Abstract
Multiplex PCR/Reverse Line Blot Hybridization assay allows the detection of up to 43 molecular targets in 43 samples using one multiplex PCR reaction followed by probe hybridization on a nylon membrane, which is re-usable. Probes are 5' amine modified to allow fixation to the membrane. Primers are 5' biotin modified which allows detection of hybridized PCR products using streptavidin-peroxidase and a chemiluminescent substrate via photosensitive film. With low setup and consumable costs, this technique is inexpensive (approximately US$2 per sample), high throughput (multiple membranes can be processed simultaneously) and has a short turnaround time (approximately 10 hours).
The technique can be utilized in a number of ways. Multiple probes can be designed to detect sequence variation within a single amplified product, or multiple products can be amplified simultaneously, with one (or more) probes used for subsequent detection. A combination of both approaches can also be used within a single assay. The ability to include multiple probes for a single target sequence makes the assay highly specific.
Published applications of mPCR/RLB include detection of antibiotic resistance genes1,2, typing of methicillin-resistant Staphylococcus aureus3-5 and Salmonella sp6, molecular serotyping of Streptococcus pneumoniae7,8, Streptococcus agalactiae9 and enteroviruses10,11, identification of Mycobacterium sp12, detection of genital13-15 and respiratory tract16 and other17 pathogens and detection and identification of mollicutes18. However, the versatility of the technique means the applications are virtually limitless and not restricted to molecular analysis of micro-organisms.
The five steps in mPCR/RLB are a) Primer and Probe design, b) DNA extraction and PCR amplification c) Preparation of the membrane, d) Hybridization and detection, and e) Regeneration of the Membrane.
Keywords: Molecular Biology, Issue 54, Typing, MRSA, macroarray, molecular epidemiology
Protocol
Careful consideration must be given to primer and probe design. All available sequences of the targets of interest from databases such as GenBank should be utilized to identify conserved areas which are suitable targets. Where a large number of targets are being amplified in a single mPCR assay, each amplified sequence should be of similar length, and should not exceed 300 base pairs to avoid competition. An alternative application of the method is to amplify one longer target using primers against conserved regions and to use multiple probes to identify sequence variation within the amplicon. In this instance, the amplified PCR product may be longer. Primer annealing temperatures should all be similar, with PCR conditions adjusted accordingly. Primers which form strong secondary structure or primer dimer should be avoided. The Sigma Aldrich DNA calculator (http://www.sigma-genosys.com/calc/DNACalc.asp) can be used to reliably predict these features.
DNA probes should be designed to have annealing temperatures close to 60°C. To maximize specificity, two probes for each target of interest can be included in the assay, one hybridizing to the forward DNA strand adjacent to the reverse primer binding site, and the other to the reverse strand adjacent to the forward primer binding site. In this case, both forward and reverse primers must be biotin-modified. If only one probe per amplified target is being used, then only one primer need be biotin modified.
When designing primers and probes for the mPCR/RLB assay it is strongly advised to use in silico methods to predict PCR products and probe hybridization to correlate with the in vitro results. Ideally this is done using isolates for which the whole genome sequence is available. Software such as FastPCR (available at http://primerdigital.com/fastpcr.html) can be used for this purpose. Weak or absent probe signal can be predicted where in silico analysis indicates base pair mismatches resulting in low probe annealing temperatures.
DNA extraction techniques will vary depending on the samples being tested, and PCR conditions will be dependent on primer design. Readers are referred to publications on individual assays for more information regarding DNA extraction and PCR reagents and conditions1-19.
Each assay is run with appropriate controls to provide at least one positive and one negative probe signal for each probe on the membrane, as well as a DNA-free control. Furthermore it is beneficial to include a probe at the top of the membrane that is expected to be positive for all samples (eg: a species or genus specific probe in the case of a micro-organism). This serves as a positive control probe, but also permits easy orientation of the results.
1. Preparation of Membrane
- Prepare the following solutions:
- 100ml 0.5M NaHCO3 (pH 8.4)
- 100 ml 0.5M NaHCO3
- 20 ml 16% EDAC
- 250 ml 0.1 M NaOH
- 250 ml 2xSSPE
- 250 ml 2xSSPE/0.1% SDS – place in waterbath at 60°C
- 250 ml 20 mM EDTA.
Pre-heat oven to 60°C.
Clean the Immunetics Miniblotter with 70% ethanol.
Dilute the oligonucleotide probes in 0.5 M NaHCO3 to a final concentration of 2 pmol/μl and a volume of 200 μl.
