Crystal structures of the human mitochondrial helicase hSuv3 in complex with AMPPNP and with a short strand of RNA are presented.
Keywords: mitochondrial helicases, human Suv3, SF2 helicases
Abstract
Suv3 is a helicase that is involved in efficient turnover and surveillance of RNA in eukaryotes. In vitro studies show that human Suv3 (hSuv3) in complex with human polynucleotide phosphorylase has RNA degradosome activity. The enzyme is mainly localized in mitochondria, but small fractions are found in cell nuclei. Here, two X-ray crystallographic structures of human Suv3 in complex with AMPPNP, a nonhydrolysable analog of ATP, and with a short five-nucleotide strand of RNA are presented at resolutions of 2.08 and 2.9 Å, respectively. The structure of the enzyme is very similar in the two complexes and consists of four domains. Two RecA-like domains form the tandem typical of all helicases from the SF2 superfamily which together with the C-terminal all-helical domain makes a ring structure through which the nucleotide strand threads. The mostly helical N-terminal domain is positioned externally with respect to the core of the enzyme. Most of the typical helicase motifs are present in hSuv3, but the protein shows certain unique characteristics, suggesting that Suv3 enzymes may constitute a separate subfamily of helicases.
1. Introduction
The human nuclear SUV3 gene (SUPV3L1) encodes an NTP-dependent RNA/DNA helicase (Suv3p, hSuv3p) which is related to the DexH/D (Ski2p) superfamily. The gene has been conserved during evolution and is present in purple bacteria and all eukaryotes (Dmochowska et al., 1999 ▶). In the yeast Saccharomyces cerevisiae the Suv3 protein is localized in mitochondria and forms a degradosome complex with the 3′–5′ exoribonuclease Dss1 (Stepien et al., 1992 ▶; Dziembowski et al., 2003 ▶; Malecki et al., 2007 ▶). Both proteins are required for the degradation and surveillance of mitochondrial RNAs.
In humans, the hSuv3 (hSuv3p) protein is predominantly localized in the mitochondrial matrix (Minczuk et al., 2002 ▶) and is necessary for the degradation of mature mtRNAs, of processing intermediates transcribed from H- and L-strands and of aberrant RNA molecules (Szczesny et al., 2010 ▶). The molecular partner of hSuv3 in human mitochondria is likely to be polynucleotide phosphorylase (PNPase), as the two proteins have been found to interact in vitro (Wang et al., 2009 ▶) and to copurify from human cells (Szczesny et al., 2010 ▶).
The human hSuv3 protein is not confined to mitochondria. It is also localized in the cell nucleus and has been found to have several interacting partners: HBXIP (Minczuk et al., 2005 ▶), BLM helicase and WRN helicase (Pereira et al., 2007 ▶). Perturbations in mammalian SUV3 gene expression have been found to lead to apoptosis, elevated levels of sister-chromatid exchange and impaired ATP production, finally resulting in cellular senescence and death (Szczesny et al., 2007 ▶; Pereira et al., 2007 ▶). Interestingly, conditional knockout of the mouse SUV3 gene results in premature-aging phenotypes, including loss of fat and muscle, kyphosis, cachexia and scaling of feet and tail (Paul et al., 2009 ▶).
In summary, the hSuv3 protein is one of the players in regulating mitochondrial RNA metabolism, chromatin maintenance and cell death.
Despite the accumulation of substantial amounts of biological data, the structure of hSuv3 has not been described to date. Any bioinformatics approach to modelling its structure has been doomed to failure since the sequence similarity of hSuv3 to any known protein structure is below 25%. In order to facilitate further studies of this important enzyme, we have initiated crystallographic studies. Here, we present two structures of human Suv3: one in complex with the nonhydrolysable analog of ATP AMPPNP (2.08 Å resolution; PDB entry 3rc3) and a second structure harboring a short piece of bound RNA oligonucleotide (PDB entry 3rc8, 2.9 Å resolution). A comparison of these structures with those of other helicases gives detailed information about the unique characteristics of this enzyme, its ATP-binding pocket and the nucleotide-binding site.
2. Materials and methods
2.1. Cloning, expression and purification of hSUV3
All of the preparation steps were performed similarly to the procedure described previously (Szczesny et al., 2007 ▶), with some modifications for crystallization trials. The cDNA fragment coding residues 47–722 of hSuv3 [hSuv3(47–722)] was amplified according to the procedure described by Tropea et al. (2007 ▶). The amplicon was inserted into entry vector pDON, generating pDON-hSuv3(47–722), followed by recombination into pDEST-His-MBP, producing pMBP-hSuv3(47–722). BL21 (DE3) CodonPlus-RIL cells (Stratagene) containing pMBP-hSuv3(47–722) were grown in M9 medium under conditions suppressing the production of methionine (Van Duyne et al., 1993 ▶). At an OD600 of about 1.0, the bacterial culture was cooled to 291 K. At this point, 0.4 ml 1 M IPTG and 75 mg selenomethionine were added per litre of culture. The culture was centrifuged after an additional 18 h of shaking at 291 K and 180 rev min−1. The cell pellet from 1 l bacterial culture was resuspended in 35 ml buffer A [50 mM Tris–HCl pH 8.0, 50 mM K2HPO4, 0.3 M KCl, 10%(v/v) glycerol, 25 mM imidazole, 5 mM β-mercaptoethanol]. After the addition of benzamidine–HCl to a final concentration of 3 mM, the cells were stored at 193 K. All further purification steps were performed at 277 K. Frozen bacteria were thawed and lysed by sonication. The crude extract was centrifuged at 40 000g for 40 min and the supernatant was applied onto a 20 ml Ni–NTA (Ni2+–nitrilotriacetic acid, Qiagen) column equilibrated with buffer A. The column was washed with ten column volumes of buffer A adjusted to 2 M KCl followed by ten column volumes of buffer A. Bound proteins were eluted with buffer A supplemented with 200 mM imidazole. The eluate was adjusted to 1 mM dithiothreitol (DTT) and 1 mM EDTA. TEV (tobacco etch virus) protease (Tropea et al., 2007 ▶) was added to a ratio of 1:100 TEV protease:total protein. After overnight digestion, the protein solution was precipitated with ammonium sulfate (0.39 g ml−1) and centrifuged at 10 000g for 20 min. The pellet was resuspended in buffer A lacking imidazole and applied onto a 25 ml Ni–NTA column. hSuv(47–722) was eluted with buffer A, which was followed by a second precipitation with ammonium sulfate (0.39 g ml−1) and centrifugation. The pellet was resuspended in 2 ml buffer B (0.25 M NaCl, 20 mM HEPES pH 7.5, 10% glycerol, 10 mM DTT, 0.02% NaN3) and applied onto a Superdex 200 size-exclusion chromatography column equilibrated with the same buffer. Fractions containing hSuv(47–722) were collected, concentrated to 10 mg ml−1 and set up for crystallization. The selenomethionine-enriched protein was used to set up the AMPPNP cocrystallization trials. Once the SeMet structure had been solved, native hSuv(47–722) was expressed in Luria–Bertani broth, purified as described above and cocrystallized with Escherichia coli crude RNA.
