhibited IL-2 promoter. Addition of both TSA and FoxP3 showed only slight further repression (Fig. 1D). Therefore the activation of LTR or inhibition of IL-2 by FoxP3 and TSA is likely through an overlapping pathway.
Figure 1. HDAC inhibitor TSA does not further enhance HIV-1 gene expression in FoxP3-expressing T cells.
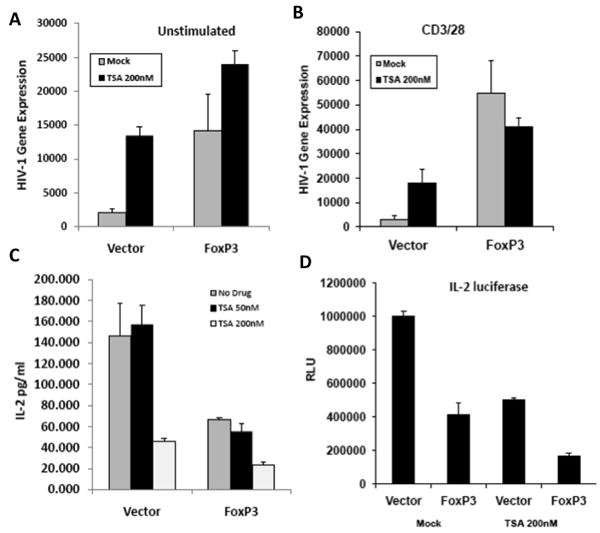
Jurkat cells transduced with control vector or FoxP3 (>95%) were infected with VSV-g pseudotyped HIV-NL4-luciferase reporter virus. At 24 hours post infection, cells were left unstimulated (A) or activated (B) with anti-CD3/CD28 mAb, and treated with mock or TSA for 24 hours. HIV-1 gene expression (luciferase activity) was determined at 48 hpi. Data represent summarized results from >3 experiments. Error bars indicate SD of triplicate samples, and * indicates p<0.05. (C) Both TSA and FoxP3 inhibit IL-2 gene expression. Jurkat cells are transfected with empty vector or FoxP3 and treated with 50 or 200uM TSA and activated with PMA and Ionomycin for 16 hours. IL-2 expression in the supernatant was measured. (D) Luciferase driven by the IL-2 promoter was co-transfected with FoxP3 or control vector in Jurkat cells. Cells were activated, lysed and luciferase was determined. Data represents multiple experiments done in triplicate.
Foxp3 expression reduces HDAC activity in human T cells
The data above suggest that FoxP3 may affect HDAC activity to modulate gene expression. Utilizing a live cell permeable HDAC substrate, we first measured HDAC activity in purified human CD4+CD25- (Th) and CD4+CD25+ (Treg) populations. In resting T cells, both Th and Treg cells expressed similar low levels of HDAC activity (data not shown). After activation with CD3 and CD28 antibodies, HDAC activity was elevated about 5 fold in Th cells. However, the HDAC activity was lower in activated Treg cells than in Th cells (20-30%, Fig. 2A). Thus primary human Treg cells had lower HDAC activity. We next determined the effect of Foxp3 on cellular HDAC deacetylase activity. Foxp3 expression in Jurkat T cells significantly decreased cellular HDAC activity (about 20%, Fig. 2B). Thus Foxp3 expression was associated with reduced HDAC activity in human T cells.
Figure 2. Foxp3 inhibits HDAC activity in human T cells.
