Abstract
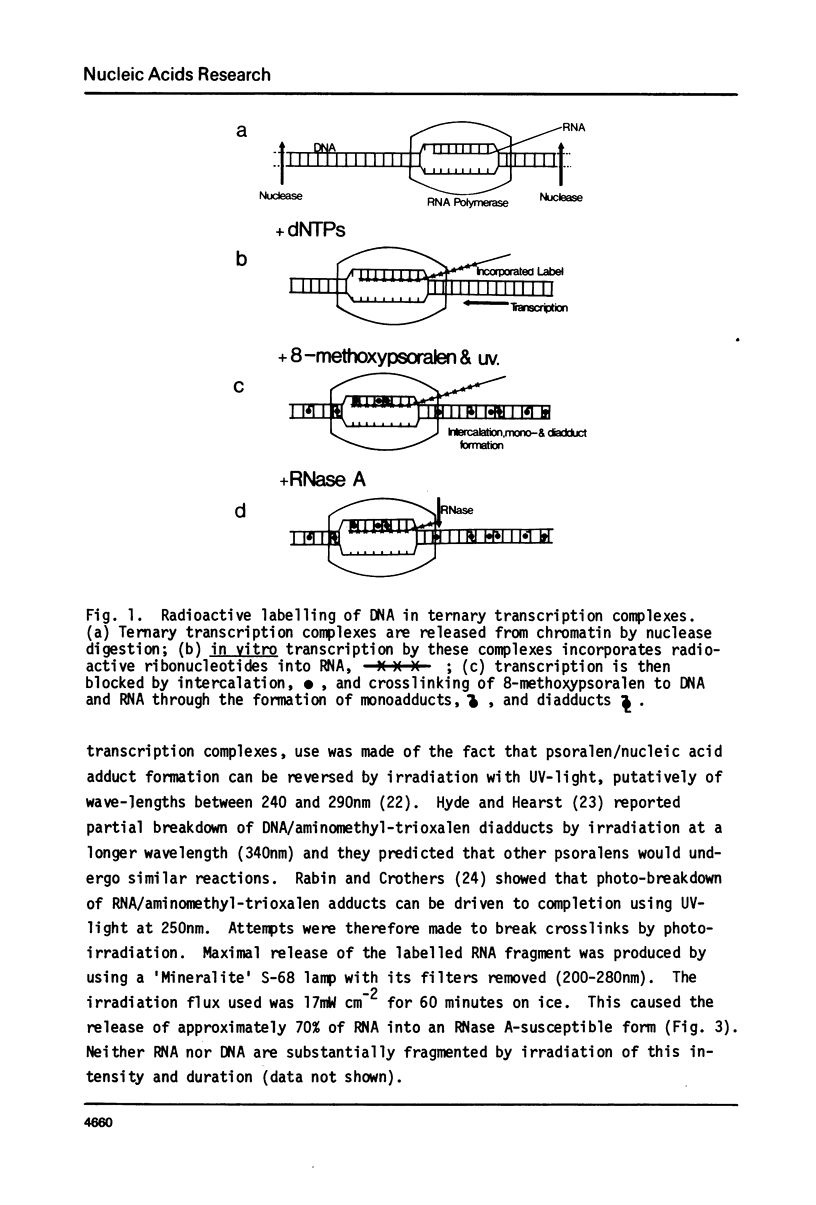
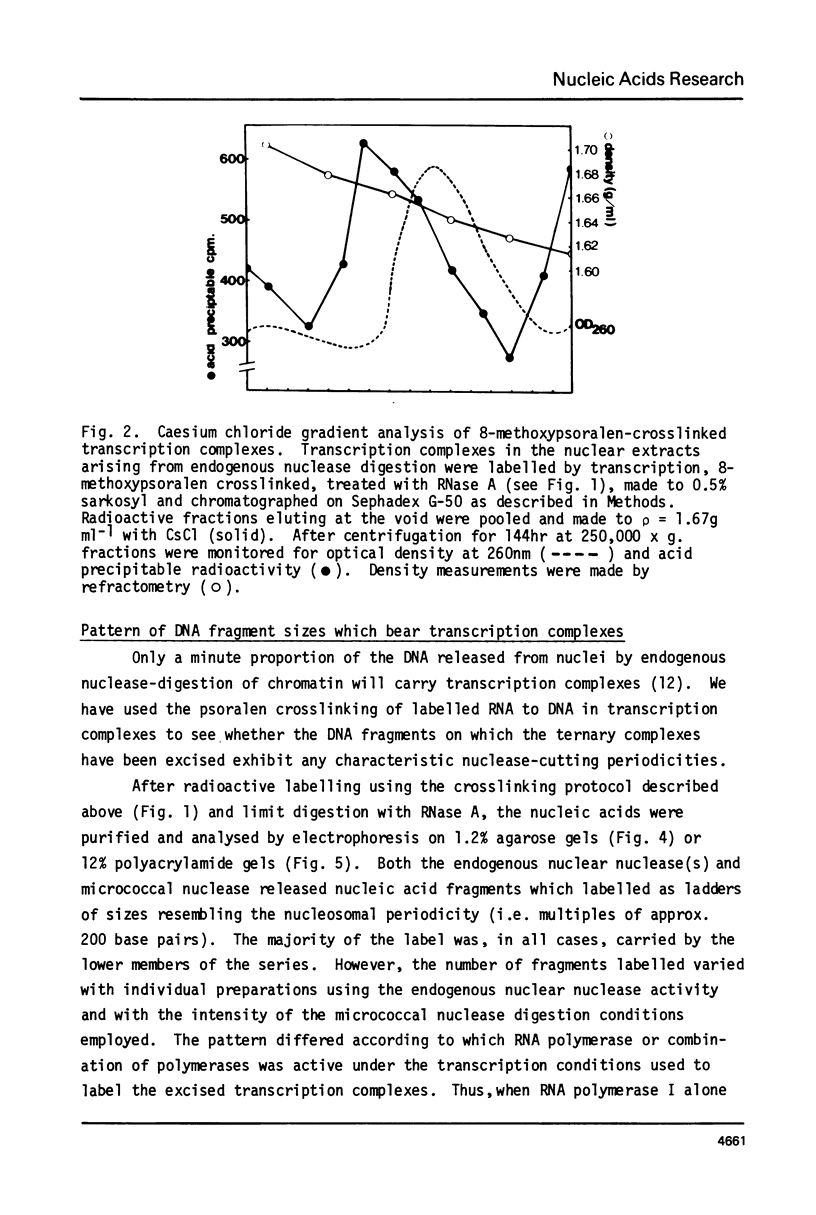
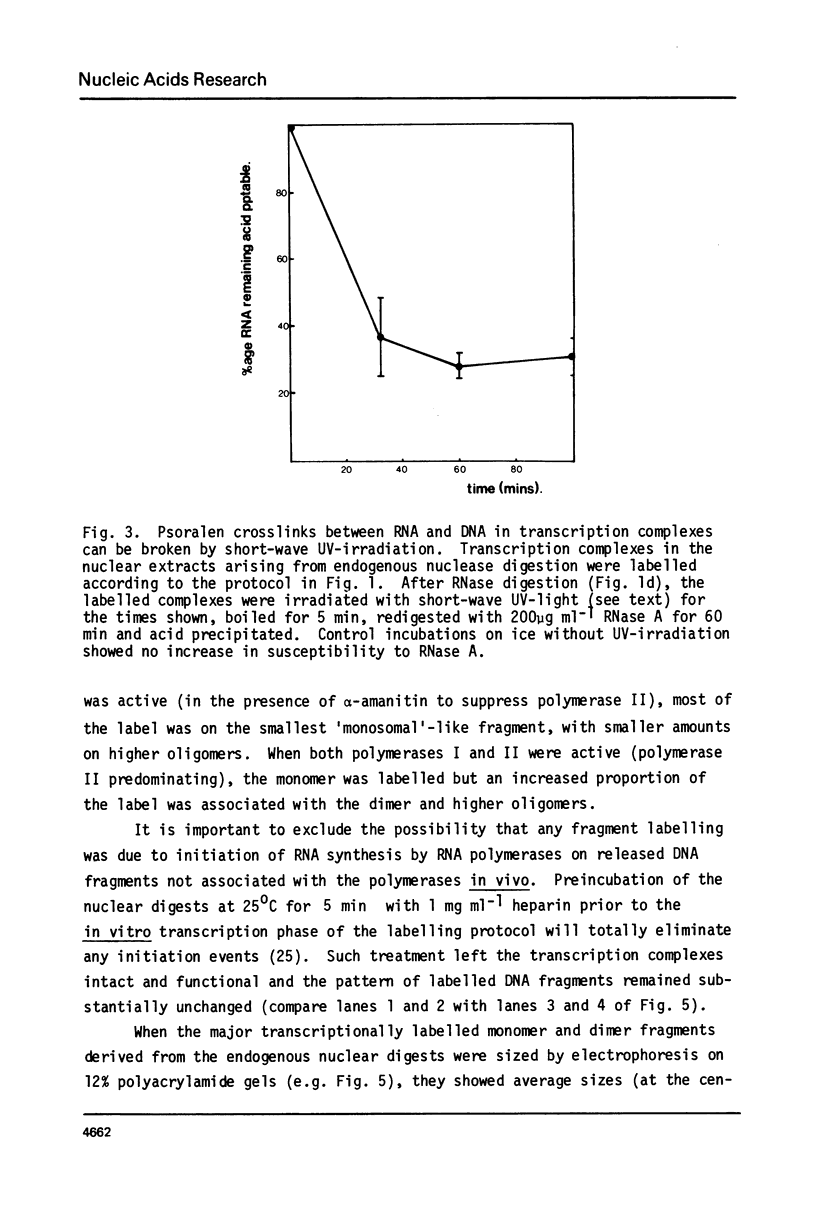
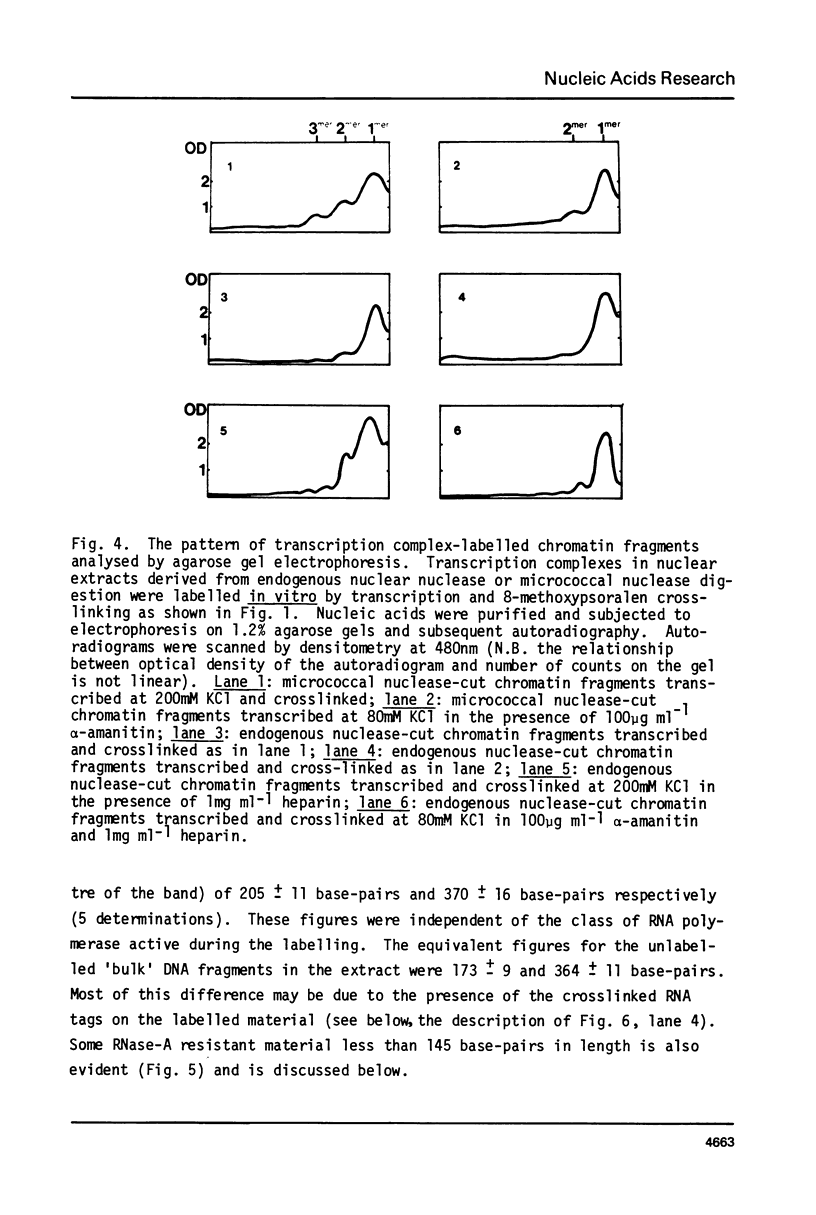
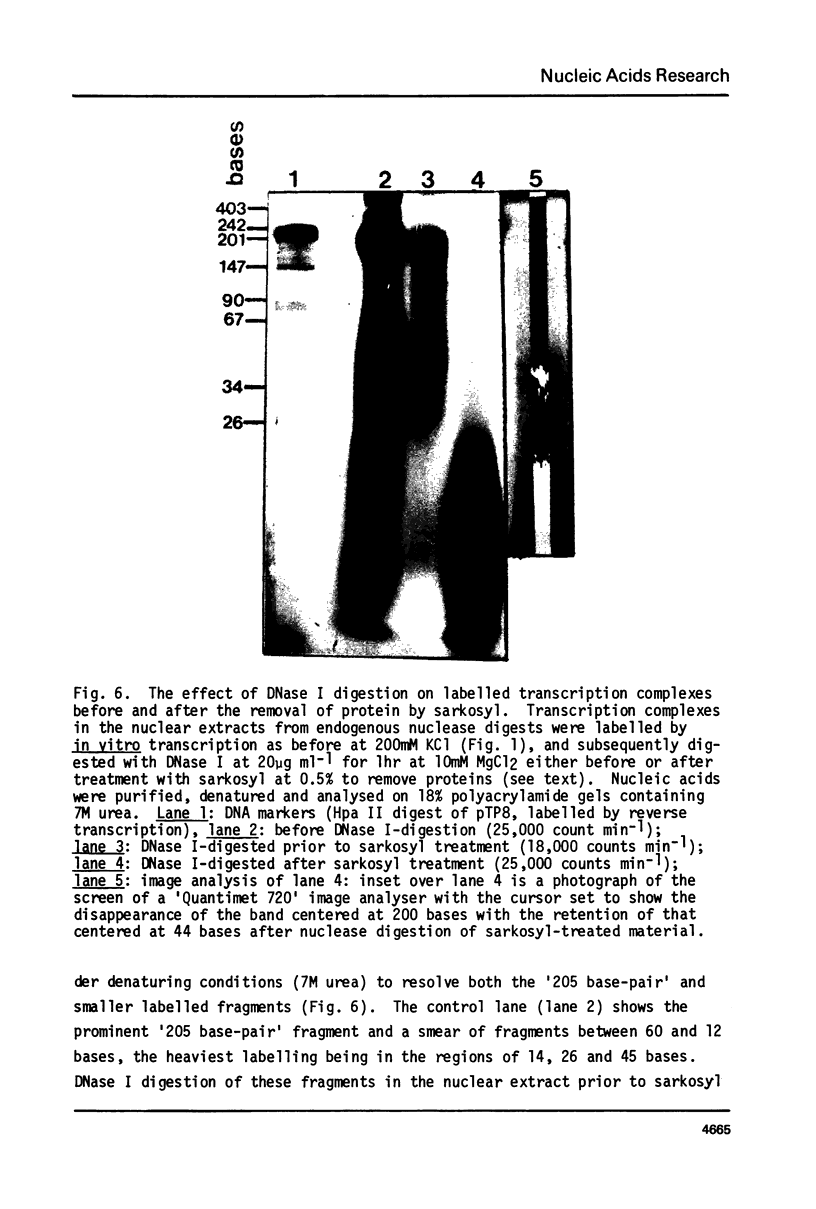
Digestion of rat liver nuclei by endogenous nucleases or micrococcal nuclease releases a chromatin fraction containing RNA polymerases I and II bound to DNA fragments in ternary transcription complexes. To label the DNA in these transcription complexes, the polymerases were allowed to add radioactively labelled ribonucleotides in vitro to in vivo-initiated RNA chains. During this transcription step, nucleic acids were photochemically cross-linked using 8-methoxypsoralen. Nucleic acids in transcription complexes were then sized by gel electrophoresis. Under conditions where RNA polymerases I and II were active in vitro, most of the labelled DNA was found in a series of fragments of sizes which were multiples of approximately 200 base-pairs. When polymerase I alone was active, the smallest member of this series carried the bulk of the label; when polymerase II also was active, a significant proportion of the label was carried on the dimer and higher oligomers. Proteins other than polymerase alone are shown to be responsible for the pattern of DNA fragments protected from nucleases. Therefore active RNA polymerases I and II in vivo are in close proximity to structures protecting DNA fragments, the sizes of which are similar to those found in nucleosomes. We have yet to establish that these structures are composed of histones.