Abstract
Protein quality control (PQC) senses and repairs misfolded/unfolded proteins and, if the repair fails, degrades the terminally misfolded polypeptides through an intricate collaboration between molecular chaperones and targeted proteolysis. Proteolysis of damaged proteins is performed primarily by the ubiquitin-proteasome system (UPS). Macroautophagy (commonly known as autophagy) may also play a role in PQC-associated proteolysis, especially when UPS function becomes inadequate. The development of a range of heart diseases, including bona fide cardiac proteinopathies and various forms of cardiac dysfunction has been linked to proteasome functional insufficiency (PFI). Both PFI and activation of autophagy have been observed in the heart of well-established mouse models of cardiac proteinopathy. A causal relationship between PFI and autophagic activation was suggested by a study using cultured cardiomyocytes but has not been established in the heart of intact animals. Taking advantage of an autophagy reporter, we demonstrated here that pharmacologically induced proteasome inhibition is sufficient to activate autophagy in cardiomyocytes in both intact animals and cell cultures, unveiling a potential cross-talk between the two major degradation pathways in cardiac PQC.
Keywords: Proteasome, autophagy, autophagic flux, cardiomyocytes, mice
Introduction
Protein quality control (PQC) is a basic cellular process that functions to assist protein (re) folding, protect nascent or unfolded polypeptides from aggregating, and selectively degrade terminally misfolded polypeptides [1]. Like in other cells, PQC plays an indispensable role in maintaining protein homeostasis (also referred to as proteostasis) in cardiomyocytes. PQC senses and repairs misfolded/unfolded proteins and, if the repair fails, degrades the terminally misfolded polypeptides through an intricate collaboration between molecular chaperones and targeted proteolysis. Proteolysis of damaged proteins is performed primarily by the ubiquitin-proteasome system (UPS) [1]. But recent studies suggest that macroautophagy (commonly known as autophagy) may also play a role in PQC-associated proteolysis, especially when UPS function becomes inadequate [2]. The development of a range of heart diseases, including bona fide cardiac proteinopathies and various forms of cardiac dysfunction has been linked to proteasome functional insufficiency (PFI) [2, 3]. Meanwhile, proteasome inhibition is clinically employed to treat certain types of cancer and has shown promising efficacy but severe adversary effects on the heart have been reported [3]. Hence, it is important to have a better understanding of how cardiomyocytes and the heart respond to proteasome inhibition/malfunction.
As the two major intracellular protein degradation pathways, the UPS and the autophagy-lysosome pathway are responsible for the clearance of proteins and organelles that are no longer needed or unwanted in eukaryotic cells [4]. Earlier studies defined proteins into two categories including “short-lived” and “long-lived” proteins based on their degradation kinetics [5]. The UPS predominantly degrades shortlived normal protein molecules after they have fulfilled their duty in the cell [6]. On the other hand, autophagy is primarily responsible for degrading long-lived proteins [7]. Notably, the distinction of substrate preference between the two proteolytic systems is relative. Recent studies indicate that the UPS can participate in the degradation of long-lived proteins while autophagy can also be involved in the degradation of short-lived proteins [5, 8].
An alternative way to categorize proteolytic mechanisms is based on function rather than the degradation kinetics. From the functional point of view, the UPS degrades two types of proteins: the fully functional proteins which are degraded as a regulatory mechanism, such as proteins involved in regulation of cell division, gene transcription, signal transduction, and endocytosis; and abnormal proteins, thereby serving as a critical step of posttranslational PQC in the cell [1, 9]. Autophagy also degrades two types of proteins: functional proteins which are degraded in a bulk fashion as a nutrient recycling mechanism during starvation, and perhaps aggregated misfolded proteins or defective organelles. It appears that these two degradation systems cooperate with each other to degrade the misfolded proteins [10]. Misfolded cytosolic proteins are partitioned into two distinct PQC compartments. Soluble misfolded proteins accumulate in a juxtanuclear compartment preferentially for degradation by the UPS [11]. In contrast, terminally aggregated proteins are partitioned in a perivacuolar inclusion and are mainly degraded by autophagy because they cannot be efficiently degraded by the proteasome [10, 11]. However, the interplay between the UPS and autophagy in normal and diseased hearts remains poorly understood.
Using a surrogate substrate reporter system, we have previously revealed PFI in the heart of two bona fide mouse models of cardiac proteinopathy [12, 13]. Aberrant protein aggregation was subsequently shown to play a critical role in the proteasome impairment induced by expression of misfolded proteins in cardiomyocytes [13, 14]. More recently, we reported that autophagy is adaptively activated in the heart of the well-established mouse model of cardiac proteinopathy and this is accompanied by upregulation of p62 [15]. The p62 is a substrate of autophagy and its increase is often used as an indicator of decreased autophagic flux. The paradoxical increase of p62 during autophagic activation in cardiac proteinopathy occurs at the transcription level and may in fact mediate the autophagic activation in the cell under a proteotoxic stress [15]. Interestingly, a previous report showed in cultured cardiomyocytes that pharmacological inhibition of the proteasome increased simultaneously autophagosomes and p62 in cultured cardiomyocytes [16]. Indeed, it has been shown in non-cardiac cells that p62 is a target gene of the Keap1-Nrf2 regulatory pathway [17]. Keap1, a BTB protein, serves as the substrate-specific adaptor in a Cullin3-based ubiquitin ligase for Nrf2. Under unstressed conditions, Keap1 constitutively binds Nrf2 and targets Nrf2 for ubiquitination and proteasome-mediated degradation [18, 19]. Upon oxidative stress, dissociation of Keap1 from Nrf2 stabilizes Nrf2 and allows Nrf2 translocate to the nucleus to activate its target genes [20]. More recent studies further demonstrated that p62 can disrupt Keap1-Nrf2 interaction, forming a feed-forward loop [21]. Hence, we sought to test the hypothesis that PFI is sufficient to activate autophagy in the heart. We found that pharmacologically induced proteasome inhibition (PSMI) increased autophagosomes in mouse hearts and this increase is due to increased autophagic flux in cardiomyocytes.
