Abstract
Structures of tert-butylcarbamate ions in the gas phase and methanol solution were studied for simple secondary and tertiary carbamates as well as for carbamate-containing products and internal standards for lysosomal enzyme assays used in newborn screening of a α-galactosidase A deficiency (GLA, Fabry disease), mucopolysaccharidosis I (Hurler disease), and mucopolysaccharidosis II (Hunter disease). Protonation of simple t-butylcarbamates can occur at the carbonyl group which is the preferred site in the gas phase. Protonation in methanol solution is more favorable if occurring at the carbamate nitrogen atom. Protonation of more complex t-butylcarbamates occurs at amide and carbamate carbonyl groups, and the ions are stabilized by intramolecular hydrogen bonding which is affected by solvation. Tertiary carbamates containing aminophenol amide groups were calculated to have substantially greater gas-phase basicities than secondary carbamates containing coumarin amide groups. The main diagnostically important ion dissociation by elimination of 2-methylpropene (isobutylene, i-C4H8) and carbon dioxide is shown by experiment and theory to proceed in two steps. Energy-resolved collision-induced dissociation of the Hurler’s disease enzymatic product ion, which is a coumarin-diamine linker-t-butylcarbamate conjugate (3a+), indicated separate energy thresholds for the loss of i-C4H8 and CO2. Computational investigation of the potential energy surface along two presumed reaction pathways indicated kinetic preference for the migration of a t-butyl hydrogen atom to the carbamate carbonyl resulting in the isobutylene loss. The consequent loss of CO2 required further proton migrations that had to overcome energy barriers.
Introduction
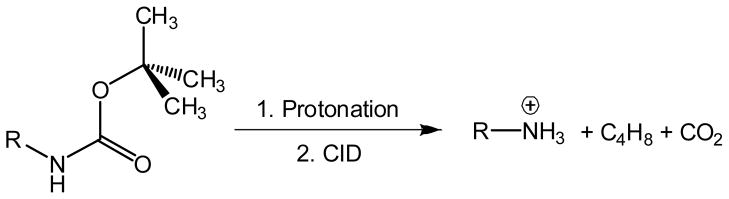
The tert-butylcarbamate (t-BOC) group serves as a useful protecting group in peptide and other syntheses that utilize active ester coupling methods.1 t-BOC groups are readily removed by a reaction with trifluoroacetic acid, indicating protonation of the carbamate functionality as the initial cleavage step.2 Dissociations of gas-phase cations containing t-butylcarbamate groups proceed by a coupled elimination of 2-methylpropene (isobutylene, i-C4H8) and carbon dioxide to result in the combined loss of 100 Da neutral fragments (Scheme 1).3 The dissociation has been extensively utilized in tandem mass spectrometry for the quantification of enzyme products and internal standards by multiplex protocols for the detection of lysosomal storage disorders4,5 caused by several enzyme deficiencies, for example, acid α-glucosidase (GAA, Pompe disease),6–8 acid α-galactosidase (GLA, Fabry disease),6–8 α-L-iduronidase (mucopolysaccharidosis MPS I, Hurler disease),9 iduronate-2-sulfatase (MPS-II, Hunter disease),10,11 N-acetylgalactosamine-6-sulfate sulfatase (MPS-IVA, Morquio A disease),12 and N-acetylgalactosamine-4-sulfate sulfatase (MPS VI, Maroteaux-Lamy disease).13
Scheme 1.
Loss of i-C4H8 and CO2 from protonated t-butylcarbamates.
