Abstract
The mTOR Complex 1 (mTORC1) protein kinase is a master growth regulator that is stimulated by amino acids. Amino acids activate the Rag guanosine triphosphatases (GTPases), which promote the translocation of mTORC1 to the lysosomal surface, the site of mTORC1 activation. We found that the vacuolar H+-adenosine triphosphatase ATPase (v-ATPase) is necessary for amino acids to activate mTORC1. The v-ATPase engages in extensive amino acid-sensitive interactions with the Ragulator, a scaffolding complex that anchors the Rag GTPases to the lysosome. In a cell-free system, ATP hydrolysis by the v-ATPase, but not the lysosomal pH gradient, was necessary for amino acids to regulate the v-ATPase-Ragulator interaction and promote mTORC1 translocation. Results obtained in vitro and within cells suggests that amino acid signaling initiates within the lysosomal lumen. These results identify the v-ATPase as a component of the mTOR pathway and delineate a lysosome-associated machinery for amino acid sensing.
Amino acids are the building blocks of proteins and intermediates in lipid and adenosine triphosphate (ATP) synthesis. They also initiate a signaling cascade in cells that leads to activation of the master growth regulator mTOR Complex 1 (mTORC1). This multi-component protein kinase integrates inputs from growth factors as well as nutrient and energy supply to control many biosynthetic and catabolic processes (1). Most signals upstream of mTORC1 converge on TSC1-TSC2, a heterodimeric tumor suppressor that negatively regulates the Rheb guanosine triphosphatase (GTPase), which is an essential activator of mTORC1 protein kinase activity (2, 3). In contrast, amino acids signal to mTORC1 by promoting its binding to a distinct family of GTPases, the Rag GTPases (4, 5). The Rags form heterodimers consisting of RagA or RagB, which are highly similar to each other, bound to RagC or RagD, which are also highly related. In an amino acid-sensitive fashion, the Rag GTPases recruit mTORC1 to the surface of lysosomes, which also contain Rheb (5). The trimeric Ragulator complex, which comprises the p18, p14, and MP1 proteins, anchors the Rag GTPases to the lysosome, and, like the Rags, is necessary for mTORC1 activation by amino acids (6). Together, the Ragulator and Rag heterodimer form an amino acid-regulated docking site for mTORC1 on the lysosomal surface.
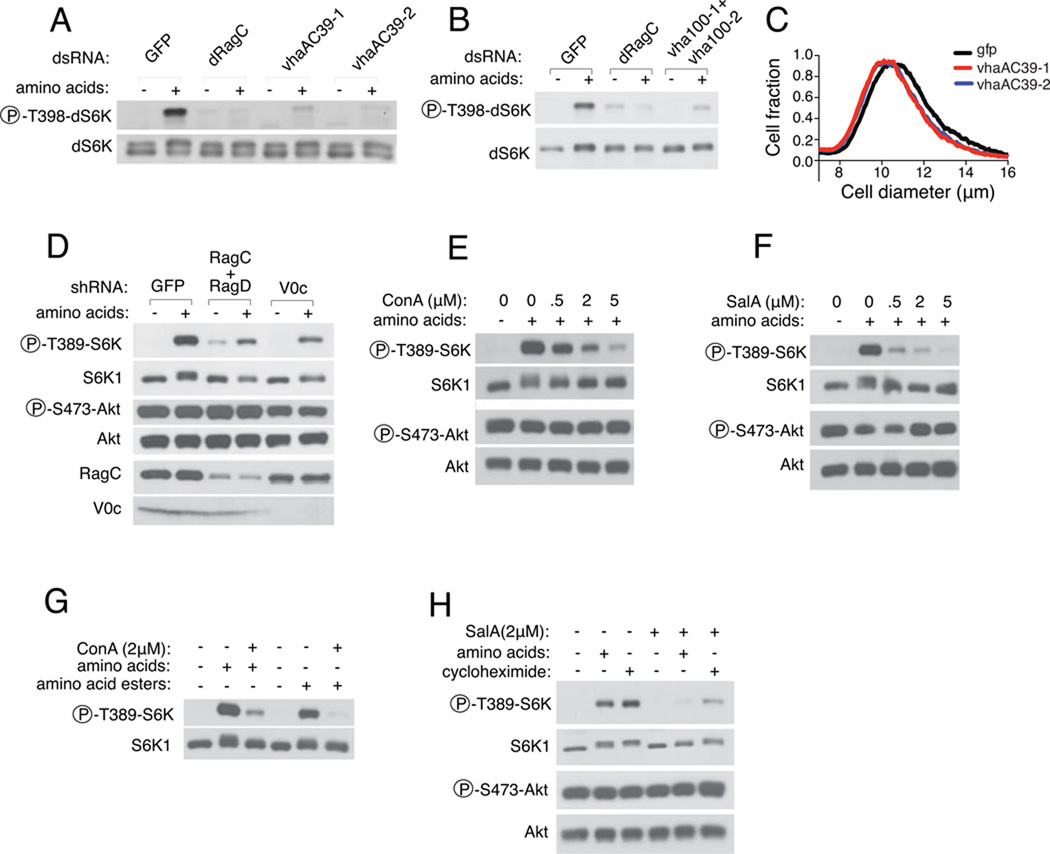
Amino acid signaling has been proposed to initiate, alternatively, at the plasma membrane or inside the cell, but this key issue remains unsettled (7–10). The localization of the Rag GTPases on lysosomes, but not other Rheb-containing endomembranes, suggests that this organelle has an important role in amino acid signaling to mTORC1. To determine if lysosome-associated processes and proteins participate in the activation of mTORC1 by