2.2. Crude RNA-extract preparation
Overnight-grown E. coli (DH5α strain) cells were treated according to Qiagen plasmid purification protocols, omitting RNase A from the P1 buffer. The cells were resuspended in P1 buffer, lysed with P2 buffer and the solution was neutralized with N3 buffer. After removal of precipitant, the supernatant was mixed with 95% ethanol in a 1:2.5 ratio. The resulting precipitant was spun down, washed with 70% ethanol, air-dried and resuspended in buffer B.
2.3. Crystallization
Two crystallization trials were performed. Initially, 1 mM AMPPNP was added to the purified protein solution and hanging drops were set up after mixing the protein solution in a 1:1 ratio with a well solution consisting of 6% PEG 3350, 0.1 M sodium citrate pH 6.5 and 10% glycerol at 277 K. Crystals of hSuv3 grew after about one week.
In the second crystallization trial hSuv3 protein was mixed with crude RNA extract from E. coli. The protein and crude RNA were incubated at 277 K overnight, which was followed by addition of RNaseA at a 1:100 RNaseA:hSuv3 ratio. The mixture was incubated for 4 h at 277 K with the purpose of ‘shaving off’ any RNA fragments sticking out of the protein. Finally, the solution of hSuv3 was concentrated to 10 mg ml−1 protein. The crystallization conditions were the same as for the AMPPNP complex.
2.4. Structure solution and refinement
A crystal of approximately 0.12 × 0.15 × 0.3 mm in size was dipped for about 5 s into a solution prepared from the well solution supplemented with 20% glycerol and transferred in a rayon loop to a stream of cold nitrogen gas at the goniostat of the SER-CAT beamline 22ID of the Advanced Photon Source, Argonne National Laboratory equipped with a MAR300 CCD detector. One set of diffraction data was collected at a wavelength of 0.9754 Å, corresponding to the high-energy remote region near the X-ray absorption edge of selenium. The structure was solved using the program HKL-3000 (Minor et al., 2006 ▶). The initial SAD phases were introduced into the ARP/wARP procedure (Perrakis et al., 1997 ▶), which built 584 residues (of the expected 675). The hSuv3 model was finalized by refinement with REFMAC (Murshudov et al., 2011 ▶) interspersed with Coot graphics sessions (Emsley & Cowtan, 2004 ▶). The final model, with an R factor of 15.96% and an R free of 20.32%, contained 607 residues of the protein, one molecule of AMPPNP and 513 water sites. The remaining expected parts of the protein chain were disordered and were not identifiable in the electron-density maps. The statistics of the diffraction data and model refinement are summarized in Table 1 ▶.
Table 1. Statistics of the diffraction data and structure refinement.
Values in parentheses are for the highest resolution shell.
| hSuv3AMPPNP | hSuv3RNA | |
|---|---|---|
| Data collection | ||
| Beamline | 22ID, SER-CAT | 22ID, SER-CAT |
| Temperature (K) | 100 | 100 |
| Wavelength () | 0.9754 | 1.0000 |
| Space group | P32 | P32 |
| Unit-cell parameters | ||
| a = b () | 89.39 | 94.43 |
| c () | 88.07 | 88.03 |
| Resolution limits () | 402.08 (2.152.08) | 302.9 (3.02.9) |
| Reflections measured | 179028 (13871) | 107954 (11070) |
| Unique reflections | 47146 (4565) | 19390 (1957) |
| Multiplicity | 3.8 (3.2) | 5.6 (5.5) |
| Completeness, native (%) | 99.6 (96.5) | 100 (100) |
| Completeness, anomalous (%) | 98.6 (91.1) | |
| R merge (%) | 5.7 (32.6) | 7.5 (40.7) |
| Average I/(I) | 26.9 (3.4) | 22.6 (4.5) |
| Wilson B factor (2) | 33.9 | 75.2 |
| Refinement | ||
| Resolution () | 402.08 | 302.9 |
| R factor (%) | 15.96 | 17.72 |
| Working reflections | 46139 | 18398 |
| R free (%) | 20.32 | 23.78 |
| Free reflections | 965 | 991 |
| Protein atoms | 4854 | 4869 |
| RNA atoms | 107 | |
| AMPPNP atoms | 31 | |
| Ion sites (Na+, Cl, azide) | 5 | |
| Water sites | 522 | |
| R.m.s.d. bonds () | 0.026 | 0.016 |
| R.m.s.d. angles () | 1.98 | 1.66 |
| Ramachandran statistics | ||
| Most favored | 517 [95.7%] | 475 [87.5%] |
| Allowed | 22 [4.1%] | 63 [11.6%] |
| Generously allowed | 0 | 3 [0.6%] |
| Disallowed | 1 [0.2%] | 2 [0.4%] |
| PDB code | 3rc3 | 3rc8 |
The crystal of the RNA complex of hSuv3 was treated according to the same cryoprocedure and diffraction data were also collected on SER-CAT beamline 22ID but using a wavelength of 1.0 Å, since in this case the protein did not contain selenomethionine but rather native methionine. The previously refined molecule of hSuv3 (stripped of solvent and ligand molecules) was positioned in the cell with MOLREP (Vagin & Teplyakov, 2010 ▶) and refined with REFMAC. The short fragment of RNA was identified in the electron density and included in refinement. It was decided that the resolution of 2.9 Å did not warrant the inclusion of water solvent. The final R factor was 17.72% and R free was 23.78%. The remaining information about the data and refinement of hSuv3–RNA is included in Table 1 ▶.
