Abstract
The ability to modify/visualize biological surfaces, and then study the modified cell/virion in a range of in vitro and in vivo environments is essential to gaining further insight into the function of specific molecules or the entire entity. Studies of biological surface modification are generally limited to genetic engineering of the organism or the covalent attachment of chemical moieties to the cell surface1,2. However these traditional techniques expose the cell to chemical reactants, or they require significant manipulation to achieve the desired outcome, making them cumbersome, and they may also inadvertently affect the viability/functionality of the modified cell. A simple method to harmlessly modify the surface of cells is required.
Recently a new technology, KODE Technology has introduced a range of novel constructs consisting of three components: a functional head group (F), a spacer (S) and a lipid tail (L) and are known as Function-Spacer-Lipid or FSL constructs3. The spacer (S) is selected to provide a construct that is dispersible in water, yet will spontaneously and stably incorporate into a membrane.
FSL construct functional moieties (F) so far include a range of saccharides including blood group-related determinants, sialic acids, hyaluronan polysaccharides, fluorophores, biotin, radiolabels, and a range of peptides3-12. FSL constructs have been used in modifying embryos, spermatozoa, zebrafish, epithelial/endometrial cells, red blood cells, and virions to create quality controls systems and diagnostic panels, to modify cell adhesion/ interaction/ separation/ immobilization, and for in vitro and in vivo imaging of cells/virions3-12.
The process of modifying cells/virions is generic and extremely simple. The most common procedure is incubation of cells (in lipid free media) with a solution for FSL constructs for 1-2 hours at 37°C4-10. During the incubation the FSL constructs spontaneously incorporate into the membrane, and the process is complete. Washing is optional. Cells modified by FSL constructs are known as kodecytes6-9, while virions are kodevirions10.
FSL constructs as direct infusions and kodecytes/kodevirions have been used in experimental animal models7,8,10. All kodecytes/kodevirions appear to retain their normal vitality and functionality while gaining the new function of the F moiety7,8,10,11.
The combination of dispersibility in biocompatible media, spontaneous incorporation into cell membranes, and apparent low toxicity, makes FSL constructs valuable research tools for the study of cells and virions.
Protocol
The following protocol describes the generic procedure for the insertion of FSL constructs (Figure 1) into biological membranes. For simplicity the protocol will only refer to cells (kodecytes), which are at a 100% concentration. However, the term cells is interchangeable with virions (kodevirions) or organisms or cellular structures and their concentration in diluent is not important, provided it is always consistent between tests, controls, and experiments. With red blood cells the packed cell volume is typically 80%, but with virions, embryos and other cells it is typically less than 1%. The method is very robust and will produce modified membranes when used by any of its variations, but refinement will be required to optimize and standardize the degree of modification required for specific applications.
1. Preparation of FSL Constructs
To prepare the FSL construct stock solution first reconstitute the dry FSL product (Table 1) by the addition of 1.0mL of diluent (or as specified in the product insert) to the product vial. Briefly sonicate (30 seconds). This will prepare a 1mg/mL solution, which can be aliquoted into 100 μL sterile containers and stored at 2-8°C for up to one week, or frozen for up to 3 months.
To prepare working FSL construct solutions for insertion, just prior to use briefly sonicate (30 seconds) the stock solution to homogenize any micelles. Dilute the FSL construct in buffer (preferably not containing lipids or highly hydrophobic material) to the concentration required or over a range if desired.
Notes:
FSL constructs will usually insert into cells in lipid containing media but will typically require as much as a 50X higher FSL working concentrations than if in PBS or other lipid free media.
The working range dilutions will depend on the application, the detection method sensitivity, the type of FSL construct, and the degree of modification required. Typically the range of dilution will be between 10-500 μg of FSL per mL of diluent for carbohydrate FSL constructs and 1-100 μg/mL for other FSL constructs such as fluorophores and biotin.
If preparing insertion diluents containing multiple FSL construct, simply add the constructs together in the same diluent at their normal working concentration. If one construct is at much higher concentration than another, some adjustment (increase) in concentration may be required for the lesser construct. Alternatively constructs may be added sequentially, preferably with the highest concentration construct first, followed by the lesser.
