Summary
AtxA, a unique regulatory protein of unknown molecular function, positively controls expression of the major virulence genes of Bacillus anthracis. The 475-amino acid sequence of AtxA reveals DNA-binding motifs and regions similar to proteins associated with the phosphoenolpyruvate:carbohydrate phosphotransferase system (PTS). We used strains producing native and functional epitope-tagged AtxA proteins to examine protein-protein interactions in cell lysates and in solutions of purified protein. Co-affinity purification, non-denaturing poly-acrylamide gel electrophoresis, and bis(maleimido)hexane (BMH) cross-linking experiments revealed AtxA homo-multimers. Dimers were the most abundant species. BMH cross-links available cysteines within 13Å. To localize interaction sites, six AtxA mutants containing distinct Cys→Ser substitutions were tested for multimerization and cross-linking. All mutants multimerized, but one mutation, C402S, prevented cross-linking. Thus, BMH uses C402 to make the inter-molecular bond between AtxA proteins, but C402 is not required for protein-protein interaction. C402 is in a region bearing amino acid similarity to Enzyme IIB proteins of the PTS. The AtxA EIIB motif may function in protein oligomerization. Lastly, cultures grown with elevated CO2/bicarbonate exhibited increased AtxA dimer/monomer ratios and increased AtxA activity, relative to cultures grown without added CO2/bicarbonate, suggesting that this host-associated signal enhances AtxA function by shifting the dimer/monomer equilibrium toward the dimeric state.
Introduction
Coordinate control of gene expression in response to niche-specific signals is a common theme for microorganisms. Often one or more trans-acting factors regulate transcription of sets of genes encoding products appropriate for physiological responses to environmental signals. Classic bacterial signal transduction systems are two-component systems comprised of a sensor/transducer protein, which has kinase activity and a response regulator protein, which has nucleic acid binding activity. Other systems include multi-component signal transduction pathways with multiple inputs and regulatory points, and apparent ‘stand-alone’ regulators that appear to mediate transcriptional changes in the absence of other factors, either because the regulator senses a signal directly or because a cognate sensor has not been identified (Bourret & Silversmith, 2010).
Coordinate control of virulence gene expression in Bacillus anthracis, the causative agent of anthrax, is mediated by AtxA, a 475-amino acid protein encoded by the virulence plasmid pXO1 (Uchida et al., 1993, Koehler et al., 1994). An atxA-null mutant of B. anthracis is attenuated in a murine model of anthrax disease (Dai et al., 1995). Deletion of atxA results in decreased transcription of the structural genes for the anthrax toxin protein genes, pagA, lef, and cya, located at distinct loci on pXO1 (182 kb), and the capsule biosynthesis operon, capBCADE, located on another virulence plasmid pXO2 (96 kb) (Uchida et al., 1993, Koehler et al., 1994, Dai et al., 1995, Fouet & Mock, 1996, Uchida et al., 1997, Guignot et al., 1997, Sirard et al., 2000, Mignot et al., 2003). In addition to controlling established virulence genes, atxA regulates at least 39 other genes on the chromosome, pXO1, and pXO2 (Hoffmaster & Koehler, 1997, Hoffmaster & Koehler, 1999, Bourgogne et al., 2003).
The molecular basis for AtxA function as a positive regulator of B. anthracis virulence genes has not been established. The amino acid sequence of AtxA reveals a number of motifs associated with potential functional domains. The amino terminal region of the protein contains adjacent winged helix and helix-turn-helix (HTH) motifs. The HTH motif is similar to that of the Streptococcus pyogenes virulence regulator Mga (Tsvetanova et al., 2007), which binds specific DNA sequences in the promoter regions of target genes (McIver et al., 1995). These motifs suggest DNA-binding activity (Uchida et al., 1997, Tsvetanova et al., 2007, Koehler, 2009), but sequence specific DNA-binding by AtxA has not been reported. The central region of the AtxA amino acid sequence reveals motifs suggesting two phosphoenolpyruvate:carbohydrate phosphotransferase system (PTS) regulation domains (PRDs). PRDs are generally found in bacterial proteins that function to activate or anti-terminate transcription of genes involved in the metabolism of specific carbohydrates and their function is typically affected by phosphorylation (Deutscher et al., 2006). Transcriptional profiling studies have not revealed AtxA targets associated with catabolism (Bourgogne et al., 2003), but Tsvetanova et al. (2007) reported that phosphorylation of specific histidine residues, H199 and H379, each within a PRD, impacts AtxA activity.
A key host-related signal associated with AtxA-regulated gene expression is CO2/bicarbonate. Growth of B. anthracis in 5% or greater atmospheric CO2 in buffered medium results in elevated transcription of the toxin, capsule, and other AtxA-regulated genes (Bartkus & Leppla, 1989, Cataldi et al., 1992, Sirard et al., 1994, Koehler et al., 1994, Dai et al., 1995, Fouet & Mock, 1996). Some studies suggest that the CO2/bicarbonate effect on toxin genes occurs in an atxA-dependent manner (Koehler et al., 1994, Dai & Koehler, 1997, Mignot et al., 2003, Chitlaru et al., 2006), yet the signal does not appear to significantly affect steady state levels of atxA transcript or AtxA protein (Dai & Koehler, 1997, Drysdale et al., 2005). Moreover, a protein-ligand relationship between CO2/bicarbonate and AtxA has not been established, nor have additional proteins associated with AtxA function been identified.
Although CO2/bicarbonate does not significantly impact atxA expression, AtxA levels are affected by a number of other signals and trans-acting proteins. Levels of atxA transcript are altered in response to temperature, redox state, and carbohydrate availability (Dai & Koehler, 1997, Wilson et al., 2009, Chiang et al., 2011). Deletion of ccpA, the carbon catabolite control protein, results in decreased transcription of atxA (Chiang et al., 2011). Mutations in genes involved in cytochrome c biogenesis increase atxA transcription in non-toxin inducing conditions (Wilson et al., 2009). Only one regulatory protein has been demonstrated to interact directly with the atxA promoter; the transition state regulator AbrB binds to the atxA promoter to repress transcription during exponential phase growth in batch culture (Saile & Koehler, 2002, Strauch et al., 2005). AtxA protein levels are also subject to post-translational control. CodY, a pleiotropic transcriptional regulator found in a number of Gram-positive bacteria (Sonenshein, 2005), influences AtxA protein levels by an unknown mechanism (van Schaik et al., 2009).
In this report we provide data indicating that AtxA exists in a homo-oligomeric state and that AtxA-AtxA contacts occur within a previously uncharacterized carboxy-terminal EIIB-like motif. We also show a correlation between growth in elevated CO2/bicarbonate, the steady state level of AtxA multimer, and AtxA activity.
Results
AtxA activity in vivo
AtxA is a cytoplasmic protein with a predicted MW of 55.6 kDa (Uchida et al., 1993). Some functional domains have been postulated (Tsvetanova et al., 2007, Koehler, 2009); however, the structure of AtxA has not been solved. Moreover, the multimeric state of AtxA in vivo and whether AtxA associates with other B. anthracis proteins, have not been discerned.
Expression of the atxA gene is highly regulated. Therefore, to investigate AtxA form and function in vivo, we developed a system to control AtxA protein levels and quantitatively assess AtxA activity in B. anthracis. Alleles of atxA encoding native, mutant, and C-terminal epitope-tagged versions of the protein were cloned such that expression was under control of the IPTG-inducible hyper-spank promoter on a low copy number plasmid, pUTE657. Recombinant epitope-tagged proteins, AtxA-His and AtxA-FLAG, facilitated immuno-detection and purification from B. anthracis. AtxA positively controls transcription of multiple genes in B. anthracis, although a direct interaction of the AtxA protein with promoters of target genes has not been demonstrated. One of the genes most strongly regulated by AtxA is lef, encoding the lethal factor protein, a component of the anthrax toxin (Bourgogne et al., 2003). Plasmids bearing IPTG-inducible atxA alleles were introduced into a B. anthracis strain (UT376) which harbors a markerless deletion of atxA and a transcriptional reporter, Plef-lacZ, at the native lef locus on pXO1. AtxA activity was quantified by measuring β-galactosidase activity following IPTG induction of the atxA alleles.
Strains were cultured in CACO3 in a 5% CO2 atmosphere, conditions shown previously to be conducive for atxA-controlled toxin gene expression (Dai et al., 1995). Figure 1 shows relative AtxA levels and activities in B. anthracis cultures expressing native AtxA, AtxA-His and AtxA-FLAG. RNA polymerase β subunit was used as a loading control. Induction with 30 μM IPTG yielded levels of epitope-tagged proteins that were comparable to untagged AtxA (Fig. 1A), indicating that protein stability was unaltered by the C-terminal His- and FLAG-tags. The cultures also displayed similar levels of β-galactosidase activity (Fig. 1B), indicating that the recombinant proteins do not exhibit altered function. The amount of induced AtxA is 8.5-fold greater than the level of AtxA expressed from its native promoter on pXO1 and results in 3-fold more β-galactosidase activity (data not shown). Notably, expression of the Plef-lacZ fusion in the atxA-null strain containing the empty vector (pUTE657) was barely detectable, confirming previous reports that transcriptional control of the lef gene is highly dependent upon AtxA (Uchida et al., 1993, Dai et al., 1995).
Fig. 1.