Several other programs from the CCP4 suite (Winn et al., 2011 ▶) were used for various purposes during structure refinement and comparison with other models of helicases. The illustrations were produced using PyMOL (DeLano, 2002 ▶; Figs. 1a, 2a, 3a, 5 and 6) and CHEMDRAW (CambridgeSoft; Figs. 2b and 3b).
3. Results and discussion
3.1. hSuv3 model
The structures of hSuv3 in complex with AMPPNP and with RNA belong to the same space group P32, but with a and b unit-cell parameters that differ by about 5 Å. The structure of the protein is shown in Fig. 1 ▶(a) and is similar in both complexes. The aspects relevant to the interaction of hSuv3 with AMPPNP or RNA will therefore be discussed based on the corresponding structures containing either AMPPNP (PDB entry 3rc3) or RNA (PDB entry 3rc8), which are referred to as hSuv3–AMPPNP or hSuv3–RNA, respectively. The AMPPNP and RNA are presented in the corresponding electron density in Figs. 2 ▶(a) and 3 ▶(a), respectively.
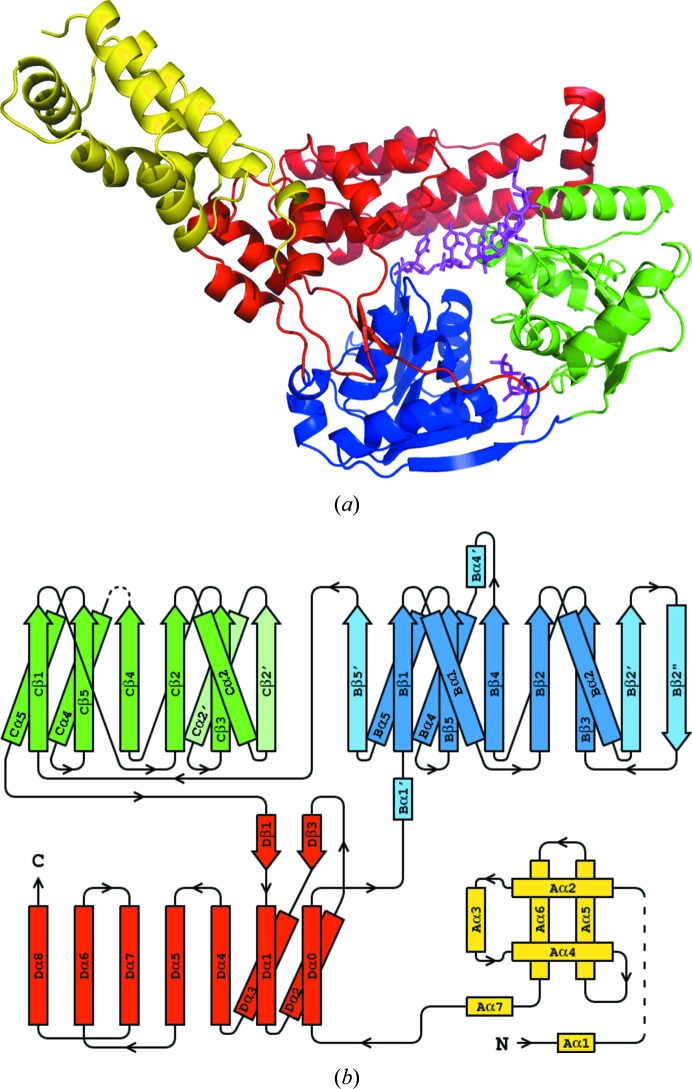
Figure 1.
(a) The structure of hSuv3 with its four domains colored as follows: the N-terminal domain A is shown in yellow, the two RecA domains B and C are shown in blue and green, respectively, and the C-terminal domain D is shown in red. Both AMPPNP and RNA are positioned as in their individual complexes and shown in magenta. (b) Topology of hSuv3. The secondary-structure elements of domains B and C which are not present in the canonical topology of RecA are represented in pale colors.
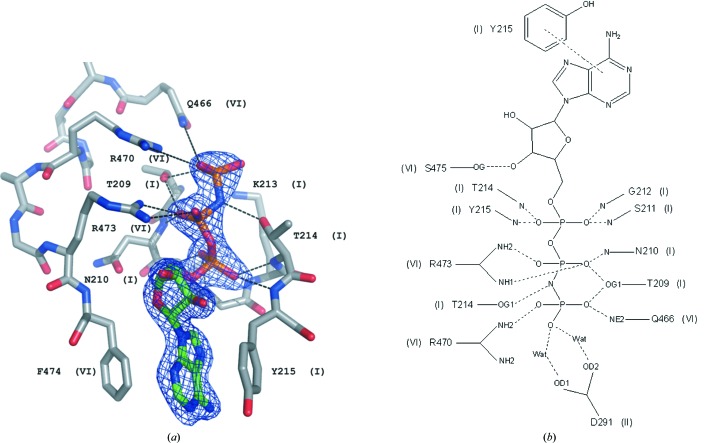
Figure 2.
(a) The molecule of AMPPNP in hSuv3–AMPPNP with the corresponding F obs electron-density map (at a 1.5σ contour level) and its surrounding residues. (b) Schematic representation of the hydrogen-bond interactions between AMPPNP and the surrounding residues of hSuv3. The side chain of Tyr215 is involved in π–π interactions with the adenine ring of AMPPNP. Roman numerals refer to the helicase motifs of particular residues.
