Abstract
Apoplastic α-glucosidases occur widely in plants but their function is unknown because appropriate substrates in the apoplast have not been identified. Arabidopsis contains at least three α-glucosidase genes; Aglu-1 and Aglu-3 are sequenced and Aglu-2 is known from six expressed sequence tags. Antibodies raised to a portion of Aglu-1 expressed in Escherichia coli recognize two proteins of 96 and 81 kD, respectively, in vegetative tissues of Arabidopsis, broccoli (Brassica oleracea L.), and mustard (Brassica napus L.). The acidic α-glucosidase activity from broccoli flower buds was purified using concanavalin A and ion-exchange chromatography. Two active fractions were resolved and both contained a 96-kD immunoreactive polypeptide. The N-terminal sequence from the 96-kD broccoli α-glucosidase indicated that it corresponds to the Arabidopsis Aglu-2 gene and that approximately 15 kD of the predicted N terminus was cleaved. The 81-kD protein was more abundant than the 96-kD protein, but it was not active with 4-methylumbelliferyl-α-d-glucopyranoside as the substrate and it did not bind to concanavalin A. In situ activity staining using 5-bromo-4-chloro-3-indolyl-α-d-glucopyranoside revealed that the acidic α-glucosidase activity is predominantly located in the outer cortex of broccoli stems and in vascular tissue, especially in leaf traces.
The α-glucosidases (EC 3.2.1.20, α-d-glucoside glucohydrolase) are a widespread and diverse group of enzymes that serve a variety of functions, depending on the subcellular location and organism in which they are found. Despite their origins in several unrelated gene families, they all share an unusually wide and overlapping substrate specificity, making identification from biochemical properties alone difficult (for review, see Frandsen and Svensson, 1998). Various forms of α-glucosidase can hydrolyze α-1,1-; α-1,2-; α-1,3-; α-1,4-; and α-1,6-linked Glc from glycoproteins or from the nonreducing ends of carbohydrates ranging in size from disaccharides to starch. They can also catalyze α-glucosyltransferase reactions (Yamasaki and Suzuki, 1980; Yamasaki and Konno, 1985).
Apoplastic forms of α-glucosidase having acidic pH optima occur widely in plants (Klis, 1971; Parr and Edelman, 1975; Yamasaki and Konno, 1987, 1992; Beers et al., 1990), but their function remains elusive because of the apparent lack of appropriate substrates in the apoplast (Fry, 1995). A chloroplastic form of α-glucosidase with a neutral pH optimum was isolated from pea and is thought to function in starch degradation, perhaps by removing rare α-1,2 or α-1,3 linkages that, if present, could block the action of the more abundant amylases and phosphorylases (Beers et al., 1990; Sun et al., 1995). Two different ER forms of α-glucosidase are also known: glucosidase I and II, which sequentially remove α-1,2- and α-1,3-linked Glc, respectively, from nascent glycoproteins as part of the quality control system that functions during glycoprotein folding (Hebert et al., 1995).
We began characterizing the α-glucosidases of Arabidopsis by searching the Arabidopsis EST collection for genes homologous to Family 31 α-glucosidases, which include mammalian lysosomal α-glucosidase (Hoefsloot et al., 1988) and intestinal sucrase/isomaltase (Chantret et al., 1992) and several fungal α-glucosidases (Dohman et al., 1990; Sugimoto and Suzuki, 1996; Nakamura et al., 1997a). The ER glucosidase II is a distant relative of this family (Trombetta et al., 1996; Arendt and Ostergaard, 1997). ESTs from several different Arabidopsis genes were identified, and one, 38A2T7 (accession no. T04333), was used as a probe to clone the Aglu-1 gene (Monroe et al., 1997). The deduced amino acid sequence of Aglu-1 contains 902 amino acids with a predicted mass of 101 kD, including the putative signal sequence. The amino acid sequence of Aglu-1 is 42% to 48% identical to those of the recently reported cDNA clones from barley (Tibbot and Skadsen, 1996), spinach (Sugimoto et al., 1997), and sugar beet (Matsui et al., 1997). Although it is not completely sequenced, a second Arabidopsis gene, Aglu-2, known from six ESTs, is similar to Aglu-1 and each of the other plant α-glucosidases.
The polypeptides of the α-glucosidases all have deduced masses of just over 100 kD, but the products of the four sequenced genes are all subjected to posttranslational modification, including proteolysis and glycosylation, probably in the secretory pathway, since all of the genes contain putative signal sequences. In barley seeds antibodies against the cloned α-glucosidase expressed in Escherichia coli recognize polypeptides of 81 and 95 kD (Tibbot et al., 1998). Active forms of barley seed α-glucosidase are also glycosylated (Sun and Henson, 1990). Four active forms of α-glucosidase from spinach seeds were separated by Sugimoto et al. (1995) and found to be 78, 78, 82, and 82 kD. The smaller forms were much more active against soluble starch than were the larger forms, but peptide sequences from three of the forms were all found within a single deduced cDNA sequence (Sugimoto et al., 1997). The active product of the sugar beet α-glucosidase is 91 kD (Chiba et al., 1978). As with spinach seeds, four forms of the sugar beet enzyme, varying both in affinity for the cell wall fraction and in substrate specificity, were separated from cultured cells (Yamasaki and Konno, 1989). Apparently, posttranslational modifications affect not only the size of the enzymes but also their substrate specificity. Other acidic α-glucosidases for which sequence information is lacking are also glycosylated, including those from soybean (Yamasaki and Konno, 1985) and banana (Konishi et al., 1991). We conclude that most if not all acidic α-glucosidases are glycosylated and proteolytically processed, but for no form of the enzyme is the extent of glycosylation or the specific proteolytic cleavage sites known.
Apoplastic α-glucosidases are well characterized biochemically, and some have been sequenced; however, little is known about their physiological function, especially in vegetative tissues. In this paper we describe the Aglu gene family from Arabidopsis and report on the isolation of a glycosylated, acidic α-glucosidase from broccoli (Brassica oleracea L.) flower buds. N-terminal sequence from the broccoli enzyme indicates that it is derived from a homolog of the Arabidopsis Aglu-2 gene that has undergone proteolysis. Cellular and tissue-level location of the activity and functional implications of that location are also described.
MATERIALS AND METHODS
Plant Materials
Arabidopsis plants were grown at 24°C under continuous illumination on a growth cart (Grower's Supply, Ann Arbor, MI). The growth medium was ProMix BX (Premier Brands, Red Hill, PA) supplemented with macronutrients and micronutrients as described by Somerville and Ogren (1982). Mustard (Brassica napus cv Southern Giant Curled) seeds were soaked in water with 0.1% Triton X-100 for 40 min, 95% ethanol for 4 min, 30% (v/v) sodium hypochlorite for 10 min, then rinsed five times with sterile water. Seeds were grown in 250-mL flasks containing 50 mL of autoclaved deionized water for 3 d. Flasks were shaken at 120 rpm under continuous illumination. Broccoli (Brassica oleracea) spears were purchased at a local market and stored at 4°C until use.
DNA Analysis
ESTs were obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus). DNA sequences from the 5′ ends of the ESTs 38A2T7 (accession no. T04333) and H8A7T7 (accession no. W43892) were obtained by cycle sequencing (Epicentre Technologies, Madison, WI) and analyzed using an automated sequencing system (Li-Cor, Lincoln, NE). Amino acid sequence alignments were conducted using the Higgins-Sharp algorithm (CLUSTAL4) in DNASIS version 3.5 (for Macintosh hardware). Identity scores were generated using default parameters.
Construction of pPW1700, Expression, and Antibody Production
A 700-bp BamHI fragment from the cDNA 38A2T7 corresponding to amino acids 388 to 605 of the deduced Aglu-1 protein was inserted into the BamHI site of pFLAG-ATS (Kodak). The resulting plasmid, pPW1700, was transformed into Escherichia coli BL21 cells for expression. Cells containing pPW1700 were grown overnight in expression broth (Luria-Bertani medium containing 50 μg/mL ampicillin and 0.4% [w/v] Glc) using a 37°C shaker. The cells were then diluted 100-fold in 5 mL of prewarmed expression broth and incubated in a 125-mL flask at 37°C with shaking. Upon reaching an A600 of about 0.2, isopropylthio-β-galactoside was added to 0.5 mm. At 2 h postinduction, only induced cells harboring pPW1700 contained the expected 24-kD polypeptide judging from Coomassie blue-stained SDS-PAGE gels. We were unable to use the anti-FLAG antibody to purify the 24-kD protein, probably because most of the induced protein was insoluble. We therefore solubilized cells containing the 24-kD polypeptide in 3 m urea with sonication and electroeluted the protein from a preparative SDS-PAGE gel (Gerhardt et al., 1994). One milligram of the purified 24-kD polypeptide was used to raise polyclonal antibodies in New Zealand White rabbits (Bethyl Labs, Montgomery, TX.) The antiserum specifically recognized the 24-kD polypeptide in immunoblots.