FSL construct concentrations above 1000 μg/mL may introduce significant amounts of lipid into a cell membrane, and may change the shape of the cell or make it more susceptible to lysis.
FSL construct working solutions should be stored at 2-8°C and used within a few days.
FSL constructs can be diluted in water but will have reduced stability and must be used within hours. If the package insert specifies water as the reconstitution diluent the product in the vial will also contain salts (as specified in the package insert).
2. Insertion of FSL Construct(s) into Membranes
First wash the cells for FSL modification free of unbound lipids by centrifugation and using PBS or lipid free cell media as wash solution.
Pack the cells or suspend in 100 μL of diluent.
Simultaneously prepare controls which ideally should be both unmodified cells (incubated with PBS rather than FSL solution) and/or cells modified with a benign but related FSL construct.
Add 100 μL of an appropriate dilution of FSL solution (containing 1 or more FSL constructs) to the cells and incubate for 1-2 hours at 37°C. A similar result can be obtained by 6 hours incubation at 25°C or overnight (18 hours) at 4°C – mixing is recommended every few hours if heavy cell suspensions are used.
Wash (optional) twice with PBS or media to remove any free FSL constructs and prepare an appropriate suspension.
Notes:
The diluent used to suspend the cells can be cell culture media, PBS, cell storage solutions, etc, but preferably without lipids ( e.g. fetal calf serum) or detergents (e.g. TWEEN).
Different volumes (than 100 μL) ratios (than 1:1 cells/FSL solution) or incubation conditions of time and temperature may be used provided the same ratio, concentration, and volume are used to obtain reproducible results.
3. Handling and Visualization
Once the FSL modification process has been completed the kodecytes can be stored at 4-8°C in lipid free media or used.
Kodecytes and kodevirions generally behave the same as unmodified cells/virions and can be handled and viewed under normal testing systems (microscopy, serology, flow cytometry, etc). They usually have no special requirements except for the avoidance of solvents/detergents/lipids that may elute the lipid constructs from membranes.
Notes:
Constructs will remain in the membrane of inactive cells, such as red cells, if stored in lipid free media for the life of the cell. In cells with active membranes the constructs will be internalized and metabolized, at a rate dependent on cell membrane activity.
Dead cells will usually contain high levels of FSL constructs; this can be used to separate nonviable cells from viable cells.
If kodecytes are stored (or used in vivo) in lipid containing media/environments, such as serum/plasma the construct will slowly elute from the membrane over a period of hours or days, depending on temperature and lipid concentrations/compositions.
4. Representative Results:
The following representative results depend on the construct used (Table 1), its inserted concentration and the relative sensitivity and specificity of the detection system(s). In general all FSL constructs will insert into cells after 1 hour but the optimal conditions will need to be established for each situation.
FSL constructs can be used to label the outside of a variety of live cells3-9,11,14. In the examples shown in Figure 2 embryos at different stages were used (as they are a fragile cell), but other cells (Figure 3) or enveloped virions (Figure 4) can also be labeled. FSL-Fluorescein allows for the direct labeling of surfaces (Figures 2-4), while FSL-biotin and other FSL constructs may require the use of a secondary detection system (usually avidin/antibodies/lectins) to show their presence (Figures 5).
Blood group markers of human or animal specificity can be attached to cells, including red cells4-9 (Figure 6). As the amount of FSL construct on a kodecyte is controllable and reproducible4-6, kodecytes for antibody quantitation can be made (Table 2 and Figure 6). The actual source of cells is not important, they can be any species, or even of the same origin as the source of antibody, thereby ensuring on the only incompatible antigen is that introduced by the FSL7,8 (Figure 6).
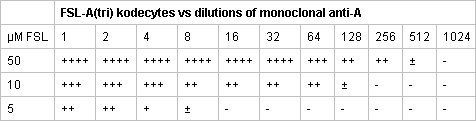 Table 2. Representative serological reactions of three different FSL-A(tri) red cell kodecytes tested against dilutions of monoclonal anti-A. The 50 μM solution of FSL-A(tri) when incubated with an equal volume of group O red cells produced strong A antigen positive cells, which could be detected by a 1:512 dilution of monoclonal anti-A. The 5- and 10 –fold lower FSL-A(tri) solutions produced kodecytes with less blood group A antigen expression.