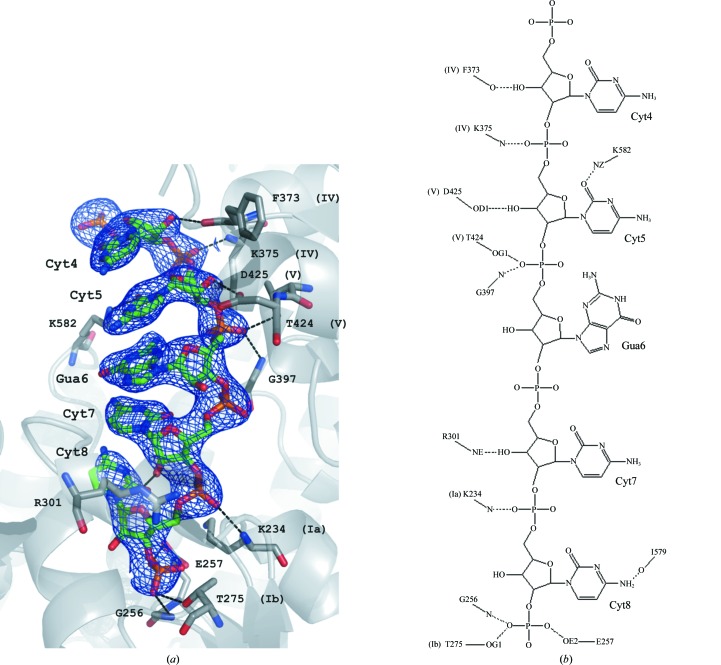
Figure 3.
(a) The identified fragment of nucleic acid in the RNA–hSuv3 structure in F obs electron density (at a 1.4σ contour level). The surrounding protein is shown in cartoon representation, with residues that interact with RNA shown in ball-and-stick representation. (b) The scheme of hydrogen bonds identified between the RNA oligonucleotide and hSuv3. Roman numerals refer to the helicase motifs of particular residues.
The construct of hSuv3 used for crystallization contained residues 47–722 of the complete genome sequence plus an additional Gly at the N-terminus. In both crystal structures fragments of the N- and C-termini (residues 46–57 and 690–722), as well as two other fragments of the chain (73–86 and 446–456), are disordered and are not visible in the electron-density map. These fragments are therefore absent from the atomic models of both structures..
The overall structure of hSuv3 can be divided into four domains, as shown in Figs. 1 ▶(a) and 1 ▶(b). Domains B, C and D form a ring structure with a tunnel at its centre. The N-terminal domain A is loosely connected to the rest of the molecule.
The helical N-terminal domain A (residues 59–170) can be approximately described as a left-handed three-helix bundle ornamented with additional helices and loops, according to the DALI server (Holm & Rosenström, 2010 ▶). No significantly similar domains were identified in the PDB.
The fold of domains B (residues 187–348) and C (residues 349–500) is typical of the SF2 family of helicases represented by the DEAD-box helicase RecA, with some variations. The B domain has eight β-strands, all parallel except for the last (Bβ2′′), and four accompanying helices (two pairs on both sides of the β-sheet) with two additional short helices. The C domain has six parallel β-strands and four helices (three on one side of the β-sheet and one on the other). In comparison with the canonical RecA fold, domain B has three additional strands and two short helices, while domain C has one strand and one additional helix. Both of them lack one helix between the third and fourth strands. The domains form a compact dimer, which is typical of many helicases.
The C-terminal domain D (residues 501–689) consists of nine helices and only one β-sheet, which consists of two short strands. The last three helices form a three-helix bundle and the preceding three are also arranged analogously to a short three-helix bundle. The DALI server identifies some three-helix bundles with low structural similarity to this domain.
In a number of multi-domain helicases, the domains additional to the two ‘standard’ α/β RecA domains constitute insertions between their strands (Caruthers & McKay, 2002 ▶). In hSuv3 the four domains are consecutive in the sequence, except for one of the helices (Dα0) linking domains A and B, which topologically belongs to the last domain D, as shown in Fig. 1 ▶(b).
The short strand of RNA, consisting of five nucleosides, identified in the hSuv3–RNA complex is located within the tunnel formed between domains B, C and D. Its localization with respect to the helicase domains B and C is analogous to the position of the short nucleic acid strands in other SF2 helicase–nucleic acid complexes (PDB entries 3pew, 3g0h, 2hyi, 2db3, 3i62, 2jlv, 2nmv, 2xgj and 3o8r; Montpetit et al., 2011 ▶; Collins et al., 2009 ▶; Andersen et al., 2006 ▶; Sengoku et al., 2006 ▶; Del Campo & Lambowitz, 2009 ▶; Luo et al., 2008 ▶; Waters et al., 2006 ▶; Weir et al., 2010 ▶; Appleby et al., 2011 ▶).
3.2. Helicase motifs in hSuv3
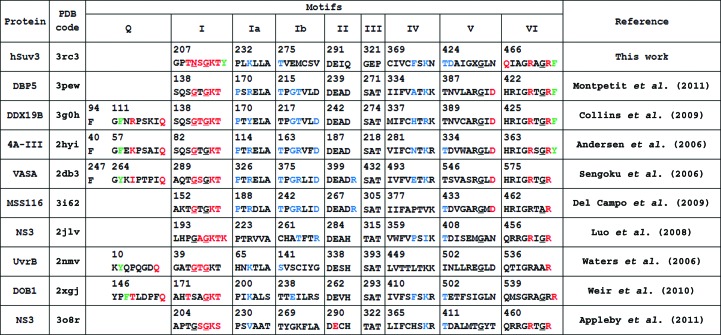
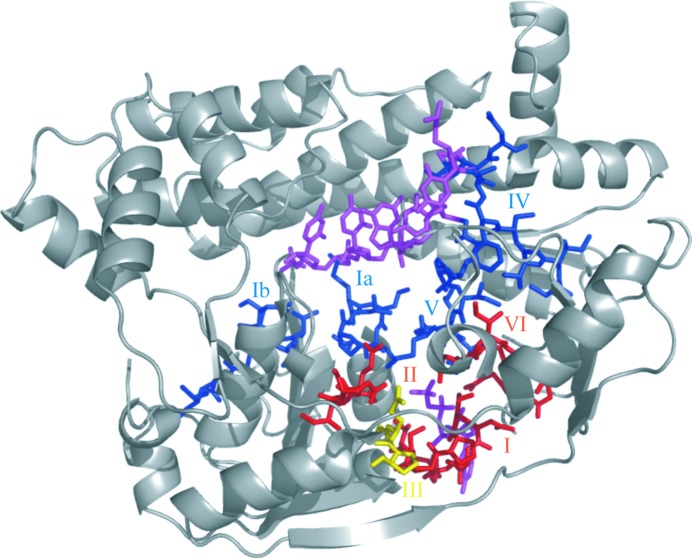
Helicases from the SF1/SF2 superfamilies possess several conserved sequence motifs, which were first identified by Gorbalenya & Koonin (1993 ▶). Fig. 4 ▶ shows the sequences of these motifs in hSuv3 compared with some other typical helicases and Fig. 5 ▶ presents their localization in the structure of hSuv3. The highest sequence similarity of hSuv3 to any known structure from the PDB is about 25% and, as shown in Fig. 4 ▶, similarity within the characteristic motifs does not seem to be higher than this.