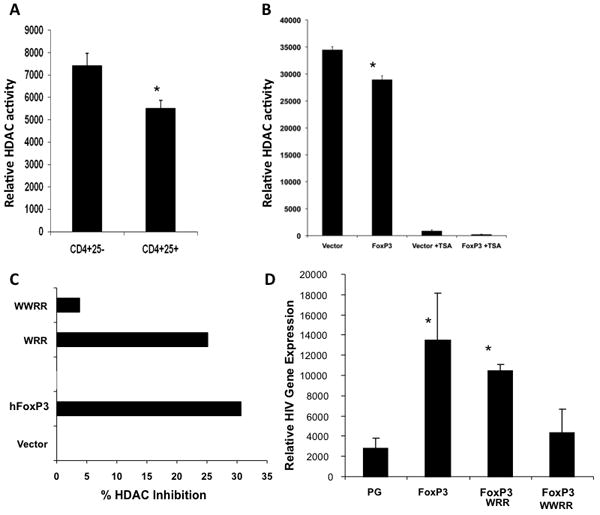
(A) Primary human Th (CD4+CD25-) and Treg (CD4+ CD25+) cells sorted from PBMC’s were activated with anti-CD3/CD28 mAb. Cells-associated HDAC activity was assessed with cell permeable HDAC substrate as described in methods. Total cellular HDAC activity in equal number of Th or Treg cells is presented. Data are representative of 3 independent experiments, each done in triplicate. Error bars indicate SD of triplicate samples, and * indicates p<0.05. (B) Jurkat cells were transduced with control or Foxp3 vector. Total cellular HDAC activity in equal number of cells is presented. TSA was added to show TSA-sensitive HDAC activity. (C) Foxp3 point mutants were expressed and HDAC activity was measured. Data represent three independent experiments. (D) FoxP3 WWRR mutant was unable to enhance HIV gene expression or to inhibit HDAC activity in human T cells. Jurkat T cells transduced (>95%) with wild type, WRR and WWRR mutant Foxp3 genes were infected with NL4-luc reporter virus and assayed for luciferase activity. Error bars indicate SD of triplicate samples, and * indicates significant differences over PG vector (p<0.05).
To define the functional domains of Foxp3 required to inhibit HDAC, a number of Foxp3 mutants with deletion and point mutations were tested. Interestingly, the WWRR mutant with mutations in the Foxp3 forkhead domain that inhibit Foxp3/NFAT interaction and Foxp3 activity (Wu et al., 2006) also was unable to inhibit HDAC activity (Fig. 2C). The Foxp3 WRR mutant, which has slightly reduced function in vivo compared to wild type Foxp3, retained HDAC inhibition activity, albeit slightly lower than wild type Foxp3 (Fig. 2C). To correlate the relative activity in activating HIV-1 gene expression in T cells, the WRR and WWRR mutants were expressed and tested in Jurkat T cells infected with HIV-luciferase reporter virus. Similar to their relative activity to inhibit HDAC activity, HIV enhancement was also impaired by the WWRR mutations but not by the WRR mutations (Fig. 2D). Therefore, critical residues in the forkhead domain of Foxp3 are required for HDAC inhibition, IL2 inhibition and HIV activation by Foxp3.
Foxp3 specifically inhibits HDAC1 activity
We next determined the specific HDAC that was inhibited by Foxp3. Since class I HDACs are responsible for most, if not all, of the TSA-sensitive HDAC enzymatic activity (Lahm et al., 2007), we expressed Foxp3 and various class I (HDAC1, 2 and 3) and one class II HDAC (HDAC4) in 293T cells. Flag-tagged HDAC1, 2, 3 and HDAC4 were immuno-precipitated from equal amounts of cell lysates in the presence or absence of Foxp3. HDAC deacetylase activity in the precipitates was assessed with an acetylated substrate of class I and II HDACs. Foxp3 was able to efficiently inhibit HDAC1 (~40% inhibition) but not HDAC 2-4 (Fig. 3A, B). The difference was not due to differences in HDAC expression or immuno-precipitation efficiency, as demonstrated by Western blot analysis of the IP’ed HDAC1-4 proteins (Fig. 3A). To confirm that inhibition of HDAC1 occurred in human T cells, Foxp3-dependent HDAC inhibition was assayed in Jurkat cells expressing HDAC1 by retroviral transduction. Ectopic expression of HDAC1 in T cells increased the cellular HDAC activity, and Foxp3 was able to inhibit HDAC1-related deacetylase activity (Fig. 3C). The Foxp3 inhibition of HDAC1 activity was not due to reduced HDAC1 expression or its nuclear translocation, because nuclear levels of HDAC1 in Jurkat cells were not affected by Foxp3 expression (Fig. 3D).
Figure 3. Foxp3 inhibits HDAC1 activity.