Purification of α-Glucosidase
All procedures were conducted at 4°C. Plant tissues were ground in a mortar and pestle with quartz sand in 2 volumes of extraction buffer (40 mm Hepes [pH 7.0], 1 m NaCl, 3 mm DTT, and 2 mm EDTA). Solid (NH4)2SO4 was added to 90% saturation and the solution was stirred for 15 min. Precipitated proteins were collected by centrifugation at 25,000g for 15 min and were dissolved in the same extraction buffer. After a centrifugation to clarify the solution, the sample was applied to a ConA-Sepharose column (3 × 0.5 cm). Nonbound proteins were washed from the column with additional extraction buffer, and then bound proteins were eluted with 15 mm methyl Glc in the same buffer. Glycosylated fractions were pooled and dialyzed overnight against 2 L of 10 mm Hepes, pH 7.0. The dialyzed sample was then applied to a CM-cellulose column (1 × 20 cm) and the nonbound proteins were washed from the column with the same buffer. Bound proteins were then eluted with a linear gradient of 0 to 1.0 m NaCl. Pooled active fractions were concentrated by placing them in a dialysis bag and laying the bag on a bed of PEG-8000 for 1 to 3 h, and/or with a Centricon-10 cartridge (Fisher Scientific).
DEAE-cellulose chromatography was carried out similarly to CM-cellulose chromatography except that the NaCl gradient was 0 to 0.3 m. For some experiments crude extracts were applied directly to the Con A column or were dialyzed overnight against 2 L of 10 mm Hepes (pH 7.0) with 150 mm NaCl before being applied to a Sephadex G-150 column.
Protein Analysis
Proteins were separated by SDS-PAGE in a minigel apparatus (Bio-Rad). The stacking and separating gels contained 4% and 12.5% acrylamide, respectively. Gels were either stained with Coomassie blue or proteins were transferred to nitrocellulose membranes. The membranes were blocked with 3% (v/v) coldwater fish gelatin in Tris-buffered saline and probed with a 1:100 dilution of anti-Aglu-1 serum or preimmune serum. Bands were visualized using peroxidase-conjugated goat anti-rabbit IgG (Sigma) diluted 1:2000 according to the manufacturer's instructions. Prestained molecular mass standards (Sigma) were used to estimate the masses of polypeptides.
For protein sequence analysis approximately 10 μg of the purified broccoli 96-kD α-glucosidase was separated by SDS-PAGE, transferred to a PVDF membrane, and stained according to the manufacturer's instructions (ABI, Columbia, MD). The N-terminal sequence was determined using Edman degradation by Midwest Analytical (St. Louis, MO).
Enzyme and Protein Assays
α-Glucosidase assays were conducted in a final volume of 0.5 or 1.0 mL of 50 mm Na acetate, pH 4.5, or 50 mm Mes, pH 7.0, and 0.1 mm 4-MUG (unless otherwise noted) at 37°C. Reactions were initiated with the addition of enzyme and were stopped with 3 mL of 0.5 m Gly, pH 10.5. Reactions stopped prior to the addition of enzyme were used as inactive controls and each assay was duplicated. Fluorescence of 4-methylumbelliferone at 450 nm was measured after excitation at 366 nm using a Spex spectrafluorimeter (Instruments S.A., Edison, NJ) and quantified using 4-methyl umbelliferone standards. One unit of activity was defined as the formation of 1 μmol product min−1 at 37°C. For pH curves 50 mm sodium acetate, Mes, or Hepes were used as buffers. For in situ α-glucosidase assays, intact Arabidopsis or mustard seedlings or hand-cut broccoli sections were incubated for 3 to 12 h at 30°C in the dark in a small volume of assay medium containing the same buffers as described above, but with 2 mm X-α-gluc (Calbiochem). The staining solution was then removed and the tissues were cleared through sequential washes with 70%, 95%, and 100% ethanol for 10 min each prior to photography. Malate dehydrogenase activity was assayed by monitoring the oxaloacetate-dependent oxidation of NADH at 340 nm in 50 mm Hepes, pH 7.5, 150 μm NADH, and 8 mm oxaloacetate, pH 6.5. Protein was measured with either Bradford reagent (Bio-Rad) or bicinchoninic acid reagent (Pierce) using BSA as the standard.
RESULTS
Arabidopsis Contains at Least Three α-Glucosidase Genes
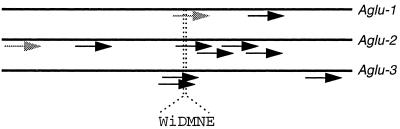
The Arabidopsis EST collection currently contains sequences from 11 unique cDNAs, each with a very high probability of matching family 31 α-glucosidases. Based on overlapping EST nucleotide sequences, alignment of deduced amino acid sequence with known α-glucosidases, and restriction mapping, we concluded that the 11 ESTs were derived from at least three different genes, which are illustrated in Figure 1. For each of the three genes, an EST was identified containing the peptide sequence WiDMNE, which is one of two signature peptides for α-glucosidases from Family 31 recognized by Nichols et al. (1998). The Asp residue in this signature peptide is located in the active site as determined by labeling with conduritol B epoxide (Sugimoto et al., 1997). We propose to name the three Arabidopsis genes Aglu-1, Aglu-2, and Aglu-3. The complete nucleotide sequences of the Aglu-1 gene (Monroe et al., 1997) and the Aglu-3 gene (Nakamura et al., 1997b) are known.
Figure 1.
Maps of the relative positions of 11 Arabidopsis α-glucosidase ESTs on the coding sequences of three α-glucosidase genes, Aglu-1, Aglu-2, and Aglu-3. Reading from left to right, ESTs (and accession numbers) from Aglu-1: 38A2T7 (T04333) and 151P15T7 (T76451); Aglu-2: H8A7T7 (W43892), G10B11T7 (N96165), 141B3T7 (T46694), 208 h21T7 (N37141), 169E22T7 (R64965), and 192B2T7 (R90271); Aglu-3: 47E11T7 (T14117), 35C12T7 (T04464), and OBO389 (ATTS5896). The relative position of the conserved active-site peptide WiDMNE is illustrated. ESTs used in this study are illustrated as shaded arrows; length of arrows corresponds roughly to the length of the known EST sequence.
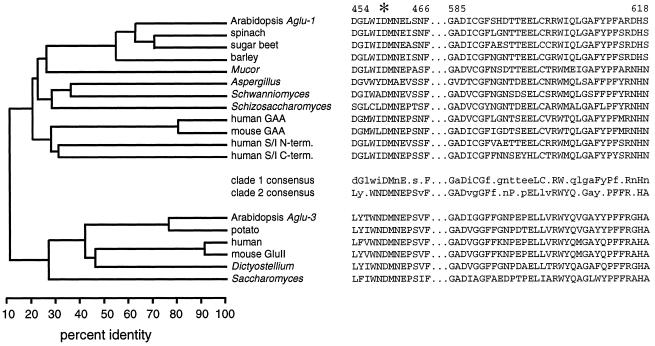
The three Arabidopsis Aglu genes fall into two clades based on amino acid sequence alignments with other sequenced genes. Figure 2 shows a phylogenetic tree and the two signature sequences for selected members of Family 31 glucoside hydrolases. For the phylogenetic analysis we included approximately 550 amino acids representing about 60% of each protein, omitting approximately 250 amino acids from each N terminus and approximately 150 amino acids from each C terminus because they were poorly conserved even in the most closely related pairs of genes. Aglu-1 fell into clade 1 with the mammalian and fungal α-glucosidases, mammalian sucrase/isomaltase, and the other apoplastic plant α-glucosidases. Sequences from ESTs derived from Aglu-2 strongly suggest that it is also a member of clade 1. Aglu-3 fell into clade 2 along with the ER glucosidase II from mouse (Arendt and Ostergaard, 1997) and several sequenced eukaryotic genes, including a cDNA from potato (Taylor et al., 1998). Partial amino acid sequence from the ER glucosidase II purified from rat liver closely matches the mouse glucosidase II sequence (Trombetta et al., 1996). Sequence identity between the two clades is only 12%, omitting the poorly conserved N- and C-terminal regions, and only 8% overall, suggesting that the two groups are very distantly related. The presence of a perfectly conserved Asp in the WiDMNE peptide and the considerable similarity among both family 31 signature regions illustrated in Figure 2 suggest a common evolutionary origin for the clades.
Figure 2.