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bellard M., Gannon F., Chambon P. Nucleosome structure III: the structure and transcriptional activity of the chromatin containing the ovalbumin and globin genes in chick oviduct nuclei. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):779–791. doi: 10.1101/sqb.1978.042.01.078. [DOI] [PubMed] [Google Scholar]
- Bloom K. S., Anderson J. N. Conformation of ovalbumin and globin genes in chromatin during differential gene expression. J Biol Chem. 1979 Oct 25;254(20):10532–10539. [PubMed] [Google Scholar]
- Bonner W. M., Laskey R. A. A film detection method for tritium-labelled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974 Jul 1;46(1):83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- Bustin M. Binding of E. coli RNA polymerase to chromatin subunits. Nucleic Acids Res. 1978 Mar;5(3):925–932. doi: 10.1093/nar/5.3.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camerini-Otero R. D., Zasloff M. A. Nucleosomal packaging of the thymidine kinase gene of herpes simplex virus transferred into mouse cells: an actively expressed single-copy gene. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5079–5083. doi: 10.1073/pnas.77.9.5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech T. R., Karrer K. M. Chromatin structure of the ribosomal RNA genes of Tetrahymena thermophila as analyzed by trimethylpsoralen crosslinking in vivo. J Mol Biol. 1980 Feb 5;136(4):395–416. doi: 10.1016/0022-2836(80)90397-6. [DOI] [PubMed] [Google Scholar]