amino acids, we used RNAi in Drosophila S2 cells to reduce the expression of a number of genes with roles in lysosomal biogenesis and function (table S1). dsRNAs targeting most of the genes did not affect the amino acid-induced phosphorylation of the ribosomal protein S6 kinase (dS6K) on T398, a readout of dTORC1 activity (table S1). In contrast, dsRNAs targeting vhaC39, vha16, vha100-1 and vha100-2, all encoding components of the vacuolar H+ATPase (v-ATPase), suppressed dS6K phosphorylation to degrees similar to that of a dsRNA targeting dRagC (table S1 and Fig. 1, A and B and fig. S1A). The dsRNAs to vhaC39 also decreased the size of S2 cells (Fig 1C). Consistent with the results in Drosophila cells, lentiviral shRNAs targeting human ATP6V0c, the orthologue of Drosophila vha16, suppressed amino acid-induced phosphorylation of S6K1 in human embryonic kidney (HEK) 293T cells (Fig. 1D and fig. S1B). These results implicate the v-ATPase in the activation of mTORC1 by amino acids.
Fig. 1.
Requirement of the v-ATPase for mTORC1 activation by amino acids. (A) Double-stranded RNA (dsRNA)-mediated depletion of vhaAC39 in Drosophila S2 cells. Cells were deprived for amino acids for 1.5 h and then stimulated with complete medium for 30 min. Proteins from cell lysates were analyzed for phosphorylation of dS6K at threonine 398 (T398). Depletion of vhaAC39 by two distinct dsRNAs is compared to that of dRagC. (B) dsRNA-mediated depletion of both vha100-1 and vha100-2 in S2 cells suppresses amino acid-induced T398 phosphorylation of dS6K. (C) Cell size measurement after depletion of vhaAC39 in S2 cells with two dsRNAs (red and blue) compared to a control dsRNA (black). (D) S6K1 phosphorylation at T389 in HEK-293T cells treated with short-hairpin RNA (shRNA) targeting GFP, RagC and RagD, and V0c. Cells were deprived of amino acids for 50 min and, where indicated, stimulated for 10 min. Immunoblotting was used to detect the indicated proteins. (E) S6K1 phosphorylation in HEK-293T cells deprived of amino acids for 50 min in the presence of the indicated concentrations of Concanamycin A (ConA) and then stimulated for 10 min with amino acids. (F) S6K1 phosphorylation in HEK-293T cells deprived of amino acids for 50 min in the presence of the indicated concentrations of Salicylihalamide A (SalA) and restimulated for 10 min with amino acids. (G) ConA blocks mTORC1 activation by alcohol esters of amino acids. HEK-293T cells were deprived of amino acids for 50 min and then stimulated for 10 min with amino acids or alcohol esters of amino acids in the presence of the indicated concentration of ConA. (H) Activation of mTORC1 by intracellular amino acids. HEK-293T cells were deprived of amino acids for 50 min and stimulated with amino acids or cycloheximide in DMSO or 2 µM SalA. Immunoblotting was used to detect the indicated proteins.