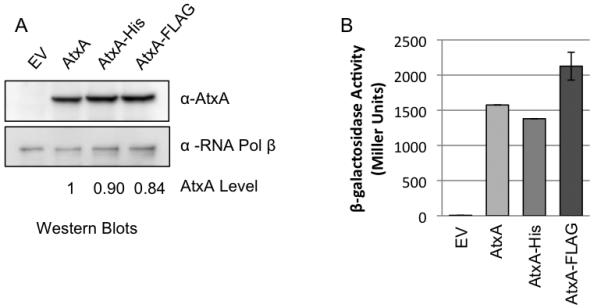
In vivo Activity of AtxA-His and AtxA-FLAG. In an atxA-null strain (UT376), production of AtxA (pUTE658), AtxA-His (pUTE991), and AtxA-FLAG (pUTE992) was induced using 30 μM IPTG during growth in CACO3. The empty vector sample (EV) was derived from UT376 (pUTE657) which lacks atxA. Samples were obtained at the transition to stationary phase (4 h; OD600 1.2 to 1.7). (A) AtxA and RNA Polymerase β subunit in soluble cell lysates were detected via immunoblotting. The AtxA levels indicated were normalized relative to the corresponding RNA polymerase β level. (B) The β-galactosidase activity of B. anthracis mutants harboring the Plef-lacZ reporter and IPTG-inducible atxA alleles was determined as described previously (Miller, 1972).
Multimerization of AtxA
In vivo AtxA – protein interactions are of interest because DNA-binding proteins and proteins containing PRDs often function as dimers (Declerck et al., 2001, Huffman & Brennan, 2002, Fux et al., 2003, Ben-Zeev et al., 2005, Graille et al., 2005). Furthermore, some investigations have suggested that an additional factor is required for AtxA function (Koehler et al., 1994, Tsvetanova et al., 2007). To explore possible protein-protein interactions, we subjected lysates from B. anthracis strains producing native AtxA to Blue Native PAGE (BN-PAGE) to determine the migration pattern of AtxA in native conditions. Electrophoresed proteins were transferred to membranes and probed with α-AtxA antibody. The mobility of immuno-reactive bands relative to molecular weight markers suggested that AtxA was in a complex (Fig. 2A).
Fig. 2.
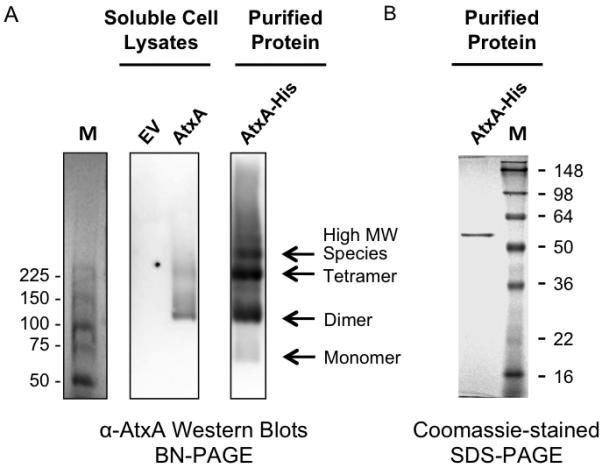
Oligomeric states of AtxA. (A) Lysates from B. anthracis strains UT376 (pUTE657) and UT376 (pUTE658), containing an empty vector (EV) and IPTG-inducible atxA respectively, and affinity-purified AtxA-His were electrophoresed on BN polyacrylamide gels. AtxA protein was detected using Western blotting. M = molecular weight markers (ProSieve Unstained Protein Marker, VWR). (B) Affinity purified AtxA-His from B. anthracis UT376 (pUTE991) was subjected to SDS-PAGE and stained with Coomassie. M = molecular weight markers (SeeBlue Plus2 Pre-Stained Standard, Invitrogen).
To test for stable interactions between AtxA and other cytosolic proteins, we subjected cell lysates of B. anthracis cells producing AtxA-His to affinity purification using NTA-Ni resin. Purified AtxA-His was analyzed using BN-PAGE and denaturing PAGE. Western hybridization using α-AtxA serum revealed three major bands migrating at approximately 110 kDa, 225 kDa, and > 225 kDa and a minor band migrating at approximately 60 kDa (Fig. 2A). In contrast, Coomassie staining after SDS-PAGE revealed only one band in the AtxA-His eluate and its gel mobility was consistent with the size of an AtxA-His monomer (56 kDa) (Fig 2B). Since no other proteins were detected in the AtxA-His eluate via SDS-PAGE and the protein mobilized as multiple species on BN-PAGE, these results suggested that AtxA forms multiple homomeric complexes in cell lysates.
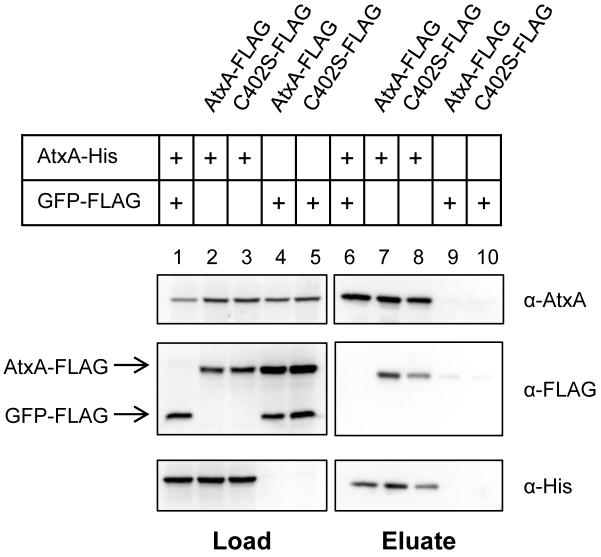
To detect association of two or more monomers of AtxA, we combined culture lysates of B. anthracis strains producing AtxA-His or AtxA-FLAG and used affinity chromatography to purify AtxA-His. A lysate from a B. anthracis strain producing GFP-FLAG was used as a control. Lysates were derived from pools of cultures containing (1) AtxA-His and GFP-FLAG, (2) AtxA His and AtxA-FLAG, and (3) AtxA-FLAG and GFP-FLAG. Proteins were captured using NTA-Ni resin, eluted with imidazole, and detected by Western blotting. As shown in Figure 3 (lanes 1-3), prior to affinity purification each tagged protein was detected in the appropriate pools. Eluates from NTA-Ni resin are shown in lanes 4-6. AtxA-FLAG was detected in the eluate only when combined with AtxA-His (lane 5). GFP-FLAG served as a negative control to ensure that non-specific proteins were washed from the NTA-Ni resin and that the His and FLAG tags did not interact. These results demonstrate that AtxA-His and AtxA-FLAG form a stable complex in vitro. Considering these data and the results from BN-PAGE, we conclude that AtxA self-associated to form dimers, tetramers, and higher MW species.
Fig. 3.
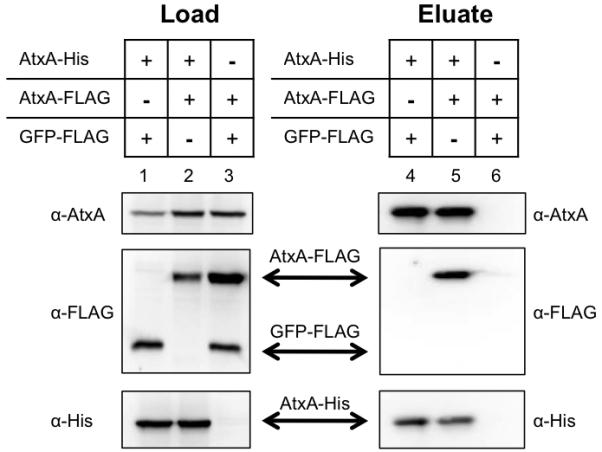
Specific binding of AtxA-His to AtxA-FLAG. Lysates from B. anthracis atxA-null strains (UT376) containing AtxA-His (pUTE991), AtxA-FLAG (pUTE992), or GFP-FLAG (pUTE1013) induced with 30 μM IPTG were subjected to co-affinity purification using NTA-Ni resin. Western blots probed with α-AtxA, α-FLAG, and α-His antibodies were performed on soluble cell lysates (Load, lanes 1-3) and purified proteins (Eluate, lanes 4-6).
Role of the Carboxy-terminal Region of AtxA in Multimerization
To further explore the properties of AtxA multimerization, we tested for the ability of the cross-linking reagent BMH to covalently fix AtxA complexes in cell lysates. BMH reacts specifically and irreversibly to link cysteine residues within 13Å. AtxA has cysteine residues at positions 96, 161, 202, 356, 370, and 402 (Fig. 5A). A cell lysate from an IPTG-induced culture expressing AtxA was treated with BMH, subjected to SDS-PAGE, and probed for AtxA via Western blot (Fig. 4). Two bands were detected slightly above the 98-kDa marker when the lysate was treated with BMH (lane 6), while a single band near the 50-kDa marker was detected in an untreated lysate (lane 2). The predicted molecular weight of AtxA is 55.6 kDa; therefore, the approximately 100-kDa bands may represent two AtxA proteins joined via BMH. The appearance of the doublet band indicates two separate AtxA complexes present in the cross-linked lysate. To investigate the composition of the protein complexes, we affinity purified AtxA His from an atxA-null strain and subjected the protein to BMH cross-linking. The migration pattern of cross-linked, purified AtxA-His was similar to that observed for AtxA and AtxA-His cross-linked in cell lysates (Fig. 4, lanes 6-8); a doublet band migrated near the 98-kDa molecular weight marker. We conclude that the doublet does not result from AtxA linked to another protein. Rather, the doublet suggests at least two conformational states of the dimeric AtxA complex.
