Abstract
The emerging field of bionanotechnology aims at revolutionizing biomedical research and clinical practice via introduction of nanoparticle-based tools, expanding capabilities of existing investigative, diagnostic, and therapeutic techniques as well as creating novel instruments and approaches for addressing challenges faced by medicine. Quantum dots (QDs), semiconductor nanoparticles with unique photo-physical properties, have become one of the dominant classes of imaging probes as well as universal platforms for engineering of multifunctional nanodevices. Possessing versatile surface chemistry and superior optical features, QDs have found initial use in a variety of in vitro and in vivo applications. However, careful engineering of QD probes guided by application-specific design criteria is becoming increasingly important for successful transition of this technology from proof-of-concept studies towards real-life clinical applications. This review outlines the major design principles and criteria, from general ones to application-specific, governing the engineering of novel QD probes satisfying the increasing demands and requirements of nanomedicine and discusses the future directions of QD-focused bionanotechnology research (critical review, 201 references).
1. Introduction
The development of materials, structures and systems with physical dimensions of 1 to 100 nanometers (nm) has a tremendous impact on the advancement of a wide range of fields including catalysis, computing, photonics, energy, and medicine. As a result, interest in nanotechnology has increased dramatically during the last decade. The National Nanotechnology Initiative budget, for example, has expanded by approximately 6 times since 2000.1 In contrast to widely used bulk counterparts, nanomaterials possess novel unusual and useful physicochemical properties that emerge at minute length scales. Metallic nanostructures in the presence of an electromagnetic field, for example, exhibit electron density oscillations which are highly sensitive to environmental perturbations. Iron oxide nanoparticles become superparamagnetic, exhibiting field-inducible magnetic dipoles. Carbon nanotubes possess remarkable tensile strength and controllable electrical conductivity. Semiconductor nanoparticles emit tunable and spectrally narrow fluorescence light upon excitation. These structures have been synthesized in a variety of shapes, sizes and configurations, and the theoretical framework explaining the unique optical, chemical and electronic properties of nanomaterials has been built. Meanwhile, nanomaterials have been incorporated in a variety of useful products ranging from stain-repellent fabrics and nanoparticle-containing sunscreens to lipid-encapsulated anticancer drugs and sensitive bioanalytical tools. With the number of nanotechnology-based patents growing exponentially,2 such items are rapidly appearing on the market. As new applications are developed, especially in such critical fields as energy generation and medicine, the impact of nanotechnology on the economy and on society will become increasingly more profound.
One of the most promising applications of nanotechnology has been in the area of biomedical research. Nanoscale sensors find their use in sensitive molecular diagnostics and high throughput bioanalytics, while nanoparticle-based drug carriers enable spatial and temporal control of drug delivery and release. Of great interest are organic and inorganic nanostructures that incorporate radiolabels and contrast agents for in vivo imaging techniques, such as Positron Emission Tomography (PET), Computed Tomography (CT), Single Photon Emission Computed Tomography (SPECT), Magnetic Resonance Imaging (MRI), sonography, and optical imaging. In combination with these macroscale modalities, nanoscale probes are important tools for molecular imaging – visualization, characterization, and quantification of biological processes at the molecular level within living systems.3,4 Fluorescent semiconductor nanoparticles, commonly referred to as quantum dots (QDs), represent a particularly interesting class of probes well-suited for advanced fluorescence imaging applications, such as multiplexed quantitative analysis of cellular phenotypes, real-time monitoring of intracellular processes, and in vivo molecular imaging.5–12 Exhibiting many supreme characteristics compared to conventional fluorophores, including size-tunable and spectrally narrow light emission along with efficient light absorption throughout a wide spectrum, improved brightness with outstanding resistance to photobleaching and degradation, and extremely large Stokes shift, QDs greatly expand the capabilities of fluorescence imaging. Furthermore, QDs provide a suitable platform for engineering of multifunctional nanodevices with capabilities of exploiting multiple imaging modalities or merging imaging and therapeutic functionalities within a single nanoparticle.
Utilization of unique photo-physical and chemical properties rendered by QDs for addressing challenging issues raised by biomedical research has promoted development of novel imaging probes, traceable drug delivery vehicles, and multifunctional nanocomposites. Active exploration of QD-based biomedical applications has resulted in more than 300% increase in related peer-reviewed publications since 2002 (based on PubMed and Nature.com searches). This review provides a synopsis of the key achievements in nanoscience that have initiated the work on utilizing QDs for biomedical applications and discusses recent developments that have converted QDs into clinically relevant tools. Brief overview of the photophysical properties and surface engineering strategies describes design principles guiding development of QDs into imaging probes and drug delivery vehicles. In-depth discussion of cell and tissue molecular profiling along with live-cell and in vivo molecular imaging presents the current state of the QD-based diagnostic and therapeutic applications and outlines potential future directions within these areas of research. Finally, review of the QD-based nanocomposites provides an introduction to an exciting emerging field of multimodal imaging and nano-therapeutics.
2. General principles for engineering of QD probes
QDs are semiconductor nanoparticles often made from hundreds to thousands of atoms of group II and VI elements (e.g. CdSe and CdTe) or group III and V elements (e.g. InP and InAs). Bulk semiconductors are materials with a relatively small band gap (less than 4 eV) between the valence and conduction bands, thus behaving like insulators at ambient conditions and exhibiting electrical conductivity only under external stimulation. Electrons in the ground state that are typically localized to individual atoms (i.e. comprising valence band) can be promoted to higher energy levels where electrons are free to move throughout the material (i.e. populate the conduction band) by supplying an amount of energy that exceeds the band gap. In certain cases, relaxation of an electron results in the release of bandgap energy in the form of light (fluorescence). QDs are crystalline particles that range from 2 to 10 nanometers in diameter. Physical size smaller that the exciton Bohr radius results in a 3-dimensional quantum confinement of charge carriers within the QD and limits the number of possible energy states that an electron can occupy (Fig. 1), thus giving nanoparticles novel properties not achievable in bulk materials.13–15 Additionally, relatively small size comparable to that of large biomolecules (e.g. antibodies) aids in engineering of biologically functional materials.
Fig. 1.
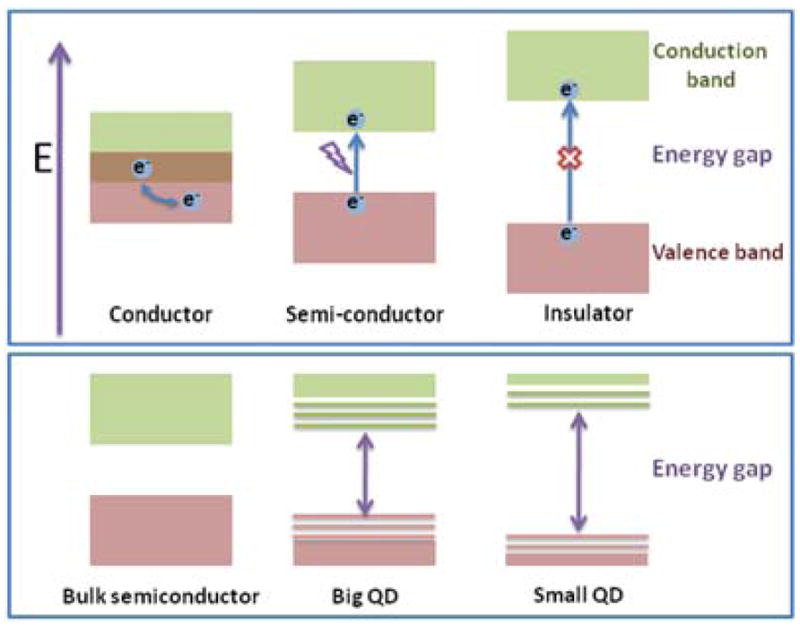
Electronic structure of bulk conductor, semiconductor, and insulator materials (top panel) and semiconductor nanoparticles (bottom panel). Bulk semiconductor materials have fully populated valence band and empty conduction band separated by a relatively small band gap. When an energy exciding the band gap is supplied, valence-band electrons acquire sufficient energy to populate conduction band and enable electric current flow. In nanoparticles, valence and conduction bands split into discrete energy levels, with the energy gap between closest possible valence and conduction levels increasing with decreasing particle size (and increasing degree of confinement of charge carriers).
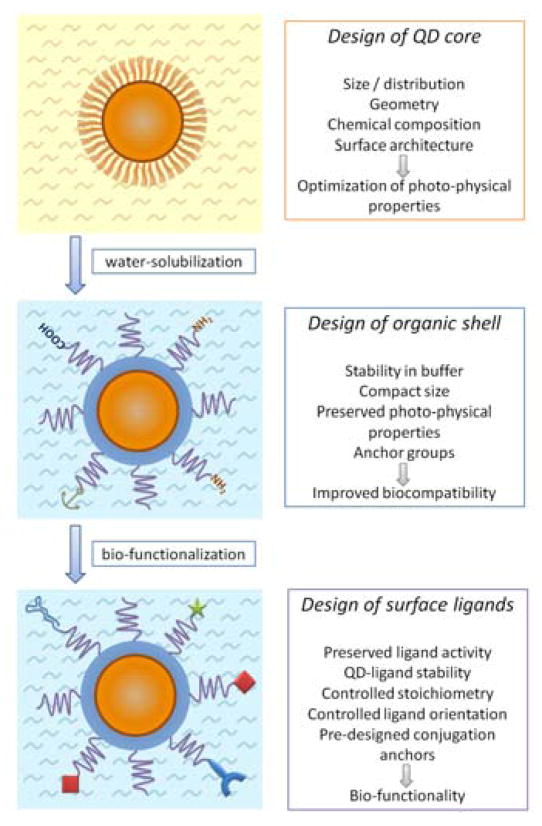
Inorganic nanoparticle core provides a rigid foundation for the development of QD probes. Manipulation of the core chemical composition, size, and structure controls the photo-physical properties of the probe. However, bare nanoparticles usually cannot interact with biological systems and do not possess any biological functionality. Careful design of coating materials that can encapsulate the QD core and shield it from the environment yields biocompatible probes with controllable physicochemical properties. Further decoration of the QDs with biomolecules imparts the bio-functionality and enables probe interaction with biological systems. Therefore, preparation of QD-based probes and nanodevices represents a multi-step process. Each step is guided by individual design principles aiming at controlling optical, physical and chemical properties of the final probe (Fig. 2).
Fig. 2.
General steps and design criteria in engineering of QD probes for biomedical applications.
2.1 Design of the quantum dot core
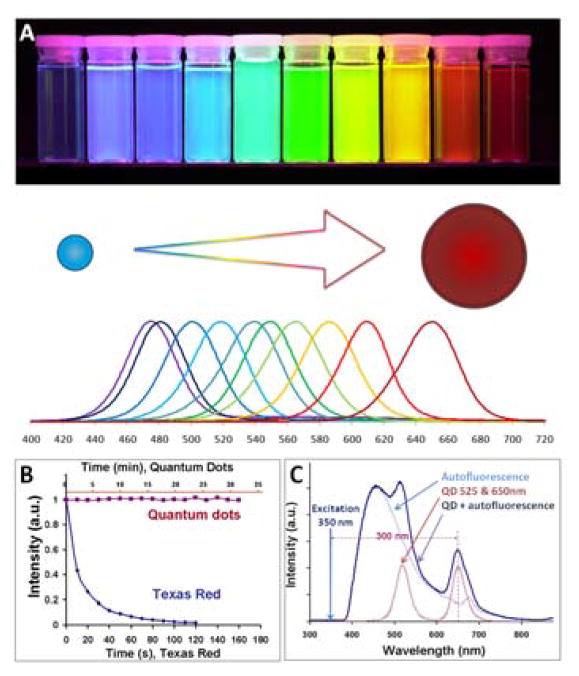
The QD core defines optical properties of the probe and represents a structural scaffold for engineering of nanodevices. In general, the QD core should be compact and highly stable with precisely controlled nanoparticle size distribution, geometry, chemical composition, and surface chemistry. Initial reports on preparation of semiconductor nanoparticles utilized QD synthesis in aqueous solutions and yielded particles with poor fluorescence efficiencies and large size variation. Advancements in synthetic procedures and surface chemistry have enabled production of water-soluble QDs with higher quantum yield (QY, up to 40–50%) and relatively narrow size distribution (exhibiting spectral emission width of ~50 nm for CdTe/CdSe particles16 and down to 19 nm for ZnSe QDs17). However, aqueous synthesis still suffers from poor control over the QD photo-physical and chemical properties. A major leap towards synthesis of highly uniform colloidal CdSe QDs was made in 1993 by Bawendi and coworkers by developing a high-temperature organometallic procedure,18 which is now widely used for synthesis of QDs for a variety of applications. In this procedure pyrolysis of organometallic precursors at high temperature yields nucleation and growth of nanocrystals, while coordination of trioctyl phosphine/trioctyl phosphine oxide (TOP/TOPO) base with unsaturated metal atoms on the QD surface prevents the formation of bulk semiconductor. Yet, utilization of a highly toxic and unstable Cd precursor (dimethyl cadmium) imposes restrictions on the equipment and reaction conditions and limits flexibility in the QD core design. A leap towards large-scale preparation of high-quality QDs has been done by Peng et al using alternative cheap precursor materials (such as CdO).19,20 Relatively mild and simple reaction conditions along with slower nucleation and growth rates offer extensive flexibility in engineering of QD chemical composition, geometry, and photo-physical properties. Precise kinetic control over a nanoparticle growth achieved with organometallic procedure enables preparation of QD populations with narrow size distribution. Therefore, as the difference in energy between the discrete ground and excited states increases with increasing degree of confinement (i.e. decreasing particle size), the size of the bandgap and, consequently, the color of emitted light can be fine-tuned by adjusting the QD size (Fig. 3A).21 With optimization of reaction conditions and utilization of size focusing via re-injection of precursors, an emission spectral width below 20 nm has been achieved.22–24 Further bandgap engineering by varying the chemical composition of nanocrystals has produced QDs emitting light from the UV, throughout the visible, and into the infrared spectra (400–4,000 nm).21,24–30
Fig. 3.
Unique photo-physical properties of QD probes. A) Narrow size-tunable light emission profile enables precise control over the probe color via varying the nanoparticle size. B) Outstanding photostability of QDs enables real-time monitoring of probe dynamics and accurate quantitative analysis, whereas quick photobleaching of organic dyes limits such applications. C) Capability of absorbing high-energy (UV-blue) light without damaging the probe and emitting fluorescence with a large Stokes shift enables efficient separation of the QD signal over the fluorescent background. Reprinted from ref. 54, Copyright (2005), with permission from Elsevier.
Narrow size-tunable light emission has proven to be highly beneficial for multiplexed molecular labeling (e.g. for phenotyping cell populations31 or detection of molecular signatures of cancer32), as little or no cross-talk between adjacent colors enables simultaneous detection and quantification of multiple fluorescence signals. Furthermore, high electron density of QDs and direct correlation between the particle size/composition and emission wavelength facilitate detailed evaluation of low-resolution fluorescence images with high-resolution imaging modalities – multiplexed imaging based on particle size can be achieved with transmission electron microscopy (TEM),33 while that based on particle chemical composition – with electron spectroscopic imaging (ESI).34 Multiplexing capability of QDs is complemented by efficient light absorption over a broad spectral range (hundreds of nanometers), as essentially any photon in UV-visible range with energy exceeding the band gap can be absorbed without damaging the nanoparticle. Unlike organic fluorophores, molar extinction coefficient of QDs gradually increases toward shorter wavelength, allowing multicolor QDs to be simultaneously excited by a single high-energy light source (e.g. UV lamp), thus eliminating the need for multiple excitation sources, reducing the cost of imaging instrumentation, and simplifying data analysis.
While providing good control over the particle size, original organometallic procedure produces QDs with low QY, compromising the utility of such particles as fluorescent probes. Moreover, TOPO-coated QDs are unstable with respect to photooxidation, resulting in effective degradation of nanocrystals and potential QD toxicity due to release of free Cd ions.13 Both issues arise from the relatively large number of atoms exposed on the surface of nanoparticles. In the nanoscale regime, surface atoms play a major role in determining the catalytic, electronic, and optical properties. As the radius of a spherical particle decreases, the ratio of its surface area to volume rapidly increases placing larger number of atoms on the surface.35 Surface atoms lack neighbors with which to form chemical bonds and thus possess unoccupied electron orbitals. Commonly referred to as dangling bonds or surface trap sites, these orbitals can trap charge carriers and either prevent or delay electron-hole recombination and subsequent photon emission, thus reducing the fluorescence QY.36, 37 Furthermore, such sites might exhibit enhanced chemical reactivity and compromise chemical stability of the nanoparticles. In order to prevent some of these undesirable characteristics, dangling bonds can be saturated by organic and inorganic capping layers.
Several groups have developed high-bandgap-energy inorganic shells (e.g. CdS and ZnS) several layers thick that effectively passivate the photoactive core of QDs.38–40 Wider band gap of the shell efficiently confines the exciton to the core, reducing nonradiative relaxation pathways and increasing QY.41 Careful choice of core and shell materials as well as optimization of the shell thickness are necessary to minimize the lattice strain between the core and shell and maximize the QD photo-physical properties. Although thin shells (1–2 monolayers) often produce the highest fluorescence yields, thicker shells (4–6 monolayers) provide more core protection from photooxidation and degradation.42 For example, Peng et al have observed confinement of the hole created during excitation within the CdSe core by a higher-bandgap CdS shell.40 As a result of such confinement, hole-dependent photo-oxidative processes that cause QD degradation and result in the loss of fluorescence is impeded. Also, a thicker shell might significantly reduce QD blinking (intermittence in light emission) associated with charge trapping and un-trapping at surface defects of a nanocrystals or due to charge ejection from the QD (Auger ionization) followed by recombination process.43–46 Since blinking might cause signal fluctuations in ultrasensitive detection, loss of distance information when movement of a single molecule is observed, and spectral jumping (change in the emission peak position), its elimination is often desirable.
Alternative approaches aim at achieving better fluorescence efficiency by optimizing the surface structure of nanocrystals and minimizing the number of surface trap sites. Some success in this direction has been observed with adjusting the precursor mixture composition and improving surface coating with multiple organic ligands (e.g. use of alkylamine surfactants, such as (hexa/octa/do)decylamine, along with TOPO).27,47–49 In one example, Talapin et al have stabilized CdSe QDs with alkylamines, achieving QY of 40–50% at room temperature (vs. 10–25% QY of as-prepared QDs).49 Qu and Peng have systematically studied the formation of photoluminescence bright point (presumably resulting from an optimal nanocrystal surface structure) during the QD synthesis, obtaining red-emitting CdSe nanoparticles with QY as high as 85% at room temperature without using inorganic cappling layer.27 However, further optimization of reaction conditions for preparation of multicolor QDs is required, and evaluation of single-core QD photo-physical properties and stability in aqueous environment is necessary in order to assess applicability of such nanoparticles for biological applications.
Both enhanced QD brightness and improved stability play a critical role in utilization of QD probes for accurate quantitative bioanalytics, single-molecule detection, real-time molecular tracking, and in vivo imaging. Having QY comparable to that of organic dyes while absorbing light more efficiently, an individual QD is 10–20 times brighter than organic fluorophores.8,50,51 Moreover, properly passivated core/shell QDs are thousands of times more photostable than organic dyes, resisting photobleaching for more than 30 minutes of continuous high-energy illumination (Fig. 3B).52–54 Unprecedented photostability renders QDs well suited for imaging when long exposure to excitation source is required, while keeping signal intensity constant and allowing for consistent analysis of samples (e.g. high-resolution 3D reconstruction,55 real-time molecule tracking,56 long-term monitoring of system response,57 etc.). Furthermore, capability to excite red QDs with high-energy blue light without damaging the probes enables utilization of the large Stokes shift for efficient separation of QD signal from predominantly blue-green autofluorescence of biological molecules (Fig. 3C).
Advances in synthesis and surface passivation technologies made QDs appealing platforms for engineering of biological probes with the advantages of enhanced photostability, improved brightness, tunable fluorescence, and single-source multicolor excitation. An ongoing work on controlling the QD surface properties and functionalization with biological ligands aims at transforming these materials into biologically compatible and bio-functional nanodevices.
2.2 Transition towards biologically compatible probes
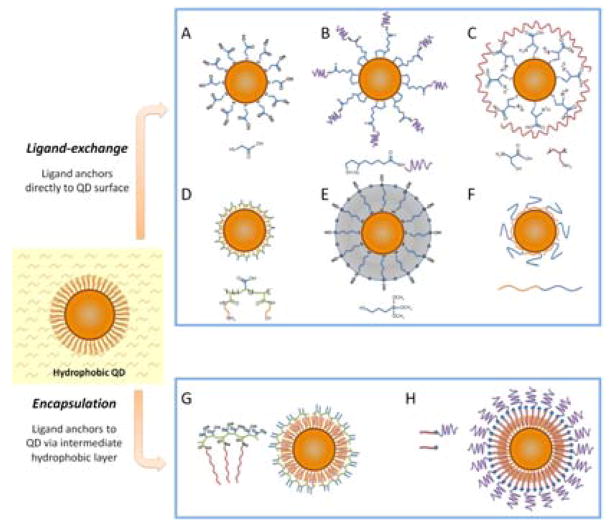
Organic phase synthesis produces high quality hydrophobic QDs soluble only in nonpolar organic solvents, such as chloroform and hexane. However, in order to be useful for biological applications QDs must be made water-soluble. In general, water-solubilization procedure should yield nanocrystals soluble and stable in biological buffers, preserve the original photo-physical properties, retain relatively small particle size, and provide reactive groups for subsequent conjugation to biomolecules. Several different approaches have been developed to produce water-soluble QDs satisfying these criteria.