The v-ATPase consists of multi-component V0 and V1 domains and operates through an incompletely understood mechanism in which each cycle of ATP hydrolysis by the V1 sector generates torque that rotates the membrane domain of V0, known as the rotor. In turn, this movement enables the transfer into the lysosomal lumen of protons, which acidify it (11). The macrolides Concanamycin A (ConA) and Salicylihalamide A (SalA) are structurally diverse inhibitors of the v-ATPase that do not have other known targets (11–14). In 293T cells, both ConA and SalA inhibited amino acid-induced phosphorylation of S6K1 in a concentration-dependent manner (Fig. 1, E and F). The inhibition of S6K1 phosphorylation occurred after short (15–50 min) treatment times (fig. S2A) and without concomitant alterations in lysosomal morphology (fig. S2, B and C) or inhibition of Akt phosphorylation, a readout of growth factor signaling (Fig. 1, E and F).
The finding that the v-ATPase and its activity are necessary for mTORC1 activation by amino acids, led us to consider potential roles for it in the pathway. One possibility is that the v-ATPase functions downstream of amino acids and is part of the amino acid-induced signaling pathway that culminates in mTORC1 activation. Another conceivable function is that the proton gradient generated by the v-ATPase is required for amino acids to be transported into the cellular compartment where an amino acid sensor is located. To examine these possibilities we tested whether the v-ATPase is required for alcohol ester derivatives of amino acids to activate mTORC1. These esters freely diffuse across membranes, and, within the cytoplasm and lysosomes, are hydrolyzed by esterases into native amino acids (15). A mixture of amino acid esters or leucine methyl ester activated mTORC1 with efficiencies comparable to that of their respective native amino acids (fig. S1, C and D). Moreover ConA also inhibited the S6K1 phosphorylation induced by amino acids esters (Fig 1G and fig. S1E). Consistent with these findings, SalA also suppressed mTORC1 activation induced by cycloheximide, which, by inhibiting translation, boosts concentrations of intracellular amino acids (7, 16) (Fig. 1H). Thus, these results are consistent with the v-ATPase having a role downstream of intracellular amino acids in the initiation or propagation of the amino acid-induced signal to mTORC1.
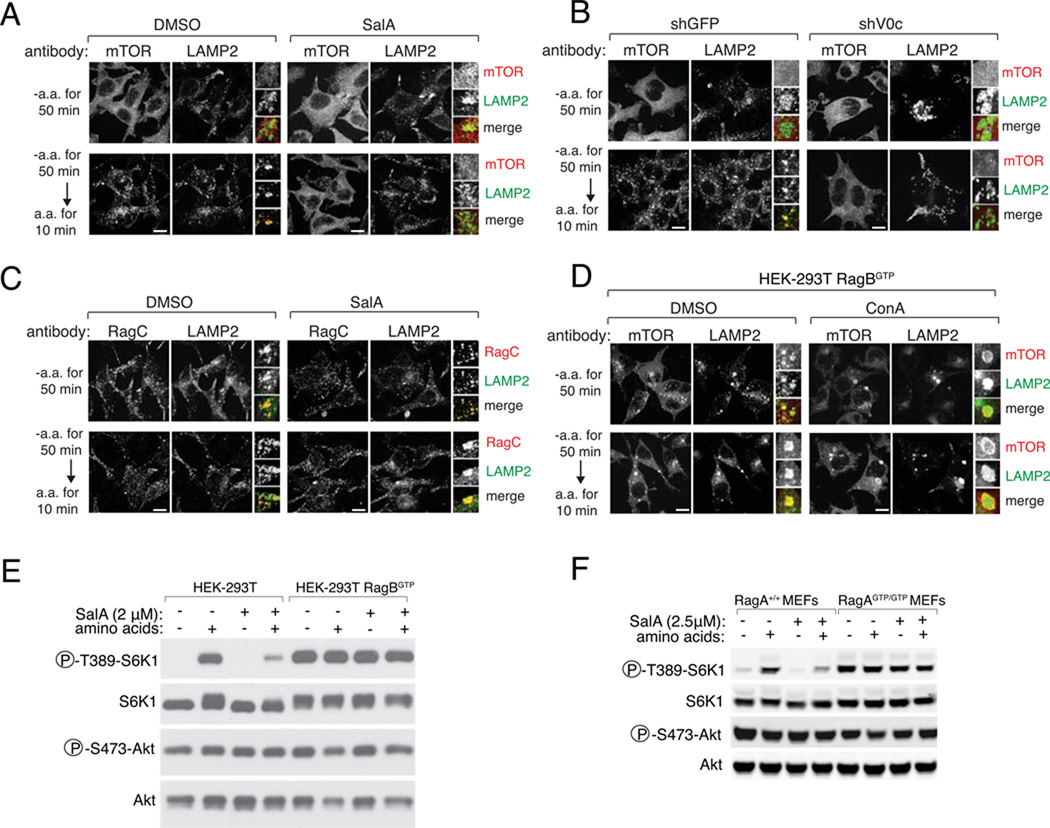