Fig. 5.
Determination of the site of BMH cross-linking. (A) The AtxA protein, which contains six cysteine residues, has five putative motifs: a winged helix-turn-helix (WH), a Mga-like helix-turn-helix (HTH), two phosphotransferase regulation domains (PRD1 and PRD2), and an Enzyme IIB-like motif (EIIB). (B) Following induction with 30 μM IPTG, cells from cultures of atxA-null strains (UT376) containing pUTE658-derived atxA point mutants were lysed and treated with BMH. SDS-PAGE (4-15%) and Western blots with AtxA-specific antibody were used to detect various forms of AtxA. (C) A similar experiment was performed using derivatives of the parent strain (UT375) expressing atxA from its native locus and pUTE992-derived FLAG-tagged versions of AtxA or the AtxA point mutants. Westerns were probed with α-FLAG and α-AtxA antibodies. EV = empty vector. Molecular weights of the protein standards are as indicated (SeeBlue Plus2, Invitrogen).
Fig. 4.
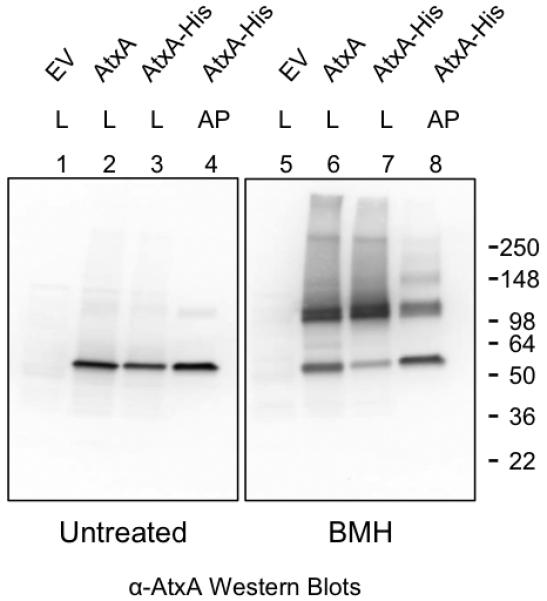
Fixation of AtxA complexes with a cysteine-reactive cross-linking agent. Cultures of the atxA-null strain UT376 containing pUTE658 (encoding AtxA) or pUTE991 (encoding AtxA-His) were induced with 40 μM IPTG. Cell lysates or affinity-purified AtxA-His were treated with the cross-linking agent BMH. SDS-PAGE (4-15%) and Western blots with AtxA-specific antibody were used to detect various forms of AtxA. EV = empty vector, L = cell lysate, and AP = affinity-purified. Molecular weights of the protein standards are listed (SeeBlue Plus2, Invitrogen).
BMH-mediated cross-linking of AtxA proteins allowed us to test for region(s) at the interface of AtxA multimers. We created strains harboring distinct atxA Cys→Ser alleles (Fig. 5A). Mutant AtxA proteins were expressed in the atxA-null background and cell lysates were reacted with BMH. The presence of the dimer band in lysates containing mutated AtxA (Fig. 5B) indicates that these proteins are capable of multimerization and are cross-linked with BMH. The lysate containing AtxA C402S differed significantly from lysates containing other AtxA mutants. The AtxA C402S sample contained a relatively low level of cross-linked protein and a high level of the apparent monomer. This result suggests that C402 is the major site of BMH crosslinking, but some other Cys residue(s) can participate in cross-linking.
To determine if the dominant BMH cross-link occurs between C402 of two AtxA monomers, AtxA C402S-FLAG was expressed in the parent strain of B. anthracis, which also expresses native AtxA from the atxA locus on pXO1. We reasoned that if BMH utilizes C402 on both proteins then, after cross-linking, FLAG-reactive protein would be detected as a monomer band (55 kDa) and not as a dimer containing AtxA C402S-FLAG and AtxA. Following BMH cross-linking, the native AtxA plus AtxA C402S-FLAG sample contained a high level of monomer when examined in Western blots probed with α-FLAG antibody (Fig. 5C, lane 5) similar to the sample containing only AtxA C402S (Fig. 5B, lane 8). In contrast, samples containing native AtxA plus Cys→Ser AtxA mutant proteins (C202S and C370S) or native AtxA plus AtxA-FLAG showed high levels of cross-linked protein reacting with α-FLAG antibody (Fig. 5C, lanes 2-4). Further, when the same samples were probed with α-AtxA antibody, dimer bands representing cross-linked AtxA (tagged and native) were present in all samples (lanes 6-10). Dimer bands detected in the AtxA C402S-FLAG plus AtxA sample probed with α-AtxA (lane 10) likely represent native AtxA because dimer bands were not detected when the same sample was probed with α-FLAG antibody (lane 5). The relatively low level of cross-linked protein in the sample containing only native AtxA (lane 6) and the sample containing native AtxA plus AtxA C402S-FLAG (lane 10) reflect low level AtxA expression from the native atxA locus. Induction of the tagged proteins (lanes 2-5 and 7-10) resulted in approximately 8.5-fold more tagged protein compared to native AtxA (Untreated lanes and data not shown). These results indicate that the major BMH cross-linked species is formed using C402 on two AtxA monomers.
To verify that AtxA C402S can form a complex with AtxA despite being defective for BMH cross-linking, we performed a co-affinity purification experiment using AtxA C402S-FLAG mixed with AtxA-His. After eluting protein from NTA-Ni resin, AtxA C402S-FLAG was present in the sample with AtxA-His, indicating that AtxA C402S has normal multimerization properties (Fig. 6). Taken together, these data demonstrate that BMH utilizes C402 to cross-link two AtxA proteins and suggests that the carboxy-terminus of AtxA is involved in oligomerization.
Fig. 6.
AtxA C402S does not have a multimerization defect. Lysates from the B. anthracis atxA-null strain UT376 expressing AtxA-His (pUTE991), GFP-FLAG (pUTE1013), AtxA-FLAG (pUTE992), or AtxA C402S-FLAG (pUTE992 C402S) were subjected to co-affinity purification using NTA-Ni resin. Samples were treated as described in Fig. 3.
Assessment of the carboxy-terminal 91-amino acid sequence of AtxA using the Conserved Domains Database (Marchler-Bauer et al., 2007) and Protein Homology/Analogy Recognition Engine (Phyre, (Kelley & Sternberg, 2009)), predicted that this region of AtxA is similar to an EIIB motif of the PTS, as depicted in Figure 5A. EIIB domains typically function enzymatically to phosphorylate carbohydrates after the sugars cross the cell membrane via an EIIC domain or protein. We performed cross-linking experiments using full length AtxA, AtxA1-385 (deletion of the EIIB-like motif), and AtxA385-475 (only the EIIB-like motif) to determine if the EIIB-like region of AtxA is involved in AtxA oligomerization. Cell lysates containing full-length and truncated FLAG-tagged proteins synthesized in parent and atxA-null backgrounds were subjected to BMH cross-linking. Bands corresponding to 56-kD, 40-kD, and 11-kD monomers of AtxA-FLAG, AtxA1-385-FLAG and AtxA385-475-FLAG, respectively, were observed in untreated lysates probed with α-FLAG antibody (Fig. 7 lanes 2-4 and 6-8). The BMH-treated samples displayed the typical dimer bands (100 kDa) for AtxA-FLAG (lanes 10 and 14). The migration of AtxA1-385-FLAG was not altered after cross-linking; only a 40-kDa band was present (lanes 11 and 15). This result is consistent with the requirement of C402 for cross-linking. The several FLAG-reactive bands present in lanes 12 and 16 represent multiple protein complexes containing AtxA385-475-FLAG and unidentified proteins. The large α-FLAG-reactive cross-linking products do not represent AtxA cross-linked to AtxA385-475-FLAG because the banding patterns of AtxA385-475-FLAG in the parent (lane 12) and atxA-null strain (lane 16) were identical. These data indicate that the EIIB-like region of AtxA is involved in AtxA oligomerization and suggest that the EIIB-like domain in the absence of other regions of the AtxA protein displays less specificity for AtxA, interacting with several other proteins in cell lysates. It is notable that the truncated AtxA proteins do not exhibit AtxA activity. B. anthracis UT376 mutants producing AtxA1-385, AtxA1-385-FLAG, AtxA385-475, or AtxA385-475-FLAG show no expression of the Plef-lacZ fusion (data now shown).
Fig. 7.