Table 2. Representative serological reactions of three different FSL-A(tri) red cell kodecytes tested against dilutions of monoclonal anti-A. The 50 μM solution of FSL-A(tri) when incubated with an equal volume of group O red cells produced strong A antigen positive cells, which could be detected by a 1:512 dilution of monoclonal anti-A. The 5- and 10 –fold lower FSL-A(tri) solutions produced kodecytes with less blood group A antigen expression.
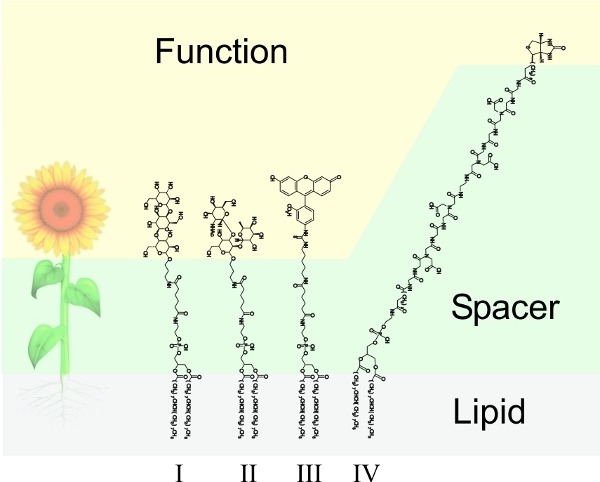 Figure 1. FSL construct overview. Like a flower FSL constructs consists of three major components. The functional head (F) in the case of the FSL construct can be a variety of different biologically functional groups, a spacer (S) designed to induce spacing of F away from the membrane, improve water dispersibility and be non-reactive with serum, while the diacyl lipid allows the construct to spontaneously incorporate into surfaces. (I) FSL-GB3 with a blood group Gb3 (or Pk) trisaccharide epitope Galα4Galβ4Glcβ. (II) FSL-A(tri) with a blood group A trisaccharide epitope GalNAcα3(Fucα2)Galβ. (III) FSL-Fluorescein. (IV) FSL-biotin with a single biotin F moiety. The functional group of structures in I-III are conjugated to an activated adipate derivative of dioleoylphosphatidylethanolamine (DOPE) while the spacer of structure IV is carboxymethylglycine-adipate based.
Figure 1. FSL construct overview. Like a flower FSL constructs consists of three major components. The functional head (F) in the case of the FSL construct can be a variety of different biologically functional groups, a spacer (S) designed to induce spacing of F away from the membrane, improve water dispersibility and be non-reactive with serum, while the diacyl lipid allows the construct to spontaneously incorporate into surfaces. (I) FSL-GB3 with a blood group Gb3 (or Pk) trisaccharide epitope Galα4Galβ4Glcβ. (II) FSL-A(tri) with a blood group A trisaccharide epitope GalNAcα3(Fucα2)Galβ. (III) FSL-Fluorescein. (IV) FSL-biotin with a single biotin F moiety. The functional group of structures in I-III are conjugated to an activated adipate derivative of dioleoylphosphatidylethanolamine (DOPE) while the spacer of structure IV is carboxymethylglycine-adipate based.