A key event in amino acid signaling is the Rag GTPase-mediated translocation of mTORC1 to the surface of lysosomes (5, 6). In cells treated with ConA or SalA or depleted of ATP6V0c, mTOR failed to cluster onto lysosomes in response to amino acids and instead was found within cells in a diffuse staining pattern (Fig. 2, A and B and fig. S3, A–C). Unlike the Ragulator (6), the v-ATPase appears not to be necessary for anchoring the Rag GTPases to the lysosomal surface because pharmacological or RNAi-mediated inhibition of the v-ATPase did not affect lysosomal localization of RagC (Fig. 2C and fig. S3, C and D). To test whether the v-ATPase might function upstream of the Rag GTPases we employed a RagB mutant that is constitutively active (RagBGTP) and renders mTORC1 signaling insensitive to amino acid starvation (4, 5). If the v-ATPase is required for the activation of the Rag GTPases, expression of RagBGTP should rescue the defects in mTOR lysosomal recruitment and S6K1 phosphorylation caused by ConA and SalA. Indeed, in cells stably expressing RagBGTP, the lysosomal localization of mTOR and the phosphorylation of S6K1 were insensitive not only to amino acid starvation but also to ConA and SalA treatment (Figs 2, D and E and fig. S3F). Consistent with these results, in knock-in mouse embryonic fibroblasts (MEFs) that express active RagAGTP from the endogenous RagA locus, SalA (Fig. 2F) and ConA (fig. S3G) did not block the constitutive S6K1 phosphorylation caused by RagAGTP. Collectively, these results place the v-ATPase downstream of amino acids but upstream of the regulation of nucleotide loading of the Rag GTPases; they also exclude its involvement in other regulatory inputs to mTORC1, such as controlling Rheb activity (6).
Fig. 2.
Requirement of the v-ATPase for lysosomal recruitment of mTORC1 by the Rag GTPases. (A) Immunofluorescence images of mTOR and LAMP1 in HEK-293T cells deprived of amino acids (top) or deprived and then stimulated (bottom) in the presence of DMSO (left) or 2.5 µM SalA (right). The mTOR and LAMP2 channels are shown separately. Inset shows a higher magnification of a selected field. In the merge, yellow indicates co-localization. (B) HEK-293T cells expressing a lentivirally-encoded shRNA targeting GFP (left) or V0c (right) were deprived of amino acids (top) or deprived and then stimulated (bottom). (C) Staining for RagC and LAMP2 in HEK-293T cells deprived of amino acids (top) or deprived and then stimulated (bottom) in the presence of DMSO (left) or 2.5 µM SalA (right). (D) HEK-293T cells stably expressing the constitutively active RagBQ99L mutant (293T RagBGTP) deprived of amino acids (top) or deprived and stimulated (bottom) in the presence of DMSO (left) or 2.5 µM ConA (right). (E) S6K1 phosphorylation in HEK-293T cells and HEK-293T RagBGTP cells deprived of amino acids for 50 min in the presence of DMSO or 2 µM SalA and stimulated for 10 min with amino acids. (F) S6K1 phosphorylation in wild-type MEFs (RagA+/+) or in MEFs homozygous for the constitutive active RagA Q66L mutant (RagAGTP/GTP); cells were deprived of amino acids for 50 min in the presence of DMSO or 2.5 µM SalA and stimulated for 10 min with amino acids.
In all images, scale bars represent 10 µm.