Figure 4.
Structurally aligned sequences of typical helicase motifs in hSuv3 and selected SF2 family helicases from the PDB containing both ATP analogs and fragments of nucleic acids. Residues hydrogen bonded to ATP are shown in red, those hydrogen bonded to nucleic acid are shown in blue and those stacked with adenine are shown in green. Residues in a left-handed α-helical conformation are underlined.
Figure 5.
Localization of typical helicase motifs in hSuv3. The N-terminal domain A is not shown. The ATP-binding motifs (I, II and VI) are presented in red, those binding RNA (Ia, Ib, IV and V) are shown in blue and motif III, with its unique sequence GEP, is shown in yellow. This figure visualizes both AMPPNP and RNA positioned as in their individual complexes. AMPPNP and RNA are shown in magenta.
According to the classification of Cordin et al. (2006 ▶), motifs Q, I, Ia, Ib, II and III reside in the first RecA domain B and motifs IV, V and VI reside in the second RecA domain C. Motifs Q, I and VI take part in binding ATP, motifs II and III take part in the hydrolysis of ATP and motifs Ia, Ib, IV and V take part in binding of the nucleic acid (Caruthers & McKay, 2002 ▶). A more recent classification, taking several additional motifs into account, has been proposed by Jankowsky and coworkers (Fairman-Williams et al., 2010 ▶; Jankowsky, 2011 ▶), but the sequence conservation of the new motifs is much lower and it is not possible to assign them unequivocally to hSuv3.
The characteristics of these motifs in hSuv3 in comparison with some other structures of helicases present in the PDB (Berman et al., 2000 ▶) and quoted by their PDB codes are discussed below.
3.3. Binding of AMPPNP: motifs Q, I, II, III and VI
Several residues around the ATP-binding pocket, mostly from motifs I, II, III and VI, form hydrogen bonds to the ligand in the structure of the AMPPNP–hSuv3 complex, as presented in Figs. 2 ▶(a) and 2 ▶(b). Inspection of the sequence and structure of hSuv3 leads to the conclusion that hSuv3 does not possess motif Q. Motif Q appears in helicases possessing the so-called ‘cap’ attached to the first RecA domain (Tanner et al., 2003 ▶) and contains residues engaged in binding ATP. When this motif exists, e.g. in 3g0h, 2hyi and 2db3 (Fig. 4 ▶), it contains an aromatic Tyr or Phe with its ring positioned parallel and interacting with the adenine aromatic ring system. The third residue after Tyr/Phe (of various types) provides its carbonyl O atom for the formation of a hydrogen bond to the adenine N6 atom. Most characteristically, the eighth residue after Tyr/Phe is Gln and forms two hydrogen bonds between its OE1 and NE2 atoms and the adenine N6 and N7 atoms, respectively.
The structure of hSuv3 does not have the insertion in the form of a ‘cap’ characteristic of the above-mentioned helicases and belongs to the group of helicases that lack motif Q altogether. In all helicase structures that contain ATP (or an analog, e.g. AMPPNP) there is always at least one aromatic residue (sometimes two) involved in π–π interactions with the adenine ring. In hSuv3 this role is taken by Tyr215, located in the sequence at the end of motif I. On the other side of the adenine moiety resides Phe474, which is located immediately after motif VI in the sequence. The aromatic ring of Phe474 is not completely parallel to the adenine, so that the π-sandwich is not ideal. The N atoms of adenine do not form hydrogen bonds to any protein atoms, but to solvent water molecules. Similarly to other helicases, if the adenine N atoms are not hydrogen bonded to protein they are surrounded by solvent. In some helicases the Tyr residue stacked with adenine is located at the end of the last β-strand of the first RecA domain. The location of an ATP-stacking Tyr or Phe at the end of motif I, as in hSuv3, is not encountered in any other helicase containing adenosine present in the PDB.
Motif I, which is also called the Walker A motif, is present in all helicases and contains residues that bind ATP. The sequence of this motif in hSuv3 is GPTNSGKTY. The last seven residues of this fragment in hSuv3 all directly bind to AMPPNP either through main-chain hydrogen bonds (N atoms of Asn210, Ser211 and Gly212 with O atoms of phosphates), side-chain hydrogen bonds (Thr209, Thr214 and, through a water molecule, Lys213) or by π-stacking with the adenine ring (Tyr215). The canonical fingerprint of SF2 helicases is GxGKS/T (Gorbalenya et al., 1989 ▶) but, in contrast to the majority of other helicases, the first Gly in this fragment is changed to Asn in hSuv3.