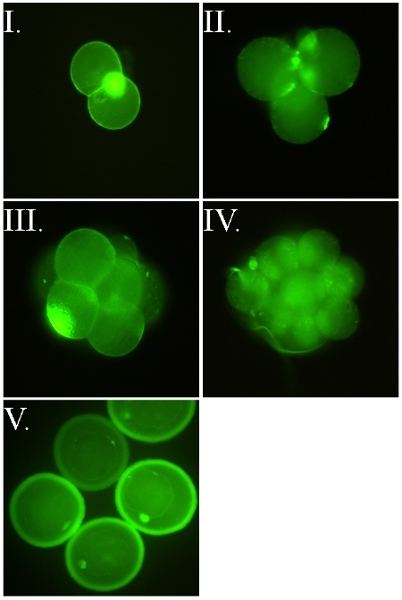 Figure 2. FSL-Fluorescein labeled murine embryos. All images are of live embryos that were labeled when their zona pellucida (ZP) was intact (i.e. the FSL construct passed through the ZP to label the embryo inside). Images I-IV are of ZP-free embryos released from their ZP with acid tyrodes post labeling with FSL-Fluorescein. Identical staining occurs regardless of whether embryos were FSL modified with ZP intact or ZP free. Embryos are directly labeled with FSL-Fluorescein, by the 2 hour 37°C method in serum free cell culture media, washed and then viewed under fluorescence microscopy. (I) ZP free two-cell murine embryo, also showing classic intense polar body staining. (II-III) ZP free four-cell and eight cell murine embryos, both showing shadowy staining of the cells which are outside the microscope plane of focus. (IV) ZP free 16 cell murine embryo. (V) ZP intact murine blastocyst embryos (d4-d5) where both the embryo and zona are labeled.
Figure 2. FSL-Fluorescein labeled murine embryos. All images are of live embryos that were labeled when their zona pellucida (ZP) was intact (i.e. the FSL construct passed through the ZP to label the embryo inside). Images I-IV are of ZP-free embryos released from their ZP with acid tyrodes post labeling with FSL-Fluorescein. Identical staining occurs regardless of whether embryos were FSL modified with ZP intact or ZP free. Embryos are directly labeled with FSL-Fluorescein, by the 2 hour 37°C method in serum free cell culture media, washed and then viewed under fluorescence microscopy. (I) ZP free two-cell murine embryo, also showing classic intense polar body staining. (II-III) ZP free four-cell and eight cell murine embryos, both showing shadowy staining of the cells which are outside the microscope plane of focus. (IV) ZP free 16 cell murine embryo. (V) ZP intact murine blastocyst embryos (d4-d5) where both the embryo and zona are labeled.
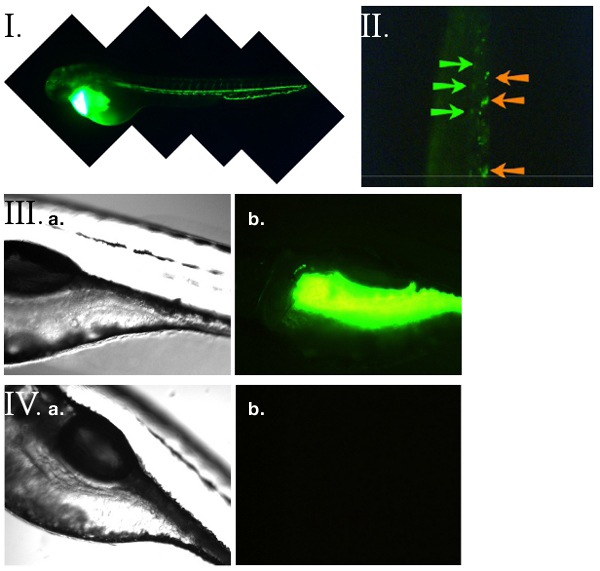 Figure 3. FSL-Fluorescein and zebrafish14. (I) Microangiography on a 52 hpf (hours post fertilisation) zebrafish larvae directly injected into the circulation with FSL-Fluorescein. The zebrafish vasculature is stained. (II) FSL-Fluorescein heterogenous Zebrafish kidney tissue cells (ZK kodecytes) were created ex vivo then micro-injected into the circulation of a 52 hpf recipient Zebrafish. In vivo observations of the ZK kodecytes were made 2 hours post injection by imaging of the vasculature under fluorescence with time-lapse microscopy. Shown is a single video frame with large slow-moving or immobile cells (indicated with orange arrows) and fast moving cells (blurred images due to movement - indicated with green arrows). Labeling allows for real-time in vivo observation of the behavior and biodistribution the ZK kodecytes. (III) Oral uptake of FSL-Fluorescein achieved by immersing zebrafish embryos in FSL-Fluorescein containing media for up to 5 days. Washing was achieved by transferring embryos into media containing no FSL constructs (at least 6 hours is required but can be for several days). (IIIa) Bright field microscopy of the FSL-Fluorescein treated zebrafish corresponding to the adjacent fluorescence image (IIIb). The fluoresence was preferentially located in the intestinal tract. No staining was observed in untreated control embryos (IVa and IVb).