Phylogenetic relationship among Family 31 α-glucosidases and aligned signature peptides indicate that there are two clades. Left, Phylogenetic tree indicating percent identity at the amino acid level using about 60% of each protein. Approximately 250 amino acids from the N termini and 150 amino acids from the C termini of each protein were omitted from this analysis. Right, Sequences of two signature peptide regions along with consensus sequences for each clade. Consensus amino acids are uppercase if perfectly conserved and lowercase if conserved in more than half of the sequences. The sequences (and accession numbers) used in the analysis from clade 1 were: Aglu-1 from Arabidopsis (AF014806), α-glucosidases from spinach (D86624), sugar beet (D89615), barley (U22450), Mucor javanicus (D67034), Aspergillus niger aglA (D45356), Schwanniomyces occidentalis glucoamylase GAM1 (M60207), Schizosaccharomyces pombe C30D11.01C (1723210), lysosomal acid α-glucosidase (GAA) from human (M34424) and mouse (P70699), and the N-terminal and C-terminal halves of the human sucrase/isomaltase (M22616). In clade 2 the sequences (and accession numbers) used were Aglu-3 from Arabidopsis (AB007646), cDNAs from potato (AJ001374) and human (AJ000332), GluII from mouse (U92793), ModA from Dictyostellium discoideum (U72236), and the Saccharomyces cerevisiae open reading frame YBR229c (Z36098).
Arabidopsis Contains Acidic and Neutral α-Glucosidases
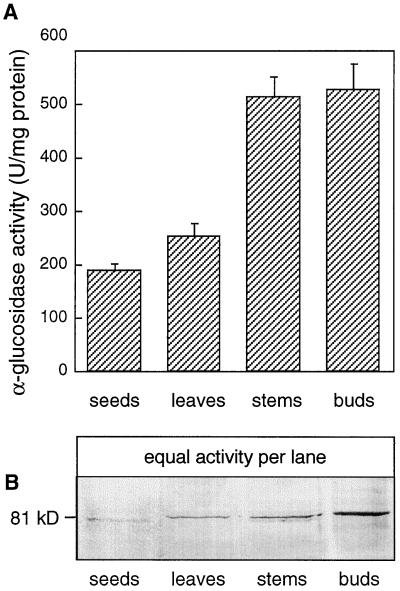
When crude extracts of Arabidopsis organs were assayed for α-glucosidase activity using 4-nitrophenyl-α-d-glucopyranoside as the substrate, very low levels of activity were observed. We therefore used 4-MUG as the substrate because of its increased sensitivity. The broad pH profile of activity in crude extracts (data not shown) suggested that several forms with optimal activity at acidic and neutral pHs existed in all tissues. Neutral forms of α-glucosidase from pea, banana, and barley have negligible activity below pH 5.0 (Beers et al., 1990; Konishi et al., 1991; Sun et al., 1995), so we assayed various extracts at pH 4.5. Figure 3A shows that stems and “buds” (including some stem tissue, flowers, flower buds, and young siliques) contained more acidic activity on a total protein basis than did leaves or seeds. To determine whether this activity was the product of any of the Aglu genes, we raised antibodies to a fragment of an EST derived from Aglu-1.
Figure 3.
α-Glucosidase activity and immunoblot of extracts from Arabidopsis tissues probed with anti-Aglu-1. A, α-Glucosidase activity in Arabidopsis tissues. Assays were conducted at pH 4.5 using 4-MUG as the substrate. Bud tissue included some stem tissue, flowers, flower buds, and young siliques. Data represent the means ± sd (n = 3). B, Detection of an 81-kD protein in Arabidopsis tissues with anti-Aglu-1 serum. Equal amounts of α-glucosidase activity (8 units) from the tissues represented in A were separated by SDS-PAGE and probed with anti-Aglu-1 serum. U, Unit.
Antibodies to Aglu-1 Recognize an 81-kD Polypeptide in Arabidopsis
A 700-bp BamHI fragment from the cDNA 38A2T7 (accession no. T04333) from Aglu-1 was expressed in E. coli and polyclonal antibodies were raised to the purified 24-kD polypeptide. This polypeptide contains the WiDMNE signature sequence and comprises about 25% of the deduced amino acid sequence of Aglu-1, beginning at about the midpoint of the protein. Equal levels of α-glucosidase activity from the same tissues represented in Figure 3A were separated by SDS-PAGE and probed with anti-Aglu-1 serum. Figure 3B shows that an 81-kD protein was recognized by the antiserum in some of the extracts, however, there was no clear relationship between the amount of the 81-kD protein and the level of α-glucosidase activity measured at pH 4.5. The bud sample contained much more of the immunoreactive protein than did other tissues (Fig. 3B). Similar blots probed with the preimmune serum did not reveal the 81-kD protein. Because of the potential for cross-reaction of the anti-Aglu-1 serum with products of the Aglu-2 gene, it could not be determined if the 81-kD Arabidopsis protein was the product of Aglu-1 or Aglu-2. Regardless, the size of this protein was similar to some of the products of the homologous spinach and barley acidic α-glucosidases (Sugimoto et al., 1995; Tibbot et al., 1998). We therefore proceeded to characterize both the 81-kD polypeptide and the major acidic α-glucosidase activity from vegetative tissues to determine whether they were related.
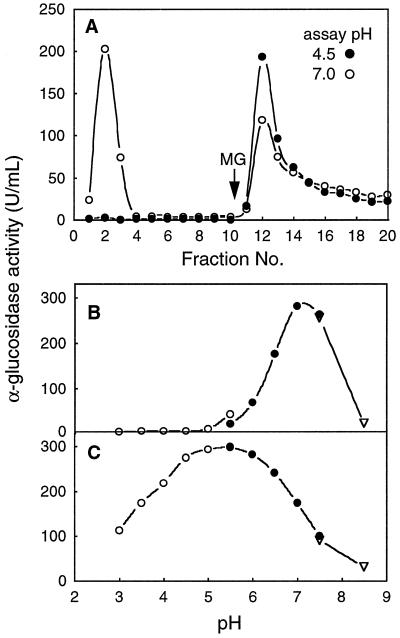
Konishi et al. (1991) were able to separate the major acidic and neutral forms of α-glucosidase from banana pulp using ConA-Sepharose affinity chromatography. A crude extract from Arabidopsis leaf tissue was prepared in a neutral extraction buffer containing 1 m NaCl, which is known to aid the release of apoplastic α-glucosidase (Parr and Edelman, 1975). Figure 4A shows that some α-glucosidase activity passed through the ConA column, whereas a fraction bound to ConA and was eluted with 15 mm methyl Glc. The nonbinding (ConA) fraction had optimal activity at pH 7.0 to 7.5 (Fig. 4B), which is typical of neutral α-glucosidases, whereas the bound (ConA+) fraction had optimal activity at pH 4.5 to 6.0 (Fig. 4C), typical of acidic α-glucosidases. As expected, the neutral form was nearly inactive below pH 5.0.
Figure 4.
Separation of neutral and acidic α-glucosidases from an Arabidopsis leaf extract using a ConA-Sepharose column, and the pH profile of each activity peak. A, ConA-Sepharose column. Bound proteins were eluted with 15 mm methyl Glc (MG). α-Glucosidase activity was measured at pH 4.5 (•) and pH 7.0 (○). B, pH profile of pooled fractions 2 and 3 from A. C, pH profile of pooled fractions 12 to 14 from A. For B and C, buffers were sodium acetate (○), Mes (•), or Hepes (▿). U, Unit.
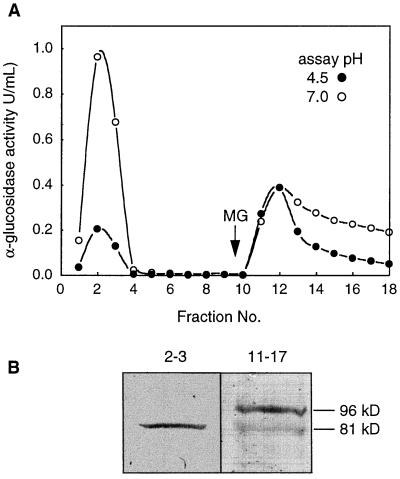
A crude extract from Arabidopsis stem and bud tissue enriched in the 81-kD protein was then prepared and passed through a ConA column to determine whether the 81-kD protein binds. Figure 5A shows that the ConA− fraction was most active at pH 7.0, but unlike the ConA− leaf fraction, which contained negligible activity at pH 4.5 (Fig. 4B), the ConA− stem+bud fraction had low but significant activity at pH 4.5. The ConA+ stem+bud fraction was active at both pH 4.5 and 7.0 (Fig. 5A). Both the ConA− and ConA+ fractions were then concentrated and probed with anti-Aglu-1 serum on an immunoblot. Surprisingly, nearly all of the 81-kD protein was in the ConA− fraction (Fig. 5B). The ConA+ fractions, which contained most of the acidic activity, contained a 96-kD immunoreactive protein that was much less abundant than the 81-kD protein since it required a greater degree of concentration to observe on immunoblots. This may explain why it was not observed in immunoblots of crude extracts.
Figure 5.