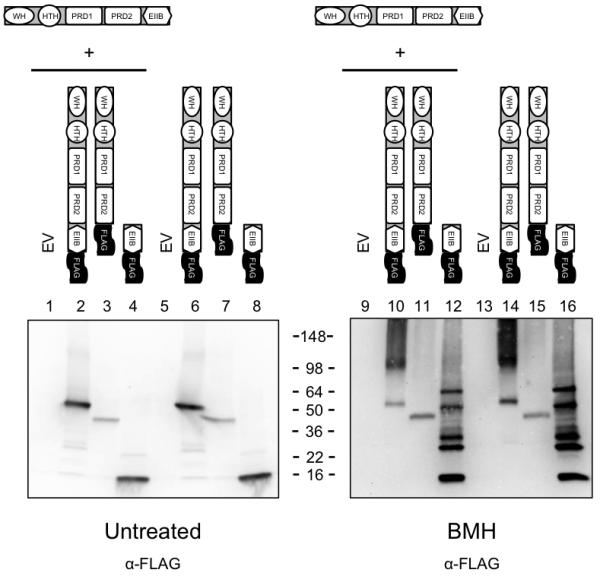
Multimerization of AtxA385-475. Cell lysates of parent (UT375) and atxA-null strains (UT376) containing AtxA-FLAG (pUTE992), AtxA1-385-FLAG (pUTE1019-FLAG), and AtxA385-475-FLAG (pUTE1022-FLAG) induced using 30 - 50 μM IPTG were treated with the cross-linking agent BMH. SDS-PAGE (4-20%) and Western blots with FLAG-specific antibody were used to detect various forms of AtxA. EV = empty vector. Molecular weights of the protein standards are listed (SeeBlue Plus2, Invitrogen).
Relationship between CO2/bicarbonate, AtxA Multimerization and Function
AtxA-dependent transcription of lef and other genes is enhanced during growth in buffered media in 5% CO2, relative to growth in air (Uchida et al., 1993, Sirard et al., 1994, Dai et al., 1995, Fouet & Mock, 1996). To address relationships between the CO2/bicarbonate signal, the oligomeric state of AtxA, and AtxA function in vivo, we compared AtxA multimerization and activity using cells cultured in CA medium with or without CO2/bicarbonate. Growth rates and IPTG-induced AtxA levels were similar for B. anthracis UT376 (pUTE658) cultures grown with or without added CO2/bicarbonate (data not shown). For each culture, AtxA function was assessed as β-galactosidase activity resulting from expression of the Plef-lacZ reporter gene, and cell lysates were subjected to BMH-mediated cross-linking to determine the levels of AtxA dimer. A representative experiment is shown in Figure 8. The AtxA dimer/monomer ratio was approximately 2-fold greater in cells cultured with elevated CO2/bicarbonate. The increased dimer level correlated with increased AtxA activity. The affect of the CO2/bicarbonate signal on the AtxA dimer/monomer ratio and AtxA activity was consistent in multiple experiments. These results show that the CO2/bicarbonate signal shifts the dimer/monomer equilibrium of AtxA toward the dimeric state and this shift is associated with enhanced protein function.
Fig. 8.
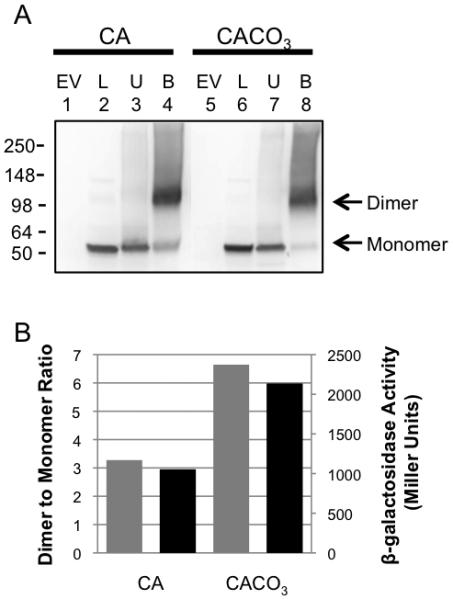
The CO2/bicarbonate effect on AtxA multimerization and activity. UT376 (pUTE658), an atxA-null strain harboring the native atxA gene driven by an IPTG-inducible promoter and the Plef-lacZ transcriptional fusion (as a reporter for AtxA activity) was cultured in CA in air and in CACO3 in a 5% CO2 atmosphere. Cells were collected two hours after induction with IPTG. (A) SDS-PAGE (4-15%) and Western blots with AtxA-specific antibody were used to detect various forms of AtxA. EV = empty vector (pUTE657), L = cell lysate, U = untreated samples, B = BMH treated samples. (B) Intensities of bands corresponding to the AtxA dimer and monomer were measured using ImageJ. Dimer/monomer ratios are shown as gray bars. β-galactosidase activities, resulting from expression of the Plef-lacZ reporter, are shown as black bars.
Discussion
We have determined that AtxA forms a multimeric protein complex independent of other proteins, and that the level of homodimeric AtxA is increased relative to monomeric AtxA in cells cultured in conditions conducive for function. Our experiments employing the BMH cross-linking reagent allow us to discern features of the AtxA multimer. BMH is specific for reduced, solvent-accessible cysteines located within 13 Å. We determined that C402 is the major site of cross-linking between two AtxA proteins, suggesting that C402 is likely surface exposed and in close proximity to the corresponding amino acid in an AtxA dimer. Since BMH cross-linking requires reduced cysteines, the C402 residues of associated AtxA proteins do not form a disulfide bond. Moreover, a C402S mutant forms multimers that are indistinguishable from those formed by the native protein. Our data indicate that C402 is located in a region of AtxA that forms protein-protein contacts, yet C402 is not required for the protein-protein interactions.
The Protein Homology/Analogy Recognition Engine “Phyre” (Kelley & Sternberg, 2009) is a homology-based, fold-recognition tool that provides three-dimensional structure predictions based on sequence similarity to proteins with known structures. Phyre reveals similarity between AtxA385-475, containing C402, and EIIB motifs of PTS proteins, leading us to propose a function for the carboxy-terminus of AtxA. As part of the PTS, EIIB proteins are the penultimate proteins in the phosphotransfer from phosphoenol-pyruvate (PEP) to imported carbohydrates (Deutscher et al., 2006). The PTS is composed of Enzyme I (EI) and HPr, which are general enzymes, and Enzyme II (EII) complexes, consisting of EIIA, EIIB, and EIIC, which recognize and transport specific carbohydrates. Conversion of PEP to pyruvate is catalyzed by EI. EI~P transfers the phosphate to HPr, which in turn phosphorylates EIIA. EIIA~P phosphorylates EIIB at a conserved cysteine residue within the P-loop, a flexible loop between β-sheet 1 and α-helix 1 (Evans et al., 1996). The phosphocysteine of EIIB transfers its phosphate to carbohydrate molecules crossing the cell membrane via the EIIC complex. The carboxy-terminus of AtxA is similar to GatB, an E. coli protein that is a member of the glucitol/galacitol family of EIIB proteins (Nobelmann & Lengeler, 1996). NMR studies of GatB with its cognate EIIA (GatA) reveal regions of GatB – GatA interaction (Volpon et al., 2006). It is thought that the GatB-GatA interface, which includes the P-loop, is also recognized by the corresponding EIIC protein, GatC (Volpon et al., 2006).
Interaction of GatB with two different proteins suggests involvement of EIIB-like motifs in protein-protein binding. In experiments, when AtxA385-475-FLAG is induced in an atxA-null strain or the parent strain containing AtxA, we detect four prominent α-FLAG-reactive products in cross-linked cell lysates. In contrast, when full-length AtxA-FLAG is induced in either strain, cross-linked lysates contain only one α-FLAG-reactive product, which corresponds to an AtxA-AtxA dimer. The presence of multiple cross-linked products in the AtxA385-475 sample suggests that the EIIB-like motif in the absence of other AtxA domains loses some specificity for protein-protein interactions. These data lead us to propose that the EIIB-like motif is involved in AtxA-AtxA interaction, yet one or more motifs within residues 1-385 of AtxA are required to confer specificity to the AtxA-AtxA interaction and to exclude interactions of the AtxA EIIB-like domain with other proteins.
The presence of an EllB-like motif adjacent to PRDs is not unique to AtxA. EIIB-like domains are found in certain other regulators, including the transcriptional activators LicR, MtlR, CelR, ManR and LevR (Deutscher et al., 2006, Joyet et al., 2010, Sun & Altenbuchner, 2010). Yet, unlike AtxA, these other regulators also contain EIIA-like motifs and the regulators control a small number of genes encoding proteins associated with use of a specific carbohydrate or group of carbohydrates. For example, in Streptococcus mutans CelR regulates the production of proteins required for cellobiose catabolism (Zeng & Burne, 2009), and MtlR induces expression of the mtl operon when Bacillus subitilis is in the presence of mannitol (Henstra et al., 1999, Watanabe et al., 2003). In contrast, AtxA is a virulence gene regulator that affects transcription of multiple genes on the chromosome and virulence plasmids within B. anthracis, none of which are known to be associated with catabolism (Bourgogne et al., 2003).
Interestingly, a cysteine residue within the EIIB-like motif in the MtlR protein of Bacillus subtilis was recently shown to be phosphorylated (Joyet et al., 2010). MtlR activity decreases when P~EIIAMtl phosphorylates MtlR at C419, the conserved cysteine within the P-loop of the EIIB-like motif. In the Phyre-produced model of the EIIB-like motif of AtxA, C402 is located in the P-loop, but the residue is not at the position typically observed for the conserved phosphocysteine in EIIB proteins of the PTS or for the EIIB-like motif in MtlR and LicT. Moreover, current data do not support phosphorylation of AtxA at this site. Phosphorylated C402 would have prevented cross-linking by BMH in our experiments. Also, Tsvetanova et al. (2007), reported that phosphorylation of AtxA occurs only within the PRDs at residues H199 and H379. 32PO4-labelled AtxA was immunoprecipitated from B. anthracis cell lysates, but no radio-labelled AtxA was evident in a H199A / H379A mutant.