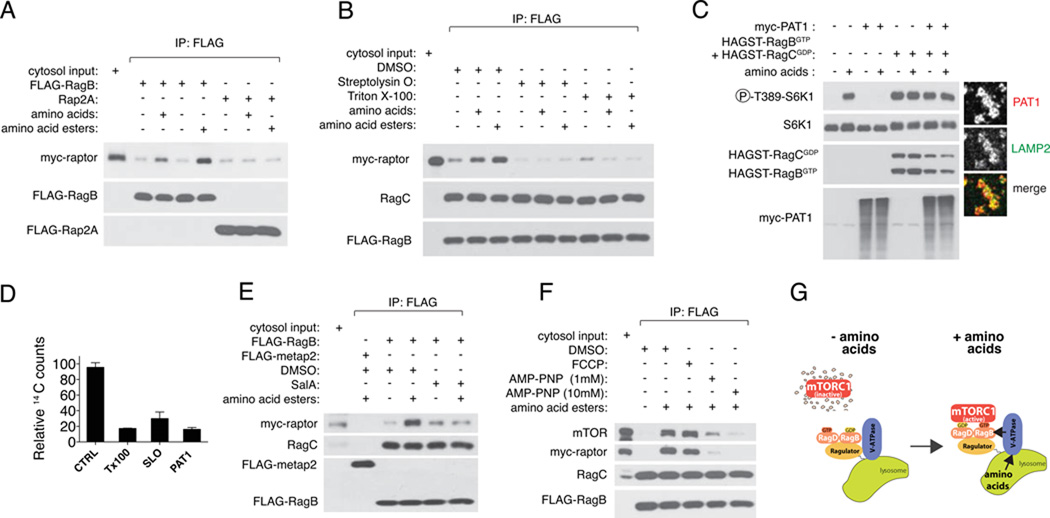
We tested whether a physical interaction exists between the v-ATPase and the Rags or Ragulator or both. Semiquantitative mass spectrometric analyses of anti-FLAG immunoprecipitates prepared from 293T cells expressing FLAG-tagged Ragulator components (p18 or p14) or RagB, revealed the presence of many subunits of the v-ATPase (Fig. 3A). Immunoblot assays with antibodies to endogenous V0 (c and d1) and V1 (A, B2, and D) subunits confirmed that Ragulator co-immunoprecipitates with the V0 and V1 domains (Fig. 3, B and C), whereas the Rags co-immunoprecipitate with V1 subunits only (fig S4, A and B). Although easily detected in immunoblot assays as co-immunoprecipitating with Ragulator, the c subunit of V0 was not detected by mass spectrometry, likely because of its highly hydrophobic nature. The v-ATPase did not co-immunoprecipitate with lysosomal (LAMP1) or cytoplasmic (Metap2) control proteins (Fig. 3, B and C). In vitro assays with purified recombinant proteins verified a direct interaction between the V0 component d1 and p18, but not p14, and between the V1 component D with p18 and, to a lesser degree, with p14 (Fig. 3D). No direct interactions were detected between the Rag GTPases and purified v-ATPase subunits (fig. S4C), consistent with the relatively low abundance of v-ATPase subunits in the RagB immunoprecipitates analyzed by mass spectrometry (Fig. 3A). Thus, in addition to scaffolding the Rag GTPases to the lysosomal surface, Ragulator provides a physical and functional link between the v-ATPase and the Rag GTPases.
Fig. 3.
Interaction of the v-ATPase with the Ragulator-Rag GTPases. (A) Cartoon summarizing mass spectrometry analyses of immunoprecipitates from FLAG-p18 (left), FLAG-p14 (center) and FLAG-RagB (right) expressing HEK-293T cells. v-ATPase subunits are color-coded according to their peptide representation (scale at the far right). (B) Binding of Ragulator to the V0 domain. HEK-293T cells stably expressing FLAG-tagged p18 and p14 were lysed and subjected to FLAG-immunoprecipitation followed by immunoblotting for V0c and V0d1. FLAG-LAMP1 and FLAG-Metap2 served as negative controls. (C) Binding of Ragulator to the V1 domain. HEK-293T cells stably expressing FLAG-tagged p18, p14, LAMP1 and Metap2 were lysed and subjected to FLAG-immunoprecipitation followed by immunoblotting for V1A, V1B2 and V1D. (D) (Top) In vitro binding assays in which purified FLAG-p18 and FLAG-p14 were incubated with recombinant V0d1 fused to glutathione S-transferase (HA-GST-V0d1), immobilized on glutathione agarose beads. Samples were subjected to immunoblotting for FLAG to detect bound Ragulator components. HA-GST-Rap2A served as a negative control. (Bottom) In vitro binding assays in which purified FLAG-p18 and FLAG-p14 were incubated with recombinant V1D fused to glutathione S-transferase (HA-GST-V1D). HA-GST-metap2 served as a negative control.