Separation of α-glucosidases from an Arabidopsis stem+bud extract by ConA chromatography and an immunoblot of the pooled active fractions probed with anti-Aglu-1. A, ConA-Sepharose column. Bound proteins were eluted with 15 mm methyl Glc (MG). α-Glucosidase activity was measured at pH 4.5 (•) and pH 7.0 (○). U, Unit. B, Immunoblot of activity peaks separated in A. Nonbinding activity (fractions 2 and 3) and bound activity (fractions 11–17) from A was concentrated 24-, and 215-fold, respectively, and equal volumes of each concentrated fraction were separated by SDS-PAGE and probed with anti-Aglu-1.
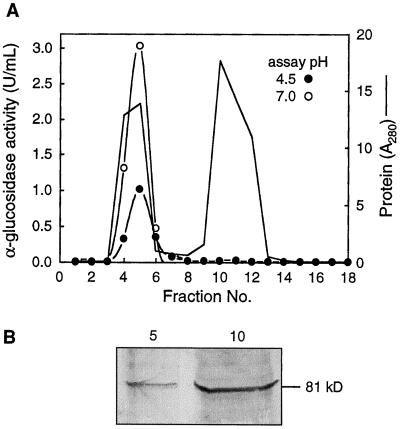
The presence of the 81-kD protein in the ConA− fraction suggested that it was either a novel neutral form of the enzyme or that the ConA− stem+bud fraction contained both the neutral form and an acidic, 81-kD polypeptide that was much less active than the ConA+ acidic form of the enzyme. To answer this question, a crude stem+bud extract from Arabidopsis was subjected to gel filtration using Sephadex 150-G to attempt to separate the neutral activity from the 81-kD polypeptide. Figure 6 shows that all of the α-glucosidase activity at both pH 4.5 and 7.0 coeluted with the first major peak of protein, but that most of the 81-kD polypeptide eluted with a second major protein peak, which had no detectable α-glucosidase activity. The 81-kD protein is therefore not the major neutral α-glucosidase, but because it had no apparent activity with 4-MUG, we did not characterize it further. It is possible that the 81-kD protein is not an α-glucosidase but that it simply shares a common epitope recognized by anti-Aglu-1. However, as noted above, other acidic plant α-glucosidases that were not assayed with 4-MUG have a mass that is similar to the 81-kD Arabidopsis protein.
Figure 6.
Separation of the Arabidopsis α-glucosidase activity from most of the 81-kD polypeptide using Sephadex G-150. A, Sephadex G-150 column. α-Glucosidase activity was assayed at pH 4.5 (•) and pH 7.0 (○). Total protein was monitored at A280 (solid line). U, Unit. B, Immunoblot of fractions 5 and 10 from A. Equal volumes (10 μL) of each fraction were separated by SDS-PAGE and probed with anti-Aglu-1.
Since the major acidic α-glucosidase activity in Arabidopsis appeared to be associated with a relatively rare 96-kD protein enriched in bud tissues, we continued our purification of the activity using broccoli flower bud tissue, which was more readily available. We anticipated that broccoli, being a crucifer, would contain α-glucosidase proteins to which our Arabidopsis Aglu-1 antiserum would bind. Indeed, crude broccoli bud extracts also contained an 81-kD protein recognized by the anti-Aglu-1 serum (data not shown).
Purification of Acidic α-Glucosidase from Broccoli
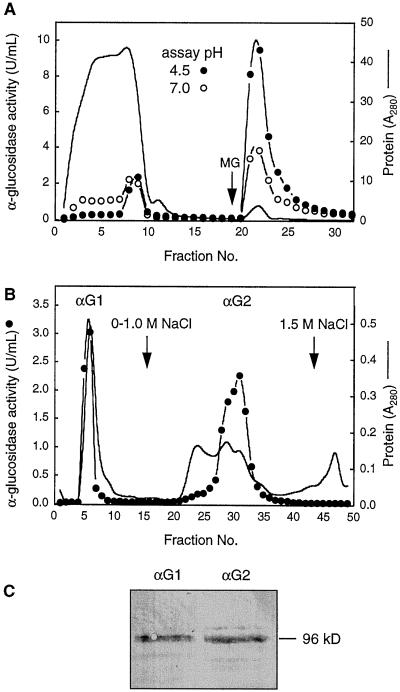
Broccoli flower bud and stem tissue was ground in extraction buffer and clarified with centrifugation. Proteins were precipitated from the crude extract with 90% (NH4)2SO4, dissolved, and passed through a ConA column. Figure 7A shows that less than 3% of the total protein was retained by the column, as measured by A280, but that over 80% of the α-glucosidase activity at pH 4.5 was retained by the column and was eluted with 15 mm methyl Glc. When probed with anti-Aglu-1, this glycoprotein fraction contained the 96-kD immunoreactive protein (data not shown).
Figure 7.
Isolation of the major acidic α-glucosidases from broccoli flower buds using ConA- and CM-cellulose chromatography and an immunoblot of pooled active fractions probed with anti-Aglu-1. A, ConA-Sepharose column. Bound proteins were eluted with 15 mm methyl Glc (MG). α-Glucosidase activity was assayed at pH 4.5 (•) and pH 7.0 (○). Total protein was monitored at A280 (solid line) in A and B. B, Separation of fractions 21 to 24 from A on a CM-cellulose column. α-Glucosidase activity was assayed at pH 4.5 (•). C, Immunoblot of activity peaks separated in B. αG1 (fractions 5–6) and αG2 (fractions 28–32) from B were pooled and concentrated. Equal amounts of activity at pH 4.5 (37 units) were separated by SDS-PAGE and probed with anti-Aglu-1. U, Unit.
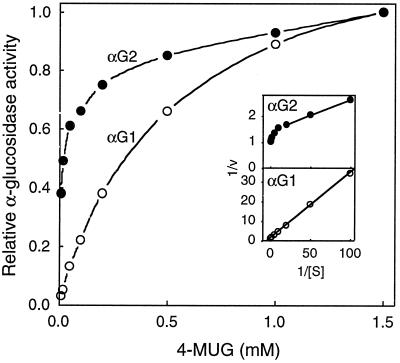
The ConA+ fractions containing the acidic activity were then pooled and dialyzed overnight against ion-exchange buffer (10 mm Hepes, pH 7.0) and applied to a CM-cellulose column. After loading the sample the column was washed with ion-exchange buffer and eluted with a linear gradient of 0 to 1.0 m NaCl in the same buffer. Two peaks of activity were separated: αG1 did not bind to the column, whereas αG2 eluted at approximately 0.5 m NaCl (Fig. 7B). The two peaks were subsequently concentrated and equal amounts of activity at pH 4.5 were separated by SDS-PAGE and probed with anti-Aglu-1 serum. Both peaks contained approximately equal amounts of the 96-kD protein (Fig. 7C), suggesting that they had similar specific activities. Figure 8 shows the effect of varying the concentration of 4-MUG on activity at pH 4.5. A Lineweaver-Burk plot of the activity in αG1 was linear and the Km for 4-MUG was approximately 0.3 mm. αG2, however, was clearly a mixture of activities since the Lineweaver-Burk plot was nonlinear. Extrapolation from the ends of the Lineweaver-Burk plot suggested that αG2 contained at least two α-glucosidases, one having a Km of approximately 0.3 mm and the other of 8.6 μm 4-MUG.
Figure 8.
Effect of 4-MUG on the activity of two broccoli α-glucosidase isoforms separated by CM-cellulose chromatography. αG1 (○) and αG2 (•) from the CM-cellulose column (Fig. 7B) were assayed at pH 4.5. Data were normalized to the level of activity at 1.5 mm 4-MUG. Inset, Lineweaver Burk plots of the same data.
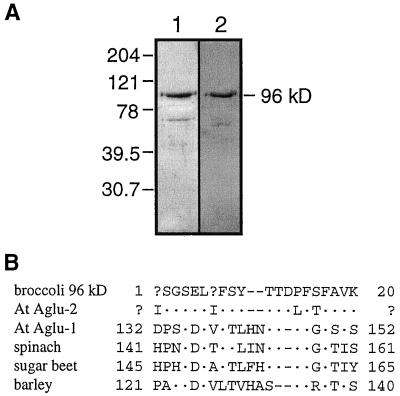
Although we succeeded in separating two 96-kD polypeptides using CM-cellulose, they were not pure, judging from Coomassie blue-stained SDS-PAGE (data not shown). Attempts to fractionate the broccoli glycoprotein fraction using a DEAE-cellulose column were disappointing because the activity eluted very broadly. However, the tail of activity eluting slowly from the DEAE-cellulose column was concentrated and found to contain a 96-kD protein that was nearly pure, based on Coomassie blue staining, and was recognized by anti-Aglu-1 serum, as illustrated in Figure 9. Approximately 10 μg of this protein fraction was separated by SDS-PAGE, transferred to a PVDF membrane, and sequenced using Edman degradation. The N-terminal sequence of this 96-kD broccoli protein was I(L)SGSELI(T)FSYTTDPFSFAVK. At positions 1 and 7 equal amounts of two different amino acids were observed, Ile and Leu, and Ile and Thr, respectively. Whether this sequence corresponds to either or both of the α-glucosidases separated by CM-cellulose is not known.