One approach involves replacing hydrophobic surface groups with hydrophilic ones by means of ligand exchange. This is usually accomplished by substitution of the native TOPO coating with bifunctional ligands, which present both a surface-anchoring group (e.g. thiol) and a hydrophilic end group (e.g. carboxyl or hydroxyl). Examples include utilization of negatively-charged carboxy-terminated thiols, such as mercaptoacetic (MAA)51 and mercaptopropionic (MPA) acids (Fig. 4A), and thiol-containing zwitterionic molecules, such as cysteine,58,59 for decoration of QD surface with hydrophilic moieties. In addition to producing ultrasmall (hydrodynamic diameter, HD, below 6 nm) and highly water-soluble nanoparticles, amine and carboxylic acid groups provide binding sites for cross-linking to proteins, peptides, and nucleic acids. Despite the simplicity of the procedure, ligand exchange with monodentate surface ligands often compromises the fluorescence efficiency, photochemical stability, and shelf life of the probes, as ligands tend to detach from the QD surface leaving behind surface trap sites and causing nanoparticle aggregation.60,61 In general, crosslinking of small ligands or substitution from mono-thio to di-thio ligands substantially improves long-term stability. For example, Liu et al have utilized di-thiol ligand dihydrolipoic acid (DHLA) conjugated to poly(ethylene glycol) (PEG) to prepare small (HD of 11.4 nm) and stable QDs with some loss of fluorescence efficiency (drop in QY from 65% to 43%) (Fig. 4B).62 In an alternative approach, Sukhanova et al have water-solubilized QDs with DL-Cysteine and further stabilized the particles with poly(allylamine), achieving improvement in QD colloidal stability and increase in QY (from 40% to 65%) (Fig. 4C).63 Jiang et al have improved the stability of mercaptoundecanoic acid shell by covalently cross-linking neighboring molecules with lysine.64 However, the dramatic increase in nanoparticle size (from 8.7 to 20.3 nm HD) induced by shell cross-linking is undesirable, and further optimization of this procedure is required. Recently, Smith and Nie have developed a new class of multidentate polymer coatings that are only 1.5–2 nm thick (Fig. 4D).65 Consisting of a poly(acrylic acid) backbone grafted with multiple anchors (thiol and amine groups), this coating renders CdTe QDs biocompatible and colloidally stable, while keeping the final HD between 5.6 and 9.7 nm.
Fig. 4.
Routes for water-solubilization of hydrophobic QDs. Ligand-exchange procedures (A–F) involve replacing the native hydrophobic surface ligands (e.g. TOPO) with hydrophilic ones by direct anchoring of ligands to the QD surface. (G–H) Encapsulation procedures preserve the native QD surface structure and over-coat QDs with amphiphilic molecules (such as polymers or lipids) via hydrophobic interactions.
A more robust ligand-exchange approach involves formation of polymerized silanol shell on the QD surface (Fig. 4E).50,66 In this procedure 3-(mercaptopropyl)trimethoxysilane (MPS) is also directly absorbed onto the nanocrystals displacing the native TOPO molecules. However, upon addition of base, silanol groups are hydrolyzed and linked with each other producing stable and compact (1–5 nm thick) silica/siloxane shell and rendering particles soluble in intermediate polar solvents (e.g. methanol or dimethyl sulfoxide). Further reaction with bifunctional methoxy compounds renders QDs soluble in aqueous buffers. Polymerized siloxane-coated nanoparticles are highly stable against flocculation. However, residual silanol groups on the QD surface often lead to precipitation and gel formation at neutral pH.41
Employing native stability and biocompatibility of biomolecules, Weiss and colleagues have demonstrated preparation of compact water-soluble QDs via ligand exchange with engineered peptides (Fig. 4F).67 With the use of phage-display libraries68 and accelerated evolution this procedure enables selection of peptide sequences that can specifically bind to any type of QDs, thus providing a universal surface coating approach. Yet, due to relatively high complexity and inaccessibility of this technique along with lack of characterization data on peptide-coated QDs such an approach is not widely used.
An alternative approach to QD water-solubilization is to retain the native TOPO coating and encapsulate the hydrophobic QDs with amphiphilic molecules such as polymers (Fig. 4G)53,69 or phospholipids (Fig. 4H).70 The hydrophobic portion of this molecule intercalates within alkyl-chain-terminated surface ligands while the hydrophilic portion (e.g. charged groups, PEG, etc.) faces outwards, interacting with the aqueous solvent and rendering the particle water-soluble. This method produces exceptionally stable water-soluble QDs with preserved optical properties, as the coating does not directly interact with the nanocrystal surface and does not disturb the surface passivation layer.71 However, deposition of several organic layers usually results in dramatic increase of the nanoparticle hydrodynamic size. For example, block copolymer coating increases the diameter of CdSe/ZnS QDs from ~4–8 nm before encapsulation to up to 30 nm HD.42,72 Size increase might be detrimental for quantitative biomarker detection in a crowded biological environment and hamper intracellular penetration of the QD probes.46,56,73 The increased thickness of polymer coating might also preclude utilization of QDs in Förster resonance energy transfer (FRET)-based applications.42,46
As new QD-based applications are being explored, more stringent requirements for QD surface coating arise. In general, the size of QDs should stay small after coating, the surface should be biocompatible, reactive groups should be available for conjugation of biomolecules and targeting ligands, and QD probes should show minimal non-specific interactions with the biological environment. With a variety of water-solubilization procedures developed, a number of QD-based biological applications have already become available. However, there is no method that satisfies all the design criteria imposed by increasing demands of biomedical research. Ligand-exchange approaches often yield compact probes at an expense of reduced stability and fluorescence efficiency, whereas polymer-encapsulation produces exceptionally stable and bright particles at an expense of increased size. Therefore, engineering of novel coatings that combine the protective features of encapsulation procedures with the compactness of small ligands represents an active area of research.
2.3 Development of bio-functional QD nanodevices
In order to utilize high quality QDs for bioimaging, detection, and drug delivery applications, bio-functionality has to be added to otherwise inert nanoparticles. This is usually achieved by decorating QDs with proteins, peptides, nucleic acids, or other biomolecules that mediate specific interactions with living systems. Surface engineering is thus crucial not only for tuning the fundamental properties of nanomaterials and rendering them stable and soluble in different environments, but also for creating nanoparticle-biomolecule hybrids capable of participating in biological processes. Such hybrids should combine useful properties of both materials involved, i.e. optical properties of the nanocrystals and biological functions of ligands attached.
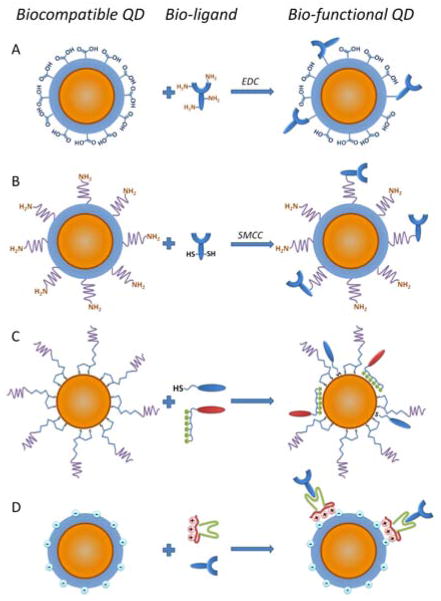
Several approaches can be used for conjugation of QDs and biological molecules. One of the most simple and popular bioconjugation methods is covalent bond formation between reactive functional groups (e.g. primary amines, carboxylic acids, alcohols, and thiols). For example, linking of proteins via primary amine groups to carboxylic acid-containing QDs can be achieved via carbodiimide-mediated amide formation (i.e. EDC, 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide, condensation reaction) (Fig. 5A). As this reaction utilizes naturally occurring amine groups it does not require additional chemical modification of proteins, preserving their natural structure; but it lacks control over the molecular orientation of the attached proteins, thus allowing attachment at a point close to ligand’s active site that might result in partial or complete loss of biological functionality of that ligand. Moreover, EDC reaction might result in QD aggregation due to crosslinking between multiple reactive sites on QDs and proteins. Another common covalent bonding procedure involves active ester maleimide-mediated amine and sulfhydryl coupling (Fig. 5B). Since free sulfhydryl groups are rare in native biomolecules, additional treatment of the ligands is often required (e.g. reduction of antibodies with dithiothreitol). This reaction yields stable QD-ligand complexes with often controlled ligand orientation. However, chemical treatment might compromise the biological activity of ligands and cause reduced sensitivity and/or specificity of the probe. Nonetheless, both approaches are widely used for variety of applications, including custom production of QD-antibody probes and preparation of QD-streptavidin conjugates. Recently, Barat et al have utilized amine-sulfhydryl coupling for preparation of compact diabody-QD probes.74 Using small cysteine-terminated antibody variable chain domains instead of complete antibodies along with site-specific conjugation of a cysteine tag, the authors have achieved decoration of QDs with fully functional antigen recognition ligands. Despite the complexity of the approach, bio-functionalization of QDs with small genetically engineered molecules carrying site-specific conjugation anchors represents a promising route for preparation of compact and highly specific QD probes.
Fig. 5.
Routes for QD bio-functionalization. Decoration of QD surface with bio-ligands can be achieved via covalent conjugation (A, B), non-covalent coordination of thiol groups or polyhistidine tags with the QD surface metal atoms (C), or electrostatic deposition of charged molecules on the QD organic shell (D).
Besides covalent bonding to organic QD shell, biomolecules can be linked directly to QD surface via coordination with metal atoms of the QD core/shell. To achieve this, QDs coated with labile small ligands are mixed with thiolated biomolecules or biomolecules containing polyhistidine (HIS) residues (Fig. 5C). As a result, small ligands are replaced on the QD surface by biomolecules. Yet, utilization of QDs with unstable displaceable surface coatings (such as mercapto compounds) and direct interaction with the QD surface might significantly reduce the brightness and stability of such bioconjugates in aqueous solutions. In a more robust variation of this approach, Medintz et al have functionalized stable DHLA-coated QDs with HIS-tagged maltose-binding protein (MBP) via coordination of oligohistidine with the QD surface at defects in DHLA surface coating. The binding event is accompanied by improved surface passivation and rise in QY (from 16% to 39%), thus enabling direct measurement of the binding stoichiometry.75 Later, this approach was successfully applied for conjugation of other HIS-tagged engineered ligands, such as enzyme sensing76 and cell penetrating77 peptides. Bio-functionalization via coordination with QD surface is attractive due to the simplicity of the reaction, control over the final bioconjugate assembly, and ability of using unmodified ligands with preserved native structure. However, custom design of ligands incorporating thiol groups or HIS-tags is often complex and suitable only for small biomolecules with relatively simple structures.
Non-covalent self-assembly of engineered proteins on the surface of QDs with preserved organic shell prevents direct access to inorganic QD core and exhibits minimal effect on the photo-physical properties (Fig. 5D). In one example a fusion protein has been utilized as an adaptor for immunoglobulin G (IgG) coupling.78,79 Electrostatic interaction between the positively charged leucine zipper domain of an adaptor protein and the negatively charged QD shell stably deposits the adaptor protein to the QD surface, while the protein G domain specifically captures the antibody Fc region. The resulting assembly features precise control over the antibody orientation and eliminates any chemical modification of IgG, thus preserving its activity. However, this procedure is often limited to conjugation of specific classes of ligands (e.g. antibodies). Moreover, the size of such bioconjugates is large due to a number of thick biomolecule layers deposited on the QD surface.
Recent achievements in merging nanoparticle encapsulation and bioconjugation steps and design of pre-functionalized surface coatings promise to provide more compact, stable, and biocompatible nanoparticles with controlled density and orientation of ligands attached. Amphiphilic polymers with a maleic anhydride backbone are being actively explored for this purpose. In organic anhydrous solvents, such polymers encapsulate TOPO-coated QDs and introduce reactive anhydride groups on the surface. In basic aqueous buffers anhydride rings are quickly hydrolyzed, yielding negatively charged carboxylic acid groups and rendering QDs water soluble.69 More importantly, anhydride groups are highly reactive towards amine-containing molecules, thus allowing covalent conjugation of a variety of biomolecules to the polymer chains without the need for post-encapsulation modification.80,81
Choice of the bio-conjugation approach depends on availability of ligands with suitable functional groups and on specific application requirements. However, common design criteria involve preserved QD photo-physical properties and ligand bio-functionality, controlled ligand orientation and binding stoichiometry, compact probe size, and good stability in physiological environment. As these criteria can be satisfied in only few specific cases, improvement of existing bioconjugation techniques and design of novel application-specific water-solubilization and bioconjugation approaches remains an active area of research. With the development of stable and bio-functional QD probes these materials will become nanoscience building blocks82 with flexible properties that could be further optimized for specific applications including biomedical imaging, detection, and nano-therapeutics.
3. QD probes for in vitro applications
In the last decade, surface engineering and bio-functionalization techniques have transformed semiconductor nanocrystals into complex cellular probes capable of interaction with biomolecules and direct participation in biological processes. In 1998, two seminal Science papers first demonstrated that semiconductor nanoparticles could be made water-soluble and used as biological imaging probes.50,51 One approach utilized silica shell encapsulation chemistry in order to produce QDs for a single-excitation dual-color cell staining.50 When derivatized with trimethoxysilylpropyl urea and acetate groups, green QDs preferentially labeled the cell nucleus, and when derivatized with biotin, red QDs labeled F-actin filaments pre-treated with phalloidin-biotin and streptavidin. The second paper was the first to demonstrate the ligand-exchange approach to QD water-solubilization.51 Subsequent conjugation of transferrin produced QD probes that were endocytosed by live HeLa cells resulting in punctate cell staining, while IgG bioconjugates were used in an aggregation-based immunoassay. Since then, a multitude of surface engineering techniques for QD solubilization and bio-functionalization have been developed, enabling application-specific design of QD probes. Such probes have found their use in a variety of in vitro applications, such as histological evaluation of cells and tissue specimens, single molecule detection and real-time tracking, long-term live-cell imaging, and study of intracellular processes.
3.1 Molecular pathology
Fluorescence microscopy is a widely used optical imaging modality for evaluation of phenotypes of healthy cells as well as for detection of molecular signatures of diseases. Histological techniques, such as fluorescence in situ hybridization (FISH) and immunohistochemistry (IHC), enable detection of nucleic acids and protein biomarkers within cells and tissue specimens with a high degree of sensitivity and spatial resolution. Organic fluorophores have been widely used in these applications, either as stains for highlighting cell structures or as specific probes for labeling biomarkers. However, applicability of organic fluorophores in multiplexed and quantitative analysis for molecular profiling, a powerful technique for study of complex molecular networks underlying physiological and pathological processes, is limited by the quick photobleaching, spectral overlap between probes, and the need to excite fluorophores at unique wavelengths. QD probes, on the other hand, exhibit photophysical properties well-suited for this application.83,84 Despite the relatively recent introduction into biomedical research, QDs have already proven to be a powerful tool for sensitive quantitative molecular profiling of cells and tissues, providing unique identification of individual cell lineages and uncovering molecular signatures of pathological processes.84,85 Utilization of QDs for staining of fixed cells and tissue specimens does not impose strict requirements on the probe biocompatibility, toxicity, or stability in biological media. However, careful design of the probe size, surface properties, and image processing algorithms are essential for this application.
The hydrodynamic size of the QD-ligand bioconjugate should be minimized in order to achieve good penetration of the probes within the cross-linked intracellular compartments of fixed cells. Membrane-bound compartments, such as nucleus and mitochondria, represent especially difficult targets for QD staining. For example, Wu et al have investigated the utility of QD-streptavidin and QD-antibody bioconjugates for simultaneous labeling of membrane-associated Her2 receptor and of a nuclear antigen in breast cancer cells (Fig. 6).53 While staining of cell surface antigens is reliable and effective, staining of cytoplasmic and nuclear markers is more variable, resulting from the relatively large size of the probes. In another example, Tholouli et al have employed the biotin-streptavidin linkage for preparation of QD-oligonucleotide probes for FISH-based studies of mRNA.86 Biotinylated DNA probes pre-incubated with QD-Streptavidin conjugates enable detection of 3 mRNA targets in a 1-step FISH procedure. Yet, pre-conjugation of multiple oligonucleotides to QDs significantly increases the overall size of the probe, thus requiring specimen permeabilization with proteinase K, which necessarily degrades cell and tissue architecture and destroys most of the protein-based biomarkers useful for IHC studies. Chan et al have resolved this issue by developing a more controlled procedure for pre-conjugation of exactly one oligonucleotide probe per QD via biotin-streptavidin linkage.87 Starting with QD-streptavidin conjugates, excess streptavidin sites are blocked with biocytin (water soluble biotin derivative), and only a few biotinylated oligonucleotides are allowed to bind. Further purification of QD-oligo conjugates with agarose gel electrophoresis yields relatively small mono-oligonucleotide FISH probes suitable for multiplexed mRNA detection under mild specimen permeabilization. As a result, a combined QD-based FISH-IHC procedure has been developed to compare cellular distribution patterns of vesicular monoamine transporter (Vmat2) mRNA and immunoreactivity of tyrosine hydroxylase in dopaminergic neurons.87 In general, with larger QD probes, stronger permeabilization of specimens with detergents and/or enzymes might be required to obtain sufficient intracellular access; however, chemical treatment might damage the target molecules, thus reducing staining sensitivity and providing inaccurate quantitative information about biomarker expression levels. Furthermore, entrapment of larger QD probes within cells hampers post-staining washing of unbound probes and reduces the specificity of staining. Therefore, engineering of more compact probes is highly beneficial.
Fig. 6.
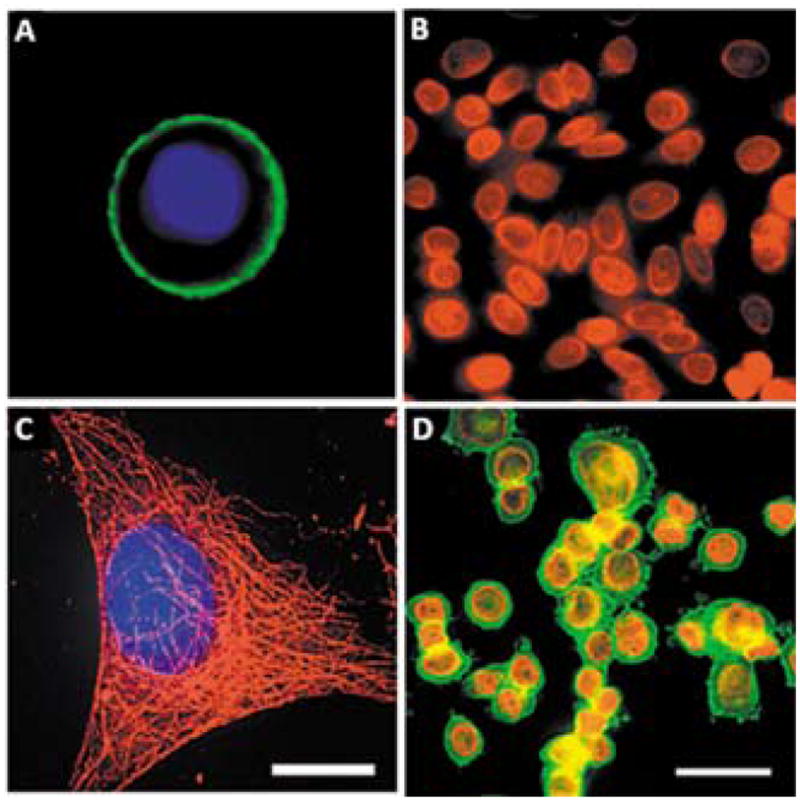
Labeling of surface and intracellular targets with QD probes. In single-color examples membrane-associated Her2 receptors are detected with primary antibodies and QD-labeled secondary IgG (A, green), while intracellular nuclear antigens (B, red) and microtubules (C, red) are visualized with primary IgG/secondary IgG-biotin/QD-Streptavidin cascade. Both labeling routes can be applied simultaneously for a two-color staining (D). The nuclei are counter-stained with Hoechst 33342 (blue) in A and C. Reprinted by permission from Macmillan Publishers Ltd.,53 copyright (2003).
QD surface engineering is critical for minimizing the non-specific binding of QD probes to biomolecules, a common reason of reduced staining signal-to-noise ratio and decreased sensitivity and specificity of the target detection. Majority of the non-specific binding results from the electrostatic interactions, when highly charged QD probes are used, and from hydrophobic interactions, when QDs with exposed hydrophobic regions or partially hydrophobic ligands are used. Decoration of QDs with uncharged hydrophilic moieties (e.g. PEG) and zwitterionic molecules produces highly water-soluble and stable probes while efficiently eliminating non-specific interactions. For example, QD probes used in the majority of published research have a layer of PEG that shields the QD core from the environment and provides anchor points for ligand attachment. Popularity of QD-PEG comes from the outstanding non-fouling properties of PEG as well as high stability of probes in a wide range of experimental conditions, which facilitates engineering of QD probes for virtually any application. However, addition of a PEG layer often results in increased particle HD leading to the detrimental size-dependent consequences described above. Zwitterionic coatings, on the other hand, become utilized more often as smaller probes are being developed. Featuring a densely packed alternating positively and negatively charged groups, these coatings do not favor electrostatic or hydrophobic interactions while providing an overall neutral well-hydrated surface. However, zwitterionic coatings tend to show high pH-sensitivity, thus imposing more stringent requirements on bioconjugation and staining conditions. Alternatively, the QD surface can be completely over-coated with large biomolecules (e.g. proteins) shielding the QD from the environment and mimicking the native functionality of the ligand; yet, possible dramatic increase in probe size renders this approach most appropriate for labeling of extracellular targets.