Cut a BiodyneC nylon membrane to 15x15 cm. Using a pencil and ruler, rule off a 0.5 cm space across top of membrane and write details here.
Seal membrane in plastic bag with 20 ml freshly made 16% EDAC solution and rock at room temperature for 10 minutes.
Wash membrane in deionised water for 30 seconds.
Place membrane in miniblotter with channels running across the membrane (parallel to pencil line). Put support cushion in place and close blotter. Aspirate fluid from channels.
Fill lanes 1 and 45 with 150 μl 0.5 M NaHCO3. Use 150 μl of each probe solution to fill lanes 2-44 in sequence, being careful to avoid air bubbles. If an air bubble does appear in the channel, keeping the pipette in place, rapidly aspirate the solution to allow the air bubble to float to the top of the pipette, then retry. Incubate at room temperature for 5 minutes.
Aspirate probe solutions, remove membrane and wash in 250 ml 0.1 M NaOH at room temperature for 9 minutes.
Wash membrane in 250 ml 2xSSPE for 30 seconds.
Wash membrane in 250 ml pre-warmed 2xSSPE/0.1% SDS in oven at 60°C for 5 minutes.
If not being used for hybridization immediately, wash membrane in 240 ml 20 mM EDTA at room temperature for 20 minutes (reserving remaining 10ml), then seal membrane in plastic bag with remaining 10 ml 20 mM EDTA and store in refrigerator at 4°C.
Wash miniblotter with Pyroneg detergent and brush. Rinse and allow to dry.
2. Hybridization and Detection
- Prepare the following solutions:
- 250 ml 2xSSPE/0.1% SDS - store in water bath at 60°C.
- 500 ml 2xSSPE/0.5% SDS – store in water bath at 60°C
- 500 ml 2xSSPE/0.5% SDS – store in oven at 42°C
- 500 ml 2xSSPE - keep at room temperature
- 500 ml 1% SDS – store in water bath at 60°C initially (for step 3)
- 250 ml 20 mM EDTA – store at room temperature (for step 3).
Turn on one oven at 60°C and one oven to 42°C. Bring water to boil in a large beaker on a hotplate.
Clean Immunetics Miniblotter with 70% ethanol.
Aliquot 10 ml of 2xSSPE/0.1% SDS into a small container. Add 20 μl of each PCR product to 150 μl 2xSSPE/0.1% SDS in numbered tubes.
Boil PCR products for 10 min at 100°C using styrofoam holders. Place on ice immediately for at least 5 minutes.
Wash membrane in remaining 240 ml 2xSSPE/0.1% SDS in 60°C oven for 5 minutes.
Place membrane in miniblotter with backing cushion. Orientate so channels run vertically (perpendicular to pencil line).
Suction excess liquid from miniblotter channels.
Add remaining 150 μl 2xSSPE/0.1% SDS to first and last channels. Add boiled PCR products to remaining channels (2-44) in sequence, being careful to avoid air bubbles.
Place miniblotter flat in 60°C oven for 1 hour to allow hybridization to take place.
Aspirate liquid from each channel then remove membrane from miniblotter.
Wash twice in prewarmed 250 ml 2xSSPE/0.5% SDS in 60°C oven for 10 minutes.
Moisten nylon separating mesh with 2xSSPE/0.5% SDS at 42°C and use it to roll up membrane.
Place rolled up membrane into roller tube, and unfurl to ensure membrane abuts the interior surface. Add 3 μl streptavidin-peroxidase conjugate (Roche Applied Science) to 15 ml 2xSSPE/0.5% SDS at 42°C and add to roller tube. Screw cap on tightly and incubate in roller oven at 42°C for 60 minutes. Ensure direction of roll is such that membrane does not tighten. Check periodically for leaks.
Wash miniblotter in Pyroneg detergent and water, rinse and allow to dry.
Remove membrane from roller tube. Wash twice in remaining 2xSSPE/0.5% SDS at 42°C for 10 minutes.
Wash twice in 2xSSPE at room temperature for 5 min.
Turn oven up to 80°C and place 500ml 1% SDS in oven to pre-warm.
Make up 15 ml Amersham ECL detection solution (7.5 ml of solution A and 7.5 ml of solution B). Discard 2xSSPE and add detection solution. Rock gently by hand for 2 minutes ensuring complete coverage of membrane with solution. Discard chemiluminescent solution.
Cut two sheets of plastic transparency to fit exposure cartridge. Place membrane between the two sheets and place into exposure cartridge.
Proceed to darkroom. Under red light, add ECL detection film to cartridge and expose for 5 minutes. Dog-ear lower right corner of film upon removal to allow correct orientation when viewing.