Figure 9.
Purified 96-kD broccoli α-glucosidase stained with Coomassie blue or probed with anti-Aglu-1 serum, and the N-terminal sequence from the same protein aligned with other plant α-glucosidase sequences. A, SDS-PAGE and immunoblot of the 96-kD broccoli α-glucosidase. Proteins (4 μg) were separated by SDS-PAGE and either stained with Coomassie blue (lane 1) or probed with anti-Aglu-1 serum (lane 2). The numbers on the left represent molecular mass standards in kilodaltons. B, N-terminal sequence from the broccoli 96-kD α-glucosidase aligned with other plant α-glucosidases. Sequences (and accession numbers) were from Arabidopsis Aglu-1 (AF014806), Arabidopsis Aglu-2 (sequence from the EST H8A7T7), spinach (D86624), sugar beet (D89615), and barley (U22450).
Alignment of the 20-residue N-terminal sequence from the broccoli 96-kD protein with Arabidopsis Aglu-1 and other plant α-glucosidases revealed that it matched a region 121 to 145 amino acids from the translation start site of each sequence (Fig. 9). Although Aglu-2 is not completely sequenced, an EST from Aglu-2, H8A7T7 (accession no. W43892), appeared to span this region. We resequenced the 5′ end of this cDNA and found that the corresponding region of Arabidopsis Aglu-2 matched the broccoli sequence at 16 of the 20 residues (Fig. 9). If the two ambiguous residues are both Ile, then identity between the broccoli 96-kD protein and Arabidopsis Aglu-2 increases to 90%. Identity between the broccoli peptide and each of the other plant α-glucosidases, including Arabidopsis Aglu-1, ranged from only 39% to 47%. The broccoli protein therefore appears to be encoded by the homolog of the Arabidopsis Aglu-2 gene. Assuming that the structures of the broccoli and Arabidopsis Aglu genes are similar, our results indicate that the 96-kD protein undergoes proteolytic processing during its synthesis, in which approximately 15 kD of the N terminus is removed. Using Aglu-1 as the model gene, removal of 15 kD would leave at most 86 kD of polypeptide, suggesting that the 96-kD protein from Arabidopsis (Fig. 5B) and broccoli (Fig. 9) may contain at least 10 kD of glycan. The deduced Aglu-1 protein contains nine potential N-glycosylation sites, only one of which would be removed with the N-terminal 15 kD if Aglu-1 and Aglu-2 are similarly processed. Our prediction that the enzyme contains at least 10 kD of glycan is thus well within the maximum potential glycan mass, assuming that the glycan is N-linked, that Aglu-2 has a similar number of N-glycosylation sites, and that all of the sites have high Man or complex glycans attached.
As noted above, amino acid sequence conservation among the sequenced plant α-glucosidases is highest in the central 60% of the proteins. It is interesting that the proposed cleavage site falls at the border of a poorly conserved region and a well-conserved region (marked by the asterisk in Fig. 10). Percent identities within the N-terminal 15-kD region of the known plant α-glucosidases range from only 13% to 34%, whereas that of the central region, omitting the last 150 amino acids, range from 52% to 70%. Sequence conservation drops to between 10% and 25% within the C-terminal 13 kD.
Figure 10.
Alignment of the known plant α-glucosidase sequences. Sequences are from Arabidopsis Aglu-1, spinach, sugar beet, and barley. For accession numbers, see legend for Figure 9. Dots represent amino acids that are identical to the Arabidopsis Aglu-1 sequence. The N terminus of the broccoli 96-kD protein is indicated with an asterisk. The 24-kD Aglu-1 polypeptide expressed in E. coli for antibody production is underlined.
Acidic α-Glucosidase Activity in Mustard Is Apoplastic
Confirming through traditional methods that the acidic α-glucosidase activity in Arabidopsis and broccoli is located in the apoplast produced equivocal results because only low levels of activity could be extracted from Arabidopsis stems by centrifugation, and these experiments necessitated breaking some cells and releasing the more active neutral form of the enzyme. Moreover, our attempts to isolate protoplasts free of the acidic α-glucosidase activity were hampered by the presence of large amounts of acidic α-glucosidase in fungal cellulase preparations. We therefore reasoned that if the acidic α-glucosidase was indeed apoplastic, as in other plants, then it should elute from uncut tissues into a buffer containing 1 m NaCl if the tissues were small and not covered by a thick cuticle. We chose to use 3-d-old mustard seedlings that were grown under water with shaking so as to retard the development of the cuticle. Seedlings grown in this manner appeared to be identical to soil-grown plants. Mustard also contained 81- and 96-kD proteins recognized by Anti-Aglu-1 serum (data not shown).
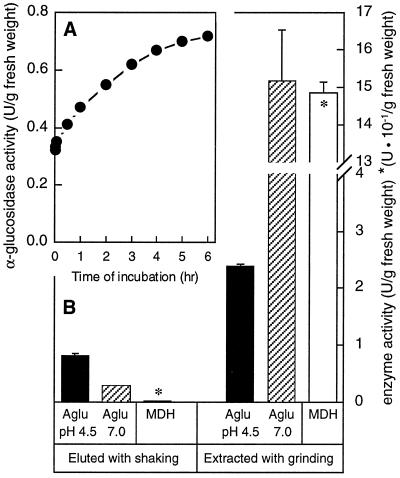
Seedlings were gently blotted dry, weighed, placed in flasks containing 10 volumes of extraction buffer, and shaken at 120 rpm. After only 1 min significant α-glucosidase activity at pH 4.5 was observed in the surrounding buffer (Fig. 11A). Over a 6-h period of shaking the level of activity in the buffer increased, indicating that the enzyme was slowly diffusing out of the seedlings. At the end of 6 h the medium contained activity that was about 3-fold higher at pH 4.5 than it was at pH 7.0, suggesting that only the acidic enzyme had eluted (Fig. 11B). Similar, untreated plants were ground with sand in the same extraction buffer to measure the total extractable activity. About 3-fold more activity at pH 4.5 was extracted from the seedlings with grinding, but activity at pH 7.0 was over 50-fold higher in the ground extract than in the elution buffer from shaken plants (Fig. 11B). Some of the activity at pH 4.5 from the ground plants could have been due to the neutral mustard enzyme, but if its pH profile is similar to that of the Arabidopsis neutral enzyme (Fig. 4B), this contribution would be low. As a marker of cell breakage, malate dehydrogenase activity in the buffer around the shaken plants was measured and found to be less than 0.2% of that from the ground plants, indicating that this method of elution was extremely gentle. These results indicate that at least one-third of the acidic α-glucosidase activity in these seedlings was located in the apoplast. The remainder of the activity could be inside the cell membrane, or it might not have been able to diffuse out of the apoplast because of the presence of a cuticle on the epidermis of some seedlings or internal barriers to apoplastic diffusion, such as the suberized root endodermis.
Figure 11.
Localization of acidic α-glucosidase activity in the apoplastic fraction of mustard seedlings. A, Time course of elution of α-glucosidase activity from intact mustard seedlings. Three-day-old mustard seedlings grown under water were shaken in extraction buffer containing 1 m NaCl for 6 h. α-Glucosidase in the medium was assayed periodically with 4-MUG at pH 4.5. B, Activity of α-glucosidase (Aglu) at either pH 4.5 or 7.0 and malate dehydrogenase (MDH) in the elution buffer from A after 6 h, and in extracts from similar but untreated plants. Data represent the means ± sd (n = 3). U, Unit.
To determine whether the apoplastic fraction contained the 96-kD immunoreactive protein, a similar salt extract of mustard seedlings was passed through a ConA column, concentrated, and analyzed by immunoblot. All of the acidic activity in the extract bound to the ConA column, and the most active fractions contained a 96-kD protein recognized by anti-Aglu-1 serum (data not shown).
The time course of elution of the acidic α-glucosidase activity from mustard seedlings suggested that two different pools of activity were eluted. One pool came out within the 1st min and was perhaps located at the surface of the seedlings, whereas the other diffused out more slowly over a period of hours (Fig. 11A). To investigate the possibility that plants contain multiple pools of α-glucosidase activity in surface and internal tissues, we used 2 mm X-α-gluc as a substrate to stain seedlings and broccoli stem sections.