(E) The Ragulator-V1 interaction, but not the Ragulator-V0, is regulated by amino acids. HEK-293T cells stably expressing FLAG-tagged p18, p14 and Metap2 were deprived of amino acids for 90 min, or deprived and then stimulated with amino acids for 15 min. Following lysis, samples were subjected to FLAG-immunoprecipitation and immunoblotting for the indicated v-ATPase subunits. (F) SalA blocks regulation of Ragulator-V1 interaction by amino acids. HEK-293T cells stably expressing FLAG-p14 were deprived of amino acids for 90 min, or deprived and then stimulated with amino acids for 15 min, in the presence of DMSO or 2 µM SalA. (G) Cartoon summarizing the Ragulator-v-ATPase interactions identified in (A)–(F). Orange denotes regulation by amino acids, blue indicates lack of regulation.
Consistent with amino acids acting upstream of the v-ATPase, amino acids regulated the interaction between the V1 domain of v-ATPase and Ragulator and Rag GTPases. Amino acid starvation and stimulation strengthened and weakened, respectively, the interaction (Fig. 3E and fig. S4D). In contrast, amino acids did not affect the interaction of Ragulator with the V0 domain of the v-ATPase (Fig. 3E) or of the V1 and V0 subunits with each other (as does glucose starvation (17)) (fig. S4D). Treatment of cells with SalA, like amino acid deprivation, strengthened the interaction between Ragulator and V1 domain subunits and, moreover, largely prevented amino acids from weakening the interaction (Fig. 3F and fig. S4E). Thus, amino acids induce a structural rearrangement of the v-ATPase-Ragulator-Rag GTPase complex that is blocked by SalA (Fig. 3G).
The v-ATPase is large, complex, and has many functions that cannot easily be teased apart in live cells (11, 18–22). Thus, to better understand its role in amino acid signaling to mTORC1 we developed a cell-free system that recapitulates the amino acid-induced binding of mTORC1 to the Rag GTPases on the lysosomal surface (see methods). We prepared a light organellar fraction from 293T cells expressing FLAG-RagB that had been deprived of amino acids. The organelles were briefly stimulated with amino acids or amino acid esters, and then incubated with cytosol containing myc-tagged raptor (Fig. 4A and fig. S5A). In this system amino acids, and especially amino acid esters, increased binding of myc-raptor to FLAG-RagB-containing vesicles but not to control vesicles (Fig. 4A and fig. S5B). As expected, in preparations containing the FLAG-RagBGTP mutant, the binding of myc-raptor was constitutively high and largely insensitive to amino acids (fig. S5C and S5D). We also prepared purified lysosomes by immunoisolating them from 293T cells (see methods) (fig. S6, A–C). Again, amino acid esters induced binding of myc-tagged raptor to isolated lysosomes (fig. S6D). Cytosol appeared to be dispensable, because highly purified, FLAG-tagged raptor showed amino acid-induced binding to organelles containing GST-tagged Rag heterodimers (fig. S6E). Thus, lysosomes contain all the machinery required for sensing amino acids and activating the Rag GTPases.
Fig. 4.
In vitro analysis of mTORC1 activation by amino acids. (A) In vitro binding of myc-raptor to FLAG-RagB- but not to FLAG-Rap2A-containing vesicles. Organelle preparations were left unstimulated, or were stimulated with amino acids or amino acid esters and incubated with myc-raptor-containing cytosol. After FLAG immunoprecipitation, bound myc-raptor was detected by immunoblotting (B) Intact FLAG-RagB lysosomes, FLAG-RagB lysosomes permeabilized with Streptolysin-O, and FLAG-RagB lysosomes permeabilized with Triton X-100, were left unstimulated, stimulated with amino acids, or stimulated with amino acid esters. Myc-raptor was detected by immunoblotting (C) S6K1 phosphorylation at T389 in HEK-293T cells transiently expressing FLAG-S6K1, FLAG-S6K1 + myc-PAT1, FLAG S6K1 + HAGST-tagged active Rag mutants, or FLAG-S6K1 + myc-PAT1 + HAGST-active Rags. Cells were deprived of amino acids for 50 min, or starved and then stimulated for 10 min (see methods). The indicated proteins were detected by immunoblotting. The band pattern of myc-PAT1 is likely due to glycosylation. (right) Immunofluorescence images of lysosomes from HEK-293T cells transiently expressing myc-PAT1 and stained for myc tag (top, red in the merge) and for LAMP2 (center, green in the merge). (D) Accumulation of 14C-labeled amino acids into lysosomes immunopurified from HEK-293T cells expressing LAMP1-mRFP-FLAGX2. Lysosomes were either left intact or permeabilized with Triton X-100 or Streptolysin O prior to measurement. Overexpression of PAT1 largely abolished amino acid accumulation inside lysosomes. Each value represents the mean ± SD of three independent samples. (E) FLAG-RagB lysosomes were treated with DMSO or SalA, activated with amino acid esters and then incubated with myc-raptor. An organellar fraction from FLAG-metap2 expressing cells served as negative control. (F) FLAG-RagB lysosomes were stimulated with amino acid esters in the presence of the proton ionophore FCCP or the non-hydrolyzable ATP analog AMP-PNP at 1 mM or 10 mM. Organelle samples were then incubated with mycraptor cytosol, followed by FLAG-IP and immunoblotting for myc-raptor and endogenous mTOR. (G) Model for inside-out activation of mTORC1 by lysosomal amino acids. Accumulation of amino acids inside the lysosomal lumen generates an activating signal that is transmitted to the Rag GTPases via the v-ATPase-Ragulator. In turn, the Rags physically recruit mTORC1 to the lysosomal surface.