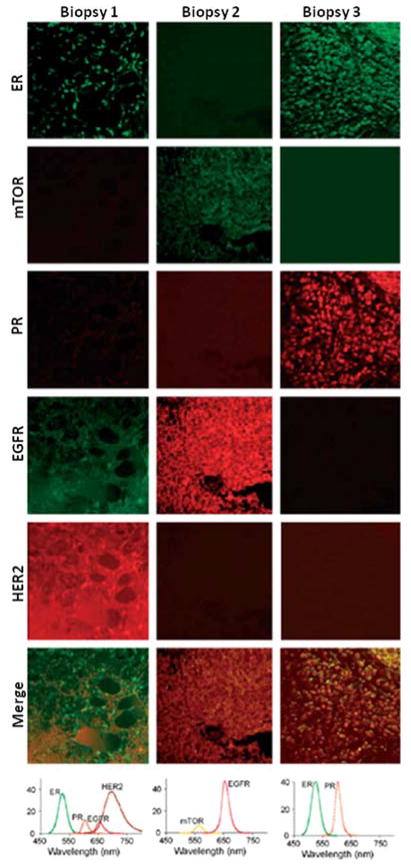
The high brightness and photostability of QD probes enables sensitive and robust measurement of the biomarker expression levels. However, accurate quantitative analysis of multiple biomarkers and comparison of their relative levels of expression within a single specimen further demand standardization of image acquisition and processing algorithms. Extraction and analysis of individual QD spectra from a composite image can be achieved with spectral imaging.84,88 Generally, spectral imaging systems incrementally apply narrow band-pass filters and collect a series of images for each wavelength band over a specified spectrum, thus providing spectral information for each pixel of an image. Deconvolution of known emission profiles from the resulting composite image separates different probe signals from each other and from the background fluorescence. However, quantitative comparison of different biomarkers in multiplexed staining might be compromised by the strong signal enhancement of larger (red) QD and reduction of smaller (green-blue) QD signals. For example, Ghazani and coworkers have demonstrated three-color staining of lung carcinoma xenografts for epidermal growth factor receptor (EGFR), E-cadherin, and cytokeratin with 655, 605, and 565 nm QD-based assays and noticed significant enhancement of 655 nm signal over 565 nm one, attributing this phenomenon to FRET from smaller to larger QDs.89 Further, the discordance in fluorescence intensity of individual probes directly relates to light absorption properties of QDs, as larger QDs possess larger absorption cross-sections and thus collect light more efficiently. The effect of FRET depends on the density and distribution of biomarkers, which is hard to predict and account for during quantitative analysis. However, differences in photo-physical properties of individual probes can be readily characterized in advance and incorporated into signal analysis algorithms. In a recent study, Yezhelyev et al have demonstrated the multiplexed labeling and quantification of three clinically significant breast cancer markers – Her2, ER, and PR – on formalin-fixed paraffin-embedded (FFPE) breast cancer cells.32 In order to account for signal enhancement of red QDs and compare expression levels of biomarkers within one sample, acquired data is adjusted according to the relative QD intensities (QD655:QD605:QD565 = 8:4:1 as measured in a separate experiment for equal QD concentrations), yielding relative biomarker abundance consistent to that obtained with conventional techniques (IHC, Western blot, and FISH). This technology has been further validated by the detection and quantification of a panel of five biomarkers on FFPE breast cancer tissue biopsies (Fig. 7).
Fig. 7.
Multiplexed labeling of breast cancer tissue biopsies. Normalization of the fluorescence according to relative QD intensities is required for accurate quantitative analysis of biomarker expression. Reproduced with permission from ref. 32. Copyright 2007 Wiley-VCH Verlag GmbH & Co. KGaA.
Future advancements in the area of QD-based molecular pathology will be centered around highly multiplexed quantitative molecular profiling. Engineering of more compact and sensitive QD probes with outstanding stability and non-fouling properties will, therefore, remain the major focus of research in this area. Decreasing the band gap by tuning the QD chemical composition, for example, might enable shifting QD emission into deep blue90 or far red30 region, while keeping the particle size constant within 4–6 nm range. However, further reduction of the QD inorganic core size below 3–4 nm might be highly challenging. Meanwhile, significant probe size reduction can be achieved via engineering of the compact organic coating layers and ligands that offer great design flexibility. Substitution of thick shells with thinner zwitterionic coatings, development of monovalent probes, and utilization of smaller targeting ligands (e.g. peptides and aptamers) will, thus, become essential for engineering of robust and stoichiometric QD probes and their translation to clinical diagnostics.
3.2 Real-time monitoring of dynamic molecular processes
Staining of fixed cells and tissue specimens provides information on biomarker expression and distribution; however, the study of intracellular molecular pathways underlying the physiological and pathological processes is limited by the static nature of this technique. Real-time imaging of live cells, on the other hand, enables the study of highly complex and dynamic biological processes that occur at molecular level. While the relatively large size of QD probes often hampers cellular entry and intracellular targeting, access to the biomarkers expressed on the cell membrane is usually readily achievable. Consequently, the majority of applications reported in the literature describe dynamics of membrane proteins (e.g. receptor diffusion) and membrane-associated processes (e.g. endocytosis and intracellular trafficking) rather than monitoring of intracellular targets. As a general guideline, QD probes for real-time live cell imaging should have compact size and high stability in biological buffers and cell culture media, exhibit high brightness and photostability for single-molecule imaging, show no toxicity or interference with cell physiology throughout the duration of experiment, and possess biological functionality for interaction with target biomolecules.
Majority of the QD probes used for live cell imaging employ a well-characterized and robust PEG coating as a universal non-fouling shield against protein adsorption. Resistance to protein binding conveys high stability in a wide range of buffers as well as cell culture media, precluding QD aggregation, non-specific interaction with cells, and off-target effects (e.g. receptor activation, enhanced endocytosys, etc.). In addition, such coating efficiently protects the QD core and preserves the beneficial photo-physical properties. Being 10–20 times brighter and orders of magnitude more photostable than organic fluorophores, QDs are well-suited for sensitive single-probe detection and long-term probe monitoring.50,51 In combination with advanced imaging techniques (e.g. 3-D tracking confocal microscopy,91 pseudo total internal reflection fluorescence microscopy,56 oblique angle fluorescence microscopy,92 etc.), QD probes enable the study of active and passive molecular transport mechanisms under high-background environments. Furthermore, distinguishing single QD probes from the small QD aggregates by the characteristic fluorescence intermittency (or blinking) improves the accuracy of measurement by eliminating the contribution of QD clusters. Outstanding resistance to photobleaching and degradation enables probe monitoring for several hours or days. For example, Jaiswal et al have utilized this property for visualization of QD endocytic uptake and specific cell-surface labeling of P-glycoprotein transporters over the course of 14 hours, acquiring images at a rate of 1 frame per minute.93 Localization of particles within the endosomes of live HeLa cells and D. discoideum amoebae could be monitored over the course of more than a week with minimal loss of QD fluorescence.
Accurate examination of physiological processes in native environment is often hard to achieve, as any chemical modification introduced to the system (e.g. labeling with a fluorophore or expression of a foreign reporter protein) might potentially change intramolecular interactions and interfere with normal cell physiology. This issue is especially keen for QD-based studies, since biomolecules must be tagged with bulky (sometimes several times larger than the studied biomolecule) probes. Therefore, design of QD probes that introduce minimal changes to the cell physiology and lack short-term cyto-toxicity is essential for the QD-based investigation of dynamic molecular processes. Much success in overcoming this challenge has been achieved in the study of cell receptor diffusion and interaction. In a single-molecule imaging study, Dahan et al have used QDs for labeling of individual glycine receptors on the surface of cultured spinal neurons and tracking the receptor diffusion in and out of synaptic cleft (Fig. 8).73 Differential 2-D diffusion coefficients of receptors have been measured over time spans 240 times longer than previously achieved using organic dyes as tags, with 4 to 8-fold better spatial resolution, and with a signal to noise ratio almost an order of magnitude higher. While the steric effect of QD probes could not be assessed through this study, relative characterization of receptor diffusion patterns within the synaptic, perisynaptic, and extrasynaptic regions was achieved. In another study, QDs have been used to reveal a previously unknown receptor diffusion mechanism for recovery from synaptic depression in neurons.94 Tracking of the rapid lateral diffusion of QD-labeled AMPA glutamate receptors have shown diffusion behavior comparable to that of organic dye-labeled receptors, while providing a robust fluorescence signal for the duration of experiment. Murcia et al have demonstrated that labeling of individual cell membrane lipids with QDs does not affect lipid diffusion (as compared to dye-labeled lipids), while enhanced brightness of the probe enables high-speed single molecule tracking at 1000 frames per second.92 Overall, it has been shown by several studies that QD probes do not significantly interfere with the diffusion of labeled biomolecules on the cell membrane, thus permitting both absolute measurement of diffusion coefficients and self-consistent relative studies of biomolecule diffusion under varying conditions.
Fig. 8.
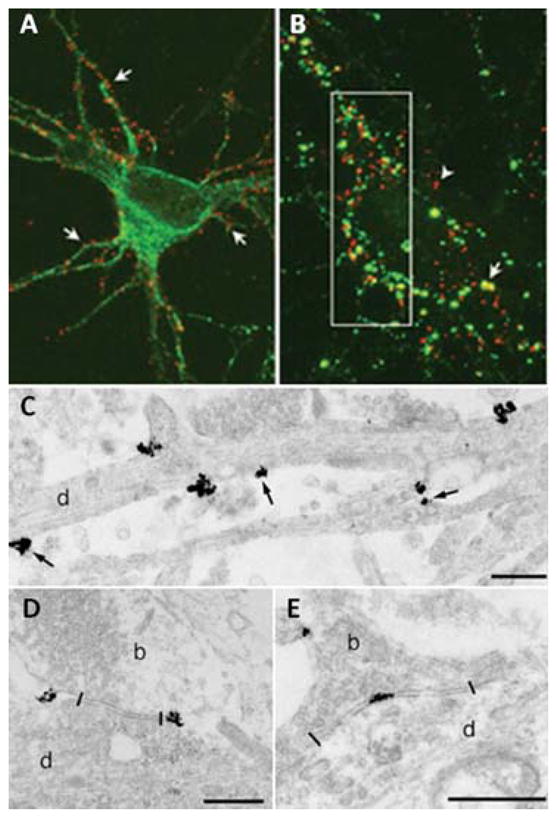
Labeling of individual glycine receptors in cultured spinal neurons. QD probes label glycine receptors throughout somatodendritic compartment (A) and can be located adjacent to (B, arrowhead) or in front of (B, arrow) inhibitory synaptic boutons. TEM examination reveals QD clustering at the extrasynaptic (C), perisynaptic (D), and synaptic (E) regions. Reprinted from ref. 73 with permission from AAAS. Copyright (2008).
Besides providing insight on the molecular dynamics of cell membrane components, QD probes facilitate the detailed study of such important processes as endocytosis and intracellular trafficking. Due to the relatively small size, individual QDs can be uptaken by the cells via endocytosis, incorporated within the endosomes, and be transported like any other endocytosed cargo without interfering with the mechanism of the process, thus representing a useful model system for the study of these phenomena. For example, Cui et al have studied the dynamics of axonal internalization and neuronal retrograde transport of the nerve growth factor (NGF) by tagging native NGF with QDs.56 While recording an average retrograde endosome movement speed consistent with previous bulk measurements of NGF transport, real-time monitoring of individual QD-NGF-containing endosomes has revealed a “stop-and-go” behavior and occasional anterograde movement, thus providing insight on the diversity in transport mechanisms. In another study Zhang et al have utilized the unique size and pH-dependent fluorescence of QDs for the study of the dynamics of synaptic vesicles during multiple rounds of neuronal transmission without perturbing the vesicle cycling.95 Monitoring of individual QD-loaded synaptic vesicles has enabled characterization of complete vesicle fusion (full-collapse fusion) and transient fusion (so-called kiss-and-run behavior) with respect to time and frequency of impulse firing, and uncovered new aspects of neurotransmitter release and replenishment mechanisms.
Efficient specific interaction with cell components requires otherwise inert QD probes to possess biological functionality, which is usually conveyed by decoration of QDs with targeting biomolecules. Often such moieties are represented by the receptor ligands attached to QD surface either covalently or through a streptavidin-biotin linker. For example, Lidke et al have decorated QDs with epidermal growth factor (EGF), a ligand for erbB/HER transmembrane receptors, to study the early steps of receptor-mediated signal transduction.57 While not interfering with receptor signaling, QDs have enabled visualization of specific EGF-receptor binding followed by heterodimerization of receptor components, endocytosis, and previously unreported retrograde transport of EGF-QDs along cell filopodia (Fig. 9). In a later report by the same group, antigen uptake and processing by dendritic cells have been studied using QDs functionalized with pathogen-specific ligands.96 Highly stable ligand-coated QDs mimicking viruses and pathogenic microorganisms provide a powerful model system for the detailed characterization of the immune response mechanisms. Yet, labeling of membrane targets via ligand-receptor binding followed by receptor activation might be undesirable, whereas labeling of non-receptor targets is impossible with this approach. Therefore, significant portion of current research is focused on the development of alternative targeting mechanisms. An interesting approach has been demonstrated by Roullier et al who have functionalized QDs with a chelator, tris-nitrilotriacetic acid (tris-NTA), pre-loaded with Ni for labeling of biomolecules with ubiquitous HIS tags.97
Fig. 9.
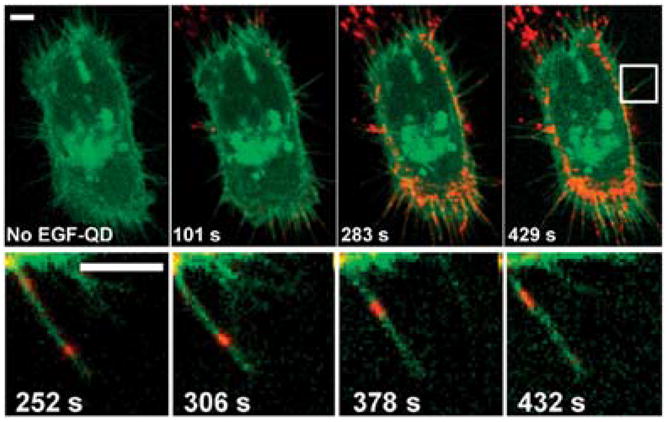
Labeling of erbB/Her transmembrane receptors with QD-EGF probes. Continuous observation of QDs in live cells enabled monitoring of receptor heterodimerization, probe endocytosis, and QD-EGF retrograde transport along cell filopodia. Reprinted by permission from Macmillan Publishers Ltd.,57 copyright (2004).
Future advances in continuous monitoring of dynamic molecular processes within living systems will rely on the expanded capabilities brought by highly bright and photostable QD probes. Having size comparable to proteins or small viruses, QDs are capable of carrying multiple biomolecules that mediate antigen recognition, receptor binding, endocytosis, and intracellular trafficking, thus facilitating the design of a variety of minimally invasive model systems for the study of cell physiology.
3.3 Labeling of intracellular targets in live cells
Like the QD-based investigation of cell physiology described above, labeling of intracellular targets in live cells examines molecular processes occurring within living systems; yet it presents a unique set of challenges and probe design requirements. As QDs cannot easily cross intact cell membrane and diffuse within the crowded intracellular environment, specific labeling of intracellular components is highly problematic. Moreover, elimination of unbound probes from intracellular environment is limited, increasing the possibility of false-positive detection. Therefore, besides being non-toxic and biocompatible, a functional QD probe for live-cell intracellular labeling should employ efficient intracellular delivery mechanism (including cell uptake and cytoplasmic release) as well as quick and complete elimination of unbound probes.
A variety of techniques have been developed for the delivery of macromolecular cargo within cells, such as microinjection, electroporation, chemical transfection, and ligand-mediated uptake.98 However, these processes, as well as subsequent intracellular distribution of internalized particles, are difficult to predict and control due to the high dependency on such factors as cell phenotype,99 nanoparticle size,100,101 and surface coating.53,102,103 Moreover, QD probes often get damaged by the transfection procedure or become sequestered within endosomes and lysosomes, being unable to reach the cytosolic molecular targets. For example, Derfus et al have shown that although both transfection using cationic liposomes and electroporation result in cytosolic QD delivery, internalized particles become aggregated by an unknown mechanism, whereas only microinjection results in diffuse cytosolic staining.104 Therefore, development of efficient QD-compatible cytosolic delivery techniques is critical for the real-time exploration of intracellular processes.
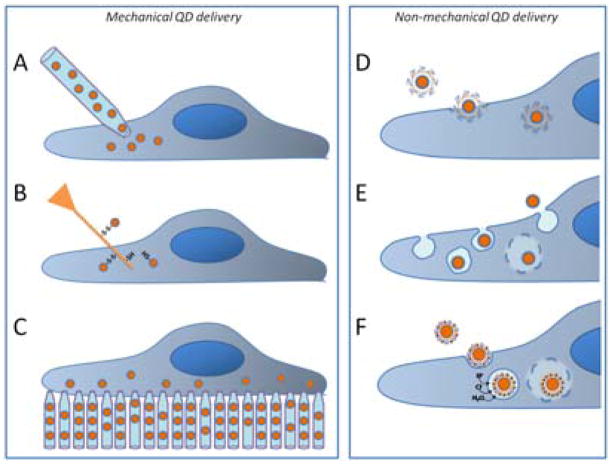
Mechanical techniques similar to traditional microinjection represent the most straight-forward approach to QD intracellular delivery, as virtually no modification of QD probes already available for extracellular labeling is required. For example, peptide-functionalized QD probes delivered to the cytoplasm via microinjection successfully exploit active peptide-specific transport mechanisms to reach target compartments, nucleus and mitochondria, within several hours after delivery (Fig. 10A).104 In another example, Yum et al have utilized gold-coated boron nitride nanotubes (with a diameter of 50 nm) to deliver QDs within the cytoplasm or nucleus of live HeLa cells with consequent 30-minute monitoring of QD diffusion within those compartments (Fig. 10B).105 Linking the ubiquitous QD-Streptavidin probes onto the nanotubes via reducible disulfide bonds enables delivery of intact QDs to a controlled intracellular location without much damage to the cell. While the QD probes used in this study did not carry targeting ligands, the technique can be expanded to deliver functionalized QDs as well. However, being quite labor-intensive and low-throughput, both techniques might only find use in limited single-cell studies. Aiming at high-throughput intracellular delivery, Park et al have engineered arrays of vertically aligned carbon nanosyringes that, upon cell growth on top of them, provide cytosolic access for injection of unmodified QDs (Fig. 10C).106 Efficient and consistent delivery of QD probes within large cell populations promises to enable studies of cell heterogeneity, inter-cellular communication, and cell population response to changing exogenous factors; yet complex manufacturing of the arrays as well as unpredictable effect of changed surface topology on the cell physiology might hamper wide use of this technique.
Fig. 10.
Mechanical (A–C) and non-mechanical (D–F) routes for intracellular delivery of bio-functional QDs within live cells. A) Microinjection enables intracellular loading of unmodified QD probes along with carrier solution on a cell-by-cell basis. B) Delivery with nanotubes offers precise control over QD delivery location, but requires QD anchoring to nanotubes via reducible linkers. C) High-throughput microinjection via nanosyringe arrays delivers unmodified QDs within large cell population, but changes the surface topology for cell growth. D) QDs functionalized with cell-penetrating peptides might employ endosome-mediated and non-endosomal pathways (depending on the peptide structure), offering flexibility in tuning the QD-cell interaction. E) Pinocytosis enables uptake of unmodified QD probes with consequent cytoplasmic distribution. F) Utilization of active receptor-mediated QD uptake via endocytosis followed by endosomal escape via proton-sponge effect represents a highly efficient non-invasive delivery method with specific targeting capabilities.
Non-mechanical approaches are gaining increasing popularity due to the potential for high-throughput robust QD intracellular delivery with minimal intrusion to cell physiology. Functionalization of QDs with engineered peptides, small versatile biomolecules, might provide great flexibility in tuning the QD interaction with cell components (Fig. 10D).107 Linking of short peptide sequences to the QD surface can be achieved by a variety of methods, including covalent conjugation to existing functional groups,108–110 electrostatic adsorption,79 biotin-streptavidin binding,111,112 and direct coordination to the nanocrystal surface via HIS sequences.76,77 In general, highly cationic peptides facilitate enhanced interaction with the cell membrane and QD internalization, whereas additional targeting moieties govern intracellular distribution. For example, Delehanty et al have modified QDs with HIS-tagged cell penetrating peptide based on the HIV-1 Tat protein motif, achieving efficient internalization of QDs via endocytosis,77 while Rozenzhak et al have added the nuclear localization sequence for nuclear targeting and apoptotic GH3 domain for triggering cell death.112 Other groups have explored similar cationic peptides, such as polyarginine111 and polylysine,113 for achieving cell entry. Despite the versatility of QD-peptide conjugates for labeling of intracellular targets, this approach still suffers from the uncontrolled probe aggregation and lysosomal sequestration inside cells.