Tissue-Level Localization of Acidic α-Glucosidase
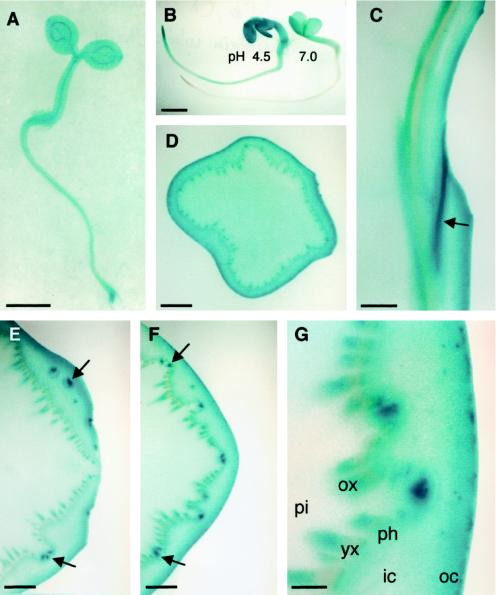
Three-day-old Arabidopsis seedlings grown on wet filter paper and stained with X-α-gluc at pH 4.5 appeared blue all over, but staining was strongest in root tips and in vascular tissues, as seen in Figure 12A. In 3-d-old mustard seedlings staining was much stronger at pH 4.5 than at 7.0, indicating that this activity was probably due to the apoplastic acidic α-glucosidase (Fig. 12B). To determine the spatial distribution of staining in a larger plant, hand sections of broccoli stems were similarly stained. Cross-sections through stems 2 to 3 cm below the apex revealed strongest staining in the cortex, especially in the outer 0.5 mm of the stem (Fig. 12D). In addition, some staining of the vascular tissue was observed in this section. Longitudinal sections through a leaf abscission zone showed strongest staining in the leaf trace (Fig. 12C). Staining of the inner xylem and outer cortex, even in the abscission zone, could also be seen. Cross-sections through and below a leaf abscission zone revealed a similar pattern (Fig. 12, E–G). The strongest staining was observed in leaf traces above and below the point of separation from the vascular cylinder. Figure 12G shows a higher magnification of the cross-section in Figure 12F, illustrating the relative lack of staining in the younger xylem, pith, and inner cortex. Staining of the abscission zone was also observed prior to leaf abscission (data not shown). The fast-eluting and slow-eluting pools of activity observed in mustard seedlings (Fig. 11A) may correspond to the epidermal and vascular activity, respectively (Fig. 12G).
Figure 12.
In situ activity of α-glucosidase assayed with α-X-gluc at pH 4.5 for 12 h, except where noted. A, Three-day-old Arabidopsis seedling. B, Three-day-old mustard seedlings stained at pH 4.5 (left) or 7.0 (right) for 3 h. C, Longitudinal section through a broccoli stem. The curved surface on the right side is a leaf abscission zone. Unstained tissue on the left is pith. Strong staining is seen in the leaf trace indicated by the arrow. D, Cross-section of a broccoli stem approximately 3 cm from the apex. E, Cross-section of a broccoli stem through a leaf abscission zone. Arrows point to strong staining in leaf traces at or above the point where they separate from the vascular cylinder. A leaf abscission zone is on the right side of the image. F, Same as E but approximately 4 mm basal. Arrows point to the same leaf traces as in E, but both leaf traces are below the point of separation from the vascular cylinder. G, Finer detail of F. pi, Pith; ox, older xylem; yx, young xylem; ph, phloem; ic, inner cortex; oc, outer cortex. Note the increased staining toward the outer cortex and in the older xylem and in the leaf traces. Bars = 1 mm (A, B, and D); 2 mm (C, E, and F); or 0.5 mm G.
Because of the relatively low activity of α-glucosidase in broccoli stem sections assayed with X-α-gluc, incubations were carried out over 12 h. This raised the possibility that some of the activity could be due to de novo synthesis induced by wounding. Inclusion of 10 μm cycloheximide in the assay solution did not affect staining, so the observed α-glucosidase activity was not due to de novo synthesis. Moreover, wounding did not appear to induce activity in broccoli stems. Adding 1 μm 1-deoxynojirimycin, an α-glucosidase inhibitor, to the assay mixture completely blocked staining with X-α-gluc, but it had no effect on staining with X-β-gluc, suggesting that the observed staining with X-α-gluc was indeed due to α-glucosidase activity.
The observed staining could be due to apoplastic α-glucosidase or, if X-α-gluc penetrated cell membranes, to a neutral α-glucosidase inside the cell, although, as mentioned above, staining was much stronger at pH 4.5, which suggests that the acidic apoplastic enzymes contributed most of the activity (Fig. 12B). When we tested the activity of crude extracts from leaf tissue with both 4-MUG and X-α-gluc at pH 4.5 and 7.0, relative activities with the two substrates were quite different. Activity with 4-MUG was about 2-fold higher at pH 7.0, whereas activity with X-α-gluc was 12-fold higher at pH 4.5, suggesting that either the neutral form of the enzyme is unstable in the presence of X-α-gluc or its solvent, or that the neutral form of the enzyme simply does not act on X-α-gluc. Regardless, in situ assays with X-α-gluc as the substrate appear to be specific for acidic α-glucosidase.
DISCUSSION
The Aglu Gene Family
Acidic α-glucosidases are a poorly understood group of enzymes, due in part to the fact that they have no known function in the apoplast, where they are located. In several plants multiple forms of the enzyme were separated and were shown to have distinct structural and catalytic properties, but no potential substrates are known to exist in the apoplast. Because of their broad substrate specificity and structural heterogeneity, identification of these enzymes from biochemical properties alone is problematic. We therefore chose to characterize the products of the Arabidopsis α-glucosidase gene family at the molecular level.
In Arabidopsis there are at least three members of this family, two of which, Aglu-1 and Aglu-2, are as similar to each other as they are to other acidic α-glucosidases that have been cloned from barley (Tibbot and Skadsen, 1996), spinach (Sugimoto et al., 1997), and sugar beet (Matsui et al., 1997). A third more distantly related α-glucosidase gene, Aglu-3, is most similar to the ER glucosidase II, which catalyzes the hydrolysis of α-1,3-linked Glc from glycoproteins as they fold. Arabidopsis also contains an abundant neutral α-glucosidase, which does not bind to ConA. It is likely that this enzyme is similar to the plastid form of α-glucosidase isolated from pea (Beers et al., 1990; Sun et al., 1995). The large size difference between the pea neutral form (24 kD) and the various acidic forms (78–96 kD) suggests that they may be encoded by different gene families.
Genes homologous to the Aglu gene family are ubiquitous in eukaryotes but are apparently absent in prokaryotes. If glycoprotein processing in the ER arose early in the evolution of eukaryotic cells, perhaps most of Family 31 α-glucosidases, including the mammalian lysosomal α-glucosidase and secreted sucrase/isomaltase, as well as the secreted fungal and plant α-glucosidases, are derived from an ancestral ER glucosidase II gene. All would have retained their signal peptides but would have then diversified in terms of their substrate specificity, location, and function.
Structure and Processing of the Major Acid α-Glucosidases in Crucifers
All of the plant α-glucosidase genes that have been sequenced encode deduced proteins of slightly more than 100 kD. The products of these genes, however, fall into two size classes, 78 to 82 kD and 91 to 96 kD, indicating that they all undergo proteolytic processing. They have optimal activity at an acidic pH and some were shown to be apoplastic glycoproteins. The most active acidic α-glucosidase in Arabidopsis and broccoli is a 96-kD glycoprotein that we believe contains at least 10 kD of sugar. Because it binds to ConA, the 96-kD protein probably contains some high Man glycans. We separated two isoforms of the 96-kD enzyme but were unable to distinguish them by size or activity, although one fraction appeared to be a mixture containing enzymes with different affinities for the substrate 4-MUG. Multiple isoforms of α-glucosidase with different catalytic properties were also separated from sugar beet and spinach (Yamasaki and Konno, 1989; Sugimoto et al., 1995). Whether the 96-kD broccoli α-glucosidase isoforms represent the products of different genes or differential posttranslational processing of a single gene product is not known, however, there is evidence that the latter can occur (Sturm, 1991). The glycan moieties of secreted glycoproteins are rapidly modified in the ER and Golgi by glycosidases and transglycosidases, so that the secreted forms are initially homogenous. After secretion, apoplastic N-acetylglucosaminidase and/or α-mannosidase slowly modify the complex glycans so that heterogeneity in glycan structure can exist, even at a single locus (Sturm, 1991). The two isoforms of broccoli α-glucosidase that we separated by CM-cellulose chromatography may represent the same enzyme with or without terminal GlcNAc on one or more complex glycans, thus affecting their affinity for the ion-exchange resin. Because the two proteins have similar specific activities, their structural differences may not be important to their function; however, they may differ in substrate specificity and/or affinity for the cell wall.