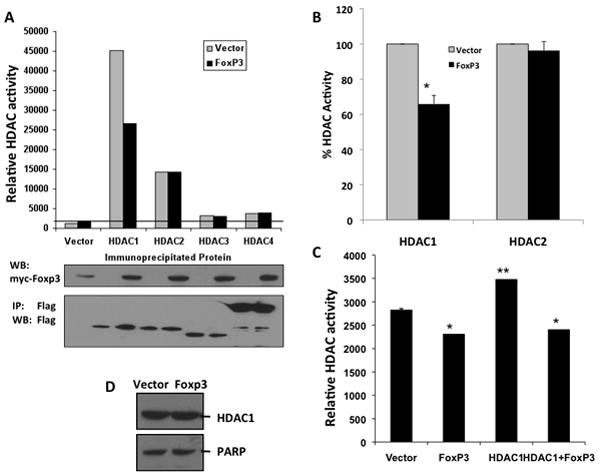
(A) Flag-tagged HDAC1-4 are cotransfected into 293T cells with vector or Foxp3. HDACs were IPed with anti-Flag mAb and HDAC activity was measured. HDAC1 (but not HDAC2-4) activity is specifically inhibited by Foxp3 coexpression. Western blots show relative IP of each HDAC and FoxP3 expression. (B) A summary of 3 independent experiments with HDAC1 and HDAC2 in 293T cells co-transfected with Foxp3. Error bars indicate SD, and * indicates significant differences (p<0.05). (C) Foxp3 inhibits HDAC1 activity in Jurkat T cells. Jurkat cells transduced with vector or Foxp3 in the presence or absence of HDAC1 expression were analyzed for HDAC activity. Error bars indicate SD. * indicates significantly lower than vector control, and ** indicates significantly higher than control (p<0.05). (D) HDAC1 nuclear levels are unchanged by Foxp3. Nuclear lysates of Jurkat cells transduced with vector or Foxp3 are assayed for HDAC1 protein levels by western blot for HDAC1, with PARP as controls.
Foxp3 interacts with HDAC1 via critical domains
There are several mechanisms of HDAC regulation, including posttranslational modification and association with a multi-subunit protein complex. We wanted to determine if Foxp3 interacts with HDAC1 to regulate its deacetylase activity. To test if Foxp3 and HDAC1 are associated in vivo, 293T cells were transfected with Foxp3 with or without HDAC1. When HDAC1 was IP’ed with an antibody, Foxp3 was co-IP’ed (Fig. 4A). Similarly, Foxp3 in Jurkat cells was associated with endogenous HDAC1 (Fig. 4B). To map the domain of Foxp3 required for HDAC1 interaction, 293T cells were transfected with Foxp3 or successive N-terminal truncation mutants of Foxp3 deleting the Proline-rich domain (Foxp3-C), the zinc-finger (Foxp3-LZ-FKH), and all domains up to the forkhead (Foxp3-FKH). The FKH domain of Foxp3 was sufficient to associate with HDAC1. Importantly, HDAC1 association with Foxp3 was inhibited by the WWRR mutations in the FKH domain, but not by the WRR mutations (Fig. 4C). Thus the WWRR mutant, which is defective in modulating gene expression, was unable to associate with HDAC1 and unable to inhibit intracellular HDAC activity. The data suggest that the interaction of HDAC1 with Foxp3 was required for Foxp3-mediated gene modulation in T cells.
Figure 4. Foxp3 interacts with HDAC1.
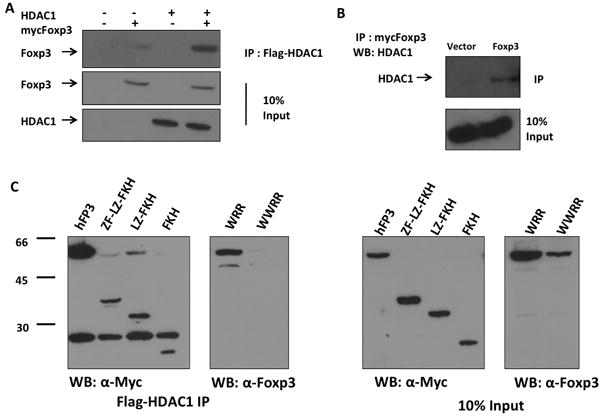
(A) 293T cells cotransfected with flag-HDAC1 and myc-tagged Foxp3 were Immunoprecipitated with anti-Flag antibody. (B) Jurkat cells transduced with myc-Foxp3 were Immunoprecipitated with anti-myc antibody and probed for endogenous HDAC1. (C) Forkhead mutation ablates Foxp3:HDAC1 interaction. 293T cells were transfected with myc-Foxp3 mutants with deletion or point mutations and Flag-HDAC1. HDAC1 was immunoprecipitated with anti-Flag antibody and Foxp3 was visualized by western blot. Total 10% of input cell lysate was included as controls. ZF-LZ-FKH, Myc-tagged FoxP3 with the N-terminal 177 amino acids deleted; LZ-FKH, Myc-tagged FoxP3 with the N-terminal 230 amino acids deleted; FKH, Myc-tagged FoxP3 with the N-terminal 311 amino acids deleted; WRR and WWRR are full-length FoxP3 mutants with point mutations in the FKH domain (5).