The fragmentation of t-BOC-derivatized primary and secondary amines proceeds as a major dissociation pathway in the presence of other groups that are usually considered to be labile, such as α and β-arylglycosides.9 However, the protonation sites in tert-butylcarbamates and the dissociation mechanisms of pertinent gas-phase cations have not been studied. Carbamic acid, which is the simplest molecule having the carbamate group, prefers protonation at the carbonyl oxygen, as established by both high-level ab initio calculations14,15 and studies of dissociation mechanisms after electron transfer.15 Gaining insight into the mechanism and energetics of dissociations involving t-BOC-containing gas-phase ions is pertinent and requisite for the further development and optimization of tandem mass spectrometric methods to be applied in metabolite analysis. Here we report a joint experimental and computational study that addresses the structures and fragmentation of tert-butylcarbamate cations relevant to tandem mass spectrometry assays of enzymes used in newborn screening of lysosomal storage disorders.
Experimental Section
Materials
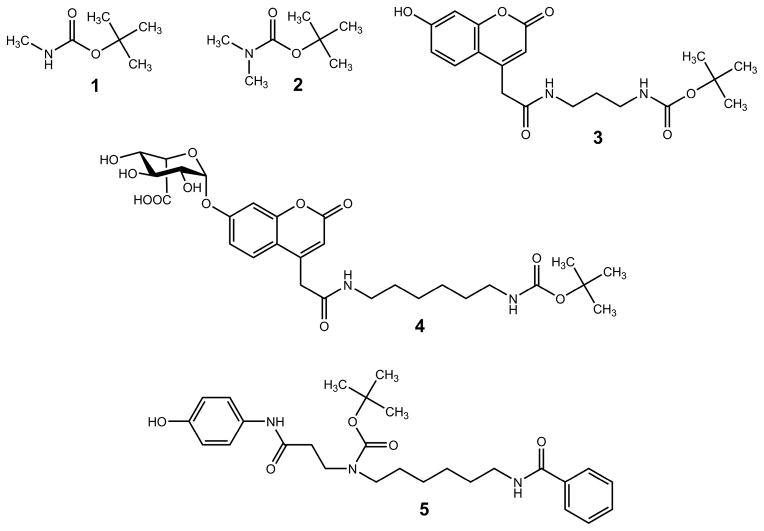
Compounds 3 (internal standard for MPS I assay),94 (product of MPS II assay)11 and 5 (internal standard for GLA assay)6 were synthesized as described previously.
Methods
Mass spectra were measured on Waters Quattro Micro API bench-top tandem quadrupole mass spectrometer (Waters, Milford, MA). Ions were produced by electrospray ionization from methanol:water:formic acid (50:50:0.1; v/v/v) solutions at concentration of 30 μM. The experimental parameters were as follows: flow rate 5 μL/min, needle voltage 3500 V, drying gas temperature 200°C and flow rate 200 L/hr, cone voltage 5.25 V. Argon was used as a collision gas at nominal pressures that were varied between 1.3 × 10−4 Torr and 5.2 × 10−4 Torr. Selected ion monitoring of m/z 377, 321, and 277 ions was performed while the acceleration voltage was ramped in 1 V steps from 0 to 20 V corresponding to nominal ion laboratory kinetic energies in the same eV range.