Both glycines in the classic motif I lie in the left-handed helical region of the Ramachandran plot in the majority of known structures of helicases. This is not unusual, since Gly residues have a similar propensity for right-handedness and left-handedness of their main chain (Nicholson et al., 1989 ▶). Non-Gly residues only rarely occur in this region of the plot owing to steric hindrance between the side chains of consecutive l-amino acids. However, the ϕ, ψ angles of Asn210 in hSuv3 locate this residue in the left-handed region of the Ramachandran plot; the conformation of the main-chain backbone is therefore the same in hSuv3 as in other helicases (Fig. 6 ▶).
Figure 6.
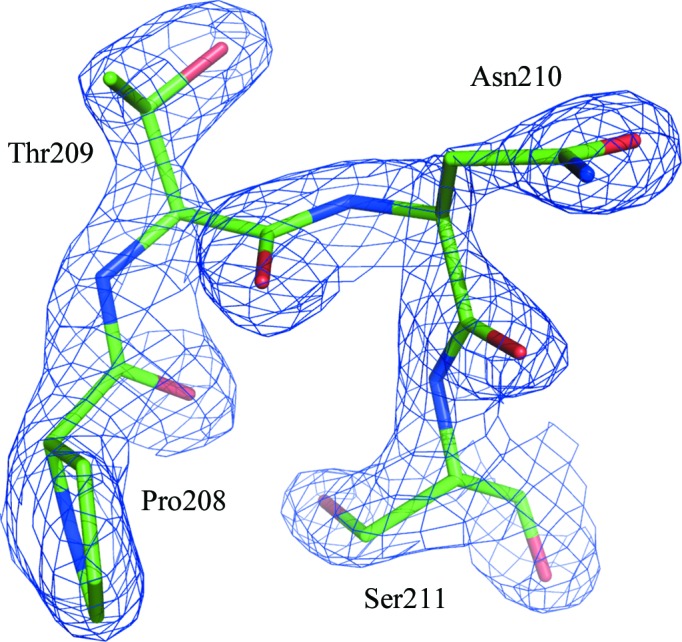
Residue Asn210 and its vicinity with the corresponding F obs electron-density map (at a 2.0σ contour level). This residue is located in the left-handed region of the Ramachandran plot.
As pointed out above, Tyr215, which forms a parallel π-stack with adenine, occurs immediately after motif I in the hSuv3 sequence. This is a unique characteristic of hSuv3, since in other helicases the aromatic residues stacking with adenine of ATP are positioned at different locations (motifs) in the protein sequence.
Motif VI is characterized by His or Gln at the beginning, two glycines at positions 4 and 7 and two arginines at positions 5 and 8; most helicases conform to this consensus. In hSuv3 this motif has the sequence QIAGRAGRF.
The side chains of Gln466, Arg470 and Arg473 form hydrogen bonds to the phosphate O atoms of AMPPNP. Immediately following the last Arg of this motif in hSuv3 is Phe474, which is located near the adenine ring of AMPPNP on the side opposite Tyr215. This is similar to some other helicase structures, in which the adenine is sandwiched between two aromatic residues (3pew, 3g0h and 2hyi).
Motif II, with its most general fingerprint, DExx, is present in all helicases. The most common amino acids in the fourth position of this motif are Asp or His, but in hSuv3 this position is occupied by Gln.
The proposed role of this motif is to indirectly (through water molecules) coordinate the Mg2+ ion of the Mg-ATP complex via the carboxyl group of Asp and to assist as a catalytic base in ATP hydrolysis through the carboxyl of Glu. In most of the available structures of helicases containing MgAMPPNP both Asp and Glu are hydrogen bonded to water molecules coordinated by the Mg2+ ion. In the hSuv3–AMPPNP structure there is no metal ion but both carboxyl OD atoms of Asp291 are connected to the terminal phosphate oxygen O2γ through two water molecules. The side chain of Glu292 is directed away and does not interact, even indirectly, with the AMPPNP. There is sufficient space available around this location to allow Glu292 to change its conformation (in concert with the nearby Met429) and to adopt a position analogous to that in structures containing Mg.
Motif III is proposed to link the processes of hydrolysis of ATP and unwinding of RNA. The characteristic sequence of this motif in SF2 helicases consists of three residues: SAT. In hSuv3 the sequence at the location of the typical SAT triplet of residues is occupied by a GEP triplet. In this respect, hSuv3 is unique among SF2 helicases and the details of the coupling of the ATPase and RNA-unwinding activities evidently differ from the majority of SF2 superfamily members. Helicases from the SF1 superfamily usually have a GDx triplet in this position, confirming that the SAT fragment is not absolutely necessary for helicase activity.
3.4. Binding of RNA: motifs Ia, Ib, IV and V
The nucleic acid-binding motifs are located on the opposite side of the double RecA dimer from the ATP-binding cleft and contain residues binding a single strand of nucleic acid. Since the RNA fragment bound to hSuv3 originated from a natural source, its sequence can only be deduced from electron density. Among the five interpretable locations of RNA bases in the map, the first and last two are best filled by cytosines and the middle one by guanine, as shown in Fig. 3 ▶(a), although their identities are not absolutely certain. Residues of hSuv3 bound to nucleic acid in the RNA complex through hydrogen bonds are shown in Figs. 3 ▶(a) and 3 ▶(b). RNA mostly interacts through its phosphate and sugar moieties; the five bases are mutually stacked but they form only two hydrogen bonds with the surrounding protein residues. Generally, the sequence conservation of these motifs is lower than in the ATP-binding motifs, but some characteristics are common to most of the SF2 superfamily members.
Among the crystal structures of SF2 helicases in the PDB, some contain fragments of single-stranded nucleic acids and are included in Fig. 4 ▶. At least five nucleotides are modeled in these structures, although additional nucleotides may be present but not visible in the electron-density map. In the hSuv3–RNA structure five nucleotides (numbered 4–8) were identified and are present in the refined model of 3rc8.
Motif Ia usually has a PxRxLA sequence; in hSuv3 it is PLKLLA. In the complexes, the carbonyl O atom of the first residue of this motif is typically hydrogen bonded to the O2′ hydroxyl group of the ribose of the third nucleotide and the peptide N atom of the third residue is hydrogen bonded to the phosphate O atom of the next nucleotide. In hSuv3 the N atom of Lys234 is hydrogen bonded to the Cyt8 phosphate O atom, but Pro232 is not bound to RNA. In some complexes containing Arg in the third position its guanidinium group makes an ion pair with the successive phosphate group of the nucleic acid, but in hSuv3–RNA the NZ atom of Lys234 is coupled to the carboxyl OE1 atom of Glu257.