In the in vitro system the alcohol esters of amino acids were more effective than native amino acids in inducing the RagB-raptor interaction (Fig. 4A). A possible reason for this is that amino acids must enter and accumulate in lysosomes to initiate signaling, and that the amino acid esters do so more easily in the in vitro preparation (15). To test the requirement for an intra-lysosomal accumulation, we used treatments that permeabilize the lysosomal membrane and thus allow amino acids to leak out. Treatment of the organellar preparation with Streptolysin-O, which introduces nanometer-size holes into the lysosomal membrane, or Triton X-100, which dissolves the membranes without disrupting the v-ATPase-Ragulator-Rag interaction, completely suppressed the effect of amino acids or their esters to promote the RagB-raptor interaction (Fig. 4B). The PAT1 transporter is a proton-coupled amino acid transporter that localizes specifically to lysosomes (fig. S7A) and exports amino acids out of the lysosomal lumen (23). In intact cells, overexpression of PAT1 completely suppressed mTORC1 activation by amino acids and this effect was fully rescued by co-expression of constitutively active RagBGTP (Fig. 4C). In contrast overexpression of PAT4, an amino acid transporter that does not localize to the lysosome, had no effect on mTORC1 activation by amino acids (fig. S7, B and C).
These results strongly suggest that amino acid signaling initiates inside the lysosome. Consistent with this possibility, stimulation of amino acid-starved 293T cells with 14C-amino acids led to the rapid appearance of labeled amino acids within lysosomes immunoisolated through a FLAG-tagged lysosomal protein (Fig. 4D and fig. S6, A and B). Amino acid accumulation was reverted by lysosome permeabilization and largely prevented by PAT1 overexpression (Fig. 4D).
In the in vitro system, disruption of the v-ATPase by SalA or by shRNA against V0c blocked the amino acid-induced interaction of raptor with RagB (Fig. 4E and Fig. S5E). SalA causes structural rearrangements in the v-ATPase that inhibit both its capacity to hydrolyze ATP and to establish the lysosomal proton gradient (13, 24). Carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP), an ionophore that dissipates the lysosomal proton gradient without interfering with the v-ATPase (25), did not prevent the amino acid-induced binding of raptor to RagB (Fig. 4F). Moreover, amino acids or their esters did not alter the lysosomal pH of intact cells, nor did they perturb lysosomal acidification in vitro (fig. S8, A–C). In contrast the non-hydrolyzable ATP analog Adenosine 5’-(β,γ-imido)triphosphate (AMP-PNP), which inhibits the ATPase activity of V1 and the consequent rotation of the stalk and the V0 proteolipid subunits, blocked the amino acid-induced interaction between raptor and RagB in a concentration-dependent manner (Fig. 4F and fig. S8D). Thus, ATP hydrolysis and the associated rotation of the v-ATPase appear to be essential to relay an amino acid signal from the lysosomal lumen to the Rag GTPases, whereas the capacity of the v-ATPase to set up the lysosomal proton gradient is dispensable.
We propose a lysosome-centric inside-out model of amino acid sensing by mTORC1 in which amino acids must accumulate in the lysosomal lumen to initiate signaling (Fig. 4G). The v-ATPase is required for amino acid signaling to mTORC1 and functions between amino acids and the nucleotide loading of the Rag GTPases. Its placement in the pathway and its amino acid-sensitive interactions with the Rag-Ragulator complex implicate it as an essential component of the amino acid sensing mechanism.