Calculations
Standard ab initio and density functional theory calculations were performed using the Gaussian 09 suite of programs.16 Gas-phase structures were optimized with B3LYP/6-31+G(d,p)17 and the local energy minima and transition states were confirmed by frequency calculations as having the appropriate number of imaginary frequencies (0 for minima, 1 for saddle points). All relative energies were corrected for zero-point vibrational energies based on harmonic frequencies that were scaled by 0.963.18 Transition states were located by scanning with B3LYP/6-31+G(d,p) the potential energy surface between the reactant and product. Intermediate structures of low energy gradients along the reaction coordinate X (dE/dX < 0.005) were analyzed by harmonic frequency calculations and the calculated Hessian matrix was used for saddle point search and optimization. Single point energies on fully optimized geometries were calculated with B3LYP/6-311++G(2d,p) and MP2/6-311++G(2d,p)19 and averaged to cancel out known errors in each method.20 Benchmark single-point energies were calculated using coupled clusters21 with single, double, and disconnected triple excitations, CCSD(T),22 and the 6-311G(d,p) basis set and expanded to effective CCSD(T)/6-311++G(3df,2p) using the standard linear formula: E[CCSD(T)/6-311++G(3df,2p)] ≈ E[CCSD(T)/6-311G(d,p)] + E[MP2/6-311++G(3df,2p)] − E[MP2/6-311G(d,p)].23 Calculations of solvated ions used the Polarizable Continuum Model (PCM)24 with standard parameters for methanol included in Gaussian 09. Solvated ion structures were fully optimized with PCM-B3LYP/6-31+G(d,p) using the pertinent gas-phase ion structures as initial guesses. Frequency calculations of solvated ions were not performed. Enthalpies and entropies were obtained from standard thermodynamic formulas using the rigid-rotor-harmonic oscillator model for the vibrational and rotational terms. Vibrational enthalpy terms for very low frequencies that exceeded 0.5kT were replaced by the 0.5kT term for free rotations.25 Vibrational entropies were not corrected. Proton affinities and gas-phase basicities were calculated as the respective 298 K reaction enthalpies and free energies for a dissociation of ion AH+ to neutral molecule A and a proton. Unimolecular rate constants were calculated using the Rice-Ramsperger-Kassel-Marcus (RRKM) theory26 and employing a modified Hase’s program27 which was recompiled for Windows XP28 and Windows 7. The RRKM rate constants were obtained by direct count of quantum states at internal energies that were increased in 2 kJ mol−1 steps from the transition state up to 400 kJ mol−1 above the reactant. Rotations were treated adiabatically and the calculated microscopic rate constants k(E,J,K) were then Boltzmann-averaged over the thermal distribution of rotational states at 298 K.
Results and Discussion
We addressed five structure types containing the t-butylcarbamate group. The chemical structures of the representative neutral compounds are shown in Figure 1. Structures 1 and 2 are simple N-methyl and N,N-dimethyl tert-butylcarbamates that were used as basic representatives of the functional groups of interest. Protonation in these systems is not affected by hydrogen bonding or interactions with other functional groups in the ion and thus represents the intrinsic property of the t-butylcarbamate moiety. Structure 3 represents the internal standard for the MPS-I assay. The molecule has a 7-hydroxycoumarin ring which is conjugated to a diamine linker capped with a t-BOC group. Structure 4 is the enzymatic product of the MPS-II assay which is an α-glycoside of α-L-iduronic acid and a coumarin conjugate aglycon. Structure 5 is the product of the GLA assay, which is chemically identical to the deuterium-labeled GLA internal standard,6 both consisting of an aminophenol, a 1,6-hexanediamine linker with a tertiary t-BOC group, and a benzamide. The common feature of structures 3–5 is that they contain several potential protonation sites in the aromatic rings, amide groups, and the carbamate group.
Figure 1.
Chemical structures of neutral t-butylcarbamates.