Develop film with automated developer or chemical baths as per manufacturer's directions.
Repeat exposure with longer or shorter exposure times if required.
3. Regeneration of Membrane
Wash membrane in 250 ml 1% SDS at 80°C for 30 minutes twice.
Wash membrane in 240 ml 20 mM EDTA at room temperature for 15 minutes.
Seal membrane in plastic bag with 10 ml 20 mM EDTA and refrigerate at 4°C for future re-use.
4. Representative Results:
The results are best viewed by placing the film over a printed grid, or else scanning the image and importing into software such as BioNumerics (Applied Maths, Sint-Martens-Latem, Belgium). Each probe result should be interpreted with reference to positive and negative control probes. The results are best graded as negative, weak or positive. Positive results are where the signal is as strong as or stronger than the positive control probe. Negative results are where the signal is absent or equal to the negative control (in the case of background signal). Weak results are where the signal is fainter than the positive control probe, but stronger than the negative control. Weak results may be the result of point mutations leading to weak probe binding, or due to non-specific signals from primer dimer formation. Using two probes per target of interest can improve the specificity, where a single weak result can safely be interpreted as non-specific signal. If any doubt remains, a single-plex PCR reaction can be performed with gel-based detection and sequencing of any amplified product to determine if the result is truly positive. A representative result is shown in figure 2.
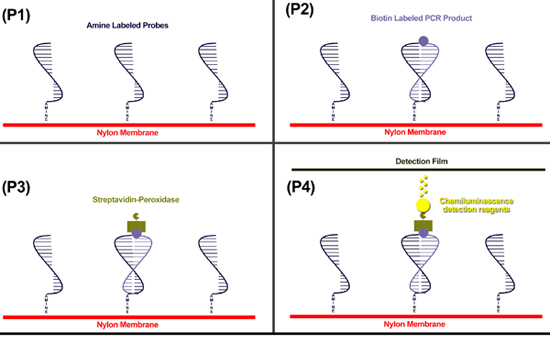 Figure 1. The mPCR/RLB principles (P1) Step 1.9. Amine modified probes are bound covalently to a nylon membrane. (P2) Step 2.10. Biotin modified PCR products are hybridized to the probes. (P3) Step 2.14. Streptavidin, labeled with peroxidase, is incubated with the membrane and binds to biotin. (P4) Steps 2.19-2.21. Peroxidase catalyses a reaction in the ECL detection reagents, producing light to which light sensitive film is exposed. The membrane is then washed for re-use.
Figure 1. The mPCR/RLB principles (P1) Step 1.9. Amine modified probes are bound covalently to a nylon membrane. (P2) Step 2.10. Biotin modified PCR products are hybridized to the probes. (P3) Step 2.14. Streptavidin, labeled with peroxidase, is incubated with the membrane and binds to biotin. (P4) Steps 2.19-2.21. Peroxidase catalyses a reaction in the ECL detection reagents, producing light to which light sensitive film is exposed. The membrane is then washed for re-use.
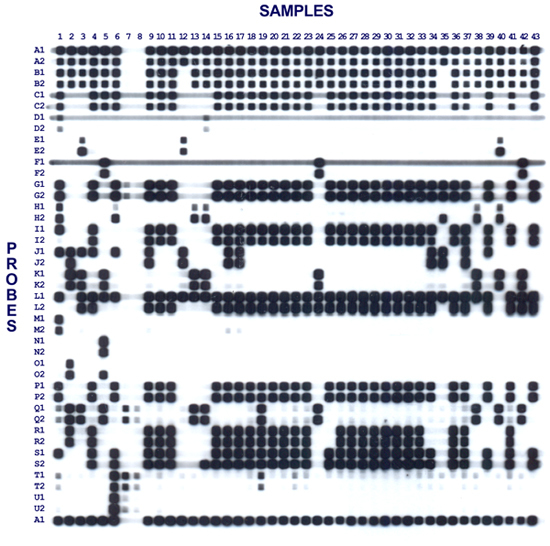 Figure 2. Representative mPCR/RLB result. Samples 1 through 6 represent positive controls – between them there is at least one positive probe signal for each of the 43 probes. Each target sequence (A through U) is detected with two different probes to maximize specificity. Probes A1 and A2 represent species-specific probes which are expected to be positive for each sample and assist with orienting the film. Probe A1 is repeated at the bottom of the membrane. Samples 7 and 8 are negative controls. Note that Probes Q1, Q2, T1 and T2 have signal in the negative controls, indicating likely nonspecific binding of primers or primer dimer product. Re-design of these probes or primers would be required. Probes C1, D1 and F1 show streaking across the membrane likely due to some non-specific uptake of the streptavidin-peroxidase conjugate or chemiluminescent substrate, however this is easily distinguished from true positive probe signals. Probes D1 and D2 have relatively weak signal compared to other probes, this may be due to larger amplicons leading to less efficient amplification. Probe J2 is negative is several samples (1, 3, 4, 6, 39) where probe J1 is positive. This most likely represents a mutation in the J2 binding site for these samples.