Materials and methods
Reagents
E64D, pepstatin A methyl ester, and MG132 were purchased from EMD Chemicals Inc. (Gibbstown, NJ). Bafilomycin A1 (BFA) and bortezomib were from LC laboratory (Woburn, MA). MG262 was purchased from BIOMOL (Plymouth Meeting, PA). DNase I and complete protease inhibitor cocktail was from Roche Applied Science (Indianapolis, IN)
Transgenic mice
The protocol for the care and use of animals in this study was approved by University of South Dakota Institutional Animal Care and Use Committee. The transgenic mouse model with ubiquitous expression of the green fluorescence protein fused microtubule associated protein 1 light chain 3 (GFP-LC3), was generously donated by Dr. N. Mizushima from National Institute for Basic Biology in Japan and maintained in the FVB/N inbred background [22]. Genotyping was carried out by PCR analysis. The primers used for PCR genotyping are as follows:
GFP-LC3: Forward 5′-ATAACTTGCTGGCCTTTCCA CT-3′; intermediate 5′-GCAGCTCATTGCTGTTCCT CAA-3′; reverse 5′-CGGGCCATTTACCGTAAGTT-AT -3′.
Heart perfusion fixation
Mice were anesthetized by inhalation of isoflurane in room air supplemented with 100% oxygen. For perfusion fixation, a 25G butterfly cannula was connected to the perfusion apparatus which was placed at 80cm above the working table and generated 80mmHg pressure by height. The chest of a mouse was opened and the butterfly cannula was then introduced into the left ventricle of the mouse. Immediately after the heart started to fill up with the fixative, an incision was made in right atrium to release fluid. The whole system was flushed with PBS for 5 minutes and then fixed with 4% paraformaldehyde at 80 mmHg for 15 minutes. After fixation, the organs of interest were removed and equilibrated overnight in 40% sucrose at 4°C.
Isolation and primary cell culture of neonatal rat ventricle myocytes (NRVMs)
NRVMs were isolated using the Cellutron Neo-myts isolation system (Cellutron Life Technology, Baltimore, MD, Cat. No. nc-6031) following the manufacturer's instructions.[23, 24] Ventricles were collected from 0- to 2-day-old rat neonates and then digested at 37°C in a beaker with stir bar stirring at a speed of 150rpm for 12 minutes. Cells were collected using 1200rpm centrifugation for 2 minutes and then resuspended in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% Fetal bovine serum (FBS), 100 μM BrdU, and 100 U/ml penicillin/streptomycin (Invitrogen, Carlsbad, CA). Several repeats of the digestion process were performed until most of the tissues were digested. To selectively enrich cardiomyocytes, the cells were preplated in 100mm non-coated dishes for 1 hour. The resulting suspended cells were counted with a hemocytometer and then plated evenly on 1% gelatin-coated plates in appropriate cell densities. The plated cells were then cultured in a 5% CO2 incubator at 37°C for at least 24 hours before the medium was changed to meet the needs of the follow-up experiments.
Recombinant adenoviruses infection of NRVMs
Recombinant adenoviruses harboring the expression cassette of GFP-LC3 (Ad-GFP-LC3) were generously provided by Dr. R. Gottlieb of the San Diego State University (San Diego, CA) [25]. Infection of cultured cardiac myocytes with the recombinant adenoviruses was generally started at 48–72 hours after myocytes were plated. Three hours after infection, the cultured mediums containing the adenovirus were replaced with fresh mediums containing 2% FBS and the cells were incubated for at least 48 hours to allow the transgene expression.
Protein extraction and western blot analysis
To prepare total proteins, ventricular myocardium tissues or cultured cells were lysed in 1 × SDS sampling buffer (50mM Tris–HCl at pH 6.8 containing 2% SDS and 10% glycerol and a complete protease inhibitor cocktail). The extracts were homogenized on ice and boiled for 5 minutes. After cell/tissue homogenates were centrifuged at 10,000×g for 10 minutes at 4°C, the supernatants were obtained as total proteins. The protein concentration was determined using Bicinchoninic acid (BCA) reagents (Pierce biotechnology, Rockford, IL). Equal amounts of samples were subjected to sodium dodecyl sulfate -polyacrylamide gel electrophoresis (SDS-PAGE), transferred to PVDF membrane using a Trans-blot apparatus (Bio-Rad, Hercules, CA). The membranes were blocked with 5% non-fat-dry milk in PBS containing 0.1% Tween-20 (PBS-T) for 1 hour at room temperature and then probed with appropriate primary antibodies overnight at 4°C. The following primary antibodies were used: anti-α-tubulin (T6199, Sigma-Aldrich; 1: 2000), anti-GAPDH (G8795, Sigma-Aldrich; 1:1000), anti-LC3 (M115-3, Medical & Biological Laboratories Co., MBL, Nagoya, Japan; 1:1000). The corresponding horseradish peroxidase-conjugated goat anti-mouse, goat anti-rabbit or goat anti-guinea secondary antibodies (Santa Cruz Biotechnology) were used respectively for chemoluminescence-based western blot analysis. The signal was detected using either enhanced chemiluminescence (ECL-Plus) reagents (GE Healthcare, Piscataway, NJ) or, for weak signals, ECL Advance Western Blotting Kit (GE Healthcare) and visualized with a VersaDoc3000 imaging system (model 3000, Bio-Rad). The signal was quantified with the Quantity One software (Bio-Rad).
Fluorescence microscopy
Perfusion-fixed mouse tissue samples were saturated with 40% sucrose solution, embedded in Tissue-Tek O.C.T. (Sakura Finetek. USA, Inc, Torrance, CA), and frozen in a -80°C ultra-low freezer until sectioning was performed. NRVMs cultured in dishes were fixed with 2% of paraformaldehyde for 10 minutes. The tissue cryosections or fixed cells were permeabilized with 1% of Triton-X100 in PBS for 1 hour, quenched with 0.1M glycine in PBS for 1 hour, and blocked with 0.5% BSA for 1 hour. The processed tissue sections were mounted in fluorescence-quenching resistant mounting medium and imaged using a fluorescence confocal microscope (Olympus Fluoview 500, Center Valley, PA). The processed cell samples were then incubated with Alexa Fluor 568-conjugated phalloidin (Invitrogen) to stain F-actin and identify cardiomyocytes. DAPI (Sigma-Aldrich) was used to stain nuclei. The fluorescence staining of cultured cells was visualized and imaged using an epi-fluorescence microscope (Zeiss Axiovert 100, Branson, MO).