Foxp3 fails to enhance HIV gene expression in human T cells expressing HDAC1-specific shRNA
To test the role of HDAC1 in FoxP3-mediated gene regulation, we first tested the effect of inhibiting HDAC1 expression in Jurkat T cells. Vector or FoxP3-expressing Jurkat T cells were stably transduced with lentiviral vectors expressing HDAC1-shRNA or control shRNA (>95% transduction), and infected with VSV-G pseudo-typed HIV-luciferase reporter virus. Under conditions where HDAC1 levels were knockdown (~50%, Fig. 5A inset), FoxP3 failed to significantly enhance HIV gene expression in Jurkat T cells expressing the HDAC1-specific shRNA (Fig. 5A). Under the same condition, Foxp3 failed to significantly inhibit IL2 gene expression in Jurkat T cells expressing the HDAC1-shRNA (data not shown). In primary human CD4 T cells, we co-transfected the plasmid DNA that express Foxp3, a control shRNA or HDAC1-specific shRNA and NL4-luciferase in purified CD4+ T cells (about 30% transfection as determined by GFP). The transfected cells were activated and analyzed for HIV gene expression. Either Foxp3 or HDAC1-shRNA significantly enhanced HIV gene expression. As in Jurkat T cells, Foxp3 failed to enhance HIV gene expression in primary human T cells with the HDAC1-shRNA (Fig. 5C). Therefore, Foxp3 interacted with HDAC1 to modulate HIV-1 gene expression in human T cells.
Figure 5. Foxp3 fails to enhance HIV gene expression in human T cells with reduced HDAC1 expression.
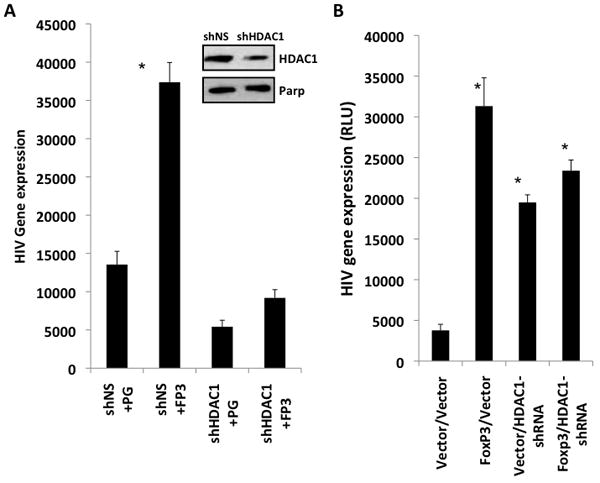
(A) Jurkat T cells were transduced with FoxP3 gene or control vector, followed with shRNA-HDAC1 (shHDAC1) or non-specific shRNA control (shNS) vector transduction (both >90% frequency). Western blot showed about 50% inhibition of HDAC1 expression (relative to PARP, inset). Cells were then infected with VSV-G pseudo-typed NL-4 luciferase virus and HIV gene expression was measured by luciferase expression. Results shown are representative of at least 3 independent experiments each done in triplicate. * p<.05, using student t-test. (B) Primary human resting CD4+ T cells were nucleofected with HDAC1 shRNA or a control shRNA, and with Foxp3 and the NL4-luciferase reporter provirus. Transfected cells were stimulated with anti-CD3/CD28 mAb at 3-4 days post transfection and HIV gene expression was measured by luciferase expression at 4-5 days post transfection. HIV-directed luciferase gene expression is shown. Data are derived from a representative experiment in triplicates. Three independent experiments were performed with similar results. * p<0.05 compared to vector controls.