Based on the position of the N terminus of the broccoli protein within the aligned α-glucosidase sequences, our data suggest that approximately 15 kD of the polypeptide was proteolytically removed from the enzyme. Several lines of evidence suggest that this processing occurred in vivo and not during enzyme purification. First, no other purified acidic α-glucosidase has a subunit size larger than 96 kD. If all of them are similarly glycosylated, then proteolytic removal of a relatively large portion of the protein is a conserved event. Second, the cleavage site occurs at the border between the poorly conserved N-terminal region and the well-conserved central region of the sequence (Fig. 10), suggesting that all of the plant α-glucosidases may be similarly processed. However, two peptide sequences from the 82-kD spinach α-glucosidase III and IV begin only 29 residues from the N-terminal Met (Sugimoto et al., 1997). These residues are well within the N-terminal 15-kD region. This would suggest that proteolysis of the spinach enzyme might occur at the C terminus. Judging from the alignment shown in Figure 10, poor sequence conservation also occurs in the C-terminal 13 kD of the cloned α-glucosidases, suggesting that both ends of the proteins may be processed. If so, then the level of glycosylation is larger than our estimate of 10 kD. Clearly, there is a large degree of structural heterogeneity among these enzymes.
Unlike barley (Tibbot et al., 1998) and spinach (Sugimoto et al., 1995), in which the smaller 78- to 82-kD acidic α-glucosidases were catalytically active, the 81-kD Arabidopsis protein was not active with 4-MUG as the substrate. It may, however, have activity toward an in vivo substrate. Because the 81-kD protein did not bind to ConA-Sepharose, it either contains all complex glycans that fail to bind to ConA, or it may be a highly deglycosylated degradation product from either Aglu-1 or Aglu-2. The function of sugar moieties on glycoproteins has been well studied, and in addition to influencing thermal stability (Kern et al., 1992), glycosylation can also influence catalytic activity. Mutational removal of each of the four N-linked glycosylation sites in lecithin-cholesterol acyltransferase decreased or increased the specific activity of the enzyme, but removal of all four sites nearly abolished activity (O et al., 1993). Further study is necessary to characterize the 81-kD cross-reacting protein and to determine if it is an active α-glucosidase.
Location of Apoplastic α-Glucosidase Activity in Vegetative Tissues and Implications for Function
Although apoplastic α-glucosidases are usually assayed using α-1,4-linked carbohydrate substrates, the lack of such linkages in the apoplastic compartment of plants has made it difficult to speculate on the potential function of these enzymes. Numerous studies of acidic α-glucosidase were conducted on cereal grain seeds, where it was assumed that the enzymes act on starch during germination. Indeed, in cereal grains an apoplastic α-glucosidase could have access to starch or its hydrolytic products, but in other seeds in which cell integrity is maintained during germination and in vegetative tissues, apoplastic α-glucosidases should not come into contact with plastidic starch except during cell lysis.
Finding that these enzymes are distantly related to the ER glucosidase II (Fig. 2) may help to shed light on this problem. During glycoprotein synthesis and folding, glucosidase II removes two α-1,3-linked Glc residues from nascent glycoproteins. Apoplastic α-glucosidases can also hydrolyze α-1,3-linkages, sometimes with more efficiency than α-1,4-linkages (Yamasaki and Konno, 1989). Perhaps the apoplastic α-glucosidases act on an apoplastic glycoprotein containing α-linked Glc. However, to our knowledge, secreted glycoproteins containing α-linked Glc have not been reported, and incorrectly processed glycoproteins are unlikely to escape the ER. Alternatively, apoplastic α-glucosidases could act on glycoproteins secreted by other organisms such as plant pathogens. The location of the enzyme activity near cuticular surfaces and especially in open leaf traces, where pathogens attempt to infect plants, makes it tempting to speculate that the enzyme plays a role in defense. Several reports have shown that exogenous application of α-glucosidase can interfere with the development of pathogenic fungi (Xaio et al., 1994; Hollenstein et al., 1995).
After landing on a plant surface, fungal conidia germinate and form a short germ tube that differentiates at the tip to produce the infection cell or appressorium (Howard and Valent, 1996). For some fungi it was shown that if the plant is not a host, an appressorium is not formed and the fungus lives saprophytically (Beckerman and Ebbole, 1996). Recognition of the host thus appears to be critical for the development of some fungi, and probably involves signaling molecules. Working with rice blast fungus, Xaio et al. (1994) observed that conidia that germinated on an artificial substrate in the presence of α-glucosidase, α-mannosidase, or protease failed to develop appressoria. No inhibition was observed when conidia were germinated in the presence of β-glucosidase, α-galactosidase, or chitinase. The authors concluded that a secreted glycoprotein involved in cell signaling was perhaps being inactivated by the enzymes before it could induce appressorium formation. Similar results were observed by Hollenstein et al. (1995) using Phytophthora megasperma f. sp. glycinae conidia on soybean hypocotyls. In that study treatment with α-glucosidase caused a delay in appressorial development and subsequent infection of soybean cells.
The location of acidic α-glucosidase activity in surface tissues and in leaf traces below the abscission zone is consistent with a role in defense, since these tissues are the first that pathogens would contact upon landing on a plant. Vessel elements of leaf traces that are exposed after leaf abscission are known to be the route for infection by some bacterial pathogens, including the causal agent of apple canker (Crowdy, 1952) and cherry canker (Crosse, 1956). Immediately after abscission the water in a vessel element retreats into the stem, carrying with it any propagules on the exposed surface. Staining of the oldest xylem tissue in broccoli stems (Fig. 12, E–G) may be due to the fact that the vessels of this xylem would have been exposed to the air through the cut base of the spear, thus representing another route for pathogen invasion. Unlike the well-studied PR proteins that are induced upon infection and constitute part of the active defense system, we could not detect any induction of acidic α-glucosidase activity by wounding or treatment of tissues with ethylene, salicylic acid, or jasmonic acid in preliminary experiments. If the enzyme is involved in defense, it probably plays a passive role.
Other possible functions for the apoplastic α-glucosidase can be envisaged that are also consistent with our localization data. For example, the enzyme may play a role in suberin or cutin synthesis since the enzyme appears to be located in places where the synthesis of those molecules takes place. Little is known about the synthesis of suberin or cutin, but perhaps glycosylated precursors are secreted to the apoplast where α-glucosidase could remove the Glc prior to polymerization or deposition. Leaf traces are filled with waxy materials after leaf abscission, so high levels of α-glucosidase activity in leaf traces is consistent with this hypothesis. The isolation of mutants should aid in testing these hypotheses.
ACKNOWLEDGMENTS
We are grateful to Kelly Poliquin for excellent technical assistance, Maarten Chrispeels for helpful suggestions, Mark Brodl and Jennifer Jenkins for critically reviewing the manuscript, and the Arabidopsis Biological Resource Center for EST clones.
Abbreviations:
- CM
carboxy methyl
- ConA
concanavalin A
- EST
expressed sequence tag
- 4-MUG
4-methylumbelliferyl-α-d-glucopyranoside
- X-α-gluc
5-bromo-4-chloro-3-indolyl-α-d-glucopyranoside
Footnotes
This work was supported by the U.S. Department of Agriculture-National Research Initiative Competitive Research Grants Program (grant nos. 94-04200 and 96-00679).