Energy-Resolved Collision Induced Dissociations
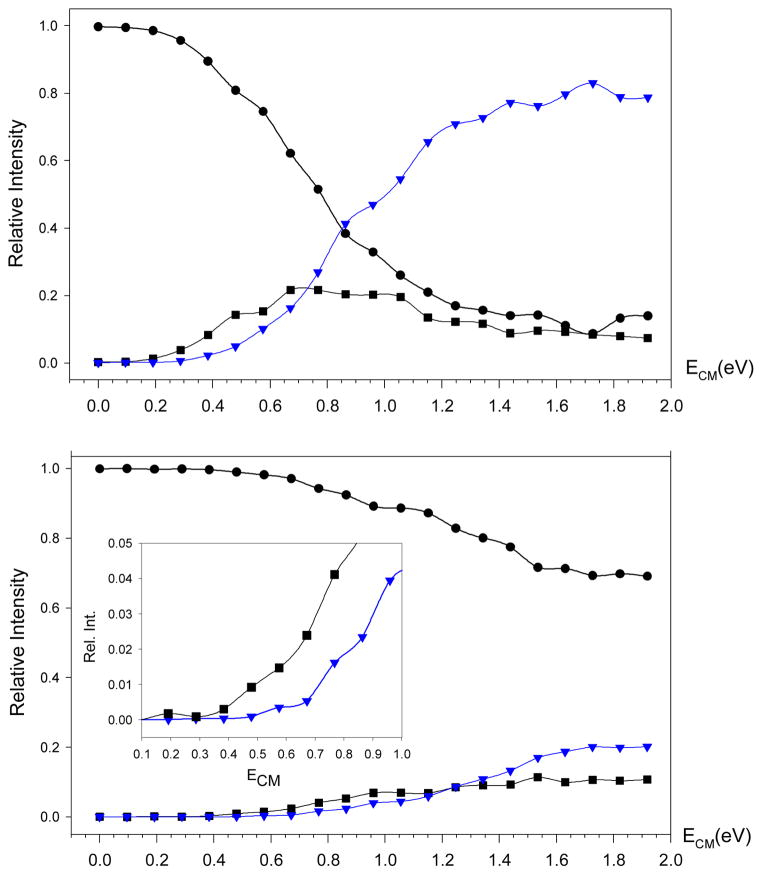
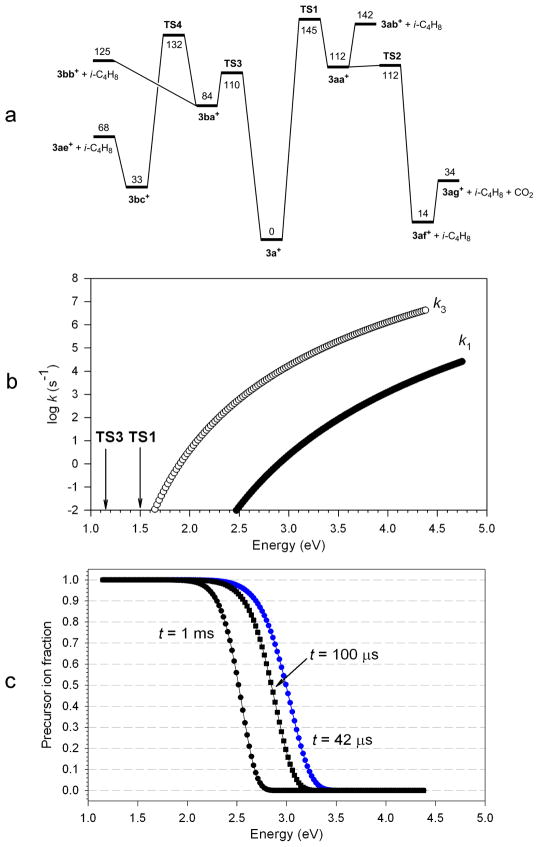
We first present experimental results from energy-resolved CID of ion 3+ formed by electrospray protonation of 3. Figure 2 shows the breakdown diagrams29 for the m/z 377 precursor ion of 3+, the m/z 321 product ion by elimination of C4H8, and the m/z 321 product ion due to a combined elimination of C4H8 and CO2, which are plotted against the center-of-mass collision energy (ECM). The breakdown diagrams were measured at three different nominal collision gas pressures; shown are the curves for the highest 5.2 × 10−4 Torr and lowest 1.3 ×10−4 Torr pressures. The curves obtained at the lowest pressure indicate a dissociation onset at ca. ECM = 0.5 eV for 1% elimination of C4H8 in the m/z 321 channel. The onset for the m/z 277 ion is shifted by 0.2–0.3 eV to higher ECM. Note that these energies have not been extrapolated to single-collision conditions and must be viewed as lower bounds for the real threshold energies.30 However, the fact that there are two separate onsets indicates that the formation of the m/z 277 ion occurs by consecutive losses of C4H8 and CO2, whereby the m/z 321 fragment ion is a stable intermediate that requires additional excitation to eliminate CO2. The relatively small difference in the estimated onset energies does not allow us to unequivocally exclude the presence of an alternative, presumably minor, pathway, which is a concomitant elimination of C4H8 and CO2 through a short-lived intermediate. Obviously, such an elimination would have a different mechanism from that forming the stable m/z 321 intermediate. Adding the estimated threshold energy for the elimination of C4H8 (0.5 eV) to the thermal energy (Eth) of the precursor ion gives a rough estimate of the critical energy (ETS) for the dissociation. Since the precursor ions entering the collision cell may not be at thermal equilibrium, their temperature and internal energy can vary from ambient (Eth = 0.58 eV at 298 K) to that of the electrospray interface (Eth = 1.36 eV at 473 K). This gives a broad range of the estimated ETS ≈ 0.5 + Eth = 1.08~1.86 eV = 104~180 kJmol−1.