Discussion
We report here that Foxp3 may function as an HDAC inhibitor to significantly enhance HIV or inhibit IL2 gene expression in T cells. HDACi TSA failed to enhance HIV-1 gene expression in Foxp3-expressing T cells. We found that human CD4+CD25+ Treg had lower levels of HDAC activity than CD4+CD25- Th cells, and ectopic expression of Foxp3 in Jurkat T cells also resulted in a decrease in cellular HDAC activity. Foxp3 specifically inhibited HDAC1 when coexpressed in 293T or Jurkat T cells, correlated with association of Foxp3 and HDAC1. Critical residues in the forkhead domain of Foxp3 were required for HDAC inhibition, association and HIV activation by Foxp3. Finally, we report that Foxp3 failed to significantly enhance HIV gene expression in human T cells with reduced HDAC1 expression.
HDAC inhibitor TSA has been proposed to relieve LTR repression by increased acetylation of histones and “relaxing” the chromatin to enhance gene expression (Laughlin et al., 1993; Steger et al., 1998; Van Lint et al., 1996). Binding of Foxp3 to target promoters is associated with changes in histone acetylation (Holmes et al., 2007; Wu et al., 2006). Thus, Foxp3 inhibition of HDAC1 may increase LTR histone acetylation. However, several reports have shown that HDACi can either activate (Chen et al., 2001; Quivy et al., 2002) or repress (Rahman et al., 2003; Yin et al., 2001) NFκB activity depending on the cell type and the context of the promoter environment. TSA-mediated activation of the HIV LTR also requires the NFκB binding sequences. We demonstrate that Foxp3 enhances HIV-1 LTR through an NFκB-dependent mechanism in T cells (Holmes et al., 2007). Foxp3 may inhibit HDAC1 to modulate the activity of NFκB at the LTR promoter, likely in cell type-, cell activation condition- and promoter-specific fashions.
It has been reported that Foxp3 can inhibit HIV LTR activity via the NFκB/NFAT sites (Grant et al., 2006; Selliah et al., 2008). The discrepancy between different reports regarding Foxp3 modulation of HIV gene expression may be due to differences in activation conditions and in specific experimental systems. Infection of resting T cells or activation following HIV-1 infection of resting T cells show Foxp3 enhancement of HIV gene expression (this report and (Holmes et al., 2007)). In contrast, Foxp3 in T cells activated prior to HIV-1 infection appears to lead to inhibition of HIV replication by Foxp3 (Grant et al., 2006; Selliah et al., 2008). This is consistent with our recent finding that HIV-1 preferentially infects and replicates in Foxp3+ T cells in humanized mouse model in vivo (Jiang et al., 2008), where human T cells are not activated prior to HIV-1 infection.
Although HDAC activity has been proposed to be involved in Treg cell function, no link has been established between Foxp3 and HDCA1. A HAT/HDAC complex including Foxp3/Tip60/HDAC7 is required for IL-2 repression by Foxp3 (Miller-Jensen et al., 2007); but it is not clear if Foxp3 modulates the activity of HDAC7. In addition, mouse CD4+CD25+ Treg cells have higher HDAC9 expression than CD4+25- cells; and Treg cells from HDAC9-/- mice are more suppressive, pointing towards a role of this class II HDAC in Treg function (Tao et al., 2007). It is likely that mouse Treg cells are distinct in HDAC expression from human Treg cells.
The biochemical mechanism by which Foxp3 inhibits HDAC1 activity is currently unclear. Our results showed that Foxp3 was associated, directly or indirectly, with HDAC1 in T cells (Fig. 4). However, co-expression of the two proteins by in vitro transcription-translation failed to demonstrate either direct binding or inhibition of HDAC1 activity by Foxp3 (D. Holmes and L. Su, Unpublished results). Since the nuclear level of HDAC1 is unchanged upon Foxp3 expression (Fig. 3D), we can rule out regulation of HDAC1 expression, stability or nuclear translocation by Foxp3. HDAC activity is regulated by posttranslational modification, association with multi-subunit complexes, and cofactor availability (Sengupta and Seto, 2004). Foxp3 is associated with a large molecular weight complex that includes Brg-1 and MBD3, components of the SWI/SNF and Mi2/NuRD complex respectively (Li et al., 2007). The Mi-2/NuRD complex also contains HDAC1, suggesting that FoxP3 may alter HDAC1 activity in association with specific complexes. It will be of future interest to elucidate the biochemical mechanism by which Foxp3 inhibits HDAC1 activity in association with HDAC1-containing complexes.