Proteasomal peptidase activities
The activity assay was done according to protocol reported by Saul R Powell [26]. Snap-frozen tissues and cultured NRVMs were homogenized on ice in 10 volumes of lysis buffer (50mM HEPES buffer at pH 7.5 containing 20 mmol/L KCl, 5 mmol/L MgCl2, 1 mmol/L DTT), and then centrifuged at 10,000×g for 10 minutes at 4 °C. The protein extracts and synthetic fluorogenic peptide substrates III Suc-LLVY- AMC (EMD Chemicals Inc.) were used for assaying the chymotrypsin-like activities. Assays were carried out using 10 μg of protein in a total volume of 200 (4.1. The peptidase activity reactions were incubated at 37 °C for 30 minutes and then quenched by adding 300 μl ice-cold ethanol. 2 ml H2O was then added into the reaction and then the fluorescence intensity was evaluated using fluorescence spectrometer (Perkin Elmer precisely LS 55, Billerica, Massachusetts) at excitation/emission wave length of 350/440 nm. Assays were carried out with and without proteasome inhibitor MG132 at a final concentration of 10 μM. The difference between the two fluorescence readings was attributed to proteasomal activity.
Statistical analysis
All quantitative data were presented as mean ± S.D. Differences between experimental groups were evaluated for significance using Student's t-test for unpaired two group comparison, non-parametric statistic if variances were unequal. One-way or two-way ANOVA followed by the posthoc test was used when appropriate. The p-value <0.05 is considered statistically significant.
Results
Pharmacological PSMI increases autophagosomes in the cardiomyocytes of intact mice
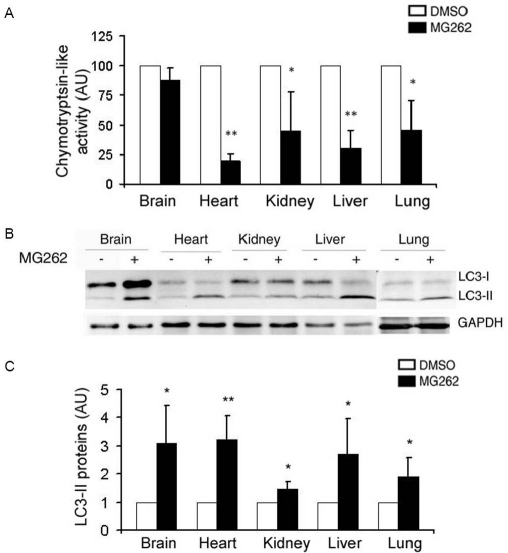
To determine whether PMSI is sufficient to activate autophagy in the heart, we first tested whether pharmacologically induced PSMI increases autophagosomes in the heart of intact mice. In this experiment, a transgenic mouse model with ubiquitous overexpression of well-established autophagy reporter GFP-LC3 was employed. To achieve systemic proteasomal inhibition, mice were intravenously injected with MG-262, a specific proteasome inhibitor [27]. MG-262 was dissolved in 60% DMSO in saline. Vehicle -treatment (CTL) used 60% DMSO in saline. Three pairs of adult GFP-LC3 mice were treated with MG-262 (5 μmol/kg) or vehicle via the tail vein. Twelve hours after the treatment, tissue samples were collected from major organs for proteasomal peptidase activity assays. Proteasomal chymotrypsin-like activity was measured using specific synthetic flurogenic peptide substrates and crude protein extracts from the tissue samples. The results showed that the proteasomal chymotrypsin-like activity in the hearts, livers, kidneys, and lungs was significantly decreased in MG262-treated mice. An 80% decrease in proteasomal chymotrypsin-like activity was achieved in MG262-treated hearts (Figure 1A).
Figure 1.
LC-II in major mouse organs is increased by systemic administration of a proteasome inhibitor. GFP-LC3 mice of 2 months of age were treated with proteasome inhibitor (MG-262, 5μmol/kg in 60% DMSO) or vehicle control (60% DMSO) via the tail vein at 12 hours before the representative organs are harvested for proteasomal chymotrypsin-like activity assays (A) and western blot analyses (B and C). A, The mean value of DMSO-treated group (vehicle control) was set as 100 AU and used to normalize the MG262-treated groups. B and C, The levels of LC3 were increased in representative organs of GFP-LC3 mice by MG-262 treatment. The representative image (B) and quantitative data of LC3 protein levels in the indicated organs of mice (C) are presented. *p<0.05, **p<0.01 vs. the DMSO group, student's t-test, n=3 mice/group.
Lipidation of LC3-I to form LC3-II allows LC3 to be incorporated into the membrane of autophagosomes; hence the abundance of LC3-II and the redistribution of LC3 from the cytosol (a diffuse pattern) to the autophagosomes (a punctate pattern) have been widely used as markers of autophagosomes [28]; To assess the impact of pharmacologically induced PSMI on autophagy activity, endogenous LC3 levels were detected by western blot analyses. The results revealed that LC3-II levels in vehicle - control mice varied substantially among different organs, but all clearly displayed significant increases upon proteasomal inhibition by MG-262. A three-fold increase in LC3-II abundance was observed in MG262-treated hearts (Figure 1B, 1C).
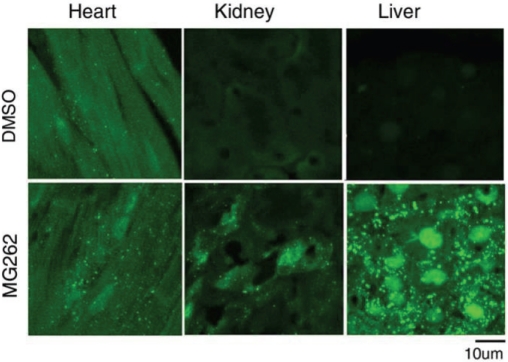
In agreement with the increase in LC3-II protein levels as revealed by western blot analyses, fluorescence confocal microscopy of sections from representative organs also revealed that GFP-LC3 puncta were increased in the hearts, kidneys, and livers of MG262-treated mice compared with control ones (Figure 2).
Figure 2.
Confocal micrographs of GFP-LC3 direct fluorescence in tissue sections of mouse hearts, kidneys and livers. GFP-LC3 mice were treated as described in Figure 1. Cryosections of ventricular myocardium, liver and kidney tissues from both DMSO-treated and MG262-treated mice were analyzed for GFP-LC3 direct fluorescence using a confocal microscope. Punctate GFP-LC3 fluorescence was significantly increased by MG262 treatment in these organs.