Figure 2.
Energy-resolved collision-induced dissociation of 3+ at nominal collision gas pressures of (top) 5.2 × 10−4 and (bottom) 1.3 ×10−4 Torr. Black circles: m/z 377; black squares: m/z 321; blue triangles: m/z 277. Inset shows the enlarged section of fragment ion relative intensities between 0.1 and 1.0 eV.
Protonation of Simple t-Butylcarbamates
The fragmentation of protonated carbamates raises questions regarding the structures and energies of the precursor ions, transition states, and intermediates, as well as the overall dissociation energies for the eliminations of C4H8 and CO2. Protonation of the carbamate group in simple t-butylcarbamates can in principle take place at one of the oxygen atoms or at the nitrogen atom. Protonated 1 (Figure 3) shows the carbonyl (1a+) and N-protonated ion structures (1b+) as the most stable tautomers. Ion 1a+ is calculated to be 20 kJ mol−1 more stable than 1b+ in the gas phase, indicating a higher proton affinity for the carbonyl oxygen in the secondary carbamate (Table 1). This preference is greatly diminished or even reversed in a methanol solution where the solvated ions 1a+ and 1b+ are practically isoenergetic. Our highest-level calculations at the effective CCSD(T)/6-311++G(3df,2p) level of theory give ΔG°298(1a+ → 1b+) = 18 and −3 kJ mol−1 in the gas phase and methanol solution, respectively. The ether-protonated tautomer (1c+) was substantially less stable than 1a+ and 1b+ in both solution and the gas phase. The methanol-solvated structure of 1c+ has long C—O bonds (Figure 3), indicating partial dissociation to a solvated complex of N-methylcarbamic acid and t-butyl cation which is again less stable than solvated 1a+ and 1b+.
Figure 3.
B3LYP/6-31+G(d,p)-optimized structures of ion tautomers of 1+ and 2+. The atoms are color-coded as follows: Green = C; red = O; blue = N, gray = H.
Table 1.
Ion Relative Energies.