Recent work on QD cell uptake and intracellular targeting has focused on employing endocytosis/pinocytosis as a universal delivery mechanism and endosome destabilization/lysis as a cytosolic access route.114 One strategy involves QD cell-loading using osmotic lysis of pinocytic vesicles (Fig. 10E). Efficient uptake is first achieved by inducing pinocytosis by incubation of cells in a hypertonic solution followed by vesicle osmotic lysis and cytoplasmic release by switching to hypotonic medium. Utilization of external control over the osmotic strength of cell medium requires no modification to QD probes and enables uniform loading of intact single QD probes to all cells within the population. For example, Courty et al have utilized this approach to load QD-tagged kinesin motors to living HeLa cells and monitor single-motor movement within the cytoplasm.115 However, drastic change in extracellular conditions is not compatible with QD loading of fragile cells, and external triggering of osmotic lysis might require extensive optimization of procedure due to wide heterogeneity in cell response to changing culture conditions. A more robust approach involves engineering of on-demand endosome- disrupting capacity within the QD probes. To achieve intracellular delivery of unmodified QD probes, Kim et al have utilized 100-nm external biodegradable delivery vesicles made of poly(D, L-lactide-co-glycolide) (PLGA) polymer.116 Functionalization of PLGA surface with antibodies enables interaction with cell surface markers, thus inducing efficient and specific cellular uptake, whereas PLGA charge reversal within low-pH endosomal environment causes membrane destabilization and endosomal escape. Finally, degradation of the polymeric vehicle within the cytoplasm releases the QD payload for specific labeling of intracellular targets. Aiming at containing all functionalities within single QD probes, Duan and Nie have coated QDs with hybrid poly(ethylene glycol)/polyethylenimine (PEG/PEI) polymers producing nanoparticles with a reasonably small HD (15–22 nm) and endosome-disrupting capacity and yet good stability and biocompatibility (Fig. 10F).117 The high amine content of PEI conveys endosomal lysis through the proton sponge effect – buffering of the endosome acidification by the amines of the polymer backbone followed by an increase in counter-ion (mostly chlorine) concentration, build-up of osmotic pressure, and eventually endosome rupture118 – while highly hydrophilic PEG layer provides QD protection from the environment, prevents aggregation, and precludes undesirable non-specific interactions with biomolecules. As a result, when incubated with live HeLa cells, such QDs are internalized, escape from the endosomes, and become distributed throughout the cytosol. However, ligand exchange and direct interaction of PEI with the QD surface necessarily causes an undesirable drop in fluorescence QY and detection sensitivity. Retention and modification of a stable coating should preclude such adverse changes in QD photo-physical properties. In one example, Yezhelyev et al have decorated negatively-charged polymer-coated QDs with tertiary amines, thus producing proton-absorbing QD probes that efficiently achieve intracellular endosomal release while featuring bright fluorescence and good colloidal stability.119
Currently, a wide range of potential techniques for QD intracellular delivery is being developed and perfected. Yet, another major obstacle – inability to remove unbound probes and determine whether QD probes have reached their intracellular targets – still remains largely unexplored. As a result, both the sensitivity and specificity of intracellular labeling suffer from the dependency on the relative number of probes that enter a cell. If too few probes are internalized, incomplete or dim labeling of targets may occur; whereas too many probes might lead to a high degree of background fluorescence and false-positive detection. Thus, effective intracellular labeling requires either active elimination of unbound probes by the cells or utilization of QD sensors that alter the wavelength or intensity of fluorescence signal upon target recognition.
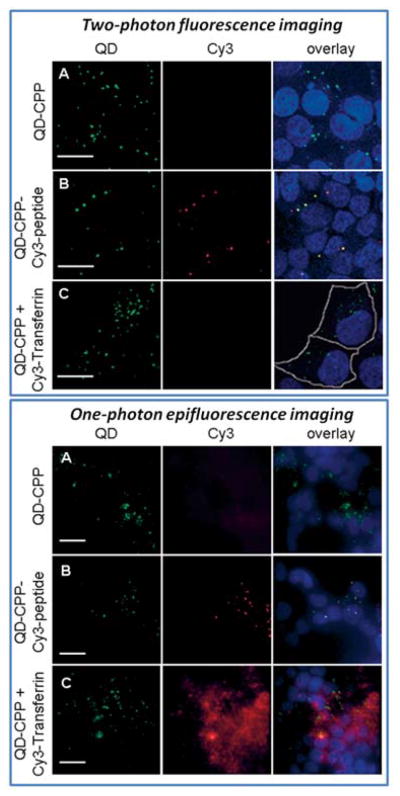
A promising technology for real-time sensing of target recognition is based on the nonradiative energy transfer (FRET) from the QD to acceptor/quencher molecules. In this approach intracellular target binding is accompanied by the change in QD-acceptor proximity and, therefore, fluorescence intensity, thus distinguishing bound probes from the background. In order to achieve efficient FRET sensing, QD probes must feature compact shell/linker structure (allowing sufficient proximity between the QD core and acceptor for nonradiative coupling), offer overlapping emission/absorption spectra for efficient energy transfer, and exploit suitable routes of excitation. Since the QD fluorescence wavelength can be tuned by adjusting the nanocrystal size and/or chemical composition, QD emission spectrum can be precisely matched with the absorption peak of an arbitrary acceptor molecule, ensuring maximum spectral overlap and efficiency of energy transfer. Broad QD absorption profile and large Stokes shift, on the other hand, enable probe excitation by wavelengths of light tens to hundreds of nanometers shorter than the emission peak, reducing non-FRET excitation of acceptor molecules and increasing the signal-to-noise ratio. Furthermore, as QDs are relatively large, multiple acceptor molecules can be attached to their surface for tuning the degree of energy transfer. Therefore, satisfying majority of the design requirements, QDs have been successfully used as FRET probes in a wide variety of sensing schemes detecting conformational changes as well as binding and cleavage events.5 For example, in order to monitor molecular interactions within live cells, McGrath et al have taken advantage of FRET between QD-transferrin probes and dye-transferrin conjugates.120 During receptor mediated endocytosis, dimerization of transferrin receptors results in increased proximity between QDs and acceptor dyes, thus enabling FRET. Yet, accidental non-FRET excitation of acceptor dye was unavoidable with standard single-photon imaging modality in this study. To minimize this artifact, Clapp et al have utilized a two-photon QD excitation route that significantly enhances the signal-to-noise ratio of intracellular FRET (Fig. 11).121 Since QDs have two-photon absorption cross sections several orders of magnitude larger than typical organic dyes, undesirable two-photon excitation (840 nm) of acceptor Cy3 dye is dramatically reduced in comparison to single-photon excitation (488 nm). Despite the promising initial labeling of cell surface markers and study of endocytosis, sensing of intracellular targeting with QD probes still remains to be shown. A number of challenges, such as probe stability, reproducibility of bioconjugation, detection sensitivity, and reliability of image acquisition and processing, need to be addressed before FRET-based QD sensors can be widely adapted for labeling of intracellular targets in living cells.
Fig. 11.
Two-photon (top panel) and one-photon (bottom panel) excitation of QD-Cy3 FRET system. QDs are efficiently excited by both methods, enabling fluorescence of conjugated Cy3 molecules via FRET. However, only two-photon excitation precludes non-FRET excitation of Cy3 dye, whereas conventional one-photon fluorescence imaging produces significant background via direct Cy3 excitation. Reproduced with permission from ref. 121. Copyright 2007 Wiley-VCH Verlag GmbH & Co. KGaA.
Employing active exocytosis of functional QD probes, on the other hand, provides an attractive route for intracellular imaging without the strict requirements of FRET sensing. However, this goal is hard to achieve due to the lack of known efficient exocytosis mechanisms and poor understanding of the intracellular behavior of QDs. The most common approach for achieving QD exocytosis is to incubate cells in starvation medium; yet poor efficiency of QD elimination and significant changes in normal cell physiology introduced by starvation might preclude from the real-time study of physiological processes. Therefore, engineering of not only intracellular delivery and targeting functionalities, but also efficient exocytosis mechanisms within the QD probes will become a major focus of future research in the area of live cell imaging.
4. QD probes for in vivo imaging
Fluorescence in vivo imaging with QD probes promises to greatly expand the capabilities of existing imaging modalities, providing access to high-resolution multiplexed vascular imaging, intraoperative image guidance, real-time cell tracking, and in vivo molecular targeting.122 MRI, CT, PET, and SPECT have become widely used imaging techniques for examination of internal structures, molecular targets, and metabolic processes in vivo. Nevertheless, PET and SPECT (which are based on detection of radioactive labels) suffer from poor spatial resolution, while MRI and CT (which are based on tissue contrast) primarily provide structural information and offer poor sensitivity. Fluorescence imaging with QD probes, on the other hand, can be performed in a multiplexed format with varying temporal and spatial resolution. For example, bulk QD measurements (e.g. in a whole-animal or whole-organ context) can be achieved via fluorescence reflectance imaging,123 whereas high-resolution examination of QD staining is available from intravital fluorescence microscopy124,125 or post-operative histological examination of excised tissues. The versatility of QDs provides vast flexibility in engineering of probes for a variety of in vivo imaging applications – blood circulation time, degradation and excretion routes, specific interaction with biomolecules and cells, and biodistribution along with QD photo-physical properties can be potentially controlled via the probe design based on the needs of a particular application. Implementation of such control, however, is not trivial as behavior of QDs in a highly heterogeneous and aggressive in vivo environment is still poorly explored. Gaining thorough understanding of the interaction between QDs and physiological systems and learning how to manipulate these interactions represent essential milestones towards benefitting from the novel in vivo imaging capabilities featured by the QD probes.
4.1 General design considerations for in vivo QD probes
Impressive progress has been achieved in engineering of bright, stable, and biocompatible probes for live cell imaging. The absence of adverse effects on cell physiology and lack of obvious short-term cytotoxicity encourage further exploration of whether QD probes can be made suitable for in vivo applications. Unlike in vitro applications, where experimental conditions can be strictly controlled by the investigator, the physiological environment presents complex and often unpredictable responses to foreign materials. Therefore, in vivo imaging with QD probes imposes another level of requirements for the probe design, most important of which are: biocompatible and non-toxic nanoparticle coatings with integrated non-fouling functionality for reduced nonspecific interactions in highly heterogeneous biological environments; reliable control over the QD biodistribution, degradation, and excretion pathways for reduced toxicity; and applicability of QD probes in non-invasive or minimally invasive intravital imaging for long-term observation of QD dynamics in vivo.
Development of novel and optimization of existing intravital imaging techniques is primarily governed by the specific application requirements and, thus, will be discussed in details later. The more important issue of QD biocompatibility and biodistribution (and associated potential toxicity), on the other hand, is relevant to all QD-based in vivo applications. Great variability in the structure and composition of the semiconductor core, particle coating, and biomolecular functionalities impede systematic investigation of QD interaction with biological systems and modes of toxicity.126–128 While early studies have shown severe cytotoxicity originating from photo-oxidation of unprotected CdSe QDs and release of Cd2+ ions,129,130 later in vitro and in vivo experiments with protected nanoparticles have not uncovered significant QD-associated adverse changes. Within different reports, potential QD toxicity has been attributed to size-based effects,100,102 Cd2+ release,129,131,132 and ROS (reactive oxygen species) induced oxidative stress.133–135 None of these mechanisms can be universally applied towards characterization of all existing or newly developed QD probes. Yet, the potential accumulation of QDs within the body and release of toxic Cd2+ ions seems to be the most prominent concern within the scientific community. Therefore, elucidation of mechanisms for preventing in vivo QD accumulation and degradation has become a priority in bio-nanotechnology research (Fig. 12).
Fig. 12.
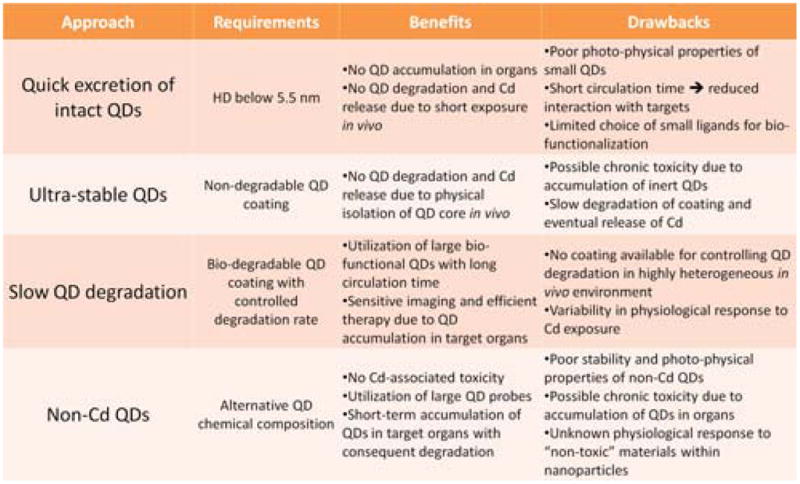
Potential routes for elimination of Cd-associated QD toxicity.
Probably the safest and most desirable approach to addressing the toxicity issue is engineering of QD probes that are quickly and completely eliminated from the body via renal or bile excretion pathways without triggering uptake by the reticulo-endothelial system (RES) and avoiding degradation pathways. This approach seems especially favorable in light of sparse information on in vivo QD degradation mechanisms and long-term effect of QD accumulation in organs. Systematic investigation of QD biodistribution performed by Choi et al in mice has identified a nanoparticle hydrodynamic size renal clearance threshold of 5.5 nm.58 When delivered systemically, small cysteine-coated QDs are readily excreted into the bladder with minimal accumulation in the liver (4.5%) and kidneys (2.6%), whereas larger particles exhibit significant liver uptake (26.5%). Further, the importance of non-fouling zwitterionic surface coatings in inhibiting the protein absorption and retaining the original nanoparticle hydrodynamic size has been emphasized. In contrast, QDs featuring charged surface undergo serum protein adsorption and increase in HD to more than 15 nm. Working towards preparation of compact QD probes, Law et al have synthesized ultrasmall (3–5 nm in diameter) cysteine-coated CdTe/ZnTe QDs and tested biodistribution of these probes in mice, finding no QDs in liver and spleen 2 weeks post- injection,59 whereas Choi et al have employed cysteine-coated CdSe(ZnCdS) QDs functionalized with cyclic-RGD peptide for tumor targeting in mouse model and observed complete elimination of 65% of injected QDs within first 4 hours, while detecting specific tumor labeling (with tumor-to-background ratio of 6.9).136 However, the few ultrasmall QDs currently available suffer from poor photo-physical properties, while preparation of better protected and bio-functionalized probes often increases the QD size, thus making renal clearance difficult. Furthermore, quick renal clearance is often undesirable, as prolonged QD circulation is required for specific targeting, high-sensitivity imaging, and therapeutic potency. Therefore, high molecular weight coatings are routinely applied to QD probes to increase their circulation time and improve bioavailability. Ballou et al have emphasized the importance of coating with high molecular weight PEG to reduce rapid clearance of QDs by liver and bone marrow,137 and Prencipe et al have achieved remarkably long blood circulation of nanomaterials encapsulated with branched PEG.138 Meanwhile, high doses of particles with positively charged amine or negatively charged carboxyl groups have been shown to initiate coagulation cascades resulting in pulmonary thrombosis and death.139 Therefore, utilization of stimulus-responsive biodegradable ligands for dynamic tuning of the QD size might become a promising design route for future in vivo probes. Such ligands would ensure prolonged QD blood circulation and/or interaction with target cells, while eventually detaching from the QD surface and releasing single nanoparticles with original size below 5.5 nm capable of efficient renal excretion.
In some cases complete elimination of QD probes from the body via renal excretion or other means might prove challenging or undesirable. Engineering of ultra-stable QDs encapsulated with inert biocompatible materials might prove helpful in this situation. If QD integrity within a human body can be retained for many years, biological systems might never be exposed to heavy metal components of the QD core. For example, Ballou et al have indicating that intact PEG-coated QDs remain in bone marrow and lymph nodes of mice for several months after injection,137 yet Fitzpatrick et al have detected signs of QD degradation (spectral blue-shifting) over a course of two years.140 While organic coatings, such as polymers and lipids, might still degrade due to exposure to biological environment, utilization of more stable inorganic materials should protect the cores of QD probes for extended periods of time. Alternatively, as cadmium poisoning results from a quick release of large amounts of this metal into a bloodstream, its preferential accumulation in kidneys, and consequent nephrotoxicity, slow degradation of QD probes within a human body followed by urinary excretion might offer a way of safe and efficient elimination of QD components over a long term. Since up to 30 ug/day of dietary Cd (coming from fish, vegetables, and other sources) can be consumed by a healthy adult without adverse effects on kidney function,141 this approach seems to be feasible. Adapting technology developed for controlled drug release towards QD encapsulation with biodegradable polymers might provide one way of achieving control over QD in vivo degradation.
Despite the proven potential for QD excretion or stable shielding, increasing safety concerns urge for complete elimination of toxic components from in vivo probes and design of biocompatible and non-toxic QDs. In one approach, Yong et al have prepared Cd-free InP/ZnS QDs and utilized these probes for targeting of pancreatic cancer cell lines;142 however, low QY (~30%) and large size (~30 nm in diameter) might limit utility of such probes for in vivo imaging. Higher-quality probes with QY of up to ~60% and HD of 17 nm have been developed by Li et al on the basis of CuInS2/ZnS QDs.143 Further, engineering of low-cost, non-toxic, and potentially biodegradable in vivo imaging probes might become available through utilization of recently developed technology for preparation of water-soluble QDs made of silicon144,145 – an inert, biocompatible, and abundant material. However, while being an attractive approach, Cd-free QDs still suffer from poor stability and inferior photo-physical properties compared to high-quality QDs made of toxic materials (such as CdSe). Therefore, improving biocompatibility of potentially toxic QD probes remains a sound and highly promising alternative, and elimination or reduction of cadmium interaction with live cells seem to be the cornerstone of such approach.
4.2 Vascular imaging
One of the most common in vivo applications of QDs is fluorescence contrast imaging of the blood vasculature and lymphatic drainage system.146 Intravenously injected QDs can highlight morphological abnormalities in vasculature, model biodistribution of nanoparticle-based drug delivery vehicles, and monitor the blood circulation dynamics, whereas intradermally delivered QDs can map the lymphatic basins along with sentinel lymph nodes (SLN) and uncover disease-related transport mechanisms (e.g. tumor metastasis pathways). Furthermore, the multicolor nature of QD probes makes it possible to investigate separate vascular systems in a multiplexed manner, providing insight into the intricate blood and lymph circulation networks within organs and tissues. In clinical practice, the ability to map tumor vasculature and lymphatic drainage pathways might not only enhance the accuracy of diagnostics, but also provide intraoperative image guidance for more effective and less invasive tumor and lymph node resection. Therefore, real-time vascular imaging with QDs has the potential to improve our understanding of vasculature-related physiological and pathological processes as well as advance clinical diagnostics and therapy.
With such a great potential, this application requires virtually no additional surface engineering of QD probes satisfying general requirements for in vivo use, as no extravasation, organ selectivity, cellular uptake, and specific target binding are necessary. However, prolonged circulation and enhanced stability in physiological conditions are often desirable for reliable data collection and long-term monitoring of probe biodistribution. Therefore, the major design focus in engineering of QD contrast agents for vascular imaging should be placed on synthesis of non-fouling and possibly biodegradable coatings that will efficiently protect the QD core, evade RES uptake and renal filtration for the duration of experiment, and then enabling eventual particle degradation and excretion. Further, engineering of fluorescence imaging systems suitable for deep-tissue in vivo imaging will be indispensable for the success of QD-based angiography.
Pioneering studies done by Larson et al have demonstrated the value of QD probes for the dynamic imaging of the blood vasculature of skin and adipose tissue in live mice.147 The relatively large size and high stability of polymer-encapsulated QDs have provided bright and persistent fluorescence contrast after intravenous injection. Performing line scans across capillaries and monitoring the propagation of QD fluorescence, the investigators have been able to measure blood flow velocities. At the same time, the large two-photon excitation action cross-section of QD probes has enabled nearly background-free vasculature imaging at tissue depths of several hundred microns with two-photon fluorescence intravital microscopy (Fig. 13). However, the surface coating used in this study was not specifically designed for prolonged QD blood circulation, and the fate of QD probes was not investigated. Ballou et al have systematically studied the effect of additional PEG coating on the circulation half-life and biodistribution of polymer-coated QDs using whole-animal real-time fluorescence reflectance imaging.137 QDs decorated with long-chain methoxy-PEG have shown significantly longer circulation half-life compared to non-modified QDs; however, the PEG shell has failed to significantly reduce the RES uptake and sequestration of particles within liver and spleen, thus limiting the blood circulation to only few hours. Moreover, extravasation of QD-PEG probes into surrounding tissues has been observed even for large particles, which might result from the non-specific interaction between QDs and endothelial cells and cause increased fluorescence background detrimental for dynamic vascular imaging. Yet, even a shorter blood circulation time is often sufficient for detailed vascular imaging. In one example Stroh et al have combined two-photon intravital microscopy, blue-emitting QDs encapsulated in PEG-phospholipid micelles, and a transgenic mice model with GFP-expressing perivascular cells to study the morphology of the tumor vasculature.148 Following systemic administration, QDs highlight the vessel boundary providing a clear picture of tumor vessel morphology while resisting extravasation for at least 30 minutes, whereas GFP fluorescence indicates the distribution of perivascular cells. Poor QD extravasation has been employed by Kim et al for studying the patho-physiology of viral infection of the central nervous system in mice.149 Using intravital two-photon microscopy, QD extravasation from brain microvasculature has been monitored as a measure of disease-associated vascular injury and blood-brain barrier breakdown.
Fig. 13.
In vivo imaging of blood vasculature with QDs. Large QDs remain within the blood vessels providing good image contrast (A), whereas FITC-labeled dextran quickly extravasates creating high background (B). Two-photon microscopy enables deeper-tissue imaging with QDs, highlighting not only the superficial vasculature (C), but also capillary network up to 250 μm deep within the tissue (D). Reprinted from ref. 147 with permission from AAAS. Copyright (2003).
Initial studies on QD-based blood vasculature imaging outline the numerous beneficial features of QD probes for this application as well as emphasize the urge for novel “stealth” coatings that would efficiently prevent interaction with biomolecules, recognition by the immune system, and extravasation, thus improving the probe circulation half-life and imaging accuracy. In addition, future coatings might feature controlled biodegradation functionality, enabling disintegration of bulky QD probes into smaller components that could be safely eliminated from the bloodstream via renal filtration.