- Cech T., Pardue M. L. Cross-linking of DNA with trimethylpsoralen is a probe for chromatin structure. Cell. 1977 Jul;11(3):631–640. doi: 10.1016/0092-8674(77)90080-0. [DOI] [PubMed] [Google Scholar]
- Chandler D. W., Gralla J. Specific binding and protection of form II SV40 deoxyribonucleic acid by ribonucleic acid polymerase II from wheat germ. Biochemistry. 1980 Apr 15;19(8):1604–1612. doi: 10.1021/bi00549a012. [DOI] [PubMed] [Google Scholar]
- Coupar B. E., Chesterton C. J. The mechanism by which heparin stimulates transcription in isolated rat liver nuclei. Polyribonucleotide elongation rates and the number of transcribing RNA polymerase molecules present. Eur J Biochem. 1977 Oct 3;79(2):525–533. doi: 10.1111/j.1432-1033.1977.tb11837.x. [DOI] [PubMed] [Google Scholar]
- Dall'Acqua F., Marciani S., Bordin F., Bevilacqua R. Studies on the photoreaction (365 nm) between psoralen and thymine. Ric Sci. 1968 Nov;38(11):1094–1099. [PubMed] [Google Scholar]
- Gottesfeld J. M. Methods for fractionation of chromatin into transcriptionally active and inactive segments. Methods Cell Biol. 1977;16:421–436. doi: 10.1016/s0091-679x(08)60117-x. [DOI] [PubMed] [Google Scholar]