| Species/reaction | Relative Energya,b |
||||
|---|---|---|---|---|---|
| B3LYP | B3LYP | MP2 | B3-MP2c | CCSD(T)d | |
| 6-31+G(d,p) | 6-311++G(2d,p) | 6-311++G(2d,p) | 6-311++G(3df,2p) | ||
| 1a+ → 1 + H+ | 882 | 883 | 865 | 874 (879)e | 877 (882)e (850)f |
| 1a+ → 1b+ | 27 | 25 | 12 | 18 (17)g (−4)h | 20 (18)g (−3)h |
| 1a+ → 1c+ | 81 | 79 | 76 | 77 (72)g (45)h | |
| 2a+ → 2 + H+ | 898 | 899 | 884 | 892 (899)e | 896 (903)e (871)f |
| 2a+ → 2b+ | 14 | 13 | 1 | 7 (9)g ( 10)h | 6 (8)g (−11)h |
| 3a → 3b | 1 | 0.5 | 15 | 8 (1.5)g | |
| 3a+ → 3a + H+ | 966 | 966 | 943 | 954 (961)e (924)f | |
| 3a+ → 3b+ | 22 | 22 | 31 | 26 (23)g (48)h | |
| 3a+ → 3c+ | 41 | 39 | 26 | 33 (29)g (18)h | |
| 3a+ → 3d+ | 79 | 78 | 66 | 72 (71)g (63)h | |
| 4a → 4b | 12 | 13 | −13 | 0 (9)g | |
| 4a+ → 4a + H+ | 960 | 958 | 967 | 962 (970)e (918)f | |
| 4a+ → 4b+ | 17 | 16 | 55 | 35 (21)d g(31)h | |
| 5a → 5b | −1 | −1 | −1 | −1 (−1)g | |
| 5a+ → 5a + H+ | 992 | 990 | 998 | 994 (1001)e (950)f | |
| 5a+ → 5b+ | 4 | 1 | 37 | 19 (3)g (−39)h | |
| 5a+ → 5c+ | 17 | 17 | 12 | 14 (10)g (−1)h | |
| 5a+ → 5d+ | 50 | 45 | 64 | 55 (36)g (−13)h | |
| 5a+ → 5e+ | 80 | 77 | 101 | 89 (71)g (−11)h | |
In units of kJ mol−1.
Including B3LYP/6-31+G(d,p) zero-point energies and referring to 0 K unless stated otherwise.
From averaged B3LYP and MP2 single-point energies
From effective single-point energies.
Proton affinities at 298 K.
Gas-phase basicities at 298 K.
Relative gas-phase free energies at 298 K.
Relative free energies in methanol solution at 298 K.
Additional N-methylation in 2 increases the overall carbamate gas-phase basicity by 21 kJ mol−1 (Table 1). A particular increase is seen for the nitrogen atom which is comparably basic as the carbonyl oxygen. The N-protonated tautomer (2b+, Figure 3) is only marginally less stable than the carbonyl protonated structure 2a+ in the gas phase. The stability order is reversed in methanol solution where solvated 2b+ is more stable than 2a+. The calculated energies indicate that simple t-butylcarbamates slightly prefer to be protonated at the nitrogen atom in methanol solution, but not in the gas phase. Considering that protonation in electrospray takes place in liquid droplets, the initially formed ions should consist of mixtures of O-carbonyl and N-protonated tautomers, with the latter being favored. However, as the gas-phase ions travel into the mass spectrometer, they can exchange protons with the residual neutral carbamate molecules to preferentially form O-carbonyl protonated tautomers. The actual composition of the gas phase ion tautomer population then may depend on the experimental conditions that determine the interactions in the solution and in the gas phase. The energy data in Table 1 further show that the relatively inexpensive B3-MP2/6-311++G(2d,p) calculations gave excellent results for the ion energetics when benchmarked against the effective CCSD(T)/6-311++G(3df,2p) data. The B3-MP2 scheme was therefore used for calculations of the larger MPS-I, MPS-II and GLA molecules and ions. These systems include several functional groups, providing multiple protonation sites and ion stabilization by intramolecular hydrogen bonding. Ion structures for these three groups are presented in the next section.