Materials and Methods
Plasmids, cells and antibodies
FoxP3 and deletion mutants in the HSPG retrovirus vector, and IL2-promoter driven luciferase were cloned as previously described (Holmes et al., 2007). Flag-tagged HDAC1-4 plasmids were provided by Dr. Tso-Pang Yao at Duke University. Foxp3 mutants WRR and WWRR were a kind gift from Dr. Ajana Rao at Harvard Medical School (Wu et al., 2006). The NL4-luc-R-E- reporter plasmid was provided by Dr. N. Landau at NYU (15). Jurkat cells were maintained in RPMI 1640 (Gibco BRL), 293T cells were maintained in DMEM and primary human T cells were maintained in Iscoves MEM, supplemented with 10% FBS (Sigma), 2mM L-glutamine, 100 U/ml penicillin, and 100 mg/ml Streptomycin. CD4+ primary T cells were purified from Ficoll separated PBMC by negative selection (>95% purity) (MACS human CD4+ T cell isolation kit) and CD4+CD25+ Regulatory T cells were purified by MACS human Regulatory T cells isolation kit (Miltenyi-Biotech, Auburn, CA). CD4+CD25+ cell purity was > 95%. T cells were activated by anti-CD3/CD28 mAb cross linked by plate-bound anti-mouse IgG as reported (Holmes et al., 2007). Antibodies used in Western Blot include anti-Flag M2 (Sigma), anti-myc monoclonal 9E10 and anti-HDAC1 polyclonal rabbit antibody (Santa Cruz).
Retrovirus production and transduction
293T cells were co-transfected with retroviral vector, VSV-G and gag/pol containing plasmids as described (Coffield et al., 2003). For retroviral transduction, Jurkat cells were spin-inoculated with retroviruses for 3 hours at 5,500 rpm with polybrene (8ug/ml). Transduction of >95% efficiency was obtained as measured by GFP expression.
Transduction of Jurkat T cells and luciferase assay
For production of single cycle HIV-Luc reporter virus, 293T cells were transfected with VSV-G and NL4-HIV-Luc (He and Landau, 1995) plasmids by Effectene (Qiagen, Valencia, CA). Jurkat cells were transduced with the VSV-g HIV-luciferase reporter viruses. Briefly, cells previously transduced with vector or Foxp3 were infected with NL4-luciferase reporter virus. At 24 hours post infection, Jurkat cells were activated with anti-CD3/CD28 for 24 hours. Cells were lysed in 1x reporter lysis buffer (promega) and luciferase expression was determined by Luciferase Assay System (Promega). Experiments were done in triplicates and repeated at least three times.
293T cell transfection, Immunoprecipitation and Western Blot
293T cells were transfected and fractionated into cytoplasmic and nuclear lysates and quantitated by BCA protein assay kit (Pierce). 500ug nuclear lysates were incubated overnight with 2ug anti-Flag antibody followed by Protein A bead incubation. For HDAC assay, the immunoprecipitated complex was washed 3X with PBS and HDAC activity was analyzed by Flour De Lys HDAC assay with HDAC substrate in HDAC assay buffer for 30 minutes at 37°C and developed (Biomol, Plymouth Meeting, PA). For Western, FoxP3 was probed with an anti-FoxP3 antiserum (Jiang et al., 2006) followed by secondary α-rabbit–IgG–HRP (1:10,000) (Amersham Biosciences) and visualized by ECL(Amersham Biosciences).
Flourimetric Cellular HDAC Activity Assay
Jurkat or primary T cells were incubated with Flour de lys intracellular HDAC substrate for 2 hours at 37°C. For HDAC assay in primary T cells, purified CD4 Th or Treg cells were stimulated for 2-5 days and assayed under similar conditions as Jurkat T cells. Experiments were done in triplicate and are repeated 3 times.
Primary T cell purification and transfection
CD4+ primary T cells were purified from Ficoll separated PBMC by MACS human CD4+ T cell isolation kit (>95% purity, Miltenyi-Biotech, Auburn, CA). Purified primary CD4+ T cells were transfected by Amaxa nucleofector kit (Amaxa Biosystems, Gaithersburg, MD). Briefly, 5×106 unstimulated CD4+ T cells were transfected with 5ug plasmid DNA (20-40% efficiency by GFP expression), and cultured for 48 hours before activation by CD3 and CD28 crosslinked with plate-bound IgG. Cells were activated for 48 hours and HIV driven or IL2 drive luciferase was measured.