Figure 2. Representative mPCR/RLB result. Samples 1 through 6 represent positive controls – between them there is at least one positive probe signal for each of the 43 probes. Each target sequence (A through U) is detected with two different probes to maximize specificity. Probes A1 and A2 represent species-specific probes which are expected to be positive for each sample and assist with orienting the film. Probe A1 is repeated at the bottom of the membrane. Samples 7 and 8 are negative controls. Note that Probes Q1, Q2, T1 and T2 have signal in the negative controls, indicating likely nonspecific binding of primers or primer dimer product. Re-design of these probes or primers would be required. Probes C1, D1 and F1 show streaking across the membrane likely due to some non-specific uptake of the streptavidin-peroxidase conjugate or chemiluminescent substrate, however this is easily distinguished from true positive probe signals. Probes D1 and D2 have relatively weak signal compared to other probes, this may be due to larger amplicons leading to less efficient amplification. Probe J2 is negative is several samples (1, 3, 4, 6, 39) where probe J1 is positive. This most likely represents a mutation in the J2 binding site for these samples.
Discussion
The mPCR/RLB method permits simultaneous detection of a large number of PCR amplicons. Because of the high sensitivity of chemiluminescent probe-based detection, a single multiplex PCR reaction with large numbers of primer pairs can be used to amplify the DNA template.
If no probe signals are obtained with one or more samples, performing gel electrophoresis with any remaining PCR product can help distinguish whether the problem is with PCR amplification or probe hybridization. Where all probe signals on the membrane are weak or absent, but gel electrophoresis indicates successful PCR amplification, possibilities include problems with labeling of the membrane, incorrect regeneration of the membrane after previous use, incorrect temperatures during hybridization or streptavidin incubation, or defective streptavidin-peroxidase conjugate or detection reagent.
If control samples indicate that individual probes may be producing false negative signals, single PCR with gel-based detection and subsequent sequencing can be performed to verify amplification of PCR product and to look for sequence variation at the probe binding site. Weak or absent probe signal may be due to large amplicon size: if possible all amplicons should be of similar length and less than 300 bp. Secondary DNA structures of amplicons may also produce poor probe binding, and may necessitate re-design of primers. Individual primer or probe concentrations may also be varied to optimize signal strength.
False positive signals due to probe binding of nonamplified primers or primer-dimer product can be investigated by performing single PCR with gel-based detection and subsequent sequencing. If this occurs, redesign of the probes may be necessitated. Point mutations in probe binding sites may lead to weak or absent signals. Allowing two probes for each amplified product makes this easy to detect. This characteristic may be exploited with careful primer and probe design to detect small sequence variations in target DNA.
In summary, while there are many methods of product detection following multiplex PCR reactions available, mPCR/RLB has the advantage of being high-throughput and inexpensive with low setup-costs. The flexibility of the method permits its use in for a wide range of applications.
Disclosures
No conflicts of interest declared.
Acknowledgments
Matthew O'Sullivan and Fei Zhou are recipients of Australian National Health and Medical Research Council Postgraduate Medical Research Scholarships.