Engineering of QD contrast agents for lymphangiography and lymph node mapping is governed by less strict and somewhat different design principles. Unlike probes for blood vessel imaging, QDs need to be small enough to get transported from interstitial space into lymph vessels, and yet large enough to be trapped in lymph nodes (in general particles with HD 5–50 nm are retained). However, neither the particle size (within 20–50 nm range) nor the surface charge has shown significant effect on the SLN mapping, providing more flexibility for probe design.150 More importantly, this is probably the only in vivo application where QD long-term toxicity and excretion routes do not present a major concern, as labeled SLNs and tissues are often removed during surgery
In an early demonstration of the clinical potential for real-time intraoperative imaging, NIR-QDs (emitting at 840–860 nm) coated with oligomeric phosphine have been injected intradermally either into the paws of mice or into the thighs of pigs and monitored with combined IR/visible reflectance videography.151 Intermediate size (15–20 nm HD) enables efficient lymphatic transport and accumulation of QDs within the SLNs, preventing further leakage to lymphatic system (Fig. 14). Importantly, labeled tissue can be clearly visualized through the skin before an incision is made, during surgery, and after node resection. In a follow-up study this technique has been utilized for mapping SLNs and identifying lymphatic drainage pathways from the lung tissue in pigs with 100% accuracy (as assessed by the conventional isosulfan blue and radioactive isotope labeling).152
Fig. 14.
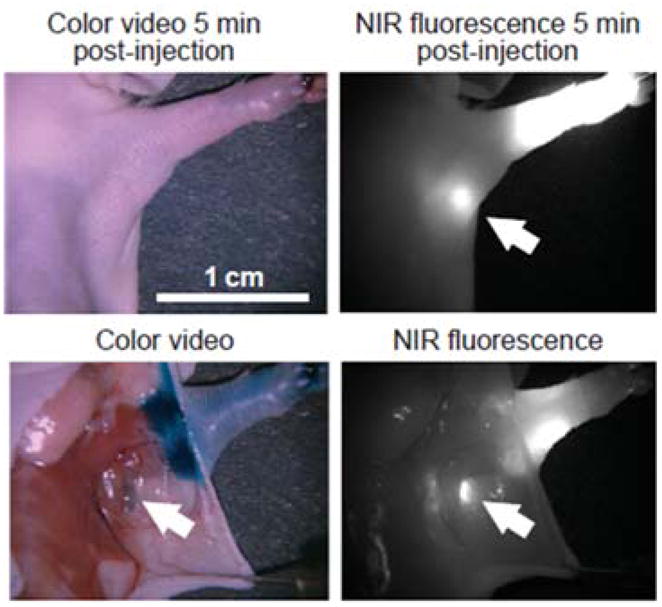
Sentinel lymph node mapping with NIR QDs. Intradermally injected QDs efficiently accumulate in SLN, enabling SLN visualization through the skin and image-guided lymph node resection. Reprinted by permission from Macmillan Publishers Ltd.,151 copyright (2004).
Aiming to image the lymphatic system beyond the SLNs, Zimmer et al have employed small (HD < 10 nm) QDs with tunable emission from 694 to 812 nm for sequential mapping of up to 5 lymph nodes following subcutaneous injection (Fig. 15).153 To achieve NIR emission from such small particles InAs/ZnSe core/shell composition (instead of common Cd-based QDs) and compact DHLA-PEG coating have been used. While decoration with short 8-unit PEG molecules increases the QD size in buffer (from 5.3 to 8.7 nm), it efficiently resists non-specific binding of proteins, thus retaining compact probe dimensions in biological environment. Labeling of distant lymph nodes beyond SLNs has also been observed for much larger polymer-coated QDs following direct injection into tumors in live mice, yet the mechanism has been attributed to bypass routes rather than to QD escape from SLNs.150 Recently, the multiplexing capability of QDs has been exploited for in vivo imaging of 5 different lymphatic basins in mice (Fig. 16). Following intracutaneous injection of 5 types of polymer-coated carboxy-QDs ranging in emission wavelength from 565 to 800 nm (HD 15–19 nm) into the paws, ears and chin of mice, Kobayashi et al have monitored the transport of QDs through lymphatic networks and accumulation in SLNs.154 Further passage of QD probes to secondary draining lymph nodes was significantly inhibited, possibly due to non-specific binding between negatively charged QD coating and proteins resulting in an increase in probe size.
Fig. 15.
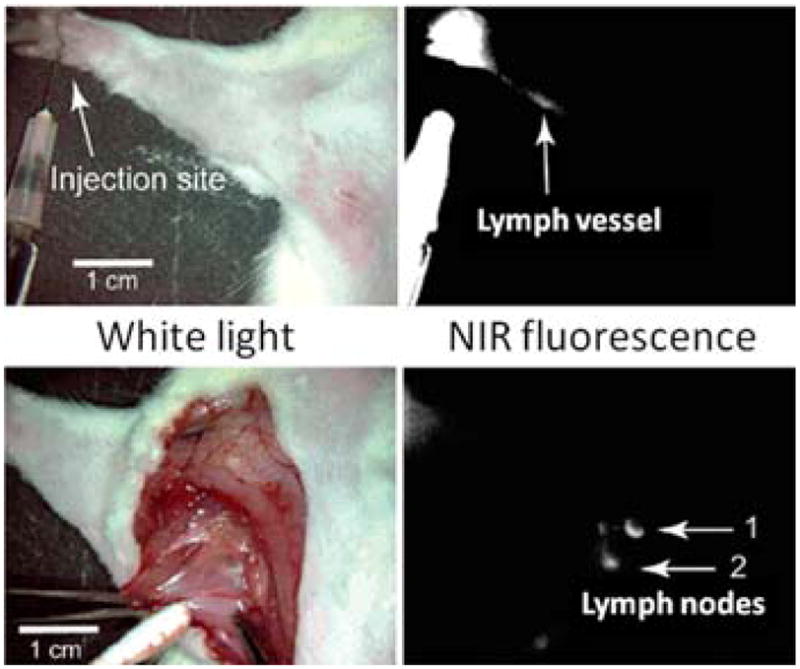
Sequential mapping of several lymph nodes with compact QDs. Small size enables QD probes to escape SLN and travel along the lymphatic system to distant lymph nodes. Reprinted with permission from ref. 153. Copyright 2006 American Chemical Society.
Fig. 16.
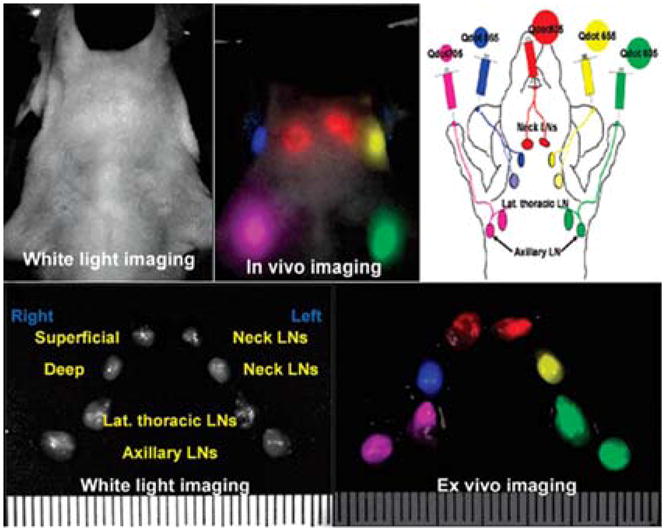
Multiplexed in vivo and ex vivo imaging of separate lymphatic networks with QD accumulation in SLNs. Reprinted with permission from ref. 154. Copyright 2007 American Chemical Society.
Cardiovascular and lymphatic angiography have been two of the most successful QD-based in vivo imaging applications. In combination with fluorescence reflectance imaging, QDs highlight macroscopic structures on a whole-animal or whole-organ scale and serve as visual tags for image-guided surgery; two-photon intravital microscopy provides high-resolution examination of superficial vessels and their surrounding tissues; and emerging advanced imaging techniques, such as multiphoton microscopy with a needle-like gradient index lens for deep-tissue imaging,155 promise to enable detailed studies of intact vasculature deep within organs. Yet, further translation of this technology into clinical practice will heavily depend on engineering of non-toxic, non-fouling, and biodegradable QD coatings as well as stable and bright QD cores.
4.3 In vivo cell tracking
Cell behavior heavily depends on the physical and chemical cues received from the local environment. Simulation of such cues with in vitro live-cell imaging studies is often limited by the inability to accurately reconstruct the complex physiological conditions, while observation of cells in their native niche in vivo is hampered by poor resolution and sensitivity of conventional imaging techniques. Even though fluorescent dyes and proteins have enabled visualization of static cancer and stem cell distributions as well as short-term cell dynamics studies,156 quick photobleaching, poor image contrast in high fluorescence background conditions, and often intrusive genetic modifications of cells greatly diminish the potential of in vivo studies over extended spatiotemporal intervals. QD probes, on the other hand, are well-suited for intracellular tagging and long-term cell tracking required for monitoring of cell migration during development, stem cell differentiation, immune system activation, oncogenesis, and tumor metastasis. For example, Dubertret et al have shown that QDs could be microinjected directly into Xenopus embryos in order to perform continuous cell lineage tracing over the course of embryonic development up to the tadpole stage, while exhibiting no evident toxicity or interference with cell division.70
Engineering of QD tags for in vivo cell tracking is based on two major design principles – efficient non-intrusive cell loading of QDs (either ex vivo or in vivo) and sensitive intravital fluorescence imaging – in addition to satisfying the general requirements for in vivo QD probes. Specific strategies for achieving QD cell loading ex vivo discussed in section 3.3 have been successfully used in the majority of cell tracking reports due to relative simplicity of procedure and availability of established cell transfection techniques. Preloaded cells can be easily introduced in vivo either via systemic administration or locally depending on the application. Yet, extraction of cells from the body for ex vivo loading might be labor-intensive and inefficient, while manipulation with cells under artificial environment might alter cell physiology. Therefore, in vivo QD cell loading techniques have recently been developed to address this issue. For example, Slotkin et al have described two novel methods for QD loading of neural stem and progenitor cells in developing mouse embryos: ultrasound-guided biomicroscopy injection, in which carboxy-QDs are directly infused into embryo brains under the control of ultrasound imaging, and in utero electroporation, in which QDs are injected intracerebrally, followed by the application of three 33-volt electrical pulses.157 Embryos appeared to develop normally post-labeling, while QD-tagged cells were able to differentiate and migrate. In another example, Jayagopal et al have systemically administered antibody-functionalized QD probes for labeling of neutrophils and leukocytes in order to continuously visualize cell dynamics (including rolling, adhesion, and extravasation) within the retinal vasculature of rats for over an hour.158 However, extracellular labeling with bulky QD-antibody probes might be only appropriate for short-term tracking of blood cells due to hampered extravasation and poor QD anchoring onto the cell surface. Despite current technical limitations, in vivo QD cell loading represents a highly promising technology for specific multiplexed tagging of cells within the physiological environment. Future advances in this field will likely employ compact QD probes with stealth coatings for efficient extravasation, non-immunogenic targeting ligands for specific cell recognition, and ligand-mediated active uptake of QDs for robust cell tagging.
Sensitive and minimally invasive intravital fluorescence imaging is another important component of QD-based in vivo cell tracking. Visible light is efficiently absorbed and scattered by tissues, thus severely limiting the depth of fluorescence imaging. Yet, similar to vascular imaging described above, several established and newly developed techniques can be successfully applied for cell tracking as well. In an early example, Voura et al have studied metastatic tumor cell extravasation into the lung tissue using DHLA-coated QDs for cell tagging and two-photon emission-scanning microscopy for post-mortem examination of excised tissue specimens.159 With the aid of lipofectamine, five groups of cells have been loaded ex vivo with QDs ranging in emission from 510 to 610 nm and then intravenously injected into live mice. High-resolution imaging of whole-mounted mouse lungs have enabled clear spectral separation of individual QD signals from each other and from tissue autofluorescence, thus facilitating study of interaction between different tumor cell populations within the same animal. However, the “snap-shot” nature of this approach conceals the dynamics of cell migration. Gao et al, on the other hand, have demonstrated the noninvasive whole-animal imaging of subcutaneously injected QD-tagged cancer cells in live mice using fluorescence reflectance imaging (Fig. 17).71 TAT or polylysine internalization peptides conjugated to the particle surface enable millions of orange QDs to be introduced into cancer cells ex vivo without affecting their ability to grow into tumors, while the high QD brightness facilitates clear visualization of implanted tumors over the background autofluorescence. In another example, Stroh et al have utilized two-photon intravital microscopy for tracking the transport of bone marrow-derived progenitor cells through tumor vessels in real time (Fig. 18).148 Cells loaded with TAT-functionalized orange-emitting QDs have been injected into the carotid artery of live mice along with blue-emitting QDs for vessel contrast. As two spectrally distinct QD types can both be excited by 800 nm two-photon light, simultaneous highlighting of tumor vasculature and continuous cell tracking at ~1 frame/second are possible. A 3-dimensional tracking of individual transplanted haematopoietic stem cells has been recently achieved by Lo Celso et al (Fig. 19).160 While fluorescent dyes have been used for cell labeling and QDs have been only utilized for microvasculature imaging, this technique can be readily expanded to QD tagging of cells and multiplexed high-resolution 3D cell tracking, thus providing access to real-time dynamic studies of cell interaction within physiological niches.
Fig. 17.
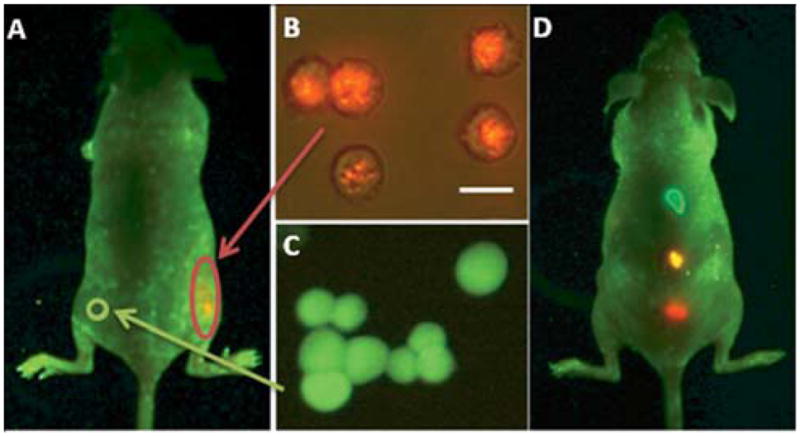
In vivo imaging of implanted QD-tagged tumor cells. A) Bright QD tags (B) enable visualization of tumor cells through skin with a non-invasive whole-animal fluorescence imaging, whereas organic dye (C) signal is indistinguishable from autofluorescence. C) Imaging of subcutaneously implanted QD-loaded microbeads shows the potential for multiplexed in vivo cell detection and tracking. Reprinted by permission from Macmillan Publishers Ltd.,71 copyright (2004).
Fig. 18.
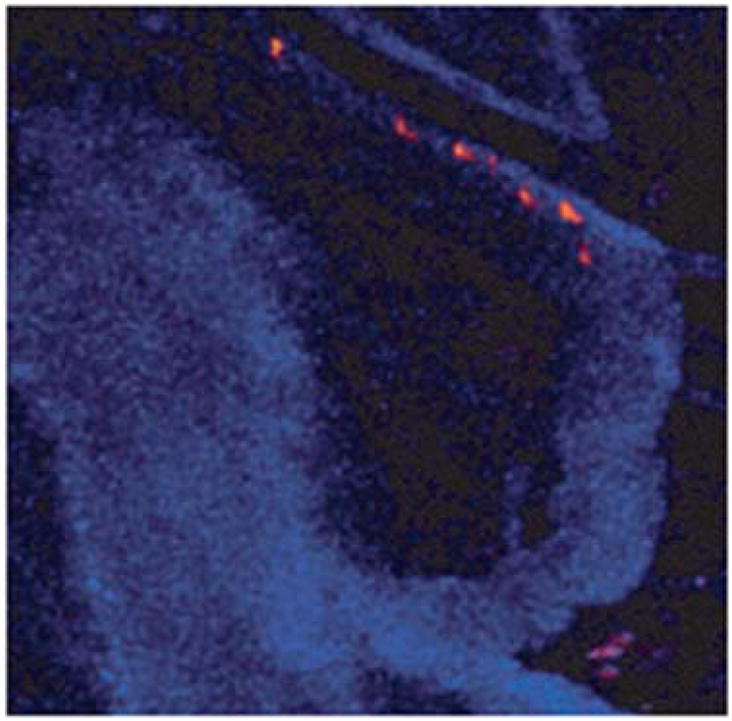
Intravital tracking of a single bone marrow-derived progenitor cell. Seven images taken with 1 second intervals are superimposed to show the movement of a QD-labeled cell (red) through a tumor blood vessel. Vasculature is highlighted with blue QDs. Reprinted by permission from Macmillan Publishers Ltd.,148 copyright (2005).
Fig. 19.
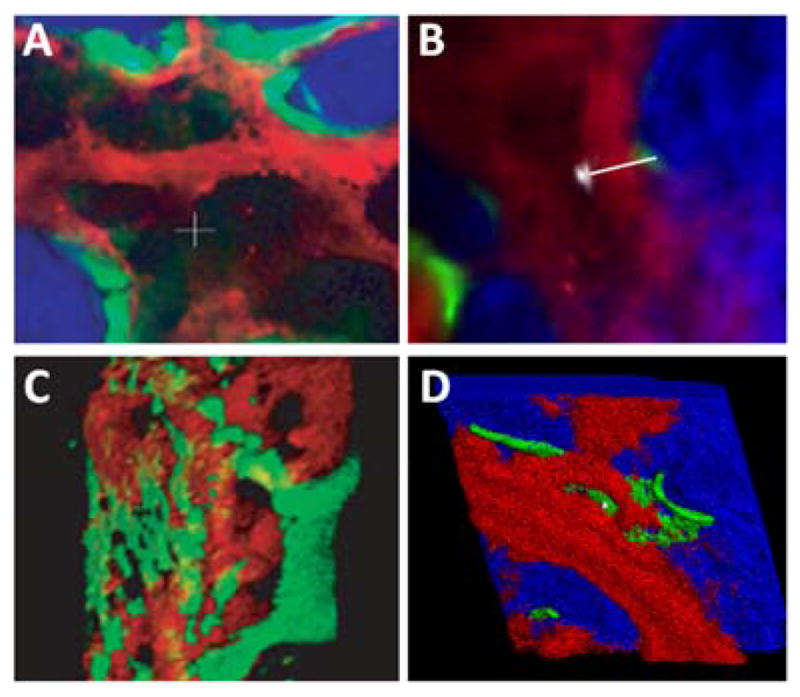
3-dimensional tracking of dye-labeled haematopoietic cells (white) within bone marrow. QDs outline the vasculature, while bone collagen is visualized with second harmonic generation (blue) and osteoblasts - via GFP fluorescence (green). 3-D reconstruction enables detailed study of bone marrow structure and precise localization of cells within their niches. Reprinted by permission from Macmillan Publishers Ltd.,160 copyright (2008).
Even in light of the promising achievements in QD-based cell tracking, the shallow depth of fluorescence imaging and significant tissue autofluorescence either limit cell tracking to subcutaneous layers or rely on intraoperative imaging if tracking within deeper tissues is required. To address this issue and improve the signal-to-background ratio of in vivo imaging So and coworkers have developed self-illuminating QD-luciferase probes for non-invasive fluorescence imaging.161 After intravenous injection of QD-loaded glioma cells, accumulation of cells within lungs is readily observed via fluorescence of QDs excited by bioluminescence resonance energy transfer process, while traditional fluorescence spectral imaging utilizing external excitation source fails to detect QD emission due to attenuation of short-wavelength excitation light and strong tissue autofluorescence (Fig. 20). However, utilization of potentially immunogenic components and requirement for supplying the substrate coelenterazine put limitations on utility of this technology for in vivo cell tracking.
Fig. 20.
In vivo imaging of self-illuminating QD probes. QDs excited via bioluminescence resonance energy transfer from conjugated luciferase are clearly visible with non-invasive whole-animal imaging (A), whereas the signal from same probes illuminated by an external short-wavelength excitation source is indistinguishable from the strong tissue autofluorescence (B). Reprinted by permission from Macmillan Publishers Ltd.,161 copyright (2006).
Steady advancements in ex vivo QD cell loading, promising initial results of in vivo QD cell tagging, and development of sensitive in vivo fluorescence imaging techniques suggest that 3-dimensional multiplexed in vivo cell tracking for the study of dynamic cell migration phenomena might become available in the near future. Achieving this objective will require engineering of novel QD surface ligands for improved cell targeting, biocompatibility, and uptake in vivo as well as employing novel intravital imaging modalities for more sensitive deep-tissue QD detection.
4.4 In vivo targeted molecular imaging
QD-based fluorescence molecular imaging represents an attractive technique for the detection of specific biomarker-expressing cells in vivo. While being simple and inexpensive in comparison to other targeted molecular imaging modalities, it provides a powerful tool for studying complex physiological phenomena (e.g. activation of immune response), detecting diseased cells and tissues (e.g. tumors), and evaluating the pharmacokinetics and biodistribution of targeted nanoparticle-based drug delivery vehicles in a whole-animal context. As discussed in previous sections, QDs have been successfully used as probes for in vitro molecular profiling of cells and tissues and as cell tags and vasculature contrast agents for in vivo imaging. However, QD-based targeted molecular imaging in vivo remains a significant challenge due to the lack of specificity in heterogeneous physiological conditions and pervasive foreign-particle clearance mechanisms. Therefore, engineering of specific targeting functionalities that efficiently exploit passive and active targeting pathways (Fig. 21), while not compromising the biocompatibility, extravasation, and blood circulation half-life of QD probes, represents the major focus of ongoing research.
Fig. 21.
Routes for in vivo QD targeting. A) Abnormal highly permeable tumor blood vasculature and poor tumor lymphatic drainage enable passive QD targeting and accumulation within the tumor via enhanced permeability and retention effect. Similar mechanism can be employed for labeling the regions with damaged or abnormal vasculature. B) Intact endothelium represents a significant barrier for QD extravasation. Labeling of biomarkers specifically expressed on the vasculature of a particular organ or tissue (e.g. tumor site) provides an efficient way of targeted in vivo imaging with bulky QD probes. C) When QD extravasation is possible, active targeting of biomarkers expressed on the cell surface enables specific labeling deep within the tissues of interest, while reducing non-specific QD accumulation in non-targeted areas.