Figure 3. FSL-Fluorescein and zebrafish14. (I) Microangiography on a 52 hpf (hours post fertilisation) zebrafish larvae directly injected into the circulation with FSL-Fluorescein. The zebrafish vasculature is stained. (II) FSL-Fluorescein heterogenous Zebrafish kidney tissue cells (ZK kodecytes) were created ex vivo then micro-injected into the circulation of a 52 hpf recipient Zebrafish. In vivo observations of the ZK kodecytes were made 2 hours post injection by imaging of the vasculature under fluorescence with time-lapse microscopy. Shown is a single video frame with large slow-moving or immobile cells (indicated with orange arrows) and fast moving cells (blurred images due to movement - indicated with green arrows). Labeling allows for real-time in vivo observation of the behavior and biodistribution the ZK kodecytes. (III) Oral uptake of FSL-Fluorescein achieved by immersing zebrafish embryos in FSL-Fluorescein containing media for up to 5 days. Washing was achieved by transferring embryos into media containing no FSL constructs (at least 6 hours is required but can be for several days). (IIIa) Bright field microscopy of the FSL-Fluorescein treated zebrafish corresponding to the adjacent fluorescence image (IIIb). The fluoresence was preferentially located in the intestinal tract. No staining was observed in untreated control embryos (IVa and IVb).
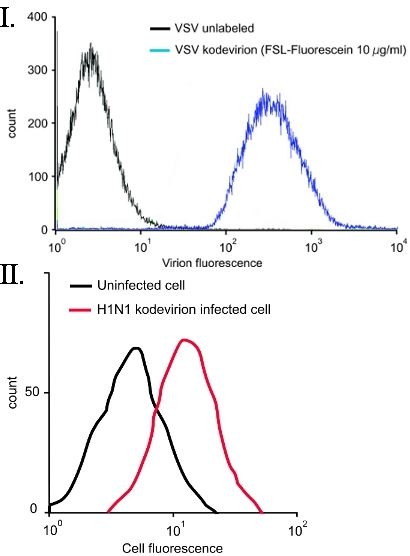 Figure 4. FSL-Fluorescein labeling of virions10 VSV and H1N1. (I) Vesicular stomatitis virus (VSV) was directly labeled with 10 μg/ml FSL-Fluorescein for 2 h at 37°C, followed by fixing with 4% paraformaldehyde and then flow cytometry. No purification of the VSV kodevirion post FSL labeling was required. (II) Flow cytometry of swine testicular cells infected with human A/Puerto Rico/8/1934 (H1N1) kodevirions labeled using FSL-Fluorescein. Uninfected cells are seen as the black line while fusion of the H1N1 kodevirion with the ST cells results in a fluorescent cell (red line).
Figure 4. FSL-Fluorescein labeling of virions10 VSV and H1N1. (I) Vesicular stomatitis virus (VSV) was directly labeled with 10 μg/ml FSL-Fluorescein for 2 h at 37°C, followed by fixing with 4% paraformaldehyde and then flow cytometry. No purification of the VSV kodevirion post FSL labeling was required. (II) Flow cytometry of swine testicular cells infected with human A/Puerto Rico/8/1934 (H1N1) kodevirions labeled using FSL-Fluorescein. Uninfected cells are seen as the black line while fusion of the H1N1 kodevirion with the ST cells results in a fluorescent cell (red line).