Motif Ib has variants of the sequence TPGRL, with the Thr at the first position being most conserved. In hSuv3 this fragment is TVEMC and the OG1 atom of the threonine is hydrogen bonded to an O atom of the last phosphate in the five-nucleotide fragment of RNA, similarly to other helicase complexes. In some helicases the N atoms of the third and fourth residues of this motif form hydrogen bonds to the O2′ hydroxyl of the fourth residue and the phosphate O atom of the fifth residue of RNA. There are no such interactions in hSuv3–RNA.
Motif IV has the consensus sequence LIFxxTxR, with the Phe at the third position being most conserved; in hSuv3 this fragment has the sequence CIVCFSKN. Typically, two residues from this fragment form hydrogen bonds to the nucleic acid. The carbonyl O atom of the fifth residue binds to the hydroxyl of the first nucleotide and the peptide N atom of the seventh residue binds to the phosphate O atom of the second nucleotide in the RNA pentamer. hSuv3–RNA conforms to this scheme, with the N atom of Phe373 and the O atom of Lys375 engaged in hydrogen bonds to RNA. If, as in some helicases, the seventh residue in this motif is Arg, it may be engaged in an ion pair with the phosphate O atom of the third RNA residue. In hSuv3–RNA the side-chain NZ atom of Lys375 resides in the vicinity but is not close enough to interact effectively with the phosphate.
In motif V, which has the typical sequence TDVxARGID, the Thr at the first position and the Gly at the seventh position are most conserved. In hSuv3 the sequence of this motif is TDAIGMGLN. The OG1 atom of the threonine is always engaged in a hydrogen bond to the phosphate of the third nucleotide of the RNA. In hSuv3–RNA the OD1 carboxyl side-chain atom of Asp425 (at the second position of the motif) is hydrogen bonded to the O2′ hydroxyl of the second RNA nucleotide, but there is no such interaction in any other helicase complexes with RNA. In some helicases the last Asp of this motif extends towards the ATP and makes a hydrogen bond to the O3′ atom of its ribose, and in hSuv3 the OD1 atom of Asn432 is connected to the corresponding ribose hydroxyl through one water molecule. Characteristically, in all helicases the Gly residue has its backbone in the left-handed α-helical conformation.
4. Conclusions
The human mitochondrial helicase hSuv3 has many characteristics typical of helicases from the SF2 superfamily, but it also has a number of features that are unique. In keeping with other SF2 helicases, hSuv3 contains two RecA-like domains and several characteristic sequence/structure motifs which are engaged in binding ATP or nucleic acids or in linking the ATPase and nucleic acid-unwinding activities. SF2 helicases often possess additional domains, but the N- and C-terminal domains of hSuv3 do not show any significant similarity to such auxiliary domains of known helicase structures apart from being almost exclusively α-helical. The C-terminal domain of hSuv3, together with the two helicase domains, forms a tunnel through which the unwound single strand of nucleic acid is threaded. The N-terminal domain of hSuv3 is located externally with respect to the enzyme core and its functional role in hSuv3 is not clear.
The typical helicase motifs in hSuv3 also have some peculiarities that are unique to this enzyme. hSuv3 does not have motif Q; rather, the tyrosine stacked with the adenine of ATP is located immediately after motif I. The first two residues of motif II in hSuv3 (DE) are absolutely conserved in SF1/SF2 helicases, but the last two residues (IQ) are rarely encountered in these positions. The residues present at the location of motif III in hSuv3 (GEP) are different from those in the usual sequence (SAT) in SF2 helicases. Motifs Ia, Ib, IV, V and VI in hSuv3, although significantly different in sequence from those in other SF2 helicases, contain certain important residues that are conserved among the whole family. Characteristically, all residues in motifs Q, I, V and VI that have a left-handed backbone conformation (not necessarily glycines) are highly conserved in all SF1/SF2 helicases.
The human Suv3 enzyme shares low sequence similarity with other structurally characterized helicases, but its sequence is 75% similar to that of the yeast enzyme. Together with the described unique structural features, this suggests that Suv3 enzymes may constitute a separate subfamily of SF2 helicases.
Supplementary Material
PDB reference: hSuv3–AMPPNP complex, 3rc3
PDB reference: hSuv3–RNA complex, 3rc8
Acknowledgments
This work was supported in part by the Polish National Centre for Research and Development (NR 13004704) and the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research and with Federal funds from the National Cancer Institute, National Institutes of Health (Contract No. HHSN261200800001). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products or organizations imply endorsement by the US Government. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. W-31-109-Eng-38.
References
- Andersen, C. B., Ballut, L., Johansen, J. S., Chamieh, H., Nielsen, K. H., Oliveira, C. L., Pedersen, J. S., Séraphin, B., Le Hir, H. & Andersen, G. R. (2006). Science, 313, 1968–1972. [DOI] [PubMed]
- Appleby, T. C., Anderson, R., Fedorova, O., Pyle, A. M., Wang, R., Liu, X., Brendza, K. M. & Somoza, J. R. (2011). J. Mol. Biol. 405, 1139–1153. [DOI] [PMC free article] [PubMed]
- Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N. & Bourne, P. E. (2000). Nucleic Acids Res. 28, 235–242. [DOI] [PMC free article] [PubMed]
- Caruthers, J. M. & McKay, D. B. (2002). Curr. Opin. Struct. Biol. 12, 123–133. [DOI] [PubMed]
- Collins, R., Karlberg, T., Lehtiö, L., Schütz, P., van den Berg, S., Dahlgren, L. G., Hammarström, M., Weigelt, J. & Schüler, H. (2009). J. Biol. Chem. 284, 10296–10300. [DOI] [PMC free article] [PubMed]
- Cordin, O., Banroques, J., Tanner, N. K. & Linder, P. (2006). Gene, 367, 17–37. [DOI] [PubMed]
- DeLano, W. L. (2002). PyMOL http://www.pymol.org.