Protonation of Coumarin and Aminophenol Conjugates
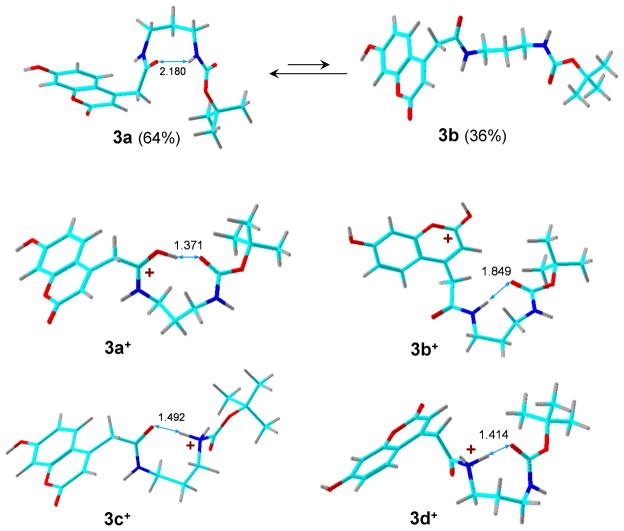
Geometry optimization of the MPS-I internal standard 3 produced two structures that differed in the conformation of the diamine linker. Structure 3a had the carbamate NH hydrogen-bonded to the amide carbonyl (Figure 4). Structure 3b had an extended linker. The calculated gas-phase free energies slightly favor 3a by 1.5 kJ mol−1 at 298 K. This order of relative free energies is reversed at temperatures >360 K because of the higher entropy of the extended-chain conformer 3b. Protonation in 3a or 3b can occur in several positions, leading to tautomers 3a+-3d+ (Figure 4). The amide carbonyl-protonated tautomer 3a+ was calculated to be the most stable cation in both the gas phase and methanol solution (Table 1) followed by the coumarin-protonated tautomer 3b+. Protonation at the amide and carbamate nitrogen atoms leads to high-energy tautomers. Interestingly, an ion tautomer protonated at the carbamate carbonyl was not a local energy minimum and upon optimization spontaneously isomerized to 3a+. Hence, structure 3a+ is expected to predominate in the gas-phase ion population. The proton affinity of 3a forming 3a+ was calculated by B3-MP2/6-311++G(2d,p) as PA = 961 kJ mol−1.
Figure 4.
B3LYP/6-31+G(d,p)-optimized structures of ion tautomers of 3+. The atoms are color-coded as follows: Green = C; red = O; blue = N, gray = H.
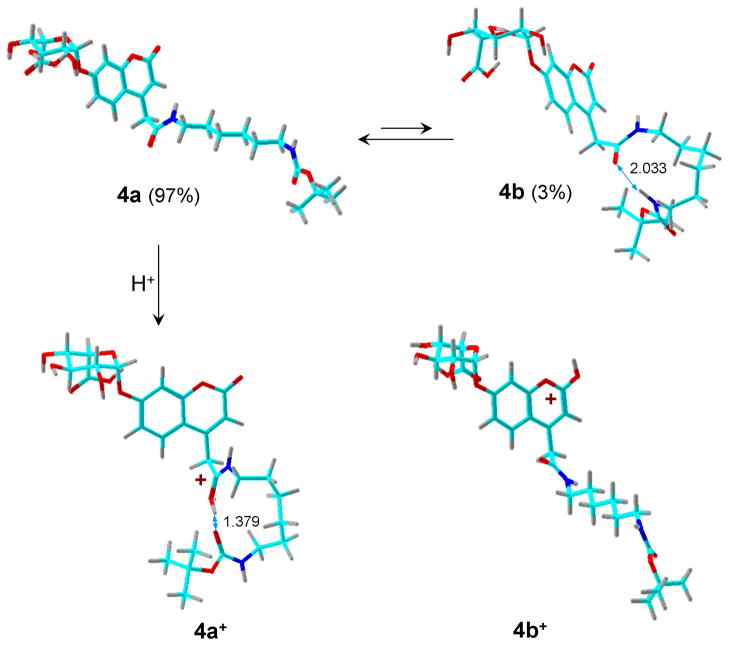
Geometry optimization of the glycoside MPS-II product 4 yielded two conformers (4a and 4b, Figure 5). The extended conformer 4a was 9 kJ mol−1 more stable (ΔG°g,298) and is expected to predominate in the gas phase. The lower ΔG°g,298 of 4a is mainly due to its higher entropy compared to that of 4b as the formation of the 11-membered ring in the latter conformer resulted in a 30 J mol−1 K−1 entropy decrease at 298 K. Protonation of 4a gave ion 4a+ in which the protonated amide group was H-bonded to the carbamate carbonyl (Figure 5). Protonation at the carbamate carbonyl resulted in an unstable ion structure that upon gradient optimization collapsed to a conformer of 4a+. Protonation at the coumarin carbonyl gave tautomer 4b+ which was less stable than 4a+. In view of the relative stabilities of 3a+-3d+, we presumed that 4a+ was the most stable ion structure and other tautomers were not studied. Table 1 data show that the proton affinity of 4a (PA = 970 kJ mol−1) was slightly higher than that of 3a. However, the entropy loss upon formation of the 13-membered H-bonding ring in 4a+ lowered the gas-phase basicity of 4a (GB = 918 kJ mol−l) below that of 3a (GB = 924 kJ mol−1).