Phosphorylation of AtxA may impact the multimeric state of the protein. Phosphorylation can stabilize or disrupt dimerization of the PRD-containing antiterminator proteins LicT and BglG (Declerck et al., 2001, Fux et al., 2003, Ben-Zeev et al., 2005, Graille et al., 2005). Dimer contacts within the PRDs of LicT are confined to α-helices 2, 3, and 5 in each PRD, and the helices are arranged in an anti-parallel orientation (van Tilbeurgh et al., 2001). Considering reports concerning LicT and BglG, it is possible that the AtxA PRDs are involved in multimerization. The Phyre-predicted structure of the PRDs of AtxA positions C202 in α-2 of PRD1, C356 in α-4 of PRD2, and C370 in α-5 of PRD2, yet in our studies, C202, C356, and C370 were not required for cross-linking of AtxA proteins by BMH. It is possible that these cysteine residues are in regions that function in multimerization. However, the sulfhydryl groups are inaccessible to BMH because they are buried in the interior of AtxA, oxidized into a disulfide bond, or are greater than 13 Å from another cysteine. More work needs to be performed to determine the protein-protein contacts existing throughout the AtxA protein and to assess potential affects of phosphorylation on multimerization of AtxA.
Our findings show for the first time a link between AtxA multimerization, function and an important signal for virulence gene synthesis by B. anthracis. Increased synthesis of poly-D-glutamic acid capsule and the anthrax toxin proteins during culture of B. anthracis in the presence of elevated CO2/bicarbonate was first reported more than sixty years ago. Yet, the mechanism by which this physiologically relevant signal affects expression of these critical virulence factors has remained largely unknown. In work presented here, we have determined that B. anthracis cultures grown in elevated CO2/bicarbonate contain higher levels of the AtxA dimer than cultures grown in air. Conditions that enhance dimerization also promote AtxA target gene expression. The positive correlation between CO2/bicarbonate level, the presence of AtxA dimers, and AtxA activity links the signal to AtxA structure and function.
Many questions remain regarding the mechanism by which CO2/bicarbonate affects AtxA function. Genetic and biochemical searches have not revealed any cellular factors that might bridge the link between the CO2/bicarbonate signal and the AtxA protein. We speculate that AtxA may bind to bicarbonate or CO2 directly. To our knowledge, only one bacterial regulatory protein has been shown to require bicarbonate to stably interact with target promoter sequences. RegA, an AraC/XylS-like regulator found in Citrobacter rodentium, binds to bicarbonate via an amino-terminal motif (Yang et al., 2008). According to the model proposed by Yang et al. (2009), the binding of bicarbonate to RegA facilitates interaction of its carboxy-terminal DNA binding motif with target sequences. AtxA does not bear apparent sequence homology to RegA, although a binding pocket may not be obvious. Multiple bicarbonate-binding proteins, with varying bicarbonate binding sites and diverse functions have been reported, including proteins associated with photosynthesis and menaquione biosynthesis (Badger & Price, 2003, Jiang et al., 2010). Direct association of bicarbonate with AtxA multimers is currently under investigation in our laboratory.
Finally, our experiments were designed to detect stable protein-protein complexes. We do not exclude the possibility of other AtxA-protein interactions. Considering that specific DNA-binding activity has not been demonstrated for purified AtxA protein, it is possible that an additional protein interacts with AtxA to allow specific DNA-binding. Moreover, a specific multimeric and/or phosphorylated form of AtxA may be required to observe this activity. Although it has been suggested that AtxA phosphorylation occurs via the HPr enzyme of the PTS, the kinase has not been identified experimentally. It is likely that the enzyme responsible for phosphorylation of AtxA makes transient protein-protein contacts and thus would not be identified by the approaches used here. Our working model is that formation of an AtxA homo-dimer is required for virulence gene activation. Future studies will focus on potential relationships between phosphorylation, multimerization, and function of AtxA.
Experimental Procedures
Culture conditions
B. anthracis strains were cultured at 37C in Luria-Bertani (LB) medium (Bertani, 1951) or Casamino Acid medium (CA) (Thorne & Belton, 1957). LB broth cultures were shaken in air. CA broth cultures were shaken in air or in 5% atmospheric CO2. For CA cultures incubated in elevated CO2, 0.8% sodium bicarbonate was added to the medium (CACO3) (Hadjifrangiskou et al., 2007). Cells from stationary phase cultures were transferred into fresh medium (CACO3, unless noted otherwise) to an initial optical density at 600nm (OD600) of approximately 0.08. Cultures were incubated for 7 h and the OD600 was determined hourly. Generally, cultures reached early-exponential phase (OD600 0.20 to 0.35) at 2 h and stationary phase (OD600 1.2 to 1.7) at 4 h. For strains harboring atxA controlled by the hyper-spank promoter (Phyper-spank) (Britton et al., 2002), expression was induced with 30-50 μM isopropyl β-D-thiogalactoside (IPTG) at 2 h and cells were harvested at 4 h.
Antibiotics were used when appropriate in the following concentrations: spectinomycin (50 μg ml−1 for E. coli and 100 μg ml−1 for B. anthracis), erythromycin (150 μg ml−1 for E. coli and 5 μg ml−1 for B. anthracis), and carbenicillin (100 μg ml−1 for E. coli). Antibiotics and other chemicals were purchased from Fisher Scientific (Fairlawn, NJ) or Sigma-Aldrich (St. Louis, MO) unless indicated otherwise.
Strain Construction
B. anthracis strains and plasmids are shown in Table 1. All B. anthracis mutants were derived from the parent strain ANR-1 (Ames non-reverting), a pXO1+ pXO2- isolate originally obtained from Ames (pXO1+ pXO2+) (Welkos et al., 2001). E. coli strains JM109, TG1, and GM2163 were employed as hosts for cloning plasmids. Amplification, manipulation, and isolation of plasmid DNA were performed according to general practices. Non-methylated plasmid DNA was obtained from E. coli GM2163 for electroporation into B. anthracis (Koehler et al., 1994, Marrero & Welkos, 1995).
Table 1.
B. anthracis strains and plasmids
| Name | Description | Reference |
|---|---|---|
| Strains | ||
| ANR-1 | Parent strain, pXO1+, pXO2− | (Welkos et al., 2001) |
| UT375 | lef promoter – lacZ fusion (Plef-lacZ) at native lef locus | This work |
| UT376 | lef promoter – lacZ fusion (Plef-lacZ) at native lef locus, atxA-null | This work |
| Plasmids | ||
| pUTE657 | Expression vector derived from pDR111 and pBC16 with IPTG-inducible Phyper-spank; Specr Ampr |
(Pflughoeft et al., 2011) |
| pUTE658 | pUTE657 - derived expression vector for AtxA; the atxA ribosome binding site and coding region controlled by Phyper-spank |
This work |
| pUTE991 | pUTE658 - derived expression vector for AtxA-His (hexa-histidine tag on the C-terminus of AtxA) |
This work |
| pUTE992 | pUTE658 - derived expression vector for AtxA-FLAG (FLAG tag on the C-terminus of AtxA) |
This work |
| pUTE1013 | pUTE657 - derived expression vector for GFP-FLAG (FLAG tag on the C-terminus of GFP); the gfpmut3a ribosome binding site, coding region, and sequence encoding FLAG controlled by Phyper-spank |
This work |
Using systems described previously (Pflughoeft et al., 2011), two strains of B. anthracis were constructed for this study: UT375, in which the lef coding sequence on pXO1 was replaced by a promoterless β-galactosidase gene (lacZ), and UT376, which contains a markerless deletion of atxA in the UT375 strain background. To construct UT375, the polymerase chain reaction (PCR) and primers TH126 and TH127 (Table S1) were used to amplify a 797-bp DNA fragment corresponding to sequence from −686 to +111 relative to the lef transcriptional start site (lefup) (Dai et al., 1995). The PCR product was cloned into pHT304-18z (Arantes & Lereclus, 1991) using PstI and BamHI restriction enzyme sites upstream of the plasmid-borne lacZ. Subsequently, a 853-bp DNA sequence corresponding to sequences +2510 to +3363 (lefdown) was amplified using primers TH128 and TH129 and cloned into the KpnI and EcoRI sites of pHT304-18z containing lefup. The lefup-lacZ-lefdown segment of DNA was removed via digestion with XhoI and EcoRI and cloned in pHY304, a temperature-sensitive E. coli – B. anthracis shuttle vector harboring an erythromycin-resistance gene (Chaffin et al., 2005). B. anthracis containing the plasmid was cultured at 41C (the temperature non-permissive for pHY304 replication) in the presence of erythromycin to select for isolates in which the plasmid inserted into the lef locus via homologous recombination. Subsequently, cultures were passaged multiple times at 30C in the absence of antibiotic to allow excision of the pHY304 derivative from the lef locus. Single colony isolates were tested using PCR and sequencing for replacement of the lef coding sequence with the promoterless β-galactosidase gene.