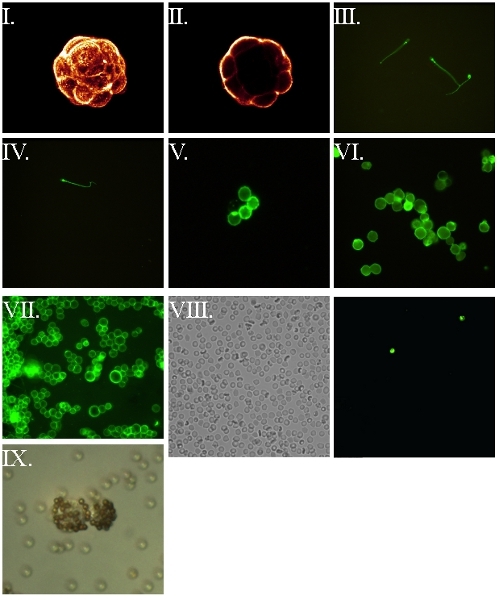 Figure 5. FSL-biotin labeled cells and subsequent visualization via labeled avidin7,8. All cells were first labeled with FSL-biotin for 1 h at 37°C, washed then reacted with fluorophore labeled avidin, washed and wet mounted for fluorescence microscopy. (I) Compiled confocal image of a murine embryo blastocyst. (II) Central confocal slice of the embryo from the preceding image. (III) Live motile human spermatozoa – blurring occurs as a consequence of their motion. (IV) Fixed (4% paraformaldehyde post insertion) human spermatozoa. (V) Human erythrocyte. (VI) Fixed (4% paraformaldehyde pre-insertion) RL95 endometrial human carcinoma. (VII) Unfixed RL95 endometrial human carcinoma cell line. (VIII) Biotin RBC kodecytes observed in a blood sample taken 2 hours post intravenous infusion of kodecytes with (VIIIa) representing a field viewed under light microscopy while (VIIIb) is the same field viewed under fluorescence, identifying the two kodecytes present in the field-of-view. Calculating the ratio of kodecytes to unlabeled cells can be use as an indicator of survival. (IX) Live human endometrial biotin kodecytes visualized by binding to avidinylated beads.
Figure 5. FSL-biotin labeled cells and subsequent visualization via labeled avidin7,8. All cells were first labeled with FSL-biotin for 1 h at 37°C, washed then reacted with fluorophore labeled avidin, washed and wet mounted for fluorescence microscopy. (I) Compiled confocal image of a murine embryo blastocyst. (II) Central confocal slice of the embryo from the preceding image. (III) Live motile human spermatozoa – blurring occurs as a consequence of their motion. (IV) Fixed (4% paraformaldehyde post insertion) human spermatozoa. (V) Human erythrocyte. (VI) Fixed (4% paraformaldehyde pre-insertion) RL95 endometrial human carcinoma. (VII) Unfixed RL95 endometrial human carcinoma cell line. (VIII) Biotin RBC kodecytes observed in a blood sample taken 2 hours post intravenous infusion of kodecytes with (VIIIa) representing a field viewed under light microscopy while (VIIIb) is the same field viewed under fluorescence, identifying the two kodecytes present in the field-of-view. Calculating the ratio of kodecytes to unlabeled cells can be use as an indicator of survival. (IX) Live human endometrial biotin kodecytes visualized by binding to avidinylated beads.
 Figure 6. FSL-Carbohydrate to add blood group markers for serologic profiling. (I) Human red cell Galili kodecytes were created with a set concentration of FSL-Galili (500 μg/mL) and tested against dilutions of human serum. Human red cells do not naturally occur with the xenoantigen Galili antigen. Such kodecytes can be used to quantitative levels of antibody in serum. In this example the donor was determined to have an anti-Galili titre of 1:32. (II) By creating kodecytes with decreasing levels of FSL ("antigen titres"), an optimal antigen level to detect antibody can be determined (in this example Lea). Cells can be created to only give a positive result when the antibody level exceeds a specific titre. The level of FSL antigen required to give a positive result depends on the quality and level of antibody being detected. For carbohydrate antigens, a FSL solution of 100 μg/mL will typically result in a strong positive reaction. (III) Human group O red cells were modified to have a specific level of A antigen (standardized A kodecytes), and used to accurately and reproducibly quantitate anti-A in human serum. In this example the anti-A titre in the group O serum tested is 1:32 and the kodecytes were prepared from the donor’s own red cells. (IV) Blood group A and B antigens simultaneously inserted into a single group O red cell sample are used to create AweakBweak kodecytes. These kodecytes can be used for ABO quality control purposes. This result shows analysis of a specifically formulated AweakBweak kodecyte tested against anti-A and anti-B reagents and gives expected weak reactions.