Autophagy is activated by PSMI in cultured cardiomyocytes
Systemic PSMI has previously shown to affect heart function and trigger pathological cardiac hypertrophy [29]. Alteration of autophagy has been observed in various forms of cardiac dysfunction [28]. Therefore, it is possible that the increase in autophagosomes in the heart by systemic PSMI may be a secondary response to cardiac dysfunction at the whole organ level. To rule out this possibility and to test whether the increase in autophagy is cardiomyocyte autonomous, we further test the effect of PSMI on cultured cardiomyocytes. In these cell culture studies, PSMI was achieved using two different types of well-documented proteasome inhibitors: MG132 and bortezomib. MG132, a peptide aldehyde, inhibits the chymotryptic activity of the proteasome by forming hemi-acetal adducts with the active site threonine nucleophile of β-subunits. It also potently inhibits thiol proteases such as cathepsin B and calpains [30-32]. In contrast, bortezomib, a peptidyl boronic acid, is a much more potent and more selective inhibitor of the chymotryptic site within the 20S proteasome. Bortezomib forms stable tetrahedral intermediates with the N-terminal threonine residues of the catalytically active proteasome β 5-subunit [31-33]. In these experiments, changes in not only the abundance of autophagosomes but also the autophagic flux were evaluated. This is important because an increase in autophagosomes can result from an increased formation or a decreased removal of autophagosomes. The former indicates autophagic activation while the latter is a sign of autophagy impairment.
Autophagic flux is increased in NRVM by MG132 treatment (Figures 3, 4)
Figure 3.
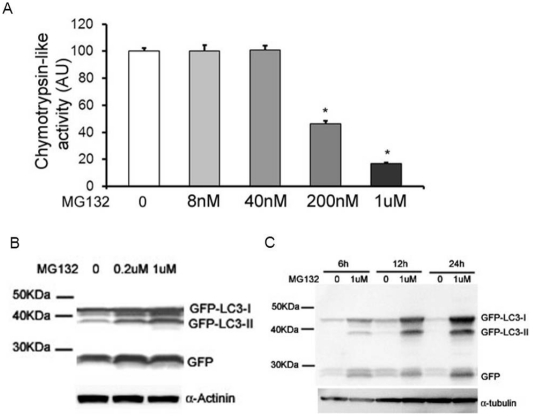
PSMI by MG132 increases GFP-LC3-II levels in NRVMs. A, MG132 treatment inhibits proteasomal chy-motrypsin-like activity in NRVMs. NRVMs were treated with the indicated doses of MG132 for 12 hours. The cells were then harvested for proteasomal chymotrypsin-like activity assay. *p<0.05 vs. CTL group, one-way ANOVA followed by the Dunnett's test, n=3. B, and C, The levels of GFP-LC3-II were increased in NRVMs by MG132 treatment in a dose- and time-dependent manner. Cultured NRVMs were infected with Ad-GFP-LC3. Three days later, NRVMs were treated with the indicated doses of MG132 for 12 hours (B) or treated with 1μM MG132 for the indicated time courses (C). The cells were then harvested for western blot analyses for the indicated proteins
Figure 4.
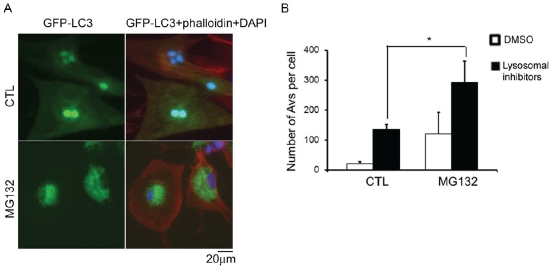
PSMI by MG132 increases autophagic flux in NRVMs. Cultured NRVMs were infected with Ad-GFP-LC3. Three days later, the cells were treated with 1μM MG-132 or DMSO for 12 hours. A, NRVMs were fixed in 2% parafor-maldehyde and stained with Alexa Fluor 568-conjugated phalloidin (red). Phalloidin stains F-actin to identify cardiomyocytes. Nuclei were stained blue with DAPI. GFP-LC3 fluorescence was diffusely distributed throughout the cytoplasm and nuclei in CTL NRVMs, whereas with MG132 treatment, GFP-LC3 formed dot-like structures in the cytoplasm, indicative of Autophagic Vacuoles (AVs). B, A mixture of lysosomal inhibitors (100nM bafilomycin A1, 5μg/mL E64D, 5ug/mL cathepsin) or DMSO was added to and remained in the cultures for another 3 hours. The quantitative analysis of the number of GFP-LC3 positive AVs per cell in each condition from 3 repeats are presented. *p<0.05, two-way ANOVA followed by the Scheffe test.
To verify the PSMI effect of MG132 on NRVMs, proteasomal peptidase activity assays were carried out with protein extracts from cells incubated with MG132 or DMSO for 12 hours. Proteasomal chymotrypsin-like activity was reduced to 46% with 200nM MG132 treatment, and to 17% with 1μM MG132 treatment (Figure 3A).
To evaluate the autophagic activity, NRVMs were infected with GFP-LC3 expressing adenoviruses (Ad-GFP-LC3) to trace autophagosomes. MG132 or DMSO was applied to NRVMs 72 hours after the Ad-GFP-LC3 infection. Protein lysates were prepared after the indicated incubation time courses and analyzed by western blot analysis using anti-GFP antibodies. On the western blot image, the positions of GFP-LC3-I (cytosolic), GFP-LC3-II (membrane-bound), and free GFP are readily identifiable. The extent of accumulation of membrane-bound GFP-LC3-II serves as a marker of autophagosome formation [34]. At 12 hours, the protein abundance of GFP-LC3-II was increased by MG132 treatment in a dose-dependent manner (Figure 3B). Moreover, the protein abundance of GFP-LC3-II was also increased by treatment with 1μM MG132 in a time-dependent manner at 6, 12, and 24 hours (Figure 3C).
To further test the autophagic activity with MG132 treatment, the subcellular localization of GFP-LC3 was examined. NRVMs were infected with Ad-GFP-LC3 and the transgene was allowed to express for 72 hours before the cells were treated with MG132 or DMSO. After 12 hours of treatment, the cells were fixed with 2% paraformaldehyde in PBS and imaged using an epifluorescence microscope. With MG132 treatment, GFP-LC3 underwent dramatic redistribution from a diffuse cytosolic signal to punctate dots (Figure 4A). The punctate dots mark autophagosomes, and the redistribution of GFP-LC3 is an indicator of autophagy induction [25].
To determine the effect of pharmacologically induced PSMI on autophagic flux in cardiomyocytes, GFP-LC3-expressing NRVMs were treated with MG132 either alone or in combination with a mixture of lysosomal inhibitors (bafilomycin A1, E64D, and pepstatin A), which inhibits autophagosome-lysosome fusion and lysosomal protease activities [25]. Fluorescence microscopy of GFP-LC3 was used to determine autophagosome prevalence in both conditions. The results showed that PSMI induced by MG132 significantly increased autophagic flux in cultured NRVMs (Figure 4B).