To generate UT376, a markerless atxA-deletion mutant of UT375, DNA fragments corresponding to approximately 1 kb upstream of the atxA translational start (−1009 to +99 from the P1 transcriptional start site) and 1 kb downstream of the atxA stop codon 435 (+1528 to +2517) were amplified using primers JR170-171 and JR172-173, respectively (Table S1), fused via splicing by overlapping extension PCR (PCR-SOE) (Horton et al., 1989), and inserted into pHY304 using the SacII and XhoI restriction enzyme sites. B. anthracis containing the Plef-lacZ reporter (UT375) was transformed with the atxA deletion construct. The strain was cultured as described above and an isolate deleted for atxA was confirmed using PCR and sequencing.
We cloned the atxA coding region into pUTE657 (Pflughoeft et al., 2011), which carries the IPTG-inducible promoter Phyper-spank from pDR111 (Britton et al., 2002) and the origin of replication from pBC16. The atxA gene was amplified using TH191 and TH192 (Table S1) to add a SalI site 5′ of the ribosome binding site (+76 from P1) and a SphI site after the translational stop codon. The resulting plasmid (pUTE658) contains atxA controlled by Phyper-spank. We used a comparable strategy to clone the gfp-mut3a sequence (Cormack et al., 1996) into pUTE657, yielding pUTE1013. Using TH193 and TH234 (Table S1), the gfp-mut3a sequence was PCR amplified from pAD123 (Dunn & Handelsman, 1999) to include the ribosome binding site and to add a C-terminal FLAG tag to gfp-mut3a.
We constructed B. anthracis strains carrying Phyper-spank –controlled atxA alleles on pUTE657 to allow expression of C-terminal epitope-tagged and mutant AtxA proteins. Mutant alleles were generated using PCR-SOE and appropriate oligonucleotide primers (Table S1). For C-terminal epitope tags (FLAG and hexa-His), primers were designed to encode the desired tag at the 3′ end of atxA prior to the stop codon. To create a 5′ deletion of atxA, oligonucleotides fused the ribosome binding site of atxA to codons 385 - 475 resulting in a construct encoding AtxA385-475. For a 3′ deletion of atxA, a stop codon was inserted after the 385th codon resulting in a construct encoding AtxA1-385. Each of the atxA alleles was cloned into the SalI and SphI sites of pUTE657.
AtxA-His purification
The recombinant epitope-tagged protein AtxA-His was purified from B. anthracis using affinity chromatography. B. anthracis UT376 (pUTE658) was cultured in 2-L baffled flasks containing 750 ml CACO3. Following induction with 50 μM IPTG, cells were harvested using centrifugation at 6200 × g for 10 min at 4C. Cells were resuspended in 3 ml Binding Buffer (5 mM imidazole, 0.5 M NaCl, 5 mM β-mercaptoethanol, 20 mM Tris pH 7.2,) supplemented with 1X EDTA-free Complete proteinase inhibitor (Roche, Indianapolis, IN), 1 mM MgCl2, and 10 units DNAse I (Ambion, Inc., Austin, TX). Cells were lysed via three passages through a French Pressure Cell Press (SLM Instruments, Inc., Urbana, IL) and soluble material was obtained following centrifugation at 30,000 × g for 20 min at 4C. Lysates were added to 1 ml NTA-Ni resin (Qiagen, Hilden, Germany) in Binding Buffer (total volume of 10 ml) and the suspension was mixed gently for 2 h at 4C. The resin was washed in batch three times with 4 ml of Binding Buffer and once with 4 ml of Wash Buffer 1 (40 mM imidazole pH 7.9, 1.0 M NaCl, 20 mM Tris pH 7.2, 5 mM β-mercaptoethanol). The resin was transferred to a gravity column and washed sequentially with (1) 10 ml Wash Buffer 1, (2) 1.5 ml Wash Buffer – High Salt (40 mM imidazole pH 7.9, 1.5 M NaCl, 20 mM Tris pH 7.2, 5 mM β-mercaptoethanol), (3) 2.5 ml Wash Buffer 1, (4) 1.5 ml Wash Buffer 2 (75 mM imidazole pH 7.9, 1.0 M NaCl, 20 mM Tris pH 7.2, 5mM β- mercaptoethanol), and (5) 2.5 ml Wash Buffer 1. Upon addition of 7.5 ml Elution Buffer (20 mM Tris pH 7.2, 800 mM imidazole pH 7.9, 500 mM NaCl, 5 mM β-mercaptoethanol), 250 μl fractions were collected in 1.5-ml tubes containing 250 μl Collection Buffer (20 mM Tris pH 7.2, 150 mM NaCl, 10% glycerol, 5 mM EDTA, 5 mM μ-mercaptoethanol). Protein concentration and purity of the fractions were assessed using the Bradford reagent (Bio-Rad, Hercules, CA) and SDS-PAGE with Coomassie staining. Fractions containing AtxA-His were pooled and dialyzed in 400 mM NaCl, 5% glycerol, 20 mM Tris pH 7.2, and 5 mM dithiothreitol (DTT).
α-AtxA Antibody and AtxA Western blots
Antiserum for Western blotting was raised in rabbits (Cocalico Biologicals, Inc., Reamstown, PA). AtxA-His, purified from E. coli using a method similar to that described above, was injected into rabbits and serum was collected on day 56. To remove non-specific antibodies, the serum was adsorbed to an immobilized E. coli lysate (Thermo Scientific) using the manufacturer’s protocol. IgG in the serum was purified using Pierce Nab Spin Columns (Thermo Scientific).
B. anthracis cell lysates for Western blotting analysis were obtained from cultures at early stationary phase (OD600 = 1.2 to 1.7). Culture samples (4 ml) were centrifuged as described above. After washing cells two times in KTE-PIC (10 mM Tris-HCl pH 8.0, 100 mM KCl, 10% ethylene glycol, and EDTA-free Complete proteinase inhibitor), cells were resuspended in KTE-PIC to a final volume of 850 μl and transferred to a 1.5-ml screw-cap tube containing 400 μl 0.1 mm Zirconia/Silica Beads (BioSpec Products, Bartlesville, OK). The samples were lysed mechanically for 2 min using a Mini BeadBeater (BioSpec Products). After centrifugation, soluble material was mixed with SDS loading buffer (final concentration of loading buffer was 5% glycerol, 100 mM DTT, 2% SDS, 40 mM Tris-Cl pH 6.8), boiled, and subjected to SDS-PAGE.
Prior to protein transfer, gels were equilibrated in CAPS Buffer (10 mM CAPS pH 11.0, 10% methanol). Using a Hoefer transfer unit (Hoefer, Inc., Holliston, MA) containing CAPS Buffer, proteins were transferred to Immobilon-P membrane (Millipore, Billerica, MA) at 50V at 4C for 1.5 to 2 h. Membranes were blocked with TBS-T (20 mM Tris base, 137 mM NaCl, 0.1% Tween 20 [pH 7.6]) containing 5% non-fat dry milk. Membranes were subsequently treated with primary antibodies (α-AtxA, α-RNA polymerase β subunit [Santa Cruz Biotechnology, Santa Cruz, CA], α-THE His [Genscript, Piscataway, NJ], and α-FLAG M2 [Sigma Aldrich]) in TBS-T. After washing with TBS-T, membranes were finally treated with the corresponding secondary antibody (goat α-rabbit–HRP conjugate or goat α-mouse-HRP conjugate [Bio-Rad]). Blots were developed using the SuperSignal West Dura Chemiluminescent Substrate (Thermo Scientific). For re-probing, membranes were stripped using Restore Western Blot Stripping Buffer (Thermo Fisher) or 62.5 mM Tris pH 6.8 containing 2% SDS and 100 mM β-mercaptoethanol.
BN-PAGE assay
To perform Blue Native PAGE (BN-PAGE), protein samples mixed with NativePAGE Sample Buffer containing 5% G-250 Sample Additive were electrophoresed using NativePAGE Novex 4-16% Bis-Tris Gels in the XCell Surelock Mini-Cell electrophoresis unit according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA). Electrophoresed proteins were transferred to PVDF at 20V overnight at 4C. Blots were fixed with 8% acetic acid for 15 min prior to probing with α–AtxA antibody as described above.
Co-affinity purification
B. anthracis UT376 containing vectors encoding AtxA-His (pUTE991), AtxA-FLAG (pUTE992), or GFP-FLAG (pUTE1013) were cultured independently. Three pools, containing 20 ml of each culture, were collected: UT376 (pUTE991) and UT376 (pUTE1013); UT376 (pUTE991) and UT376 (pUTE992); and UT376 (pUTE992) and UT376 (pUTE1013). The cells were washed with 10 ml Binding Buffer containing EDTA-free Complete proteinase inhibitor (BB-PIC). Pellets were flash frozen in an ethanol-dry ice bath and stored at −80C.
To create cleared soluble cell lysates, samples were thawed in an ice bath, resuspended in 2.5 ml BB-PIC, and transferred to two 1.5-ml screw cap tubes, each containing 500 μl 0.1 mm Zirconia/Silica beads. The samples were lysed mechanically for 1 min using a Mini BeadBeater, placed on ice for 5 min, and then subjected to an additional 1 min of bead beating. Following centrifugation at 10,000 x g for 5 min at 4C, the soluble material was incubated at 37C for 20 min. NTA-Ni resin (70 μl) was added and the samples were incubated at 4C 539 for 2 h. The resin was transferred to spin filters (EMB Chemicals, Darmstadt, Germany) and several 500-μl washes were performed in the following sequence: 2x Binding Buffer, 2x Wash Buffer 1, 1x Wash Buffer-High Salt, 2x Wash Buffer 1, 1x Wash Buffer 2, and 2x Wash Buffer 1. Proteins were eluted from the column using 125 μl Elution Buffer and then denatured with SDS loading buffer. Samples were probed with α-AtxA, α-His, and α-FLAG antibodies using Western blotting.