LITERATURE CITED
- Arendt CW, Ostergaard HL. Identification of the CD45-associated 116-kDa and 80-kDa proteins as the alpha- and beta-subunits of alpha-glucosidase II. J Biol Chem. 1997;272:13117–13125. doi: 10.1074/jbc.272.20.13117. [DOI] [PubMed] [Google Scholar]
- Beckerman JL, Ebbole DJ. MPG1, a gene encoding a fungal hydrophobin of Magnaporthe grisea, is involved in surface recognition. Mol Plant Microbe Interact. 1996;9:450–456. doi: 10.1094/mpmi-9-0450. [DOI] [PubMed] [Google Scholar]
- Beers EP, Duke SH, Henson CA. Partial characterization and subcellular localization of three α-glucosidase isoforms in pea (Pisum sativumL.) seedlings. Plant Physiol. 1990;94:738–744. doi: 10.1104/pp.94.2.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantret I, Lacasa M, Chevalier G, Ruf J, Islam I, Mantei N, Edwards Y, Swallow D, Rousset M. Sequence of the complete cDNA and the 5′ structure of the human sucrase-isomaltase gene: possible homology with a yeast glucoamylase. Biochem J. 1992;285:915–923. doi: 10.1042/bj2850915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba S, Inomata S, Matsui H, Shimomura T. Purification and properties of an α-glucosidase (glucoamylase) in sugar beet seed. Agric Biol Chem. 1978;42:241–245. [Google Scholar]
- Crosse JE. Bacterial canker of stone fruits. II. Leaf-scar infection of cherry. J Hortic Sci. 1956;31:212–224. [Google Scholar]
- Crowdy SH. Observations on apple canker. IV. The infection of leaf scars. Ann Appl Biol. 1952;39:569–580. [Google Scholar]
- Dohman RJ, Strasser AWM, Dahlems UM, Hollenberg CP. Cloning of the Schwaniomyces occidentalis glucoamylase gene (GAM1) and its expression in Saccharomyces cerevisiae. Gene. 1990;95:111–121. doi: 10.1016/0378-1119(90)90421-m. [DOI] [PubMed] [Google Scholar]
- Frandsed TP, Svensson B. Plant α-glucosidases of the glucoside hydrolase family 31: molecular properties, substrate specificity, reaction mechanism, and comparison with family members of different origin. Plant Mol Biol. 1998;37:1–13. doi: 10.1023/a:1005925819741. [DOI] [PubMed] [Google Scholar]
- Fry SC. Polysaccharide-modifying enzymes in the plant cell wall. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:497–520. [Google Scholar]
- Gerhardt P, Murray RGE, Wood WA, Krieg NR, eds (1994) Methods for General and Molecular Biology. American Society for Microbiology, Washington, DC
- Hebert DN, Foellmer B, Helenius A. Glucose trimming and reglucosylation determine glycoprotein association with calnexin in the endoplasmic reticulum. Cell. 1995;81:425–433. doi: 10.1016/0092-8674(95)90395-x. [DOI] [PubMed] [Google Scholar]
- Hoefsloot LH, Hoogeveen-Westerveld M, Kroos MA, van Beeumen J, Reuser AJJ, Oostra BA. Primary structure and processing of lysosomal α-glucosidase: homology with the intestinal sucrase-isomaltase complex. EMBO J. 1988;7:1697–1704. doi: 10.1002/j.1460-2075.1988.tb02998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenstein L, Balsiger S, Hohl R. The influence of IgG, proteases and glycosidases on adhesion to, and infection of soybean plants by Phytophthora megasperma f.sp. glycinae. Bot Helv. 1995;105:221–232. [Google Scholar]
- Howard RJ, Valent B. Annu Rev Microbiol. 1996;50:491–512. doi: 10.1146/annurev.micro.50.1.491. [DOI] [PubMed] [Google Scholar]
- Kern G, Schulke N, Schmid FX, Jaenicke R. Stability, quaternary structure, and folding of internal, external, and core-glycosylated invertase from yeast. Protein Sci. 1992;1:120–131. doi: 10.1002/pro.5560010112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klis FM. α-Glucosidase activity located at the cell surface in callus of Convolvulus arvensis. Physiol Plant. 1971;25:253–257. [Google Scholar]
- Konishi Y, Kitazato S, Asano R, Nakatani N. Polymorphism of acid and neutral α-glucosidase in banana pulp: changes in apparent pIs and affinity to con A of the enzymes during ripening. Agric Biol Chem. 1991;55:1089–1094. [Google Scholar]
- Matsui H, Iwanami S, Ito H, Mori H, Honma M, Chiba S. Cloning and sequencing of a cDNA encoding α-glucosidase from sugar beet. Biosci Biotechnol Biochem. 1997;61:875–880. doi: 10.1271/bbb.61.875. [DOI] [PubMed] [Google Scholar]
- Monroe JD, Hall BD, Gough CM, Stephen AL. Nucleotide sequence of an α-glucosidase gene (accession no. AF014806) from Arabidopsis thaliana (PGR 97-141) Plant Physiol. 1997;115:863. [Google Scholar]
- Nakamura A, Nishimura I, Yokoyama A, Lee DG, Hidaka M, Masaki H, Kimura A, Chiba S, Uozumi T. Cloning and sequencing of an α-glucosidase gene from Aspergillus niger and its expression in A. nidulans. J Biotechnol. 1997a;53:75–84. doi: 10.1016/s0168-1656(97)01664-7. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Sato S, Kaneko T, Kotani H, Asamizu E, Miyajima N, Tabata S. Structural analysis of Arabidopsis thalianachromosome 5. III. Sequence features of the regions of 1,191,918 bp covered by seventeen physically assigned P1 clones. DNA Res. 1997b;4:401–414. doi: 10.1093/dnares/4.6.401. [DOI] [PubMed] [Google Scholar]
- Nichols BL, Eldering J, Avery S, Hahn D, Quaroni A, Sterchi E. Human small intestinal maltase-glucoamylase cDNA cloning: homology to sucrase-isomaltase. J Biol Chem. 1998;273:3076–3081. doi: 10.1074/jbc.273.5.3076. [DOI] [PubMed] [Google Scholar]
- O K, Hill JS, Wang X, McCleod R, Pritchard PH. Lecithin:cholesterol acyltransferase: role of N-linked glycosylation in enzyme function. Biochem J. 1993;294:879–884. doi: 10.1042/bj2940879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parr DR, Edelman J. Release of hydrolytic enzymes from the cell walls of intact and disrupted carrot callus tissue. Planta. 1975;127:111–119. doi: 10.1007/BF00388372. [DOI] [PubMed] [Google Scholar]
- Somerville CR, Ogren WL (1982) Isolation of photorespiration mutants of Arabidopsis thaliana. In M Edelman, RB Hallick, NH Chua, eds, Methods in Chloroplast Biology. Elsevier Biomedical Press, Amsterdam, The Netherlands, pp 129–138
- Strum A. Heterogeneity of the complex N-linked oligosaccharides at specific glycosylation sites of two secreted carrot glycoproteins. Eur J Biochem. 1991;199:169–179. doi: 10.1111/j.1432-1033.1991.tb16106.x. [DOI] [PubMed] [Google Scholar]
- Sugimoto M, Furui S, Suzuki Y. Multiple molecular forms of α-glucosidase from spinach seeds, Spinacia oleraceaL. Biosci Biotech Biochem. 1995;59:673–677. [Google Scholar]
- Sugimoto M, Furui S, Suzuki Y. Molecular cloning and characterization of a cDNA encoding alpha-glucosidase from spinach. Plant Mol Biol. 1997;33:765–768. doi: 10.1023/a:1005766003923. [DOI] [PubMed] [Google Scholar]
- Sugimoto M, Suzuki Y. Molecular cloning, sequencing, and expression of a cDNA encoding α-glucosidases from Mucor javanicus. J Biochem. 1996;119:500–505. doi: 10.1093/oxfordjournals.jbchem.a021269. [DOI] [PubMed] [Google Scholar]
- Sun Z, Duke SH, Henson CA. The role of pea chloroplast α-glucosidase in transitory starch degradation. Plant Physiol. 1995;108:211–217. doi: 10.1104/pp.108.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Henson C. Degradation of native starch granules by barley α-glucosidases. Plant Physiol. 1990;94:320–327. doi: 10.1104/pp.94.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MA, George LA, Ross HA, Davies HV. cDNA cloning and characterization of an α-glucosidase gene from potato (Solanum tuberosumL.) Plant J. 1998;13:419–424. doi: 10.1046/j.1365-313x.1998.00051.x. [DOI] [PubMed] [Google Scholar]
- Tibbot BK, Henson CA, Skadsen RW. Expression of enzymatically active, recombinant barley α-glucosidase in yeast and immunological detection of α-glucosidase from seed tissue. Plant Mol Biol. 1998;38:379–391. doi: 10.1023/a:1006006203372. [DOI] [PubMed] [Google Scholar]
- Tibbot BK, Skadsen RW. Molecular cloning and characterization of a gibberellin-inducible α-glucosidase gene from barley. Plant Mol Biol. 1996;30:229–241. doi: 10.1007/BF00020110. [DOI] [PubMed] [Google Scholar]
- Trombetta ES, Simons JF, Helenius A. Endoplasmic reticulum glucosidase II is composed of a catalytic subunit, conserved from yeast to mammals, and a tightly bound noncatalytic HDEL-containing subunit. J Biol Chem. 1996;271:27509–27516. doi: 10.1074/jbc.271.44.27509. [DOI] [PubMed] [Google Scholar]
- Xiao J, Ohshima A, Kamakura T, Ishiyama T, Yamaguchi I. Extracellular glycoprotein(s) associated with cellular differentiation in Magnaporthe grisea. Mol Plant Microbe Interact. 1994;7:639–644. [Google Scholar]
- Yamasaki Y, Konno H. Two forms of α-glucosidase from soybean callus. Agric Biol Chem. 1985;49:849–850. [Google Scholar]
- Yamasaki Y, Konno H. Wall-bound α-glucosidase of suspension-cultured rice cells. Phytochemistry. 1987;26:711–713. [Google Scholar]
- Yamasaki Y, Konno H. α-Glucosidases of suspension-cultured sugar-beet cells. Phytochemistry. 1989;28:2583–2585. [Google Scholar]
- Yamasaki Y, Konno H. Wall-bound α-glucosidase of suspension-cultured sugar-beet cells. Phytochemistry. 1992;31:2605–2607. [Google Scholar]
- Yamasaki Y, Suzuki Y. Two forms of α-glucosidase from sugar beet seeds. Planta. 1980;148:354–361. doi: 10.1007/BF00388123. [DOI] [PubMed] [Google Scholar]