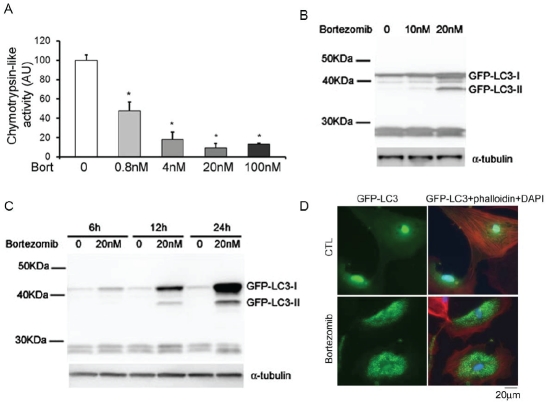
Autophagosomes are increased by bortezomib treatment in NRVMs (Figure 5)
Figure 5.
PSMI by bortezomib increases autophagosomes in NRVMs. A. Bortezomib treatment inhibited proteasomal chymotrypsin-like activities in NRVMs. NRVMs were treated with the indicated doses of bortezomib for 12 hours. The cells were then harvested for the proteasomal chymotrypsin-like activity assay. *p<0.05 vs. CTL group, one-way ANOVA followed by the Dunnett's test, n=3. B and C. The levels of GFP-LC3-II were increased in NRVMs by bortezomib treatment in a dose- and time-dependent manner. Cultured NRVMs were infected with Ad-GFP-LC3. Three days later, NRVMs were treated with the indicated doses of bortezomib for 12 hours (B) or treated with 20nM bortezomib for the indicated time courses (C). The cells were then harvested for western blot analyses for GFP and α- tubulin. D. Three days after infection with Ad-GFP-LC3, NRVMs were treated with 20nM bortezomib or DMSO for 12 hours. NRVMs then were fixed in 2% paraformaldehyde and stained with phalloidin (red). Nuclei were stained blue with DAPI. GFP-LC3 fluorescence was diffusely distributed in CTL NRVMs, whereas GFP-LC3 was found in dot-like structures in bortezomib-treated NRVMs.
To determine the PSMI effect of bortezomib on cultured NRVMs, proteasomal peptidase activity assays were carried out with protein extracts from cells incubated with bortezomib or DMSO for 12 hours. Proteasomal chymotrypsin-like activity was reduced to 47% by 0.8nM bortezomib treatment, to 18% by 4nM bortezomib treatment (Figure 5A).
To evaluate the autophagic activity with bortezomib treatment in NRVMs, Ad-GFP-LC3 was applied to mark autophagosomes. Three days after infection with Ad-GFP-LC3, NRVMs were incubated with bortezomib or DMSO. The cells were harvested after the indicated incubation hours and analyzed by western blot analysis using anti-GFP antibodies. The result revealed that GFP-LC3-II levels were increased after 12 hours of treatment with 10nM bortezomib. At 12 hours, the protein abundance of GFP-LC3-II was increased by bortezomib treatment in a dose-dependent manner (Figure 5B). Moreover, GFP-LC3-II is increased after a minimum of 6 hours of incubation with 20nM bortezomib. The protein abundance of GFP-LC3-II was also increased by bortezomib treatment in a time-dependent manner (Figure 5C).
To further test the autophagic activity with bortezomib treatment, the subcellular localization of GFP-LC3 was examined in NRVMs. After 12 hours of treatment with bortezomib, GFP-LC3 fluorescence underwent dramatic redistribution from a diffuse cytosolic signal to punctate dots, indicative of autophagy induction (Figure 5D).
Taken together, these data demonstrate that pharmacologically induced PSMI is capable of inducing autophagy both in cultured cardiomyocytes and in intact mice.
Discussion
The present study demonstrates that pharmacologically induced PSMI is sufficient to activate autophagy in cardiomyocytes in both intact animals and cell culture. We have demonstrated this by assessing the changes in the endogenous LC3-II and the abundance and distribution of transgenic GFP-LC3-II. Notably, systemic administration of a proteasome inhibitor (MG262) increases autophagosomes in major mouse organs, as indicated by increased LC3-II levels and increased GFP-LC3-labeled puncta in MG262-treated mice (Figure 1, 2). Previous studies have shown that proteasome inhibitors increase autophagy in cell lines including cortical neurons and HEK293 cells [35, 36]. One recent study showed that pharmacologically induced PSMI by 5μM MG-132 elicited modest increases in LC3-II levels in cultured cardiomyocytes [16]. The current study extends previous findings in several important ways. First, MG132 and bortezomib, belonging to two mechanistically different groups of proteasome inhibitors, are sufficient to trigger autophagy in cardiomyocytes. Moreover, to minimize the non-specific effects of proteasome inhibitors, low doses of proteasome inhibitor were tested in this study and found to be able to activate autophagy. Meanwhile, peptidase activity has been examined to monitor the proteasome inhibition effect. Our results have revealed that 200 nM of MG132, which inhibits chymotrypsin-like activity to 46%, is able to increase autophagy in cardiomyocytes. In contrast, in previous studies, MG132 at much higher doses (5μM or even higher) was used to activate autophagy [16, 36]. Furthermore, MG132 treatment combined with lysosomal inhibition increased accumulation of autophagic vacuoles and suggested an increased autophagic flux. Compared with using the sole parameter of the steady levels of LC3-II, the parameter of autophagic flux better reflects autophagic activity. More strikingly, for the first time, our study reveals that systemic proteasomal inhibition increased autophagosomes in major mouse organs.
These experiments demonstrate that pharmacologically induced PSMI is capable of elevating autophagy in cardiomyocytes and intact mice. Therefore, a direct causal link between PFI and autophagy enhancement is suggested by our study.
Mammalian cells are endowed with the ability to defend against the potentially toxic effects of misfolded proteins. The UPS and autophagy are important players for maintaining protein homeostasis in cells. Along with our another recent report [15], We have found that autophagy is upregulated in bona fide mouse models of cardiac proteinopathy as well as in response to pharmacologically induced PSMI. Given that UPS proteolytic function is inadequate in the cardiac proteinopathy mice [12, 13], this leads to the proposition that autophagy is activated to compensate for the impaired or insufficient UPS function in order to protect cells from misfolded protein stresses.
Autophagy has been generally characterized as a non-selective degradation pathway. Recently, there is a growing body of evidence suggesting that autophagy also selectively degrades various cellular structures, including protein aggregates, damaged mitochondria, and invading microbes [37, 38]. It has been found that the removal of aggregate-prone proteins related to neurodegenerative diseases is largely dependent on autophagy [39, 40]. Moreover, protein aggregates are accumulated in autophagy-deficient mice [41-43].