BMH Cross-linking
Protein cross-linking was performed as described elsewhere (Eswaramoorthy et al., 2009). Briefly, cultures containing UT376 (pUTE658) were induced with 30 μM IPTG for 2 h. Cells were washed twice with 5 ml phosphate-buffered saline (PBS) containing 10 mM EDTA and adjusted to pH 7.2. Cells were then resuspended in 1 ml PBS 10 mM EDTA pH 7.2 and lysed. For each experiment, 250 μl of cleared soluble cell lysate, representing 5 ml of culture, was mixed with 5 μl of 20 mM bis(maleimido)hexane (BMH, Thermo Scientific, prepared freshly in DMSO) and incubated at 4C for 2 h with end-over-end mixing. Control reactions lacking BMH contained the DMSO solvent only. Reactions were quenched by adding 40 mM cysteine, vortexing for 10 min, and boiling in the presence of 1X SDS loading buffer containing 100 mM DTT. The samples were analyzed on 4-15% polyacrylamide SDS gels (Bio-Rad) and AtxA was detected via Western blotting. ImageJ software (Rasband, 1997) was used to determine relative intensities of cross-reactive bands.
AtxA-His for cross-linking experiments was affinity purified using NTA-Ni resin and washes were performed with solutions made without β-mercaptoethanol. AtxA-His was eluted from the resin, and subjected to BMH cross-linking as described above.
Supplementary Material
Acknowledgements
We thank Maria Hadjifrangiskou for the construction of the pUTE657 and pUTE658 vectors, and Jason Rall for producing pUTE937. We also acknowledge Jennifer Dale and Kathryn Pflughoeft for valuable discussions.
This work was supported by Award Number R01AI033537 from the National Institute of Allergy and Infectious Diseases to T.M.K. T.G.H. was supported during a portion of this work by a training fellowship from the Keck Center Computational and Structural Biology in Biodefense Training Program of the Gulf Coast Consortia (NIH Grant No.1 T32 AI065396-01). The content of this publication is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy And Infectious Diseases or the National Institutes of Health.
References
- Arantes O, Lereclus D. Construction of cloning vectors for Bacillus thuringiensis. Gene. 1991;108:115–119. doi: 10.1016/0378-1119(91)90495-w. [DOI] [PubMed] [Google Scholar]
- Badger MR, Price GD. CO2 concentrating mechanisms in cyanobacteria: molecular components, their diversity and evolution. Journal of Experimental Botany. 2003;54:609–622. doi: 10.1093/jxb/erg076. [DOI] [PubMed] [Google Scholar]
- Bartkus JM, Leppla SH. Transcriptional regulation of the protective antigen gene of Bacillus anthracis. Infection and Immunity. 1989;57:2295–2300. doi: 10.1128/iai.57.8.2295-2300.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Zeev E, Fux L, Amster-Choder O, Eisenstein M. Experimental and computational characterization of the dimerization of the PTS-regulation domains of BglG from Escherichia coli. Journal of Molecular Biology. 2005;347:693–706. doi: 10.1016/j.jmb.2005.01.068. [DOI] [PubMed] [Google Scholar]
- Bertani G. STUDIES ON LYSOGENESIS I. Journal of Bacteriology. 1951;62:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgogne A, Drysdale M, Hilsenbeck SG, Peterson SN, Koehler TM. Global effects of virulence gene regulators in a Bacillus anthracis strain with both virulence plasmids. Infection and Immunity. 2003;71:2736–2743. doi: 10.1128/IAI.71.5.2736-2743.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourret RB, Silversmith RE. Two-component signal transduction. Current Opinion in Microbiology. 2010;13:113–115. doi: 10.1016/j.mib.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton RA, Eichenberger P, Gonzalez-Pastor JE, Fawcett P, Monson R, Losick R, Grossman AD. Genome-Wide Analysis of the Stationary-Phase Sigma Factor (Sigma-H) Regulon of Bacillus subtilis. J. Bacteriol. 2002;184:4881–4890. doi: 10.1128/JB.184.17.4881-4890.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldi A, Fouet A, Mock M. Regulation of pag gene expression in Bacillus anthracis: use of a pag-lacZ transcriptional fusion. FEMS Microbiology Letters. 1992;77:89–93. doi: 10.1016/0378-1097(92)90137-d. [DOI] [PubMed] [Google Scholar]
- Chaffin DO, Mentele LM, Rubens CE. Sialylation of group B streptococcal capsular polysaccharide is mediated by cpsK and is required for optimal capsule polymerization and expression. Journal of Bacteriology. 2005;187:4615–4626. doi: 10.1128/JB.187.13.4615-4626.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C, Bongiorni C, Perego M. Glucose-dependent activation of Bacillus anthracis toxin gene expression and virulence requires the carbon catabolite protein CcpA. Journal of Bacteriology. 2011;193:52–62. doi: 10.1128/JB.01656-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitlaru T, Gat O, Gozlan Y, Ariel N, Shafferman A. Differential proteomic analysis of the Bacillus anthracis secretome: distinct plasmid and chromosome CO2-dependent cross talk mechanisms modulate extracellular proteolytic activities. Journal of Bacteriology. 2006;188:3551–3571. doi: 10.1128/JB.188.10.3551-3571.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- Dai Z, Koehler TM. Regulation of anthrax toxin activator gene (atxA) expression in Bacillus anthracis: temperature, not CO2/bicarbonate, affects AtxA synthesis. Infection and Immunity. 1997;65:2576–2582. doi: 10.1128/iai.65.7.2576-2582.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z, Sirard JC, Mock M, Koehler TM. The atxA gene product activates transcription of the anthrax toxin genes and is essential for virulence. Molecular Microbiology. 1995;16:1171–1181. doi: 10.1111/j.1365-2958.1995.tb02340.x. [DOI] [PubMed] [Google Scholar]
- Declerck N, Dutartre H, Receveur V, Dubois V, Royer C, Aymerich S, van Tilbeurgh H. Dimer stabilization upon activation of the transcriptional antiterminator LicT. Journal of Molecular Biology. 2001;314:671–681. doi: 10.1006/jmbi.2001.5185. [DOI] [PubMed] [Google Scholar]
- Deutscher J, Francke C, Postma PW. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiology and Molecular Biology Reviews: MMBR. 2006;70:939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drysdale M, Bourgogne A, Koehler TM. Transcriptional analysis of the Bacillus anthracis capsule regulators. Journal of Bacteriology. 2005;187:5108–5114. doi: 10.1128/JB.187.15.5108-5114.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn AK, Handelsman J. A vector for promoter trapping in Bacillus cereus. Gene. 1999;226:297–305. doi: 10.1016/s0378-1119(98)00544-7. [DOI] [PubMed] [Google Scholar]
- Eswaramoorthy P, Guo T, Fujita M. In vivo domain-based functional analysis of the major sporulation sensor kinase, KinA, in Bacillus subtilis. Journal of Bacteriology. 2009;191:5358–5368. doi: 10.1128/JB.00503-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans B, Tishmack PA, Pokalsky C, Zhang M, Van Etten RL. Site-directed mutagenesis, kinetic, and spectroscopic studies of the P-loop residues in a low molecular weight protein tyrosine phosphatase. Biochemistry. 1996;35:13609–13617. doi: 10.1021/bi9605651. [DOI] [PubMed] [Google Scholar]
- Fouet A, Mock M. Differential influence of the two Bacillus anthracis plasmids on regulation of virulence gene expression. Infection and Immunity. 1996;64:4928–4932. doi: 10.1128/iai.64.12.4928-4932.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fux L, Nussbaum-Shochat A, Amster-Choder O. Interactions between the PTS regulation domains of the BglG transcriptional antiterminator from Escherichia coli. The Journal of Biological Chemistry. 2003;278:46203–46209. doi: 10.1074/jbc.M306506200. [DOI] [PubMed] [Google Scholar]
- Graille M, Zhou C-Z, Receveur-Brechot V, Collinet B, Declerck N, van Tilbeurgh H. Activation of the LicT transcriptional antiterminator involves a domain swing/lock mechanism provoking massive structural changes. The Journal of Biological Chemistry. 2005;280:14780–14789. doi: 10.1074/jbc.M414642200. [DOI] [PubMed] [Google Scholar]
- Guignot J, Mock M, Fouet A. AtxA activates the transcription of genes harbored by both Bacillus anthracis virulence plasmids. FEMS Microbiology Letters. 1997;147:203–207. doi: 10.1111/j.1574-6968.1997.tb10242.x. [DOI] [PubMed] [Google Scholar]
- Hadjifrangiskou M, Chen Y, Koehler TM. The alternative sigma factor sigmaH is required for toxin gene expression by Bacillus anthracis. Journal of Bacteriology. 2007;189:1874–1883. doi: 10.1128/JB.01333-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henstra SA, Tuinhof M, Duurkens RH, Robillard GT. The Bacillus stearothermophilus mannitol regulator, MtlR, of the phosphotransferase system. A DNA-binding protein, regulated by HPr and IICBmtl-dependent phosphorylation. The Journal of Biological Chemistry. 1999;274:4754–4763. doi: 10.1074/jbc.274.8.4754. [DOI] [PubMed] [Google Scholar]