Statistical analysis
For statistical analysis, a two-tailed Student t-test was employed where p<0.05 was considered significant.
Acknowledgments
We thank Drs. T.P. Yao (Duke University) and A. Rao (Harvard University) for providing critical reagents; Drs. Y. Zhang, A. Baldwin and B. Damania for critical discussions; G. Hench for critical reading of the manuscript; and Dedeke Brouwer, Liqun Chi and T. Anthony Curtis for technical support.
The project was supported by a grant from NIH (AI077454 to L. S.).
Footnotes
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Chen L, Fischle W, Verdin E, Greene WC. Duration of nuclear NF-kappaB action regulated by reversible acetylation. Science. 2001;293:1653–1657. doi: 10.1126/science.1062374. [DOI] [PubMed] [Google Scholar]
- Coffield VM, Jiang Q, Su L. A genetic approach to inactivating chemokine receptors using a modified viral protein. Nat Biotechnol. 2003;21:1321–1327. doi: 10.1038/nbt889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- Fontenot JD, Rasmussen JP, Williams LM, Dooley JL, Farr AG, Rudensky AY. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 2005;22:329–341. doi: 10.1016/j.immuni.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Grant C, Oh U, Fugo K, Takenouchi N, Griffith C, Yao K, Newhook TE, Ratner L, Jacobson S. Foxp3 represses retroviral transcription by targeting both NF-kappaB and CREB pathways. PLoS Pathog. 2006;2:e33. doi: 10.1371/journal.ppat.0020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Landau NR. Use of a novel human immunodeficiency virus type 1 reporter virus expressing human placental alkaline phosphatase to detect an alternative viral receptor. J Virol. 1995;69:4587–4592. doi: 10.1128/jvi.69.7.4587-4592.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes D, Knudsen G, Mackey-Cushman S, Su L. FoxP3 enhances HIV-1 gene expression by modulating NFkappaB occupancy at the long terminal repeat in human T cells. J Biol Chem. 2007;282:15973–15980. doi: 10.1074/jbc.M702051200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q, Su H, Knudsen G, Helms W, Su L. Delayed functional maturation of natural regulatory T cells in the medulla of postnatal thymus: role of TSLP. BMC Immunol. 2006;7:6. doi: 10.1186/1471-2172-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q, Zhang L, Wang R, Jeffrey J, Washburn ML, Brouwer D, Barbour S, Kovalev GI, Unutmaz D, Su L. FoxP3+CD4+ regulatory T cells play an important role in acute HIV-1 infection in humanized Rag2-/-gammaC-/- mice in vivo. Blood. 2008;112:2858–2868. doi: 10.1182/blood-2008-03-145946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi A, Garg H, Tompkins MB, Tompkins WA. Preferential feline immunodeficiency virus (FIV) infection of CD4+ CD25+ T-regulatory cells correlates both with surface expression of CXCR4 and activation of FIV long terminal repeat binding cellular transcriptional factors. J Virol. 2005;79:4965–4976. doi: 10.1128/JVI.79.8.4965-4976.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khattri R, Cox T, Yasayko SA, Ramsdell F. An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nat Immunol. 2003;4:337–342. doi: 10.1038/ni909. [DOI] [PubMed] [Google Scholar]
- Lahm A, Paolini C, Pallaoro M, Nardi MC, Jones P, Neddermann P, Sambucini S, Bottomley MJ, Lo Surdo P, Carfi A, et al. Unraveling the hidden catalytic activity of vertebrate class IIa histone deacetylases. Proc Natl Acad Sci U S A. 2007;104:17335–17340. doi: 10.1073/pnas.0706487104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughlin MA, Zeichner S, Kolson D, Alwine JC, Seshamma T, Pomerantz RJ, Gonzalez-Scarano F. Sodium butyrate treatment of cells latently infected with HIV-1 results in the expression of unspliced viral RNA. Virology. 1993;196:496–505. doi: 10.1006/viro.1993.1505. [DOI] [PubMed] [Google Scholar]