References
- O'Sullivan MV, Cai Y, Kong F, Zeng X, Gilbert GL. Influence of disk separation distance on accuracy of the disk approximation test for detection of inducible clindamycin resistance in Staphylococcus spp. J Clin Microbiol. 2006;44:4072–4076. doi: 10.1128/JCM.01632-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Kong F, Wang H, Darbar A, Gilbert GL. Simultaneous detection of nine antibiotic resistance-related genes in Streptococcus agalactiae using multiplex PCR and reverse line blot hybridization assay. Antimicrob Agents Chemother. 2006;50:204–209. doi: 10.1128/AAC.50.1.204-209.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y. Comparison of single- and multilocus sequence typing and toxin gene profiling for characterization of methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2007;45:3302–3308. doi: 10.1128/JCM.01082-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L. A new multiplex PCR-based reverse line-blot hybridization (mPCR/RLB) assay for rapid staphylococcal cassette chromosome mec (SCCmec) typing. J Med Microbiol. 2009;58:1045–1057. doi: 10.1099/jmm.0.007955-0. [DOI] [PubMed] [Google Scholar]
- O'Sullivan MV, Kong F, Sintchenko V, Gilbert GL. Rapid identification of methicillin-resistant Staphylococcus aureus transmission in hospitals by use of phage-derived open reading frame typing enhanced by multiplex PCR and reverse line blot assay. J Clin Microbiol. 2010;48:2741–2748. doi: 10.1128/JCM.02201-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Kong F, Jelfs P, Gilbert GL. Extended phage locus typing of Salmonella enterica serovar Typhimurium, using multiplex PCR-based reverse line blot hybridization. J Med Microbiol. 2008;57:827–838. doi: 10.1099/jmm.0.47766-0. [DOI] [PubMed] [Google Scholar]
- Zhou F, Kong F, Tong Z, Gilbert GL. Identification of less-common Streptococcus pneumoniae serotypes by a multiplex PCR-based reverse line blot hybridization assay. J Clin Microbiol. 2007;45:3411–3415. doi: 10.1128/JCM.01076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong F, Brown M, Sabananthan A, Zeng X, Gilbert GL. Multiplex PCR-based reverse line blot hybridization assay to identify 23 Streptococcus pneumoniae polysaccharide vaccine serotypes. J Clin Microbiol. 2006;44:1887–1891. doi: 10.1128/JCM.44.5.1887-1891.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong F, Ma L, Gilbert GL. Simultaneous detection and serotype identification of Streptococcus agalactiae using multiplex PCR and reverse line blot hybridization. J Med Microbiol. 2005;54:1133–1138. doi: 10.1099/jmm.0.46244-0. [DOI] [PubMed] [Google Scholar]
- Zhou F. Identification of 20 common human enterovirus serotypes by use of a reverse transcription-PCR-based reverse line blot hybridization assay. J Clin Microbiol. 2009;47:2737–2743. doi: 10.1128/JCM.00823-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F. Molecular identification and analysis of nonserotypeable human enteroviruses. J Clin Microbiol. 2010;48:1276–1282. doi: 10.1128/JCM.02384-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Kong F, Yang Y, Cheng J, Gilbert GL. Use of PCR and reverse line blot hybridization macroarray based on 16S-23S rRNA gene internal transcribed spacer sequences for rapid identification of 34 mycobacterium species. J Clin Microbiol. 2006;44:3544–3550. doi: 10.1128/JCM.00633-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Kong F, Wang B, McKechnie ML, Gilbert GL. Multiplex polymerase chain reaction-based reverse line blot hybridization assay to detect common genital pathogens. Int J STD AIDS. 2010;21:320–325. doi: 10.1258/ijsa.2009.008481. [DOI] [PubMed] [Google Scholar]
- McKechnie ML. Simultaneous identification of 14 genital microorganisms in urine by use of a multiplex PCR-based reverse line blot assay. J Clin Microbiol. 2009;47:1871–1877. doi: 10.1128/JCM.00120-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Kong F, Zhou H, Gilbert GL. Use of PCR and reverse line blot hybridization assay for rapid simultaneous detection and serovar identification of Chlamydia trachomatis. J Clin Microbiol. 2006;44:1413–1418. doi: 10.1128/JCM.44.4.1413-1418.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Kong F, Yang Y, Gilbert GL. A multiplex PCR-based reverse line blot hybridization (mPCR/RLB) assay for detection of bacterial respiratory pathogens in children with pneumonia. Pediatr Pulmonol. 2008;43:150–159. doi: 10.1002/ppul.20749. [DOI] [PubMed] [Google Scholar]
- Wang Y. Use of a multiplex PCR-based reverse line blot (mPCR/RLB) hybridisation assay for the rapid identification of bacterial pathogens. Clin Microbiol Infect. 2008;14:155–160. doi: 10.1111/j.1469-0691.2007.01890.x. [DOI] [PubMed] [Google Scholar]
- Wang H, Kong F, Jelfs P, James G, Gilbert GL. Simultaneous detection and identification of common cell culture contaminant and pathogenic mollicutes strains by reverse line blot hybridization. Appl Environ Microbiol. 2004;70:1483–1486. doi: 10.1128/AEM.70.3.1483-1486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong F, Gilbert GL. Multiplex PCR-based reverse line blot hybridization assay (mPCR/RLB) - a practical epidemiological and diagnostic tool. Nat Protoc. 2006;1:2668–2680. doi: 10.1038/nprot.2006.404. [DOI] [PubMed] [Google Scholar]