- Gould H. J., Cowling G. J., Harborne N. R., Allan J. An examination of models for chromatin transcription. Nucleic Acids Res. 1980 Nov 25;8(22):5255–5266. doi: 10.1093/nar/8.22.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatayama T., Omori K., Inoue A., Yukioka M. Partial characterization of RNA polymerase II complex released by micrococcal nuclease digestion of rat liver nuclei. Biochim Biophys Acta. 1981 Feb 26;652(2):245–255. doi: 10.1016/0005-2787(81)90113-1. [DOI] [PubMed] [Google Scholar]
- Hodo H. G., 3rd, Sahasrabuddhe C. G., Plishker M. F., Saunders G. F. Use of RNA polymerase as an enzymatic probe of nucleosomal structure. Nucleic Acids Res. 1980 Sep 11;8(17):3851–3864. doi: 10.1093/nar/8.17.3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde J. E., Hearst J. E. Binding of psoralen derivatives to DNA and chromatin: influence of the ionic environment on dark binding and photoreactivity. Biochemistry. 1978 Apr 4;17(7):1251–1257. doi: 10.1021/bi00600a019. [DOI] [PubMed] [Google Scholar]
- Isaacs S. T., Shen C. K., Hearst J. E., Rapoport H. Synthesis and characterization of new psoralen derivatives with superior photoreactivity with DNA and RNA. Biochemistry. 1977 Mar 22;16(6):1058–1064. doi: 10.1021/bi00625a005. [DOI] [PubMed] [Google Scholar]
- Levy A., Noll M. Chromatin fine structure of active and repressed genes. Nature. 1981 Jan 15;289(5794):198–203. doi: 10.1038/289198a0. [DOI] [PubMed] [Google Scholar]