- Li B, Samanta A, Song X, Iacono KT, Brennan P, Chatila TA, Roncador G, Banham AH, Riley JL, Wang Q, et al. FOXP3 is a homo-oligomer and a component of a supramolecular regulatory complex disabled in the human XLAAD/IPEX autoimmune disease. Int Immunol. 2007;19:825–835. doi: 10.1093/intimm/dxm043. [DOI] [PubMed] [Google Scholar]
- Miller-Jensen K, Janes KA, Brugge JS, Lauffenburger DA. Common effector processing mediates cell-specific responses to stimuli. Nature. 2007;448:604–608. doi: 10.1038/nature06001. [DOI] [PubMed] [Google Scholar]
- Moreira JM, Scheipers P, Sorensen P. The histone deacetylase inhibitor Trichostatin A modulates CD4+ T cell responses. BMC Cancer. 2003;3:30. doi: 10.1186/1471-2407-3-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono M, Yaguchi H, Ohkura N, Kitabayashi I, Nagamura Y, Nomura T, Miyachi Y, Tsukada T, Sakaguchi S. Foxp3 controls regulatory T-cell function by interacting with AML1/Runx1. Nature. 2007;446:685–689. doi: 10.1038/nature05673. [DOI] [PubMed] [Google Scholar]
- Oswald-Richter K, Grill SM, Shariat N, Leelawong M, Sundrud MS, Haas DW, Unutmaz D. HIV Infection of Naturally Occurring and Genetically Reprogrammed Human Regulatory T-cells. PLoS Biol. 2004;2:E198. doi: 10.1371/journal.pbio.0020198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivy V, Adam E, Collette Y, Demonte D, Chariot A, Vanhulle C, Berkhout B, Castellano R, de Launoit Y, Burny A, et al. Synergistic activation of human immunodeficiency virus type 1 promoter activity by NF-kappaB and inhibitors of deacetylases: potential perspectives for the development of therapeutic strategies. J Virol. 2002;76:11091–11103. doi: 10.1128/JVI.76.21.11091-11103.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman MM, Kukita A, Kukita T, Shobuike T, Nakamura T, Kohashi O. Two histone deacetylase inhibitors, trichostatin A and sodium butyrate, suppress differentiation into osteoclasts but not into macrophages. Blood. 2003;101:3451–3459. doi: 10.1182/blood-2002-08-2622. [DOI] [PubMed] [Google Scholar]
- Sakaguchi S, Sakaguchi N, Asano M, Itoh M, Toda M. Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor alpha-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J Immunol. 1995;155:1151–1164. [PubMed] [Google Scholar]
- Selliah N, Zhang M, White S, Zoltick P, Sawaya BE, Finkel TH, Cron RQ. FOXP3 inhibits HIV-1 infection of CD4 T-cells via inhibition of LTR transcriptional activity. Virology. 2008;381:161–167. doi: 10.1016/j.virol.2008.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta N, Seto E. Regulation of histone deacetylase activities. J Cell Biochem. 2004;93:57–67. doi: 10.1002/jcb.20179. [DOI] [PubMed] [Google Scholar]
- Steger DJ, Eberharter A, John S, Grant PA, Workman JL. Purified histone acetyltransferase complexes stimulate HIV-1 transcription from preassembled nucleosomal arrays. Proc Natl Acad Sci U S A. 1998;95:12924–12929. doi: 10.1073/pnas.95.22.12924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao R, de Zoeten EF, Ozkaynak E, Chen C, Wang L, Porrett PM, Li B, Turka LA, Olson EN, Greene MI, et al. Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat Med. 2007;13:1299–1307. doi: 10.1038/nm1652. [DOI] [PubMed] [Google Scholar]
- Van Lint C, Emiliani S, Verdin E. The expression of a small fraction of cellular genes is changed in response to histone hyperacetylation. Gene Expr. 1996;5:245–253. [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Borde M, Heissmeyer V, Feuerer M, Lapan AD, Stroud JC, Bates DL, Guo L, Han A, Ziegler SF, et al. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell. 2006;126:375–387. doi: 10.1016/j.cell.2006.05.042. [DOI] [PubMed] [Google Scholar]
- Yin L, Laevsky G, Giardina C. Butyrate suppression of colonocyte NF-kappa B activation and cellular proteasome activity. J Biol Chem. 2001;276:44641–44646. doi: 10.1074/jbc.M105170200. [DOI] [PubMed] [Google Scholar]
- Ziegler SF. FOXP3: of mice and men. Annu Rev Immunol. 2006;24:209–226. doi: 10.1146/annurev.immunol.24.021605.090547. [DOI] [PubMed] [Google Scholar]