When cells are subjected to misfolded protein stress, the accumulation of unfolded/misfolded proteins induces endoplasmic reticulum (ER) stress. The unfolded protein response (UPR) triggered by ER stress is an integral part of intra-cellular PQC [44]. Terminally misfolded ER proteins are retrogradely transported out of the ER and immediately subjected to ubiquitination and proteasomal degradation via ER-associated protein degradation (ERAD) in the cytosol [45]. However, sustained ER stress causes accumulation of UPS reporter substrates, which indicates that sustained ER stress has an inhibitory effect on the UPS [46]. When ERAD is overloaded by ER inhibitors or blocked by proteasome inhibitors, autophagy is mobilized to degrade terminally misfolded ER proteins via the ER-activated autophagy (ERAA) pathway [47-50]. In autophagy-defective tumor cells, although no accumulation of polyubiquitinated proteins occurs, the accumulation of ER chaperones and the oxidative protein folding machinery in autophagy-deficient cells and tumors indicates a defect in the management of protein turnover [51].
Several reports have suggested that autophagy is activated by misfolded protein stress via upregulated ER signaling. In a recent study, Hill's group reported that accumulation and aggregation of ubiquitinated proteins upregulated the UPR regulator Bip and triggered activation of autophagy in a mouse model of load-induced heart failure [16]. Additionally, accumulation of misfolded proteins, such as polyQ72 aggregates, in the ER stimulated LC3 conversion from LC3-I to LC3-II through phosphorylation of PERK (RNA-dependent protein kinase-like ER kinase) and eukaryotic initiation factor 2α (eIF2α) [48]. A related study with human tumor cells revealed that UPR protects against hypoxic tumor cells by inducing LC3 and Atg5 gene expression via the PERK/elF2α signaling branch [52].
Previously, ubiquitination was generally considered as a signal for a protein to be degraded by the proteasome. Misfolded proteins, which are often ubiquitinated, were thought to be mainly degraded by UPS. Recent findings suggest that ubiquitinated proteins are also a major class of substrate for selective autophagy. This is suggested by the elevated ubiquitinated protein levels in tissue-specific knockout of Atg7 or Atg5 in mouse brains, livers and hearts [41-43]. Therefore, attachment of ubiquitin to various cellular cargos constitutes a universal degradation signal recognized by both UPS and the autophagy-lysosome pathways. The question arises as to what determines if a ubiquitin-labeled protein substrate will enter one or the other pathway. Recent studies suggest that K48-linked polyubiquitination is associated with the UPS, whereas K63-linked polyubiquitination chains provide a signal for selective autophagic degradation [38, 53].
A potential working model is that under the circumstance of PFI or PSMI, polyubiquitinated proteins accumulate. This accumulation of polyubiquitinated proteins may provide a signal that leads to activation of selective autophagy. Under this condition, autophagy functions as a compensatory mechanism to eliminate proteins that have escaped the surveillance of the UPS. The p62 and other ubiquitin binding protein may mediate this cross-talk between the UPS and autophagy. To date, for at least cardiomyocytes, this remains an attractive hypothesis to be formerly tested.
The p62 is a multi-functional adaptor protein that has been implicated in homeostatic cell function. As an adaptor molecule, p62 links ubiquitinated proteins to the autophagic machinery [54]. The C-terminal portion of p62 binds polyubiquitinated substrates through its ubiquitin-associated (UBA) domain and directly binds to LC3 via the LC3 interacting region (LIR) motif [55]. p62 can also polymerize via its N-terminal Phox/Bem1p (PB1) domain and interact with the proteasome via an N-terminal ubiq-uitin-like (UBL) domain. Interaction of the UBL domain with proteasomes may be involved in shuttling substrates for proteasomal degradation [56]. In cultured neuronal cells, the increase in both transcript and protein levels of p62 in response to PSMI has been reported and p62 was purported to sense proteolytic stress with PSMI and be involved in mediating the alternative degradation pathway to alleviate proteolytic stress [57]. PFI, upregulation of both the transcript and protein levels of p62, and increased autophagic flux have been found to coexist in mouse hearts overexpressing human disease-linked misfolded proteins. It will be very important to sort out the inter-relationship among these derangements. The findings of the present study favor the model that PFI accumulates Nrf2; increased Nrf2 in turn increases the transcription of p62; and p62 further activates Nrf2 through a positive feedback on one hand and activates autophagy on the other hand. Increased autophagy will attempt to compensate for PFI by selectively removing misfolded proteins in the cell, thereby helping the cell to survive.
In support of the above model, reduction of p62 protein levels or interference with p62 function significantly increases cell death induced by the expression of mutant huntingtin proteins [54].
However, one study using mice with genetic inactivation of p62 and Atg7 found that loss of p62 markedly attenuated liver injury caused by autophagy deficiency; whereas p62 deficiency had little effect on neuronal degeneration [58]. The reason for this apparent discrepancy is not yet clear. With the loss of Atg7 in the liver, accumulated p62 might be above the level of optimum cell survival and the detrimental function of p62 might be activated [59]. Furthermore, when autophagy is inhibited, the accumulation of p62 was shown to account for the impaired degradation of UPS substrates [60]. Moreover, p62 overexpression at levels similar to those in autophagy-deficient cells increases polyQ aggregation and toxicity. This effect is proteasome-dependent and autophagy-independent [60]. Therefore, it appears that p62 is a double-edge sword and the consequence of its upregulation in the cell depends heavily upon the functional status of the autophagic-lysosomal pathway and may also be cell-type specific. Further investigation of the cross-talk between the UPS and autophagy in the heart is clearly warranted.
Acknowledgments
Dr. X. Wang is a recipient of the Established Investigator Award of the American Heart Association. This work was in part supported by NIH grants R01HL085629 and R01HL072166 and American Heart Association grants 0740025N (to X. W.), 0625738Z (to H.S.), and 0815571G (to Q. Z.).
Disclosure
None.