- Lilley D. M., Jacobs M. F., Houghton M. The nature of the interaction of nucleosomes with a eukaryotic RNA polymerase II. Nucleic Acids Res. 1979 Sep 25;7(2):377–399. doi: 10.1093/nar/7.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M., Pardon J. F. Structure and function of chromatin. Annu Rev Genet. 1979;13:197–233. doi: 10.1146/annurev.ge.13.120179.001213. [DOI] [PubMed] [Google Scholar]
- MASTER R. W. POSSIBLE SYNTHESIS OF POLYRIBONUCLEOTIDES OF KNOWN BASE-TRIPLET SEQUENCES. Nature. 1965 Apr 3;206:93–93. doi: 10.1038/206093b0. [DOI] [PubMed] [Google Scholar]
- Mathis D., Oudet P., Chambon P. Structure of transcribing chromatin. Prog Nucleic Acid Res Mol Biol. 1980;24:1–55. doi: 10.1016/s0079-6603(08)60670-4. [DOI] [PubMed] [Google Scholar]
- Matsui S. I., Fuke M., Busch H. Fidelity of ribosomal ribonucleic acid synthesis by nucleoli and nucleolar chromatin. Biochemistry. 1977 Jan 11;16(1):39–45. doi: 10.1021/bi00620a007. [DOI] [PubMed] [Google Scholar]
- Paoletti J. Relaxation of chromatin structure induced by ethidium binding: 1--Micrococcal nuclease digestion of the ethidium--chromatin complex. Biochem Biophys Res Commun. 1978 Mar 15;81(1):193–198. doi: 10.1016/0006-291x(78)91648-0. [DOI] [PubMed] [Google Scholar]
- Pathak M. A., Krämer D. M. Photosensitization of skin in vivo by furocoumarins (psoralens). Biochim Biophys Acta. 1969 Nov 19;195(1):197–206. doi: 10.1016/0005-2787(69)90616-9. [DOI] [PubMed] [Google Scholar]