In a pioneering study performed by Akerman et al, green and red MAA-coated QDs coupled to peptides with affinity for lung endothelium and tumor vasculature have been intravenously injected into live mice.162 Post mortem evaluation of tissue sections revealed remarkably specific QD targeting. Certainly, utilization of unstable and dim QD probes might not satisfy current requirements for in vivo QD probes, but the idea of using targeting peptides for preparation of compact and biocompatible probes holds a great potential, as long as enzyme-mediated peptide degradation and possible immunogenicity are carefully evaluated. For example, Cai et al have used polymer-encapsulated NIR CdTe/ZnS core/shell QDs functionalized with cyclic RGD (arginine-glycine-aspartic acid) peptides for targeting integrin αvβ3 (a biomarker up-regulated in cancerous tissue during proliferation, metastasis, and angiogenesis) following systemic administration in tumor-bearing mice.163 Specific in vivo tumor labeling was clearly detectable with a whole-animal hyper-spectral imaging. However, significant RES uptake of QD probes even with a 2000 MW PEG spacer calls for thorough investigation of the underlying cause of interaction with immune system and demonstrates the need for further improvements in QD surface functionalization.
Functionalization of QDs with antibodies or antibody fragments represents an alternative strategy for preparation of targeted in vivo QD probes. Utilization of antibodies is probably more straightforward compared to targeting peptides, as an extensive library of specific antibodies is widely available. However, significant increase in probe size and the potential immunogenicity of foreign proteins puts this approach under scrutiny. For example, Gao et al have successfully utilized bulky PEG-coated QD-antibody conjugates for detection of human prostate tumors grown in mice with non-invasive whole-animal fluorescence spectral imaging.71 Employing both passive targeting (i.e. accumulation of QDs within the tumor via enhanced permeability and retention effect) and active targeting (i.e. QD binding to tumor cells with antibodies against prostate-specific membrane antigen, PSMA), QDs efficiently labeled the tumor tissue. Notably, particles solubilized by an amphiphilic triblock copolymer and modified with PEG have demonstrated remarkable stability in physiological conditions (with circulation half-life of 5–8 hours). However, poor extravasation characteristics limit the use of large QD probes primarily for imaging of tumors with leaky vasculature.
Immunogenicity of targeting antibodies has been emphasized by Jayagopal et al.158 In order to prevent phagocyte recognition and uptake of IgG fragment-conjugated QD probes when administered in immune-competent rats, the authors have passivated the Fc domains on the QD surface with anti-Fc F(ab)2 fragments. Recently, single-chain Fv antibody fragments with high affinity for EGF receptors have been conjugated to QDs for specific labeling of pancreatic tumors.164 These ligands eliminate the problem of Fc binding and uptake in vivo and can be conjugated to nanoparticles at higher surface densities than whole antibodies due to their relatively small size (~26 vs. ~160 kDa). While this approach is not as accessible as targeting with whole antibodies and care is required to preserve the specificity of antigen recognition during Fv fragment preparation and conjugation, engineering of compact non-immunogenic QD probes remains highly attractive.
One of the major advantages of QDs as in vivo molecular imaging probes is their compatibility with both whole-animal imaging as well as high-resolution intravital microscopy modalities, thus permitting examination of biological processes at multiple length scales in live animals. In a whole-animal context QDs can highlight diseased tissue and organ uptake, while at the cellular level single particles can be tracked as they are transported through tissue and interact with molecular targets on the surface of cells. For example, single-particle tracking in vivo has also been demonstrated by Tada et al through the use of high resolution 3-D confocal real-time intravital microscopy technique.165 QDs conjugated to trastuzumab (a monoclonal antibody against the HER2/neu receptor) have been visualized within HER2-overexpressing human breast cancer tumors grown in mice. Following injection through the tail vein, vascular transport, extravasation, specific binding, and cellular internalization of single QD probes can be followed over the course of 24 hours. Specific interactions between probes and target cells have been observed in real time at a sampling rate of 30 frames/sec with 30 nm spatial resolution, and particle trajectories have been quantitatively analyzed in order to determine six rate-associated stages of nanoparticle transport.
QD-based in vivo targeted molecular imaging has been demonstrated with different targeting ligands and QD coatings, at different spatial resolutions and time scales, and for different target localizations. With all the heterogeneity of used probe designs, it is still not clear which route will yield the best QD probe for this application. Yet, the likely candidate probe will feature outstanding biomimetic capabilities, compact size, improved imaging depth, and small, stable, and non-immunogenic targeting ligands. Having achieved this, further probe development will address the long-term fate of QDs within the body and explore the possible degradation and excretion mechanisms.
5. Engineering of multifunctional nanodevices
The emerging field of nanomedicine seeks to revolutionize medical diagnostics and therapy through the development of multifunctional nanodevices. Recent advancements in the engineering of QD probes and promising benefits this technology can bring have dictated a shift of focus from the synthesis of single-component probes towards the design of complex nanostructures composed of multiple targeting, imaging, and therapeutic modules. For example, QDs integrated with MRI contrast agents or radionuclides can be used for dual-mode imaging, whereas when combined with drugs or nucleic acid therapeutics, QDs can serve as traceable delivery vehicles. Like single-component QD probes, these nanocomposites can be potentially targeted to specific disease biomarkers using antibodies, affinity peptides, or aptamers. In general, QDs are used as universal scaffolds for the attachment of extra components and targeting ligands due to their large surface area and modular surface chemistry. At the same time, nanoparticles are small enough to be incorporated into larger delivery vehicles, which have higher loading capacity and can be used for multistage targeting and payload release. The numerous combinations in possible composition and structure of QD-based multifunctional nanodevices make systematic analysis and formulation of generic design principles challenging. While the general and application-specific criteria outlined in previous sections remain relevant, a number of additional nuances related to extra components and expanded probe functionality must be addressed on a case-by-case basis.
5.1 QD probes for multimodal imaging
Fluorescence imaging with QDs features high sensitivity and resolution, multiplexed and real-time detection, and quantitative analysis. With such a degree of utility, QDs greatly expand the capabilities of MRI and PET probes highly suitable for non-invasive in vivo imaging. For example, QD-based nanocomposites containing magnetic nanoparticles (MNPs) and gadolinium (Gd) chelates are useful for combined fluorescence/MRI imaging, while probes containing radionuclides enable fluorescence/PET imaging. With utilization of integrated imaging techniques, such nanocomposites synergistically combine the strengths of individual modalities, facilitating the correlation of images with different resolution and tissue/molecular specificity. Among these, fluorescence imaging can be especially useful for post-operative histological evaluation of excised tissues or for intraoperative image guidance,166 which are not possible using MRI or PET imaging agents alone. Enhanced functionality of nanocomposite probes for multimodal imaging promises to help clinicians to better understand, diagnose and treat diseases, such as cancer167 and cardiovascular disorders,168 and allows for accurate collection and analysis of structural and metabolic data.169,170 For this application, QDs are unique from other fluorophores in their high brightness and photostability, which permit long-term continuous imaging in photon-limited conditions, along with modular surface chemistry, which enables conjugation of QDs with additional imaging components. The design of nanodevice architectures that preserve the imaging capabilities of individual components (especially the QD fluorescence), while featuring compact structure with desirable surface chemistry and targeting functionality represents the major focus of current research in the area of multimodal imaging.
QDs and MRI contrast agents have been incorporated in several types of multimodal imaging probes including hybrid fluorescent/superparamagnetic nanocrystals,171–173 polymeric nanobeads,174,175 and lipid-based nanocomposites.176,177 Due to their high relaxivity, MNP-based imaging probes are well suited for high-contrast MRI imaging.170 Moreover, MNPs are efficiently attracted by the magnetic field, thus facilitating quick separation of probes from complex solutions (e.g. blood). However, potential quenching of QD fluorescence by closely located MNPs represents a major concern. For example, superparamagnetic MNP-QD heterostructures prepared by either growing a semiconductor shell on top of the magnetic core (Fig. 22A)171 or by growing a separate QD attached directly to the magnetic core172 show relatively poor fluorescence properties (QY 2.3–9.7% for blue-emitting core/shell particles and 38% for red-emitting dimmers). Therefore, functionalization of QDs with alternative MRI agents or utilization of rigid linkers physically separating the QD and MNP cores to reduce quenching might yield brighter probes.
Fig. 22.
Engineering of QD-based nanocomposites for dual-modal imaging. Fluorescence/MRI probes can be prepared by synthesizing QD-MNP heterostructures (A) or incorporating QDs and paramagnetic compounds (e.g. Gd chelates) either within larger nanostructures (B) or on a single-QD platform (C). Fluorescence/PET probes can be made by functionalizing the QD surface with radionuclides (e.g. 64Cu) along with other targeting and therapeutic moieties (D). Plasmonic fluorescent probes incorporate gold nanoparticles and nanospheres either directly attached to the QD surface or grown around the QD in the form of shell (E).
In an alternative approach, QDs and chelated Gd contrast agents have been incorporated within the polymer-based nanobeads (Fig. 22B).174 Self-assembly of negatively charged MPA-coated QDs, Gd chelates and chitosan (a cationic biopolymer) via electrostatic interactions produce ~50 nm HD nanobeads that not only preserve the QD photo-physical properties, but also improve the QY, possibly due to better passivation of the nanocrystal surface. Moreover, incorporation of multiple QDs and Gd chelates within the single nanobead significantly improves the sensitivity of dual fluorescence/MRI imaging. However, the increased size of the probes often hampers extravasation and interaction with cells when administered systemically in vivo. Preparation of a single-QD dual-imaging probe has been demonstrated by Koole et al.176 QD cores protected by the biocompatible silica shell are further coated by a lipid bilayer containing PEGylated, Gd-linked, and RGD peptide-linked lipids (Fig. 22C). Resulting probes with dual fluorescence/paramagnetic functionalities feature ~60 nm HD, ~25–30% QY, and specific targeting for αvβ3 integrin. Besides providing a flexible platform for attachment of additional imaging and targeting modules, silica-lipid encapsulation efficiently prevents the Cd leakage and physically separates MRI agents from the QD core. When incubated with endothelial cells in vitro nanoparticles are readily internalized by the cells; however, the signal strength of both fluorescence detection and MRI are limited by the low content of imaging agents within each particle and significantly reduced QY.
Recently, Bruns et al have generated ~250 nm nanosomes by separately incorporating hydrophobic QDs and MNPs into the core of targeted lipoproteins.178 Real-time MRI of live mice has enabled quantitative analysis of the rapid liver clearance kinetics with high spatiotemporal resolution, whereas post-mortem fluorescence microscopy of tissue sections has revealed lipoprotein biodistribution in the liver tissue and specific uptake of nanosomes by hepatocytes. Incorporation of both QD and MNP reporters within single nanosomes will enable dual-modal imaging with these probes. Despite the promising results, this technology is not yet suitable for dual-modal in vivo imaging as liver uptake is highly undesirable for most of the applications. Further incorporation of non-fouling surface coatings and targeting ligands, however, will enable highly sensitive MR/fluorescence vascular imaging and cell tracking with nanosomes.
PET represents another non-invasive in vivo imaging modality that might benefit from added QD-based fluorescence detection. At the same time, QD biodistribution and deep-tissue imaging can be achieved with a significantly lower (10–20 fold) QD dose when done in combination with PET agents.179,180 In one example, Cai et al have quantified the ability of intravenously injected QDs to target tumors in mice by both fluorescence and PET in vivo.179 NIR QDs have been conjugated to 64Cu radionuclides and to RGD peptides for integrin αvβ3 targeting (Fig. 22D). Ex vivo PET and NIR fluorescence imaging of excised tumors and organs was strongly correlated (R2 = 0.93), demonstrating efficacy of combined preoperative/intraoperative PET/fluorescence for image-guided surgery. However, the relatively large probe size hampers extravasation and accumulation in tumor, while the extensive modification with targeting peptides enhances RES uptake. Nevertheless, this technology promises to enable sensitive quantitative evaluation of QD biodistribution and tumor targeting efficacy using non-invasive PET imaging. Recently, Schipper et al have further employed the power of real-time 3D-PET imaging to measure the uptake kinetics of non-targeted 64Cu-QDs of various sizes (5, 10, and 20 nm) and surface coatings (polymer, peptide, PEG) injected into live mice.181 Minimal changes in QD surface chemistry or physical dimensions introduced by the Cu modification ensure the accuracy of the model system. Utilization of well-characterized sensitive PET contrast agents and bright versatile QDs in compact nanocomposites makes dual PET/fluorescence imaging a valuable tool for the quantitative analysis of nanoparticle biodistribution at different resolutions and time scales.
Engineering of plasmonic fluorescent probes for optoelectronics and nanophotonics represents a new and exciting research direction in the field of multimodal imaging. In a pioneering study, Mokari et al have grown gold tips on the CdSe quantum rods, thus obtaining dual-modal optical probes with unique capability of site-specific self-assembly and biomolecule attachment.182 However, direct coupling between two materials significantly quenched the QD fluorescence. A systematic investigation of the effect of nanocomposite architecture on optical properties has been done by Jin and Gao.183 They were the first to synthesize QD-Au nanocomposites that retained fluorescence while exhibiting plasmonic features, thus being potentially useful multimodal imaging probes. Stable and highly bright (QY 75%) lipid-PEG-encapsulated QDs have been used as a platform for probe preparation. Concurrent gold shell growth on the QD surface introduces the plasmonic properties (Fig. 22E). However, fluorescence QY drops to only 18% following the Au shell deposition. The thickness of the shell as well as physical separation between the QD and Au has been determined to be responsible for fluorescence quenching. In fact it has been shown that an increase in the QD-Au gap from 3.1 to 4.8 nm improves the fluorescence (QY 39%), while increase in the Au shell thickness from 2 to 5 nm nearly completely quenches the QDs. Therefore, due to the high quenching capacity of plasmonic materials, design of plasmonic fluorescent probes in general requires physical separation of the materials as well as careful optimization of the probe architecture.
A wide variety of radioactive, optically, and magnetically-responsive agents can be incorporated into composite nanostructures. Engineering of multi-component nanodevices is opening the door to novel multimodal imaging and sensing applications. Currently, such probes are being incorporated into variety of imaging applications in laboratory settings, while in the near future QD-based nanocomposites may enable preoperative diagnostics, intraoperative image guidance, and high-resolution post-operative histological evaluation of excised tissues.
5.2 QD-based therapeutic nanocomposites
Multifunctional QD-based nanocomposites for drug delivery represent an extremely beneficial tool for nanomedicine, as functionalities for drug loading, targeting, controlled release, and monitoring of pharmacokinetics and biodistribution can be incorporated within a single unit.184 Moreover, nanodevices with integrated sensing modality can be used to monitor the drug release in real time. However, the extensive capabilities of therapeutic nanocomposites require strict design criteria guiding the development of this class of nanoparticles. In general, nanotherapeutics must be made of biocompatible and non-fouling materials that ensure long circulation time in the bloodstream and optimized pharmacokinetics, feature functionalities for overcoming multiple physiological barriers, possess sufficient drug loading capacity, and balance the carrier’s affinity for the therapeutic agent with its tendency for release. Many of the criteria related to the QD intracellular delivery and in vivo targeting have been discussed in sections 3.3, 4.3, and 4.4, while this section focuses on drug loading, release, and sensing.
QDs feature versatile surface chemistry that enables attachment of various ligands and loading of both hydrophilic and hydrophobic therapeutics. However, being a relatively new technology, QD-based drug delivery is not as robust and well-studied as other liposome or polymer-based techniques. Therefore, a more common and straightforward approach involves utilization of QDs as fluorescent markers for tagging conventional drug carriers. Liposomes and micelles, for example, are widely used for drug delivery, featuring great flexibility in size, charge, rigidity, permeability, and surface functionality.185 For visualization of these vesicles, QDs have been either linked to the surface or incorporated inside of the liposomes. To demonstrate external labeling, Weng et al have conjugated hydrophilic QDs and targeting ligands (anti-HER2 single-chain Fv antibody fragments) to PEG-phospholipids, which are incorporated into the lipid layer during liposome hydration (Fig. 23A).186 Following loading with anticancer drug doxorubicin (Dox), targeted, traceable, and fairly stable (15–30% Dox loss during 2-month storage) drug delivery vehicles are obtained. As determined by flow-cytometry, functionalized liposomes show remarkably selective targeting and potent cytotoxicity in vitro, with 900 to 1800-fold higher uptake compared to non-targeted QD-liposomes, and 16 to 30-fold higher uptake compared to HER2-negative cells. When administered systemically in vivo, QD-liposomes have ~3 hour circulation half-life and accumulate in implanted tumors. In an alternative approach, liposomes have been internally loaded with QDs, thus eliminating the need for chemical conjugation while shielding the QD tags from the physiological environment and improving the blood circulation half-life (up to 5 hours) (Fig. 23B).187,188 Despite the relative simplicity of the procedure and minimal requirements for QD tag design, both external and internal QD loading suffer from reduced drug loading capacity and poor fluorescence intensity, as surface attachment of more than 2 QDs per liposome impedes cell internalization, whereas incorporation of QDs inside of liposomes is sparse.
Fig. 23.
Engineering of QD-based therapeutic nanocomposites. QDs can be used as tags for labeling of larger drug carriers, such as liposomes (A, B) and polymeric nanoparticles (C), or as single-QD platforms for traceable drug loading and delivery (D–F).
Higher QD loading capacity has been achieved with the polymeric drug delivery vehicles. For example, Kim et al have incorporated MitoTracker dye (a model drug) and QDs (fluorescent tag) into 100 nm biodegradable PLGA nanocomposites using a microemulsion procedure and functionalized particles with trastuzumab for HER2 targeting (Fig. 23C).116 Upon ligand mediated endocytosis the vehicles undergo pH-dependent charge reversal, which leads to endosomal membrane destabilization and cytoplasmic release of the particles followed by PLGA hydrolysis, dye release, and clear mitochondria staining. Other methods of producing drug- and QD-loaded polymeric composites include microfluidic emulsification189 and electrostatic assembly of polyplexes.190,191 Therefore, established synthesis protocols enable preparation of nanocomposites with improved loading capacity that facilitate sensitive fluorescence detection (via incorporation of multiple QD tags) and enhanced therapeutic potency (via high drug content) essential for in vivo studies. However, large particles often suffer from short circulation half-life (due to enhanced RES uptake192,193) and poor tissue penetration,148 both of which significantly impede the therapeutic potential.
Individual QDs, having high surface area to volume ratio and modular surface chemistry, provide a suitable platform for the engineering of smaller targeted and traceable drug delivery vehicles. As significant progress has already been done in engineering QD probes for targeted in vivo imaging, a major focus is been placed on developing novel drug loading routes and drug loading/release monitoring capabilities with minimal effect on the overall QD properties. In one example, Bagalkot et al have covalently linked hydrophilic QDs to aptamers targeted against PSMA and loaded Dox via intercalation within the double-stranded CG portion of the aptamers at ~1:1 ratio (Fig. 23D).194 Drug loading/release sensing has been achieved with carefully designed bi-FRET arrangement. By matching the QD emission with Dox absorption the drug loading can be monitored via QD fluorescence quenching. At the same time, as Dox fluorescence is also efficiently quenched by the aptamer, the overall Dox-loaded targeted QD probes remain in a non- emitting state. Upon specific delivery into PSMA-expressing prostate cancer cells, slow Dox release can be monitored by recovery of both green QD and red Dox fluorescence. Yet, the drug loading is strictly limited by the number of aptamers conjugated to each QD, thus requiring administration of higher QD dose in order to reach a therapeutic response.
The low drug loading capacity of single QDs can be employed for delivery of highly potent therapeutics, such as siRNA, a 22 bp-long double-stranded RNA that efficiently suppresses expression of specific genes by initiating cleavage of corresponding mRNA inside cells.195 Most often siRNA is loaded on the hydrophilic surface of QDs either via covalent conjugation or electrostatic interaction. For example, Derfus et al have functionalized QDs with tumor-homing peptides and covalently conjugated anti-GFP siRNA via a cleavable disulfide linker to suppress GFP expression in HeLa cells (Fig. 23E).196 Utilization of a cleavable linker ensures high stability of a QD-siRNA complex, while providing efficient intracellular siRNA release essential for effective interference. Walther et al, on the other hand, have absorbed 80–100 bp cyanine-labeled RNAs via electrostatic interaction with cationic internalization peptides coated on the QD surface, achieving intracellular siRNA delivery.197 However, both delivery approaches suffer from endosomal sequestration of the particles and require additional treatment of cells with endosome-disrupting agents (cationic liposomes196 or chloroquine197) for efficient cytosolic release.
QD carriers that have intrinsic endosomal escape functionality along with targeting and siRNA carrying capacity have recently gained increasing attention due to their capability of one-step efficient and high-throughput siRNA delivery. To achieve this goal, fusogenic peptides109,198 that disrupt membranes via hydrophobic interactions and tertiary amine-rich polymers117,119 that elicit the proton sponge effect have been utilized. In one example, Yezhelyev et al have modified amphiphilic polymer-coated QDs with tertiary amines and tuned the surface charge to balance the competing electrostatic effects of QD-siRNA binding and intracellular siRNA release (Fig. 23F).119 In a similar report, Qi and Gao have used a pre-modified amphiphilic polymer for QD encapsulation and siRNA loading, demonstrating improved knockdown of the HER2/neu gene in SK-BR-3 cells.199 Importantly, QD carriers protect adsorbed siRNA from enzymatic degradation and enable real-time monitoring of FITC-labeled siRNA loading/release via FRET quenching of FITC signal by QDs and restoration of green fluorescence upon cytoplasmic siRNA release.