Figure 6. FSL-Carbohydrate to add blood group markers for serologic profiling. (I) Human red cell Galili kodecytes were created with a set concentration of FSL-Galili (500 μg/mL) and tested against dilutions of human serum. Human red cells do not naturally occur with the xenoantigen Galili antigen. Such kodecytes can be used to quantitative levels of antibody in serum. In this example the donor was determined to have an anti-Galili titre of 1:32. (II) By creating kodecytes with decreasing levels of FSL ("antigen titres"), an optimal antigen level to detect antibody can be determined (in this example Lea). Cells can be created to only give a positive result when the antibody level exceeds a specific titre. The level of FSL antigen required to give a positive result depends on the quality and level of antibody being detected. For carbohydrate antigens, a FSL solution of 100 μg/mL will typically result in a strong positive reaction. (III) Human group O red cells were modified to have a specific level of A antigen (standardized A kodecytes), and used to accurately and reproducibly quantitate anti-A in human serum. In this example the anti-A titre in the group O serum tested is 1:32 and the kodecytes were prepared from the donor’s own red cells. (IV) Blood group A and B antigens simultaneously inserted into a single group O red cell sample are used to create AweakBweak kodecytes. These kodecytes can be used for ABO quality control purposes. This result shows analysis of a specifically formulated AweakBweak kodecyte tested against anti-A and anti-B reagents and gives expected weak reactions.
Discussion
Modifying cells and virions with FSL constructs is a very simple and robust technique3-11. To describe and distinguish FSL construct modified cells and virions from unmodified cells, they are termed kodecytes and kodevirions respectively6-10, but only when the introduced functional group can be shown to be present on their membrane. When kodecytes are made with different concentrations of FSL constructs, they can be referred to by the concentration of the FSL construct solution used to create them, e.g. 15 μg/mL A kodecytes, or if more than one FSL is used then a combined term, e.g. A+biotin kodecytes7,8. When different cells are to be compared, including the cell type in the description is recommended, e.g. red cell biotin kodecytes or endometrial biotin kodecytes7,8.
Although the process of insertion is very robust and insertion (albeit with different rates) will occur over a large temperature range from 4 - 37°C and within minutes to hours, to obtain reproducible insertion strict control of contact time, temperature, FSL concentration (including diluent formulation), and to a lesser extent cell concentration are required. To date, all FSL constructs can be used to modify cells by the same technique4-11, however it is expected that certain conditions will be more favorable to work-flow and/or the optimal handling requirements of the cells/virions to be modified. The exact FSL concentration to achieve the desired effect needs to be determined by the user4. The resultant FSL construct modified cells/virions can usually be used in the same manner as an unmodified cell/virion in biological or analytical systems4-11. As the FSL constructs will slowly elute from modified surfaces when in contact with lipid solutions7,8, or be consumed by active cells11, these issues must be considered with respect to the level of signal or activity obtained over periods of time. Kodecytes can be fixed (e.g. glutaraldehyde/formalin) after insertion provided the fixative does not contain lipid-eluting solvents and the functional group is compatible with the fixative. Alternatively fixed cells can be modified with FSL constructs.
In addition to the modification of cell and virion surfaces FSL constructs can also be infused directly into the circulation of laboratory animals causing in vivo modification of circulating cells7 and to inhibit viruses12, toxins,12 and antibodies7. FLS constructs have also been used to decorate liposomes and can be printed onto paper surfaces13, where they immobilize, and can then be used in diagnostic assays.
The ability to easily modify live cells with an increasing range of FSL constructs should prove to be a useful research and development tool for the study of cell surface biology.
Disclosures
Deborah A. Blank and Stephen M. Henry are employees and stockholders of KODE Biotech Limited the patent owner of KODE technology. Dan Bess is an employee of Sigma-Aldrich, which has sponsored the production of this video-article.
Acknowledgments
The authors thank Dan Bess from Sigma-Aldrich for presenting the video. We also thank Don Branch, Evgenii Cherny, Scott Chesla, Elizabeth Hadac, Amanda Harrison, Damien Heathcote, Annika Hult, Chuan-Ching Lan, Susannah McIntosh, Sarvani Komarraju, Elena Korchagina, Caroline Oliver, Martin Olsson, Stephen Parker, Igor Rodinov, Alexander Tuzikov, and Eleanor Williams for their results and contributions to the design, synthesis of FSL constructs and determining biological activity.