- Rabin D., Crothers D. M. Analysis of RNA secondary structure by photochemical reversal of psoralen crosslinks. Nucleic Acids Res. 1979 Oct 10;7(3):689–703. doi: 10.1093/nar/7.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargan D. R., Butterworth P. H. A transcriptionally enriched chromatin fraction released by a Mg++ -dependent endogenous nuclease from rat liver. Cell Biol Int Rep. 1981 Jan;5(1):59–67. doi: 10.1016/0309-1651(81)90158-2. [DOI] [PubMed] [Google Scholar]
- Scheer U. Changes of nucleosome frequency in nucleolar and non-nucleolar chromatin as a function of transcription: an electron microscopic study. Cell. 1978 Mar;13(3):535–549. doi: 10.1016/0092-8674(78)90327-6. [DOI] [PubMed] [Google Scholar]
- Shen C. K., Hearst J. E. Photochemical crosslinking of transcription complexes with psoralen. I. Covalent attachment of in vitro SV40 nascent RNA to its double-stranded DNA template. Nucleic Acids Res. 1978 Apr;5(4):1429–1441. doi: 10.1093/nar/5.4.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C. K., Hsieh T. S., Wang J. C., Hearst J. E. Photochemical cross-linking of DNA-RNA helices by psoralen derivatives. J Mol Biol. 1977 Nov;116(4):661–679. doi: 10.1016/0022-2836(77)90265-0. [DOI] [PubMed] [Google Scholar]
- Siebenlist U., Simpson R. B., Gilbert W. E. coli RNA polymerase interacts homologously with two different promoters. Cell. 1980 Jun;20(2):269–281. doi: 10.1016/0092-8674(80)90613-3. [DOI] [PubMed] [Google Scholar]