Drug carriers based on single QDs are often small (< 20 nm) relative to conventional delivery vehicles, thus facilitating improved extravasation from vasculature and accumulation in target organs.184,200,201 At the same time, compact probes with zwitterionic surfaces are much more efficient in avoiding opsonization and RES uptake.58 However, the amount of drug delivered per QD is limited by the particle’s surface area, which must be shared with other ligands for molecular targeting and/or cell internalization. Therefore, the most promising in vivo application of single-QD nanotherapeutics will be in monitoring the delivery of highly potent drugs (such as siRNA) and modeling the biodistribution of other nanoparticle-based drug carriers with similar surface properties and physical dimensions, yet higher drug loading capacities (e.g. polymeric nanoparticles). Future work will likely focus on incorporation of environmentally responsive materials for controlled drug release, non-fouling surface coatings for improved biodistribution, and multistage targeting functionality for enhanced therapeutic specificity.
6. Conclusions
The application of nanoparticles for the multi-parameter comprehensive study of physiological and pathological processes in biomedical research as well as for advanced diagnostics and therapy in clinical practice presents a surging trend in nanomedicine. QDs, in particular, have emerged as one of the most promising classes of nanoparticles for biomedical imaging, drug delivery, and sensing due to their unique photo-physical properties and versatile surface chemistry. Biofunctionalization of inorganic QD cores facilitates interaction of nanoparticles with biological systems and enables direct participation in biological processes. As a result, QD probes have been utilized in a wide variety of applications spanning in vitro molecular pathology, live cell imaging, and in vivo drug delivery and tracing. With each application QDs have opened new horizons of multiplexed quantitative detection, sensitive high-resolution fluorescence imaging, and long-term real-time monitoring of probe dynamics. Aiming at expanding QD functionality even further, early steps have been made towards engineering of QD-based multi-functional nanodevices that promise to combine the benefits of multiple imaging modalities and incorporate the imaging, drug loading, and sensing capacities within a single nanoparticle. Yet, currently available QD probes are far from being ideal, leaving plenty of room for improvement of existing and development of novel nanoparticle designs. In this review we have discussed the future directions of QD-based bio-nanotechnology research and outlined the major design principles and criteria, from general ones to application-specific, governing the engineering of novel QD probes satisfying increasing demands and requirements of nanomedicine.
Acknowledgments
This work was supported in part by NIH (R01CA131797, R01CA140295), NSF, and the Department of Bioengineering at the University of Washington. X.G. thanks the NSF for a Faculty Early Career Development award (CAREER). P.Z. thanks the UW Center for Nanotechnology for a UIF fellowship and the National Science Foundation for Graduate Research Fellowship. M.S. thanks Mary Gates Endowment for Students for Research Scholarship, and Amgen Scholars Program. We are also grateful to Shivang Dave for critical reading of the manuscript.
Notes and references
- 1.Nanoscale Science Engineering and Technology Subcommittee (NSET), Committee on Technology (CT) and National Science and Technology Council (NSTC) Vol. 44 2009. [Google Scholar]
- 2.Chen H, Roco MC, Li X, Lin Y. Nat Nanotechnol. 2008;3:123–125. doi: 10.1038/nnano.2008.51. [DOI] [PubMed] [Google Scholar]
- 3.Massoud TF, Gambhir SS. Genes & development. 2003;17:545–580. doi: 10.1101/gad.1047403. [DOI] [PubMed] [Google Scholar]
- 4.Mankoff DA. J Nucl Med. 2007;48:18N–21N. [PubMed] [Google Scholar]
- 5.Medintz IL, Mattoussi H. Phys Chem Chem Phys. 2009;11:17–45. doi: 10.1039/b813919a. [DOI] [PubMed] [Google Scholar]
- 6.Medintz IL, Mattoussi H, Clapp AR. Int J Nanomedicine. 2008;3:151–167. doi: 10.2147/ijn.s614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Misra RD. Nanomed. 2008;3:271–274. doi: 10.2217/17435889.3.3.271. [DOI] [PubMed] [Google Scholar]
- 8.Resch-Genger U, Grabolle M, Cavaliere-Jaricot S, Nitschke R, Nann T. Nat Methods. 2008;5:763–775. doi: 10.1038/nmeth.1248. [DOI] [PubMed] [Google Scholar]
- 9.Smith AM, Duan H, Mohs AM, Nie S. Adv Drug Deliv Rev. 2008;60:1226–1240. doi: 10.1016/j.addr.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tholouli E, Sweeney E, Barrow E, Clay V, Hoyland JA, Byers RJ. J Pathol. 2008;216:275–285. doi: 10.1002/path.2421. [DOI] [PubMed] [Google Scholar]
- 11.Xing Y, Rao J. Cancer Biomark. 2008;4:307–319. doi: 10.3233/cbm-2008-4603. [DOI] [PubMed] [Google Scholar]
- 12.Zrazhevskiy P, Gao X. Nano Today. 2009;4:414–428. doi: 10.1016/j.nantod.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alivisatos AP. J Phys Chem. 1996;100:13226–13239. [Google Scholar]
- 14.Brus LE. J Chem Phys. 1984;80:4403–4409. [Google Scholar]
- 15.Rossetti R, Nakahara S, Brus LE. J Chem Phys. 1983;79:1086–1088. [Google Scholar]
- 16.Zeng RS, Zhang TT, Liu JC, Hu S, Wan Q, Liu XM, Peng ZW, Zou BS. Nanotechnology. 2009;20 doi: 10.1088/0957-4484/20/9/095102. [DOI] [PubMed] [Google Scholar]
- 17.Zheng YG, Yang ZC, Ying JY. Adv Mater. 2007;19:1475. [Google Scholar]
- 18.Murray CB, Norris DJ, Bawendi MG. J Am Chem Soc. 1993;115:8706–8715. [Google Scholar]
- 19.Peng ZA, Peng XG. J Am Chem Soc. 2001;123:183–184. doi: 10.1021/ja003633m. [DOI] [PubMed] [Google Scholar]
- 20.Peng ZA, Peng X. J Am Chem Soc. 2002;124:3343–3353. doi: 10.1021/ja0173167. [DOI] [PubMed] [Google Scholar]
- 21.Alivisatos AP. Science. 1996;271:933–937. [Google Scholar]
- 22.Li LS, Pradhan N, Wang Y, Peng X. Nano Lett. 2004;4:2261–2264. [Google Scholar]
- 23.Peng X, Wickham J, Alivisatos AP. J Am Chem Soc. 1998;120:5343–5344. [Google Scholar]
- 24.Zhong XH, Feng YY, Knoll W, Han MY. J Am Chem Soc. 2003;125:13559–13563. doi: 10.1021/ja036683a. [DOI] [PubMed] [Google Scholar]
- 25.Kim S, Fisher B, Eisler HJ, Bawendi M. J Am Chem Soc. 2003;125:11466–11467. doi: 10.1021/ja0361749. [DOI] [PubMed] [Google Scholar]
- 26.Pietryga JM, Schaller RD, Werder D, Stewart MH, Klimov VI, Hollingsworth JA. J Am Chem Soc. 2004;126:11752–11753. doi: 10.1021/ja047659f. [DOI] [PubMed] [Google Scholar]
- 27.Qu L, Peng X. J Am Chem Soc. 2002;124:2049–2055. doi: 10.1021/ja017002j. [DOI] [PubMed] [Google Scholar]
- 28.Smith AM, Gao X, Nie S. Photochem Photobiol. 2004;80:377–385. doi: 10.1562/0031-8655(2004)080<0377:QDNFIV>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 29.Zhong XH, Han MY, Dong ZL, White TJ, Knoll W. J Am Chem Soc. 2003;125:8589–8594. doi: 10.1021/ja035096m. [DOI] [PubMed] [Google Scholar]
- 30.Bailey RE, Nie SM. J Am Chem Soc. 2003;125:7100–7106. doi: 10.1021/ja035000o. [DOI] [PubMed] [Google Scholar]
- 31.Chattopadhyay PK, Price DA, Harper TF, Betts MR, Yu J, Gostick E, Perfetto SP, Goepfert P, Koup RA, De Rosa SC, Bruchez MP, Roederer M. Nat Med. 2006;12:972–977. doi: 10.1038/nm1371. [DOI] [PubMed] [Google Scholar]
- 32.Yezhelyev MV, Al-Hajj A, Morris C, Marcus AI, Liu T, Lewis M, Cohen C, Zrazhevskiy P, Simons JW, Rogatko A, Nie S, Gao X, Regan RMO. Adv Mater. 2007;19:3146–3151. [Google Scholar]
- 33.Giepmans BN, Deerinck TJ, Smarr BL, Jones YZ, Ellisman MH. Nat Methods. 2005;2:743–749. doi: 10.1038/nmeth791. [DOI] [PubMed] [Google Scholar]
- 34.Nisman R, Dellaire G, Ren Y, Li R, Bazett-Jones DP. J Histochem Cytochem. 2004;52:13–18. doi: 10.1177/002215540405200102. [DOI] [PubMed] [Google Scholar]
- 35.Oberdorster G, Oberdorster E, Oberdorster J. Environ Health Perspect. 2005;113:823–839. doi: 10.1289/ehp.7339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bryant GW, Jaskolski W. J Phys Chem B. 2005;109:19650–19656. doi: 10.1021/jp0535543. [DOI] [PubMed] [Google Scholar]
- 37.Gomez DE, Califano M, Mulvaney P. Phys Chem Chem Phys. 2006;8:4989–5011. doi: 10.1039/b607661k. [DOI] [PubMed] [Google Scholar]
- 38.Dabbousi BO, Rodriguez-Viejo J, Mikulec FV, Heine JR, Mattoussi H, Ober R, Jensen KF, Bawendi MG. The Journal of Physical Chemistry B. 1997;101:9463–9475. [Google Scholar]
- 39.Hines MA, Guyot-Sionnest P. J Phys Chem. 1996;100:468–471. [Google Scholar]
- 40.Peng X, Schlamp MC, Kadavanich AV, Alivisatos AP. J Am Chem Soc. 1997;119:7019–7029. [Google Scholar]
- 41.Chan WC, Maxwell DJ, Gao X, Bailey RE, Han M, Nie S. Curr Opin Biotechnol. 2002;13:40–46. doi: 10.1016/s0958-1669(02)00282-3. [DOI] [PubMed] [Google Scholar]
- 42.Medintz IL, Uyeda HT, Goldman ER, Mattoussi H. Nat Mater. 2005;4:435–446. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- 43.Efros AL, Rosen M. Phys Rev Lett. 1997;78:1110–1113. [Google Scholar]
- 44.Michalet X, Pinaud F, Lacoste TD, Dahan M, Bruchez MP, Alivisatos AP, Weiss S. Single Molecules. 2001;2:261–276. [Google Scholar]
- 45.Nirmal M, Dabbousi BO, Bawendi MG, Macklin JJ, Trautman JK, Harris TD, Brus LE. Nature. 1996;383:802–804. [Google Scholar]
- 46.Pinaud F, Michalet X, Bentolila LA, Tsay JM, Doose S, Li JJ, Iyer G, Weiss S. Biomaterials. 2006;27:1679–1687. doi: 10.1016/j.biomaterials.2005.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Comparelli R, Zezza F, Striccoli M, Curri ML, Tommasi R, Agostiano A. Materials Science and Engineering: C. 2003;23:1083–1086. [Google Scholar]
- 48.Li JJ, Wang YA, Guo W, Keay JC, Mishima TD, Johnson MB, Peng X. J Am Chem Soc. 2003;125:12567–12575. doi: 10.1021/ja0363563. [DOI] [PubMed] [Google Scholar]
- 49.Talapin DV, Rogach AL, Kornowski A, Haase M, Weller H. Nano Lett. 2001;1:207–211. doi: 10.1021/nl0155126. [DOI] [PubMed] [Google Scholar]
- 50.Bruchez M, Jr, Moronne M, Gin P, Weiss S, Alivisatos AP. Science. 1998;281:2013–2016. doi: 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- 51.Chan WC, Nie S. Science. 1998;281:2016–2018. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- 52.Gao X, Nie S. Trends Biotechnol. 2003;21:371–373. doi: 10.1016/S0167-7799(03)00209-9. [DOI] [PubMed] [Google Scholar]
- 53.Wu X, Liu H, Liu J, Haley KN, Treadway JA, Larson JP, Ge N, Peale F, Bruchez MP. Nat Biotechnol. 2003;21:41–46. doi: 10.1038/nbt764. [DOI] [PubMed] [Google Scholar]
- 54.Gao X, Yang L, Petros JA, Marshall FF, Simons JW, Nie S. Curr Opin Biotechnol. 2005;16:63–72. doi: 10.1016/j.copbio.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 55.Tokumasu F, Dvorak J. J Microsc. 2003;211:256–261. doi: 10.1046/j.1365-2818.2003.01219.x. [DOI] [PubMed] [Google Scholar]
- 56.Cui B, Wu C, Chen L, Ramirez A, Bearer EL, Li WP, Mobley WC, Chu S. Proc Natl Acad Sci U S A. 2007;104:13666–13671. doi: 10.1073/pnas.0706192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lidke DS, Nagy P, Heintzmann R, Arndt-Jovin DJ, Post JN, Grecco HE, Jares-Erijman EA, Jovin TM. Nat Biotechnol. 2004;22:198–203. doi: 10.1038/nbt929. [DOI] [PubMed] [Google Scholar]
- 58.Choi HS, Liu W, Misra P, Tanaka E, Zimmer JP, Itty Ipe B, Bawendi MG, Frangioni JV. Nat Biotechnol. 2007;25:1165–1170. doi: 10.1038/nbt1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Law WC, Yong KT, Roy I, Ding H, Hu R, Zhao W, Prasad PN. Small. 2009;5:1302–1310. doi: 10.1002/smll.200801555. [DOI] [PubMed] [Google Scholar]
- 60.Aldana J, Wang YA, Peng X. J Am Chem Soc. 2001;123:8844–8850. doi: 10.1021/ja016424q. [DOI] [PubMed] [Google Scholar]
- 61.Algar WR, Krull UJ. ChemPhysChem. 2007;8:561–568. doi: 10.1002/cphc.200600686. [DOI] [PubMed] [Google Scholar]
- 62.Liu W, Howarth M, Greytak AB, Zheng Y, Nocera DG, Ting AY, Bawendi MG. J Am Chem Soc. 2008;130:1274–1284. doi: 10.1021/ja076069p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sukhanova A, Devy J, Venteo L, Kaplan H, Artemyev M, Oleinikov V, Klinov D, Pluot M, Cohen JH, Nabiev I. Anal Biochem. 2004;324:60–67. doi: 10.1016/j.ab.2003.09.031. [DOI] [PubMed] [Google Scholar]
- 64.Jiang W, Mardyani S, Fischer H, Chan WCW. Chem Mater. 2006;18:872–878. [Google Scholar]
- 65.Smith AM, Nie S. J Am Chem Soc. 2008;130:11278–11279. doi: 10.1021/ja804306c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gerion D, Pinaud F, Williams SC, Parak WJ, Zanchet D, Weiss S, Alivisatos AP. J Phys Chem B. 2001;105:8861–8871. [Google Scholar]
- 67.Pinaud F, King D, Moore HP, Weiss S. J Am Chem Soc. 2004;126:6115–6123. doi: 10.1021/ja031691c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Whaley SR, English DS, Hu EL, Barbara PF, Belcher AM. Nature. 2000;405:665–668. doi: 10.1038/35015043. [DOI] [PubMed] [Google Scholar]
- 69.Pellegrino T, Manna L, Kudera S, Liedl T, Koktysh D, Rogach AL, Keller S, Radler J, Natile G, Parak WJ. Nano Letters. 2004;4:703–707. [Google Scholar]
- 70.Dubertret B, Skourides P, Norris DJ, Noireaux V, Brivanlou AH, Libchaber A. Science (New York, NY. 2002;298:1759–1762. doi: 10.1126/science.1077194. [DOI] [PubMed] [Google Scholar]
- 71.Gao X, Cui Y, Levenson RM, Chung LW, Nie S. Nat Biotechnol. 2004;22:969–976. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- 72.Smith AM, Duan H, Rhyner MN, Ruan G, Nie S. Phys Chem Chem Phys. 2006;8:3895–3903. doi: 10.1039/b606572b. [DOI] [PubMed] [Google Scholar]
- 73.Dahan M, Levi S, Luccardini C, Rostaing P, Riveau B, Triller A. Science. 2003;302:442–445. doi: 10.1126/science.1088525. [DOI] [PubMed] [Google Scholar]
- 74.Barat B, Sirk SJ, McCabe KE, Li J, Lepin EJ, Remenyi R, Koh AL, Olafsen T, Gambhir SS, Weiss S, Wu AM. Bioconjug Chem. 2009;20:1474–1481. doi: 10.1021/bc800421f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Medintz IL, Clapp AR, Mattoussi H, Goldman ER, Fisher B, Mauro JM. Nat Mater. 2003;2:630–638. doi: 10.1038/nmat961. [DOI] [PubMed] [Google Scholar]
- 76.Medintz IL, Clapp AR, Brunel FM, Tiefenbrunn T, Uyeda HT, Chang EL, Deschamps JR, Dawson PE, Mattoussi H. Nat Mater. 2006;5:581–589. doi: 10.1038/nmat1676. [DOI] [PubMed] [Google Scholar]
- 77.Delehanty JB, Medintz IL, Pons T, Brunel FM, Dawson PE, Mattoussi H. Bioconjug Chem. 2006;17:920–927. doi: 10.1021/bc060044i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Goldman ER, Anderson GP, Tran PT, Mattoussi H, Charles PT, Mauro JM. Anal Chem. 2002;74:841–847. doi: 10.1021/ac010662m. [DOI] [PubMed] [Google Scholar]
- 79.Mattoussi H, Mauro JM, Goldman ER, Anderson GP, Sundar VC, Mikulec FV, Bawendi MG. J Am Chem Soc. 2000;122:12142–12150. [Google Scholar]
- 80.Fernandez-Arguelles MT, Yakovlev A, Sperling RA, Luccardini C, Gaillard S, Medel AS, Mallet JM, Brochon JC, Feltz A, Oheim M, Parak WJ. Nano Lett. 2007;7:2613–2617. doi: 10.1021/nl070971d. [DOI] [PubMed] [Google Scholar]
- 81.Lin CAJ, Sperling RA, Li JK, Yang TY, Li PY, Zanella M, Chang WH, Parak WGJ. Small. 2008;4:334–341. doi: 10.1002/smll.200700654. [DOI] [PubMed] [Google Scholar]
- 82.Alivisatos AP. ACS nano. 2008;2:1514–1516. doi: 10.1021/nn800485f. [DOI] [PubMed] [Google Scholar]
- 83.Byers RJ, Di Vizio D, O’Connell F, Tholouli E, Levenson RM, Gossage K, Twomey D, Yang Y, Benedettini E, Rose J, Ligon KL, Finn SP, Golub TR, Loda M. J Mol Diagn. 2007;9:20–29. doi: 10.2353/jmoldx.2007.060119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.True LD, Gao X. J Mol Diagn. 2007;9:7–11. doi: 10.2353/jmoldx.2007.060186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jain KK. Expert Opin Pharmacother. 2005;6:1463–1476. doi: 10.1517/14656566.6.9.1463. [DOI] [PubMed] [Google Scholar]
- 86.Tholouli E, Hoyland JA, Di Vizio D, O’Connell F, Macdermott SA, Twomey D, Levenson R, Yin JA, Golub TR, Loda M, Byers R. Biochem Biophys Res Commun. 2006;348:628–636. doi: 10.1016/j.bbrc.2006.07.122. [DOI] [PubMed] [Google Scholar]
- 87.Chan P, Yuen T, Ruf F, Gonzalez-Maeso J, Sealfon SC. Nucleic Acids Res. 2005;33:e161. doi: 10.1093/nar/gni162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Levenson RM. Lab Medicine. 2004;35:244–251. [Google Scholar]
- 89.Ghazani AA, Lee JA, Klostranec J, Xiang Q, Dacosta RS, Wilson BC, Tsao MS, Chan WC. Nano Lett. 2006;6:2881–2886. doi: 10.1021/nl062111n. [DOI] [PubMed] [Google Scholar]
- 90.Zhong XH, Zhang ZH, Liu SH, Han MY, Knoll W. J Phys Chem B. 2004;108:15552–15559. [Google Scholar]
- 91.Wells NP, Lessard GA, Werner JH. Anal Chem. 2008;80:9830–9834. doi: 10.1021/ac8021899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Murcia MJ, Minner DE, Mustata GM, Ritchie K, Naumann CA. J Am Chem Soc. 2008;130:15054–15062. doi: 10.1021/ja803325b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jaiswal JK, Mattoussi H, Mauro JM, Simon SM. Nature biotechnology. 2003;21:47–51. doi: 10.1038/nbt767. [DOI] [PubMed] [Google Scholar]