- Del Campo, M. & Lambowitz, A. M. (2009). Mol. Cell, 35, 598–609. [DOI] [PMC free article] [PubMed]
- Dmochowska, A., Kalita, K., Krawczyk, M., Golik, P., Mroczek, K., Lazowska, J., Stepień, P. P. & Bartnik, E. (1999). Acta Biochim. Pol. 46, 155–162. [PubMed]
- Dziembowski, A., Piwowarski, J., Hoser, R., Minczuk, M., Dmochowska, A., Siep, M., van der Spek, H., Grivell, L. & Stepien, P. P. (2003). J. Biol. Chem. 278, 1603–1611. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Fairman-Williams, M. E., Guenther, U. P. & Jankowsky, E. (2010). Curr. Opin. Struct. Biol. 20, 313–324. [DOI] [PMC free article] [PubMed]
- Gorbalenya, A. E. & Koonin, E. V. (1993). Curr. Opin. Struct. Biol. 3, 419–429.
- Gorbalenya, A. E., Koonin, E. V., Donchenko, A. P. & Blinov, V. M. (1989). Nucleic Acids Res. 17, 4713–4730. [DOI] [PMC free article] [PubMed]
- Holm, L. & Rosenström, P. (2010). Nucleic Acids Res. 38, W545–W549. [DOI] [PMC free article] [PubMed]
- Jankowsky, E. (2011). Trends Biochem. Sci. 36, 19–29. [DOI] [PMC free article] [PubMed]
- Luo, D., Xu, T., Watson, R. P., Scherer-Becker, D., Sampath, A., Jahnke, W., Yeong, S. S., Wang, C. H., Lim, S. P., Strongin, A., Vasudevan, S. G. & Lescar, J. (2008). EMBO J. 27, 3209–3219. [DOI] [PMC free article] [PubMed]
- Malecki, M., Jedrzejczak, R., Stepien, P. P. & Golik, P. (2007). J. Mol. Biol. 372, 23–36. [DOI] [PubMed]
- Minczuk, M., Dmochowska, A., Palczewska, M. & Stepien, P. P. (2002). Yeast, 19, 1285–1293. [DOI] [PubMed]
- Minczuk, M., Mroczek, S., Pawlak, S. D. & Stepien, P. P. (2005). FEBS J. 272, 5008–5019. [DOI] [PubMed]
- Minor, W., Cymborowski, M., Otwinowski, Z. & Chruszcz, M. (2006). Acta Cryst. D62, 859–866. [DOI] [PubMed]
- Montpetit, B., Thomsen, N. D., Helmke, K. J., Seeliger, M. A., Berger, J. M. & Weis, K. (2011). Nature (London), 472, 238–242. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Nicholson, H., Söderlind, E., Tronrud, D. E. & Matthews, B. W. (1989). J. Mol. Biol. 210, 181–193. [DOI] [PubMed]
- Paul, E., Cronan, R., Weston, P. J., Boekelheide, K., Sedivy, J. M., Lee, S.-Y., Wiest, D. L., Resnick, M. B. & Klysik, J. E. (2009). Mamm. Genome, 20, 92–108. [DOI] [PMC free article] [PubMed]
- Pereira, M., Mason, P., Szczesny, R. J., Maddukuri, L., Dziwura, S., Jedrzejczak, R., Paul, E., Wojcik, A., Dybczynska, L., Tudek, B., Bartnik, E., Klysik, J., Bohr, V. A. & Stepien, P. P. (2007). Mech. Ageing Dev. 128, 609–617. [DOI] [PubMed]
- Perrakis, A., Morris, R. & Lamzin, V. S. (1997). Nature Struct. Biol. 6, 458–463. [DOI] [PubMed]
- Sengoku, T., Nureki, O., Nakamura, A., Kobayashi, S. & Yokoyama, S. (2006). Cell, 125, 287–300. [DOI] [PubMed]
- Stepien, P. P., Margossian, S. P., Landsman, D. & Butow, R. A. (1992). Proc. Natl Acad. Sci. USA, 89, 6813–6817. [DOI] [PMC free article] [PubMed]
- Szczesny, R. J., Borowski, L. S., Brzezniak, L. K., Dmochowska, A., Gewartowski, K., Bartnik, E. & Stepien, P. P. (2010). Nucleic Acids Res. 38, 279–298. [DOI] [PMC free article] [PubMed]
- Szczesny, R. J., Obriot, H., Paczkowska, A., Jedrzejczak, R., Dmochowska, A., Bartnik, E., Formstecher, P., Polakowska, R. & Stepien, P. P. (2007). Biol. Cell. 99, 323–332. [DOI] [PubMed]
- Tanner, N. K., Cordin, O., Banroques, J., Doère, M. & Linder, P. (2003). Mol. Cell, 11, 127–138. [DOI] [PubMed]
- Tropea, J. E., Cherry, S., Nallamsetty, S., Bignon, C. & Waugh, D. S. (2007). Methods Mol. Biol. 363, 1–19. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Van Duyne, G. D., Standaert, R. F., Karplus, P. A., Schreiber, S. L. & Clardy, J. (1993). J. Mol. Biol. 229, 105–124. [DOI] [PubMed]
- Wang, D. D.-H., Shu, Z., Lieser, S. A., Chen, P.-L. & Lee, W.-H. (2009). J. Biol. Chem. 284, 20812–20821. [DOI] [PMC free article] [PubMed]
- Waters, T. R., Eryilmaz, J., Geddes, S. & Barrett, T. E. (2006). FEBS Lett. 580, 6423–6427. [DOI] [PubMed]
- Weir, J. R., Bonneau, F., Hentschel, J. & Conti, E. (2010). Proc. Natl Acad. Sci. USA, 107, 12139–12144. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: hSuv3–AMPPNP complex, 3rc3
PDB reference: hSuv3–RNA complex, 3rc8