- Spindler S. R. Deoxyribonucleic acid dependent ribonucleic acid polymerase II specific initiation and elongation factors from calf thymus. Biochemistry. 1979 Sep 4;18(18):4042–4048. doi: 10.1021/bi00585a031. [DOI] [PubMed] [Google Scholar]
- Tien Kuo M., Sahasrabuddhe C. G., Saunders G. F. Presence of messenger specifying sequences in the DNA of chromatin subunits. Proc Natl Acad Sci U S A. 1976 May;73(5):1572–1575. doi: 10.1073/pnas.73.5.1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trendelenburg M. F., McKinnell R. G. Transcriptionally active and inactive regions of nucleolar chromatin in amplified nucleoli of fully grown oocytes of hibernating frogs, Rana pipiens (Amphibia, Anura). A quantitative electron microscopic study. Differentiation. 1979;15(2):73–95. doi: 10.1111/j.1432-0436.1979.tb01037.x. [DOI] [PubMed] [Google Scholar]
- Wasylyk B., Thevenin G., Oudet P., Chambon P. Transcription of in vitro assembled chromatin by Escherichia coli RNA polymerase. J Mol Biol. 1979 Mar 5;128(3):411–440. doi: 10.1016/0022-2836(79)90095-0. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Woodcock C. L., Frado L. L., Hatch C. L., Ricciardiello L. Fine structure of active ribosomal genes. Chromosoma. 1976 Oct 12;58(1):33–39. doi: 10.1007/BF00293438. [DOI] [PubMed] [Google Scholar]
- Wu C., Wong Y. C., Elgin S. C. The chromatin structure of specific genes: II. Disruption of chromatin structure during gene activity. Cell. 1979 Apr;16(4):807–814. doi: 10.1016/0092-8674(79)90096-5. [DOI] [PubMed] [Google Scholar]