- Hoffmaster AR, Koehler TM. The anthrax toxin activator gene atxA is associated with CO2-enhanced non-toxin gene expression in Bacillus anthracis. Infection and Immunity. 1997;65:3091–3099. doi: 10.1128/iai.65.8.3091-3099.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmaster AR, Koehler TM. Control of virulence gene expression in Bacillus anthracis. Journal of Applied Microbiology. 1999;87:279–281. doi: 10.1046/j.1365-2672.1999.00887.x. [DOI] [PubMed] [Google Scholar]
- Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- Huffman JL, Brennan RG. Prokaryotic transcription regulators: more than just the helix-turn-helix motif. Current Opinion in Structural Biology. 2002;12:98–106. doi: 10.1016/s0959-440x(02)00295-6. [DOI] [PubMed] [Google Scholar]
- Jiang M, Chen M, Guo Z-F, Guo Z. A bicarbonate cofactor modulates 1,4-dihydroxy-2-naphthoyl-coenzyme a synthase in menaquinone biosynthesis of Escherichia coli. The Journal of Biological Chemistry. 2010;285:30159–30169. doi: 10.1074/jbc.M110.147702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyet P, Derkaoui M, Poncet S, Deutscher J. Control of Bacillus subtilis mtl operon expression by complex phosphorylation-dependent regulation of the transcriptional activator MtlR. Molecular Microbiology. 2010;76:1279–1294. doi: 10.1111/j.1365-2958.2010.07175.x. [DOI] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJE. Protein structure prediction on the Web: a case study using the Phyre server. Nature Protocols. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Koehler TM. Bacillus anthracis physiology and genetics. Molecular Aspects of Medicine. 2009;30:386–396. doi: 10.1016/j.mam.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler TM, Dai Z, Kaufman-Yarbray M. Regulation of the Bacillus anthracis protective antigen gene: CO2 and a trans-acting element activate transcription from one of two promoters. Journal of Bacteriology. 1994;176:586–595. doi: 10.1128/jb.176.3.586-595.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Derbyshire MK, DeWeese-Scott C, Gonzales NR, Gwadz M, et al. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Research. 2007;35:D237-240–D237-240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrero R, Welkos SL. The transformation frequency of plasmids into Bacillus anthracis is affected by adenine methylation. Gene. 1995;152:75–78. doi: 10.1016/0378-1119(94)00647-b. [DOI] [PubMed] [Google Scholar]
- McIver KS, Heath AS, Green BD, Scott JR. Specific binding of the activator Mga to promoter sequences of the emm and scpA genes in the group A streptococcus. Journal of Bacteriology. 1995;177:6619–6624. doi: 10.1128/jb.177.22.6619-6624.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignot T, Mock M, Fouet A. A plasmid-encoded regulator couples the synthesis of toxins and surface structures in Bacillus anthracis. Molecular Microbiology. 2003;47:917–927. doi: 10.1046/j.1365-2958.2003.03345.x. [DOI] [PubMed] [Google Scholar]
- Miller JH. Experiments in molecular genetics. Cold Spring Harbor Laboratory; New York, NY: 1972. pp. 352–355. [Google Scholar]
- Nobelmann B, Lengeler JW. Molecular analysis of the gat genes from Escherichia coli and of their roles in galactitol transport and metabolism. Journal of Bacteriology. 1996;178:6790–6795. doi: 10.1128/jb.178.23.6790-6795.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflughoeft KJ, Sumby P, Koehler TM. Bacillus anthracis sin Locus and Regulation of Secreted Proteases. J. Bacteriol. 2011;193:631–639. doi: 10.1128/JB.01083-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasband WS. ImageJ. U. S. National Institutes of Health; Bethesda, Maryland, USA: 1997. [Google Scholar]
- Saile E, Koehler TM. Control of anthrax toxin gene expression by the transition state regulator abrB. Journal of Bacteriology. 2002;184:370–380. doi: 10.1128/JB.184.2.370-380.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirard JC, Guidi-Rontani C, Fouet A, Mock M. Characterization of a plasmid region involved in Bacillus anthracis toxin production and pathogenesis. International Journal of Medical Microbiology: IJMM. 2000;290:313–316. doi: 10.1016/S1438-4221(00)80030-2. [DOI] [PubMed] [Google Scholar]
- Sirard JC, Mock M, Fouet A. The three Bacillus anthracis toxin genes are coordinately regulated by bicarbonate and temperature. Journal of Bacteriology. 1994;176:5188–5192. doi: 10.1128/jb.176.16.5188-5192.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenshein AL. CodY, a global regulator of stationary phase and virulence in Gram-positive bacteria. Current Opinion in Microbiology. 2005;8:203–207. doi: 10.1016/j.mib.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Strauch MA, Ballar P, Rowshan AJ, Zoller KL. The DNA-binding specificity of the Bacillus anthracis AbrB protein. Microbiology (Reading, England) 2005;151:1751–1759. doi: 10.1099/mic.0.27803-0. [DOI] [PubMed] [Google Scholar]
- Sun T, Altenbuchner J. Characterization of a Mannose Utilization System in Bacillus subtilis. J. Bacteriol. 2010;192:2128–2139. doi: 10.1128/JB.01673-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne CB, Belton FC. An agar-diffusion method for titrating Bacillus anthracis immunizing antigen and its application to a study of antigen production. Journal of General Microbiology. 1957;17:505–516. doi: 10.1099/00221287-17-2-505. [DOI] [PubMed] [Google Scholar]
- Tsvetanova B, Wilson AC, Bongiorni C, Chiang C, Hoch JA, Perego M. Opposing effects of histidine phosphorylation regulate the AtxA virulence transcription factor in Bacillus anthracis. Molecular Microbiology. 2007;63:644–655. doi: 10.1111/j.1365-2958.2006.05543.x. [DOI] [PubMed] [Google Scholar]
- Uchida I, Hornung JM, Thorne CB, Klimpel KR, Leppla SH. Cloning and characterization of a gene whose product is a trans-activator of anthrax toxin synthesis. Journal of Bacteriology. 1993;175:5329–5338. doi: 10.1128/jb.175.17.5329-5338.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida I, Makino S, Sekizaki T, Terakado N. Cross-talk to the genes for Bacillus anthracis capsule synthesis by atxA, the gene encoding the trans-activator of anthrax toxin synthesis. Molecular Microbiology. 1997;23:1229–1240. doi: 10.1046/j.1365-2958.1997.3041667.x. [DOI] [PubMed] [Google Scholar]
- van Schaik W, Chateau A, Dillies M-A, Coppee J-Y, Sonenshein AL, Fouet A. The global regulator CodY regulates toxin gene expression in Bacillus anthracis and is required for full virulence. Infection and Immunity. 2009;77:4437–4445. doi: 10.1128/IAI.00716-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Tilbeurgh H, Le Coq D, Declerck N. Crystal structure of an activated form of the PTS regulation domain from the LicT transcriptional antiterminator. The EMBO Journal. 2001;20:3789–3799. doi: 10.1093/emboj/20.14.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpon L, Young CR, Matte A, Gehring K. NMR structure of the enzyme GatB of the galactitol-specific phosphoenolpyruvate-dependent phosphotransferase system and its interaction with GatA. Protein Science: A Publication of the Protein Society. 2006;15:2435–2441. doi: 10.1110/ps.062337406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Hamano M, Kakeshita H, Bunai K, Tojo S, Yamaguchi H, et al. Mannitol-1-Phosphate Dehydrogenase (MtlD) Is Required for Mannitol and Glucitol Assimilation in Bacillus subtilis: Possible Cooperation of mtl and gut Operons. J. Bacteriol. 2003;185:4816–4824. doi: 10.1128/JB.185.16.4816-4824.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welkos S, Little S, Friedlander A, Fritz D, Fellows P. The role of antibodies to Bacillus anthracis and anthrax toxin components in inhibiting the early stages of infection by anthrax spores. Microbiology. 2001;147:1677–1685. doi: 10.1099/00221287-147-6-1677. [DOI] [PubMed] [Google Scholar]
- Wilson AC, Hoch JA, Perego M. Two small c-type cytochromes affect virulence gene expression in Bacillus anthracis. Molecular Microbiology. 2009;72:109–123. doi: 10.1111/j.1365-2958.2009.06627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Dogovski C, Hocking D, Tauschek M, Perugini M, Robins-Browne RM. Bicarbonate-mediated stimulation of RegA, the global virulence regulator from Citrobacter rodentium. Journal of Molecular Biology. 2009;394:591–599. doi: 10.1016/j.jmb.2009.10.033. [DOI] [PubMed] [Google Scholar]
- Yang J, Hart E, Tauschek M, Price GD, Hartland EL, Strugnell RA, Robins-Browne RM. Bicarbonate-mediated transcriptional activation of divergent operons by the virulence regulatory protein, RegA, from Citrobacter rodentium. Molecular Microbiology. 2008;68:314–327. doi: 10.1111/j.1365-2958.2008.06171.x. [DOI] [PubMed] [Google Scholar]
- Zeng L, Burne RA. Transcriptional Regulation of the Cellobiose Operon of Streptococcus mutans. Journal of Bacteriology. 2009;191:2153–2162. doi: 10.1128/JB.01641-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.