References
- Waehler R, Russell SJ, Curiel DT. Engineering targeted viral vectors for gene therapy. Nat. Rev. Genet. 2007;8:573–587. doi: 10.1038/nrg2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strable E, Finn MG. Chemical modification of viruses and virus-like particles. Curr. Top. Microbiol. Immunol. 2009;327:1–21. doi: 10.1007/978-3-540-69379-6_1. [DOI] [PubMed] [Google Scholar]
- Henry S, Bovin N. The development of synthetic peptidolipids, glycolipids and other lipid-linked structures to create designer red cells. Transfusion. 2008;48(2S Suppl):194A–194A. [Google Scholar]
- Frame T, Carroll T, Korchagina E, Bovin N, Henry S. Synthetic glycolipid modification of red blood cell membranes. Transfusion. 2007;47:876–882. doi: 10.1111/j.1537-2995.2007.01204.x. [DOI] [PubMed] [Google Scholar]
- Hult AK, Frame T, Henry S, Olsson ML. Flow cytometry evaluation of red blood cells transformed with variable amounts of synthetic A and B glycolipids. Vox Sang. 2008;95(Suppl 1):180–180. [Google Scholar]
- Henry S. Modification of red blood cells for laboratory quality control use. Curr. Opin. Hematol. 2009;16:467–472. doi: 10.1097/MOH.0b013e328331257e. [DOI] [PubMed] [Google Scholar]
- Oliver C, Blake D, Henry S. In vivo neutralization of anti-A and successful transfusion of A antigen incompatible red cells in an animal model. Transfusion. 2011 doi: 10.1111/j.1537-2995.2011.03184.x. Forthcoming. [DOI] [PubMed] [Google Scholar]
- Oliver C, Blake D, Henry S. Modeling transfusion reactions and predicting in vivo cell survival with kodecytes. Transfusion. 2011 doi: 10.1111/j.1537-2995.2010.03034.x. Forthcoming. [DOI] [PubMed] [Google Scholar]
- Heathcote D, Carroll T, Wang JJ, Flower R, Rodionov I, Tuzikov A, Bovin N, Henry S. Novel antibody screening cells, MUT+Mur kodecytes, created by attaching peptides onto erythrocytes. Transfusion. 2010;50:635–641. doi: 10.1111/j.1537-2995.2009.02480.x. [DOI] [PubMed] [Google Scholar]
- Hadac EM, Federspiel MJ, Chernyy E, Tuzikovb A, Korchagina E, Bovin NV, Russell SJ, Henry SM. Fluorescein and radiolabeled Function-Spacer-Lipid constructs allow for simple in vitro and in vivo bioimaging of enveloped virions. J Virol Meth. 2011;10 doi: 10.1016/j.jviromet.2011.06.005. Forthcoming. [DOI] [PubMed] [Google Scholar]
- Blake D, Lan A, Love D, Bovin N, Henry S. Fluorophore-kodecytes - fluorescent function-spacer-lipid (FSL) modified cells for in vitro and in vivo analyses. FEBS J. 2010;277(Suppl 1):199–199. [Google Scholar]
- Harrison AL, Olsson ML, Brad Jones R, Ramkumar S, Sakac D, Binnington B, Henry S, Lingwood CA, Branch DR. A synthetic globotriaosylceramide analogue inhibits HIV-1 infection in vitro by two mechanisms. Glycobiology. 2010;27:515–524. doi: 10.1007/s10719-010-9297-y. [DOI] [PubMed] [Google Scholar]
- Barr K, Diegel O, Parker S, Bovin N, Henry S. Function-Spacer-Lipid (FSL) constructs enable inkjet printing of blood group antigens. FEBS J. 2010;277(Suppl 1):235–235. [Google Scholar]
- Lan C-C. Modeling inflammatory bowel disease [dissertation] Auckland: University of Auckland; 1988. [Google Scholar]