References
- 1.Wang X, Robbins J. Heart failure and protein quality control. Circ Res. 2006;99:1315–1328. doi: 10.1161/01.RES.0000252342.61447.a2. [DOI] [PubMed] [Google Scholar]
- 2.Su H, Wang X. The ubiquitin-proteasome system in cardiac proteinopathy: a quality control perspective. Cardiovasc Res. 2010;85:253–262. doi: 10.1093/cvr/cvp287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li YF, Wang X. The role of the proteasome in heart disease. Biochim Biophys Acta. 2011;1809:141–149. doi: 10.1016/j.bbagrm.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuertes G, Villarroya A, Knecht E. Role of proteasomes in the degradation of short-lived proteins in human fibroblasts under various growth conditions. Int J Biochem Cell Biol. 2003;35:651–664. doi: 10.1016/s1357-2725(02)00382-5. [DOI] [PubMed] [Google Scholar]
- 6.Hochstrasser M. Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Curr Opin Cell Biol. 1995;7:215–223. doi: 10.1016/0955-0674(95)80031-x. [DOI] [PubMed] [Google Scholar]
- 7.Yoshimori T. Autophagy: a regulated bulk degradation process inside cells. Biochem Biophys Res Commun. 2004;313:453–458. doi: 10.1016/j.bbrc.2003.07.023. [DOI] [PubMed] [Google Scholar]
- 8.Fuertes G, Martin De Llano JJ, Villarroya A, Rivett AJ, Knecht E. Changes in the proteolytic activities of proteasomes and lysosomes in human fibroblasts produced by serum withdrawal, amino-acid deprivation and confluent conditions. Biochem J. 2003;375:75–86. doi: 10.1042/BJ20030282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gomes AV, Zong C, Ping P. Protein degradation by the 26S proteasome system in the normal and stressed myocardium. Antioxid Redox Signal. 2006;8:1677–1691. doi: 10.1089/ars.2006.8.1677. [DOI] [PubMed] [Google Scholar]
- 10.Ding WX, Yin XM. Sorting, recognition and activation of the misfolded protein degradation pathways through macroautophagy and the proteasome. Autophagy. 2008;4:141–150. doi: 10.4161/auto.5190. [DOI] [PubMed] [Google Scholar]
- 11.Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454:1088–1095. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J, Chen Q, Huang W, Horak KM, Zheng H, Mestril R, Wang X. Impairment of the ubiquitin-proteasome system in desminopathy mouse hearts. Faseb J. 2006;20:362–364. doi: 10.1096/fj.05-4869fje. [DOI] [PubMed] [Google Scholar]
- 13.Chen Q, Liu JB, Horak KM, Zheng H, Kumarapeli AR, Li J, Li F, Gerdes AM, Wawrousek EF, Wang X. Intrasarcoplasmic amyloidosis impairs proteolytic function of proteasomes in cardiomyocytes by compromising substrate uptake. Circ Res. 2005;97:1018–1026. doi: 10.1161/01.RES.0000189262.92896.0b. [DOI] [PubMed] [Google Scholar]
- 14.Liu J, Tang M, Mestril R, Wang X. Aberrant protein aggregation is essential for a mutant desmin to impair the proteolytic function of the ubiquitin-proteasome system in cardiomyocytes. J Mol Cell Cardiol. 2006;40:451–454. doi: 10.1016/j.yjmcc.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 15.Zheng Q, Su H, Ranek MJ, Wang X. Autophagy and p62 in Cardiac Proteinopathy. Circ Res. 2011;109:296–308. doi: 10.1161/CIRCRESAHA.111.244707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tannous P, Zhu H, Nemchenko A, Berry JM, Johnstone JL, Shelton JM, Miller FJ, Jr, Rothermel BA, Hill JA. Intracellular protein aggregation is a proximal trigger of cardiomyocyte autophagy. Circulation. 2008;117:3070–3078. doi: 10.1161/CIRCULATIONAHA.107.763870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jain A, Lamark T, Sjottem E, Larsen KB, Awuh JA, Overvatn A, McMahon M, Hayes JD, Johansen T. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem. 2010;285:22576–22591. doi: 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol. 2004;24:8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, Kim M, Nishito Y, Iemura S, Natsume T, Ueno T, Kominami E, Motohashi H, Tanaka K, Yamamoto M. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 22.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saucerman JJ, Zhang J, Martin JC, Peng LX, Stenbit AE, Tsien RY, McCulloch AD. Systems analysis of PKA-mediated phosphorylation gradients in live cardiac myocytes. Proc Natl Acad Sci U S A. 2006;103:12923–12928. doi: 10.1073/pnas.0600137103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gomez-Ospina N, Tsuruta F, Barreto-Chang O, Hu L, Dolmetsch R. The C terminus of the L-type voltage-gated calcium channel Ca(V)1.2 encodes a transcription factor. Cell. 2006;127:591–606. doi: 10.1016/j.cell.2006.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamacher-Brady A, Brady NR, Gottlieb RA. Enhancing macroautophagy protects against ischemia/reperfusion injury in cardiac myocytes. J Biol Chem. 2006;281:29776–29787. doi: 10.1074/jbc.M603783200. [DOI] [PubMed] [Google Scholar]
- 26.Powell SR, Davies KJ, Divald A. Optimal determination of heart tissue 26S-proteasome activity requires maximal stimulating ATP concentrations. J Mol Cell Cardiol. 2007;42:265–269. doi: 10.1016/j.yjmcc.2006.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumarapeli AR, Horak KM, Glasford JW, Li J, Chen Q, Liu J, Zheng H, Wang X. A novel transgenic mouse model reveals deregulation of the ubiquitin-proteasome system in the heart by doxorubicin. Faseb J. 2005;19:2051–2053. doi: 10.1096/fj.05-3973fje. [DOI] [PubMed] [Google Scholar]
- 28.Zheng Q, Wang X. Autophagy and the ubiquitin-proteasome system in cardiac dysfunction. Panminerva Med. 2010;52:9–25. [PMC free article] [PubMed] [Google Scholar]
- 29.Tang M, Li J, Huang W, Su H, Liang Q, Tian Z, Horak KM, Molkentin JD, Wang X. Proteasome functional insufficiency activates the calcineurin-NFAT pathway in cardiomyocytes and promotes maladaptive remodelling of stressed mouse hearts. Cardiovasc Res. 2010;88:424–433. doi: 10.1093/cvr/cvq217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee DH, Goldberg AL. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 1998;8:397–403. doi: 10.1016/s0962-8924(98)01346-4. [DOI] [PubMed] [Google Scholar]
- 31.Adams J, Behnke M, Chen S, Cruickshank AA, Dick LR, Grenier L, Klunder JM, Ma YT, Plamondon L, Stein RL. Potent and selective inhibitors of the proteasome: dipeptidyl boronic acids. Bioorg Med Chem Lett. 1998;8:333–338. doi: 10.1016/s0960-894x(98)00029-8. [DOI] [PubMed] [Google Scholar]