- 94.Heine M, Groc L, Frischknecht R, Beique JC, Lounis B, Rumbaugh G, Huganir RL, Cognet L, Choquet D. Science. 2008;320:201–205. doi: 10.1126/science.1152089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang Q, Li Y, Tsien RW. Science. 2009;323:1448–1453. doi: 10.1126/science.1167373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cambi A, Lidke DS, Arndt-Jovin DJ, Figdor CG, Jovin TM. Nano Lett. 2007;7:970–977. doi: 10.1021/nl0700503. [DOI] [PubMed] [Google Scholar]
- 97.Roullier V, Clarke S, You C, Pinaud F, Gouzer GG, Schaible D, Marchi-Artzner V, Piehler J, Dahan M. Nano Lett. 2009;9:1228–1234. doi: 10.1021/nl9001298. [DOI] [PubMed] [Google Scholar]
- 98.Delehanty JB, Mattoussi H, Medintz IL. Anal Bioanal Chem. 2009;393:1091–1105. doi: 10.1007/s00216-008-2410-4. [DOI] [PubMed] [Google Scholar]
- 99.Barua S, Rege K. Small. 2009;5:370–376. doi: 10.1002/smll.200800972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jiang W, Kim BY, Rutka JT, Chan WC. Nat Nanotechnol. 2008;3:145–150. doi: 10.1038/nnano.2008.30. [DOI] [PubMed] [Google Scholar]
- 101.Nabiev I, Mitchell S, Davies A, Williams Y, Kelleher D, Moore R, Gun’ko YK, Byrne S, Rakovich YP, Donegan JF, Sukhanova A, Conroy J, Cottell D, Gaponik N, Rogach A, Volkov Y. Nano Lett. 2007;7:3452–3461. doi: 10.1021/nl0719832. [DOI] [PubMed] [Google Scholar]
- 102.Lovric J, Bazzi HS, Cuie Y, Fortin GR, Winnik FM, Maysinger D. J Mol Med. 2005;83:377–385. doi: 10.1007/s00109-004-0629-x. [DOI] [PubMed] [Google Scholar]
- 103.Kairdolf BA, Mancini MC, Smith AM, Nie S. Anal Chem. 2008;80:3029–3034. doi: 10.1021/ac800068q. [DOI] [PubMed] [Google Scholar]
- 104.Derfus AM, Chan WCW, Bhatia SN. Adv Mater. 2004;16:961–966. [Google Scholar]
- 105.Yum K, Na S, Xiang Y, Wang N, Yu MF. Nano Lett. 2009;9:2193–2198. doi: 10.1021/nl901047u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Park S, Kim YS, Kim WB, Jon S. Nano Lett. 2009;9:1325–1329. doi: 10.1021/nl802962t. [DOI] [PubMed] [Google Scholar]
- 107.Zhou M, Ghosh I. Biopolymers. 2007;88:325–339. doi: 10.1002/bip.20655. [DOI] [PubMed] [Google Scholar]
- 108.de la Fuente JM, Fandel M, Berry CC, Riehle M, Cronin L, Aitchison G, Curtis AS. ChemBioChem. 2005;6:989–991. doi: 10.1002/cbic.200500071. [DOI] [PubMed] [Google Scholar]
- 109.Koshman YE, Waters SB, Walker LA, Los T, de Tombe P, Goldspink PH, Russell B. J Mol Cell Cardiol. 2008;45:853–856. doi: 10.1016/j.yjmcc.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sewell SL, Giorgio TD. Materials Science and Engineering: C. 2009;29:1428–1432. [Google Scholar]
- 111.Lagerholm BC, Wang M, Ernst LA, Ly DH, Liu H, Bruchez MP, Waggoner AS. Nano Letters. 2004;4:2019–2022. [Google Scholar]
- 112.Rozenzhak SM, Kadakia MP, Caserta TM, Westbrook TR, Stone MO, Naik RR. Chem Commun. 2005:2217–2219. doi: 10.1039/b418454h. [DOI] [PubMed] [Google Scholar]
- 113.Silver J, Ou W. Nano Lett. 2005;5:1445–1449. doi: 10.1021/nl050808n. [DOI] [PubMed] [Google Scholar]
- 114.Qi L, Gao X. Expert Opin Drug Deliv. 2008;5:263–267. doi: 10.1517/17425247.5.3.263. [DOI] [PubMed] [Google Scholar]
- 115.Courty S, Luccardini C, Bellaiche Y, Cappello G, Dahan M. Nano Lett. 2006;6:1491–1495. doi: 10.1021/nl060921t. [DOI] [PubMed] [Google Scholar]
- 116.Kim BY, Jiang W, Oreopoulos J, Yip CM, Rutka JT, Chan WC. Nano Lett. 2008;8:3887–3892. doi: 10.1021/nl802311t. [DOI] [PubMed] [Google Scholar]
- 117.Duan H, Nie S. J Am Chem Soc. 2007;129:3333–3338. doi: 10.1021/ja068158s. [DOI] [PubMed] [Google Scholar]
- 118.Sonawane ND, Szoka FC, Jr, Verkman AS. J Biol Chem. 2003;278:44826–44831. doi: 10.1074/jbc.M308643200. [DOI] [PubMed] [Google Scholar]
- 119.Yezhelyev MV, Qi L, O’Regan RM, Nie S, Gao X. J Am Chem Soc. 2008;130:9006–9012. doi: 10.1021/ja800086u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.McGrath N, Barroso M. J Biomed Opt. 2008;13:9. doi: 10.1117/1.2939417. [DOI] [PubMed] [Google Scholar]
- 121.Clapp AR, Pons T, Medintz IL, Delehanty JB, Melinger JS, Tiefenbrunn T, Dawson PE, Fisher BR, Rourke BO, Mattoussi H. Adv Mater. 2007;19:1921–1926. [Google Scholar]
- 122.Bentolila LA, Ebenstein Y, Weiss S. J Nucl Med. 2009;50:493–496. doi: 10.2967/jnumed.108.053561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Graves EE, Weissleder R, Ntziachristos V. Curr Mol Med. 2004;4:419–430. doi: 10.2174/1566524043360555. [DOI] [PubMed] [Google Scholar]
- 124.Jain RK, Munn LL, Fukumura D. Nat Rev Cancer. 2002;2:266–276. doi: 10.1038/nrc778. [DOI] [PubMed] [Google Scholar]
- 125.Makale M. Methods Enzymol. 2007;426:375–401. doi: 10.1016/S0076-6879(07)26016-1. [DOI] [PubMed] [Google Scholar]
- 126.Hardman R. Environ Health Perspect. 2006;114:165–172. doi: 10.1289/ehp.8284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lee J, Lilly GD, Doty RC, Podsiadlo P, Kotov NA. Small. 2009;5:1213–1221. doi: 10.1002/smll.200801788. [DOI] [PubMed] [Google Scholar]
- 128.Monteiro-Riviere NA, Inman AO, Zhang LW. Toxicol Appl Pharmacol. 2009;234:222–235. doi: 10.1016/j.taap.2008.09.030. [DOI] [PubMed] [Google Scholar]
- 129.Derfus AM, Chan WCW, Bhatia SN. Nano Lett. 2004;4:11–18. doi: 10.1021/nl0347334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhang T, Stilwell JL, Gerion D, Ding L, Elboudwarej O, Cooke PA, Gray JW, Alivisatos AP, Chen FF. Nano Lett. 2006;6:800–808. doi: 10.1021/nl0603350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hoshino A, Manabe N, Fujioka K, Suzuki K, Yasuhara M, Yamamoto K. J Artif Organs. 2007;10:149–157. doi: 10.1007/s10047-007-0379-y. [DOI] [PubMed] [Google Scholar]
- 132.Su Y, He Y, Lu H, Sai L, Li Q, Li W, Wang L, Shen P, Huang Q, Fan C. Biomaterials. 2008 doi: 10.1016/j.biomaterials.2008.09.029. [DOI] [PubMed] [Google Scholar]
- 133.Cho SJ, Maysinger D, Jain M, Roder B, Hackbarth S, Winnik FM. Langmuir. 2007;23:1974–1980. doi: 10.1021/la060093j. [DOI] [PubMed] [Google Scholar]
- 134.King-Heiden TC, Wiecinski PN, Mangham AN, Metz KM, Nesbit D, Pedersen JA, Hamers RJ, Heideman W, Peterson RE. Environ Sci Technol. 2009;43:1605–1611. doi: 10.1021/es801925c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Nel A, Xia T, Madler L, Li N. Science. 2006;311:622–627. doi: 10.1126/science.1114397. [DOI] [PubMed] [Google Scholar]
- 136.Choi HS, Liu W, Liu F, Nasr K, Misra P, Bawendi MG, Frangioni JV. Nat Nanotechnol. 2009 doi: 10.1038/nnano.2009.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ballou B, Lagerholm BC, Ernst LA, Bruchez MP, Waggoner AS. Bioconjug Chem. 2004;15:79–86. doi: 10.1021/bc034153y. [DOI] [PubMed] [Google Scholar]
- 138.Prencipe G, Tabakman SM, Welsher K, Liu Z, Goodwin AP, Zhang L, Henry J, Dai H. J Am Chem Soc. 2009;131:4783–4787. doi: 10.1021/ja809086q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Geys J, Nemmar A, Verbeken E, Smolders E, Ratoi M, Hoylaerts MF, Nemery B, Hoet PH. Environ Health Perspect. 2008;116:1607–1613. doi: 10.1289/ehp.11566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Fitzpatrick JA, Andreko SK, Ernst LA, Waggoner AS, Ballou B, Bruchez MP. Nano Lett. 2009;9:2736–2741. doi: 10.1021/nl901534q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Satarug S, Haswell-Elkins MR, Moore MR. Br J Nutr. 2000;84:791–802. [PubMed] [Google Scholar]
- 142.Yong KT, Ding H, Roy I, Law WC, Bergey EJ, Maitra A, Prasad PN. ACS Nano. 2009;3:502–510. doi: 10.1021/nn8008933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Li L, Daou TJ, Texier I, Kim Chi TT, Liem NQ, Reiss P. Chem Mater. 2009;21:2422–2429. [Google Scholar]
- 144.Kang ZH, Liu Y, Tsang CHA, Ma DDD, Fan X, Wong NB, Lee ST. Adv Mater. 2009;21:661. [Google Scholar]
- 145.Park JH, Gu L, von Maltzahn G, Ruoslahti E, Bhatia SN, Sailor MJ. Nat Mater. 2009;8:331–336. doi: 10.1038/nmat2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ballou B, Ernst LA, Andreko S, Fitzpatrick JA, Lagerholm BC, Waggoner AS, Bruchez MP. Methods Mol Biol. 2009;574:63–74. doi: 10.1007/978-1-60327-321-3_6. [DOI] [PubMed] [Google Scholar]
- 147.Larson DR, Zipfel WR, Williams RM, Clark SW, Bruchez MP, Wise FW, Webb WW. Science. 2003;300:1434–1436. doi: 10.1126/science.1083780. [DOI] [PubMed] [Google Scholar]
- 148.Stroh M, Zimmer JP, Duda DG, Levchenko TS, Cohen KS, Brown EB, Scadden DT, Torchilin VP, Bawendi MG, Fukumura D, Jain RK. Nat Med. 2005;11:678–682. doi: 10.1038/nm1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kim JV, Kang SS, Dustin ML, McGavern DB. Nature. 2009;457:191–195. doi: 10.1038/nature07591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ballou B, Ernst LA, Andreko S, Harper T, Fitzpatrick JA, Waggoner AS, Bruchez MP. Bioconjug Chem. 2007;18:389–396. doi: 10.1021/bc060261j. [DOI] [PubMed] [Google Scholar]
- 151.Kim S, Lim YT, Soltesz EG, De Grand AM, Lee J, Nakayama A, Parker JA, Mihaljevic T, Laurence RG, Dor DM, Cohn LH, Bawendi MG, Frangioni JV. Nat Biotechnol. 2004;22:93–97. doi: 10.1038/nbt920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Soltesz EG, Kim S, Laurence RG, DeGrand AM, Parungo CP, Dor DM, Cohn LH, Bawendi MG, Frangioni JV, Mihaljevic T. Ann Thorac Surg. 2005;79:269–277. doi: 10.1016/j.athoracsur.2004.06.055. discussion 269–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zimmer JP, Kim SW, Ohnishi S, Tanaka E, Frangioni JV, Bawendi MG. J Am Chem Soc. 2006;128:2526–2527. doi: 10.1021/ja0579816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kobayashi H, Hama Y, Koyama Y, Barrett T, Regino CA, Urano Y, Choyke PL. Nano Lett. 2007;7:1711–1716. doi: 10.1021/nl0707003. [DOI] [PubMed] [Google Scholar]
- 155.Levene MJ, Dombeck DA, Kasischke KA, Molloy RP, Webb WW. J Neurophysiol. 2004;91:1908–1912. doi: 10.1152/jn.01007.2003. [DOI] [PubMed] [Google Scholar]
- 156.Sutton EJ, Henning TD, Pichler BJ, Bremer C, Daldrup-Link HE. Eur Radiol. 2008;18:2021–2032. doi: 10.1007/s00330-008-0984-z. [DOI] [PubMed] [Google Scholar]
- 157.Slotkin JR, Chakrabarti L, Dai HN, Carney RS, Hirata T, Bregman BS, Gallicano GI, Corbin JG, Haydar TF. Dev Dyn. 2007;236:3393–3401. doi: 10.1002/dvdy.21235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Jayagopal A, Russ PK, Haselton FR. Bioconjug Chem. 2007;18:1424–1433. doi: 10.1021/bc070020r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Voura EB, Jaiswal JK, Mattoussi H, Simon SM. Nat Med. 2004;10:993–998. doi: 10.1038/nm1096. [DOI] [PubMed] [Google Scholar]
- 160.Lo Celso C, Fleming HE, Wu JW, Zhao CX, Miake-Lye S, Fujisaki J, Cote D, Rowe DW, Lin CP, Scadden DT. Nature. 2009;457:92–96. doi: 10.1038/nature07434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.So MK, Xu C, Loening AM, Gambhir SS, Rao J. Nat Biotechnol. 2006;24:339–343. doi: 10.1038/nbt1188. [DOI] [PubMed] [Google Scholar]
- 162.Akerman ME, Chan WC, Laakkonen P, Bhatia SN, Ruoslahti E. Proc Natl Acad Sci U S A. 2002;99:12617–12621. doi: 10.1073/pnas.152463399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Cai W, Shin DW, Chen K, Gheysens O, Cao Q, Wang SX, Gambhir SS, Chen X. Nano Lett. 2006;6:669–676. doi: 10.1021/nl052405t. [DOI] [PubMed] [Google Scholar]
- 164.Yang L, Mao H, Wang YA, Cao Z, Peng X, Wang X, Duan H, Ni C, Yuan Q, Adams G, Smith MQ, Wood WC, Gao X, Nie S. Small. 2009;5:235–243. doi: 10.1002/smll.200800714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Tada H, Higuchi H, Wanatabe TM, Ohuchi N. Cancer Res. 2007;67:1138–1144. doi: 10.1158/0008-5472.CAN-06-1185. [DOI] [PubMed] [Google Scholar]
- 166.Kircher MF, Mahmood U, King RS, Weissleder R, Josephson L. Cancer Res. 2003;63:8122–8125. [PubMed] [Google Scholar]
- 167.Cai W, Chen X. J Nucl Med. 2008;49(Suppl 2):113S–128S. doi: 10.2967/jnumed.107.045922. [DOI] [PubMed] [Google Scholar]
- 168.Mulder WJ, Cormode DP, Hak S, Lobatto ME, Silvera S, Fayad ZA. Nat Clin Pract Cardiovasc Med. 2008;5(Suppl 2):S103–111. doi: 10.1038/ncpcardio1242. [DOI] [PubMed] [Google Scholar]
- 169.Cai W, Chen X. Small. 2007;3:1840–1854. doi: 10.1002/smll.200700351. [DOI] [PubMed] [Google Scholar]
- 170.Cheon J, Lee JH. Acc Chem Res. 2008;41:1630–1640. doi: 10.1021/ar800045c. [DOI] [PubMed] [Google Scholar]
- 171.Gao J, Zhang B, Gao Y, Pan Y, Zhang X, Xu B. J Am Chem Soc. 2007;129:11928–11935. doi: 10.1021/ja0731017. [DOI] [PubMed] [Google Scholar]
- 172.Gao J, Zhang W, Huang P, Zhang B, Zhang X, Xu B. J Am Chem Soc. 2008;130:3710–3711. doi: 10.1021/ja7103125. [DOI] [PubMed] [Google Scholar]
- 173.Gu H, Zheng R, Zhang X, Xu B. J Am Chem Soc. 2004;126:5664–5665. doi: 10.1021/ja0496423. [DOI] [PubMed] [Google Scholar]
- 174.Tan WB, Zhang Y. Advanced Materials. 2005;17:2375–2380. [Google Scholar]
- 175.Sathe TR, Agrawal A, Nie S. Anal Chem. 2006;78:5627–5632. doi: 10.1021/ac0610309. [DOI] [PubMed] [Google Scholar]
- 176.Koole R, van Schooneveld MM, Hilhorst J, Castermans K, Cormode DP, Strijkers GJ, de Mello Donega C, Vanmaekelbergh D, Griffioen AW, Nicolay K, Fayad ZA, Meijerink A, Mulder WJ. Bioconjug Chem. 2008;19:2471–2479. doi: 10.1021/bc800368x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Mulder WJ, Strijkers GJ, van Tilborg GA, Cormode DP, Fayad ZA, Nicolay K. Acc Chem Res. 2009 doi: 10.1021/ar800223c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Bruns OT, Ittrich H, Peldschus K, Kaul MG, Tromsdorf UI, Lauterwasser J, Nikolic MS, Mollwitz B, Merkel M, Bigall NC, Sapra S, Reimer R, Hohenberg H, Weller H, Eychmuller A, Adam G, Beisiegel U, Heeren J. Nat Nanotechnol. 2009;4:193–201. doi: 10.1038/nnano.2008.405. [DOI] [PubMed] [Google Scholar]
- 179.Cai W, Chen K, Li ZB, Gambhir SS, Chen X. J Nucl Med. 2007;48:1862–1870. doi: 10.2967/jnumed.107.043216. [DOI] [PubMed] [Google Scholar]
- 180.Schipper ML, Cheng Z, Lee SW, Bentolila LA, Iyer G, Rao J, Chen X, Wu AM, Weiss S, Gambhir SS. J Nucl Med. 2007;48:1511–1518. doi: 10.2967/jnumed.107.040071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Schipper ML, Iyer G, Koh AL, Cheng Z, Ebenstein Y, Aharoni A, Keren S, Bentolila LA, Li J, Rao J, Chen X, Banin U, Wu AM, Sinclair R, Weiss S, Gambhir SS. Small. 2009;5:126–134. doi: 10.1002/smll.200800003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Mokari T, Rothenberg E, Popov I, Costi R, Banin U. Science. 2004;304:1787–1790. doi: 10.1126/science.1097830. [DOI] [PubMed] [Google Scholar]
- 183.Jin Y, Gao X. Nat Nanotechnol. 2009;4:571–576. doi: 10.1038/nnano.2009.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Farokhzad OC, Langer R. ACS Nano. 2009;3:16–20. doi: 10.1021/nn900002m. [DOI] [PubMed] [Google Scholar]
- 185.Immordino ML, Dosio F, Cattel L. Int J Nanomedicine. 2006;1:297–315. [PMC free article] [PubMed] [Google Scholar]
- 186.Weng KC, Noble CO, Papahadjopoulos-Sternberg B, Chen FF, Drummond DC, Kirpotin DB, Wang D, Hom YK, Hann B, Park JW. Nano Lett. 2008;8:2851–2857. doi: 10.1021/nl801488u. [DOI] [PubMed] [Google Scholar]
- 187.Al-Jamal KT, Tian B, Cakebread A, Halket JM, Kostarelos K. Mol Pharm. 2009;6:520–530. doi: 10.1021/mp800187d. [DOI] [PubMed] [Google Scholar]
- 188.Al-Jamal WT, Al-Jamal KT, Bomans PH, Frederik PM, Kostarelos K. Small. 2008;4:1406–1415. doi: 10.1002/smll.200701043. [DOI] [PubMed] [Google Scholar]
- 189.Yang CH, Huang KS, Lin YS, Lu K, Tzeng CC, Wang EC, Lin CH, Hsu WY, Chang JY. Lab Chip. 2009;9:961–965. doi: 10.1039/b814952f. [DOI] [PubMed] [Google Scholar]
- 190.Chen HH, Ho YP, Jiang X, Mao HQ, Wang TH, Leong KW. Mol Ther. 2008;16:324–332. doi: 10.1038/sj.mt.6300392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Tan WB, Jiang S, Zhang Y. Biomaterials. 2007;28:1565–1571. doi: 10.1016/j.biomaterials.2006.11.018. [DOI] [PubMed] [Google Scholar]
- 192.Rensen PC, Sliedregt LA, Ferns M, Kieviet E, van Rossenberg SM, van Leeuwen SH, van Berkel TJ, Biessen EA. J Biol Chem. 2001;276:37577–37584. doi: 10.1074/jbc.M101786200. [DOI] [PubMed] [Google Scholar]
- 193.Ogawara K, Yoshida M, Higaki K, Kimura T, Shiraishi K, Nishikawa M, Takakura Y, Hashida M. J Control Release. 1999;59:15–22. doi: 10.1016/s0168-3659(99)00015-2. [DOI] [PubMed] [Google Scholar]
- 194.Bagalkot V, Zhang L, Levy-Nissenbaum E, Jon S, Kantoff PW, Langer R, Farokhzad OC. Nano Lett. 2007;7:3065–3070. doi: 10.1021/nl071546n. [DOI] [PubMed] [Google Scholar]
- 195.Whitehead KA, Langer R, Anderson DG. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Derfus AM, Chen AA, Min DH, Ruoslahti E, Bhatia SN. Bioconjug Chem. 2007;18:1391–1396. doi: 10.1021/bc060367e. [DOI] [PubMed] [Google Scholar]
- 197.Walther C, Meyer K, Rennert R, Neundorf I. Bioconjugate chemistry. 2008;19:2346–2356. doi: 10.1021/bc800172q. [DOI] [PubMed] [Google Scholar]
- 198.Moore NM, Sheppard CL, Barbour TR, Sakiyama-Elbert SE. J Gene Med. 2008;10:1134–1149. doi: 10.1002/jgm.1234. [DOI] [PubMed] [Google Scholar]
- 199.Qi L, Gao X. ACS Nano. 2008;2:1403–1410. doi: 10.1021/nn800280r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Moghimi SM, Hunter AC, Murray JC. Pharmacol Rev. 2001;53:283–318. [PubMed] [Google Scholar]
- 201.Perrault SD, Walkey C, Jennings T, Fischer HC, Chan WC. Nano Lett. 2009;9:1909–1915. doi: 10.1021/nl900031y. [DOI] [PubMed] [Google Scholar]