Supplementary Material
Acknowledgments
We thank members of the Sabatini Lab for helpful suggestions, E. Spooner for the mass spectrometric analysis, J. De Brabander (UT Southwestern) for Salicylihalamide A. Supported by grants from the National Institutes of Health (CA103866 and AI47389 and Department of Defense (W81XWH-07-0448) to D.M.S., awards from the W.M. Keck Foundation and LAM Foundation to D.M.S, and fellowship support from the Jane Coffin Childs Memorial Fund for Medical Research and the LAM Foundation to R.Z, from the Human Frontier Science Program to A.E., and from the Medical Scientist Training Program to S.W.
D.M.S. is an investigator of the Howard Hughes Medical Institute.
Footnotes
-
-Materials and Methods; Table S1 legend; Supplemental Figure S1–S8 legends
-
-Table S1
-
-Supplemental Figures 1–8
REFERENCES
- 1.Zoncu R, Efeyan A, Sabatini DM. Nat Rev Mol Cell Biol. Jan;12:21. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Inoki K, Li Y, Xu T, Guan KL. Genes Dev. 2003 Aug 1;17:1829. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J. Curr Biol. 2003 Aug 5;13:1259. doi: 10.1016/s0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- 4.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Nat Cell Biol. 2008 Aug;10:935. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sancak Y, et al. Science. 2008 May 22; [Google Scholar]
- 6.Sancak Y, et al. Cell. Apr 16;141:290. [Google Scholar]
- 7.Beugnet A, Tee AR, Taylor PM, Proud CG. Biochem J. 2003 Jun 1;372:555. doi: 10.1042/BJ20021266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bohe J, Low A, Wolfe RR, Rennie MJ. J Physiol. 2003 Oct 1;552:315. doi: 10.1113/jphysiol.2003.050674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christie GR, Hajduch E, Hundal HS, Proud CG, Taylor PM. J Biol Chem. 2002 Mar 22;277:9952. doi: 10.1074/jbc.M107694200. [DOI] [PubMed] [Google Scholar]
- 10.Wu B, et al. J Cell Biol. 2006 May 8;173:327. doi: 10.1083/jcb.200602089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forgac M. Nat Rev Mol Cell Biol. 2007 Nov;8:917. doi: 10.1038/nrm2272. [DOI] [PubMed] [Google Scholar]
- 12.Bowman BJ, McCall ME, Baertsch R, Bowman EJ. J Biol Chem. 2006 Oct 20;281:31885. doi: 10.1074/jbc.M605532200. [DOI] [PubMed] [Google Scholar]
- 13.Boyd MR, et al. J Pharmacol Exp Ther. 2001 Apr;297:114. [PubMed] [Google Scholar]
- 14.Huss M, et al. J Biol Chem. 2002 Oct 25;277:40544. doi: 10.1074/jbc.M207345200. [DOI] [PubMed] [Google Scholar]
- 15.Reeves JP. J Biol Chem. 1979 Sep 25;254:8914. [PubMed] [Google Scholar]
- 16.Price DJ, Nemenoff RA, Avruch J. J Biol Chem. 1989 Aug 15;264:13825. [PubMed] [Google Scholar]
- 17.Bond S, Forgac M. J Biol Chem. 2008 Dec 26;283:36513. doi: 10.1074/jbc.M805232200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cruciat CM, et al. Science. Jan 22;327:459. [Google Scholar]
- 19.Hiesinger PR, et al. Cell. 2005 May 20;121:607. [Google Scholar]
- 20.Hurtado-Lorenzo A, et al. Nat Cell Biol. 2006 Feb;8:124. doi: 10.1038/ncb1348. [DOI] [PubMed] [Google Scholar]
- 21.Peters C, et al. Nature. 2001 Feb 1;409:581. doi: 10.1038/35054500. [DOI] [PubMed] [Google Scholar]
- 22.Yan Y, Denef N, Schupbach T. Dev Cell. 2009 Sep;17:387. doi: 10.1016/j.devcel.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sagne C, et al. Proc Natl Acad Sci U S A. 2001 Jun 19;98:7206. doi: 10.1073/pnas.121183498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie XS, et al. J Biol Chem. 2004 May 7;279:19755. doi: 10.1074/jbc.M313796200. [DOI] [PubMed] [Google Scholar]
- 25.Steinberg BE, et al. J Cell Biol. Jun 28;189:1171. doi: 10.1083/jcb.200911083. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.