- 32.Palombella VJ, Conner EM, Fuseler JW, Destree A, Davis JM, Laroux FS, Wolf RE, Huang J, Brand S, Elliott PJ, Lazarus D, McCormack T, Parent L, Stein R, Adams J, Grisham MB. Role of the proteasome and NF-kappaB in streptococcal cell wall-induced polyarthritis. Proc Natl Acad Sci U S A. 1998;95:15671–15676. doi: 10.1073/pnas.95.26.15671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lightcap ES, McCormack TA, Pien CS, Chau V, Adams J, Elliott PJ. Proteasome inhibition measurements: clinical application. Clin Chem. 2000;46:673–683. [PubMed] [Google Scholar]
- 34.Kuballa P, Huett A, Rioux JD, Daly MJ, Xavier RJ. Impaired autophagy of an intracellular pathogen induced by a Crohn's disease associated ATG16L1 variant. PLoS One. 2008;3 doi: 10.1371/journal.pone.0003391. e3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rideout HJ, Lang-Rollin I, Stefanis L. Involvement of macroautophagy in the dissolution of neuronal inclusions. Int J Biochem Cell Biol. 2004;36:2551–2562. doi: 10.1016/j.biocel.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 36.Iwata A, Riley BE, Johnston JA, Kopito RR. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J Biol Chem. 2005;280:40282–40292. doi: 10.1074/jbc.M508786200. [DOI] [PubMed] [Google Scholar]
- 37.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 38.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 39.Yamada M, Tsuji S, Takahashi H. Involvement of lysosomes in the pathogenesis of CAG repeat diseases. Ann Neurol. 2002;52:498–503. doi: 10.1002/ana.10328. [DOI] [PubMed] [Google Scholar]
- 40.Rubinsztein DC, DiFiglia M, Heintz N, Nixon RA, Qin ZH, Ravikumar B, Stefanis L, Tolkovsky A. Autophagy and its possible roles in nervous system diseases, damage and repair. Autophagy. 2005;1:11–22. doi: 10.4161/auto.1.1.1513. [DOI] [PubMed] [Google Scholar]
- 41.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 42.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 43.Nakai A, Yamaguchi O, Takeda T, Higuchi Y, Hikoso S, Taniike M, Omiya S, Mizote I, Matsumura Y, Asahi M, Nishida K, Hori M, Mizushima N, Otsu K. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13:619–624. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 44.Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 45.Hebert DN, Molinari M. In and out of the ER: protein folding, quality control, degradation, and related human diseases. Physiol Rev. 2007;87:1377–1408. doi: 10.1152/physrev.00050.2006. [DOI] [PubMed] [Google Scholar]
- 46.Menendez-Benito V, Verhoef LG, Masucci MG, Dantuma NP. Endoplasmic reticulum stress compromises the ubiquitin-proteasome system. Hum Mol Genet. 2005;14:2787–2799. doi: 10.1093/hmg/ddi312. [DOI] [PubMed] [Google Scholar]
- 47.Fujita E, Kouroku Y, Isoai A, Kumagai H, Misutani A, Matsuda C, Hayashi YK, Momoi T. Two endoplasmic reticulum-associated degradation (ERAD) systems for the novel variant of the mutant dysferlin: ubiquitin/proteasome ERAD(I) and autophagy/lysosome ERAD(II) Hum Mol Genet. 2007;16:618–629. doi: 10.1093/hmg/ddm002. [DOI] [PubMed] [Google Scholar]
- 48.Kouroku Y, Fujita E, Tanida I, Ueno T, Isoai A, Kumagai H, Ogawa S, Kaufman RJ, Kominami E, Momoi T. ER stress (PERK/eIF2alpha phosphorylation) mediates the polyglutamineinduced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 2007;14:230–239. doi: 10.1038/sj.cdd.4401984. [DOI] [PubMed] [Google Scholar]
- 49.Ding WX, Ni HM, Gao W, Hou YF, Melan MA, Chen X, Stolz DB, Shao ZM, Yin XM. Differential effects of endoplasmic reticulum stressinduced autophagy on cell survival. J Biol Chem. 2007;282:4702–4710. doi: 10.1074/jbc.M609267200. [DOI] [PubMed] [Google Scholar]
- 50.Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, Murakami T, Taniguchi M, Tanii I, Yoshinaga K, Shiosaka S, Hammarback JA, Urano F, Imaizumi K. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, Dipaola RS, Karantza-Wadsworth V, White E. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rouschop KM, van den Beucken T, Dubois L, Niessen H, Bussink J, Savelkouls K, Keulers T, Mujcic H, Landuyt W, Voncken JW, Lambin P, van der Kogel AJ, Koritzinsky M, Wouters BG. The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. J Clin Invest. 2010;120:127–141. doi: 10.1172/JCI40027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tan JM, Wong ES, Kirkpatrick DS, Pletnikova O, Ko HS, Tay SP, Ho MW, Troncoso J, Gygi SP, Lee MK, Dawson VL, Dawson TM, Lim KL. Lysine 63-linked ubiquitination promotes the formation and autophagic clearance of protein inclusions associated with neurodegenerative diseases. Hum Mol Genet. 2008;17:431–439. doi: 10.1093/hmg/ddm320. [DOI] [PubMed] [Google Scholar]
- 54.Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pankiv S, Hoyvarde Clausen T, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of Ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;19:19. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 56.Seibenhener ML, Babu JR, Geetha T, Wong HC, Krishna NR, Wooten MW. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol Cell Biol. 2004;24:8055–8068. doi: 10.1128/MCB.24.18.8055-8068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuusisto E, Suuronen T, Salminen A. Ubiquitin-binding protein p62 expression is induced during apoptosis and proteasomal inhibition in neuronal cells. Biochem Biophys Res Commun. 2001;280:223–228. doi: 10.1006/bbrc.2000.4107. [DOI] [PubMed] [Google Scholar]
- 58.Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata JI, Ezaki J, Murata S, Hamazaki J, Nishito Y, Iemura SI, Natsume T, Yanagawa T, Uwayama J, Warabi E, Yoshida H, Ishii T, Kobayashi A, Yamamoto M, Yue Z, Uchiyama Y, Kominami E, Tanaka K. Homeostatic Levels of p62 Control Cytoplasmic Inclusion Body Formation in Autophagy-Deficient Mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 59.Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 2009;137:1001–1004. doi: 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Korolchuk VI, Mansilla A, Menzies FM, Rubinsztein DC. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol Cell. 2009;33:517–527. doi: 10.1016/j.molcel.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]