Figure 5.
B3LYP/6-31+G(d,p)-optimized structures of ion tautomers of 4+. The atoms are color-coded as follows: Green = C; red = O; blue = N, gray = H.
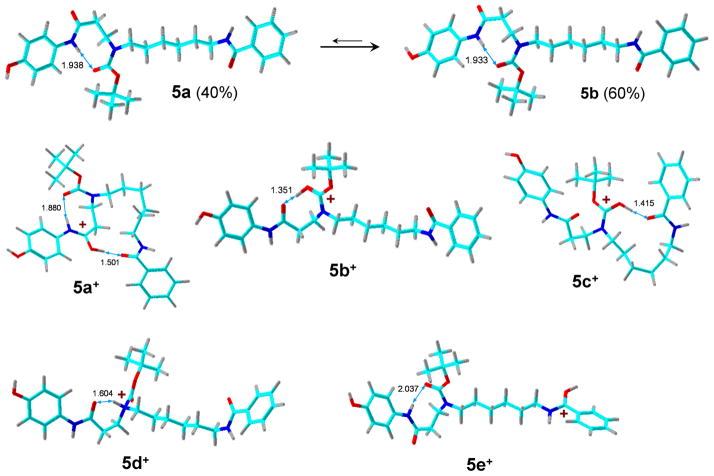
The optimized structures of the GLA product show an intramolecular H bond between the carbamate carbonyl and the aminophenol N—H bond. The phenol OH group rotamers 5a and 5b were nearly isoenergetic. Protonation of 5a/5b was studied for several ion tautomers (5a+-5e+). The lowest-energy ion structure was protonated at the aminophenol amide carbonyl which was H-bonded to the benzamide carbonyl (5a+, Figure 6). The second most stable structure (5b+) was protonated at the carbamate carbonyl which was tightly H-bonded to the aminophenol amide (Figure 6). Attempts to move the proton in 5b+ to the aminophenol amide carbonyl resulted in spontaneous collapse back to structure 5b+, indicating that the tertiary carbamate group was more basic. Structures 5a+ and 5b+ were very close in energy; 5a+ was preferred by MP2 calculations, whereas 5b+ was preferred by B3LYP calculations. Ion 5b+ was slightly more stable than its conformer 5c+ which had an H-bond between the protonated carbamate carbonyl and the benzamide group. Both 5a+ and 5b+ were substantially more stable than the N-protonated structure 5d+. We note that a definite assignment of the global energy minimum for this group of ions would require an exhaustive conformational search for each tautomer, which we have not performed.
Figure 6.
B3LYP/6-31+G(d,p)-optimized structures of ion tautomers of 5+. The atoms are color-coded as follows: Green = C; red = O; blue = N, gray = H.
The GB of the tertiary carbamate 5a (950 kJ mol−1) is notably higher than those of the secondary carbamates 3a and 4a. The relatively high basicity of the amide group in 5a is presumably due to π-conjugation with the electron-rich aminophenol system that stabilizes the cation. This is consistent with an experimental observation that protonation of 5 in electrospray is notably more efficient than protonation of coumarin conjugates and results in lower limits of detection for the enzyme products and their related internal standards.8
Dissociation Mechanisms for the Elimination of Isobutylene and Carbon Dioxide
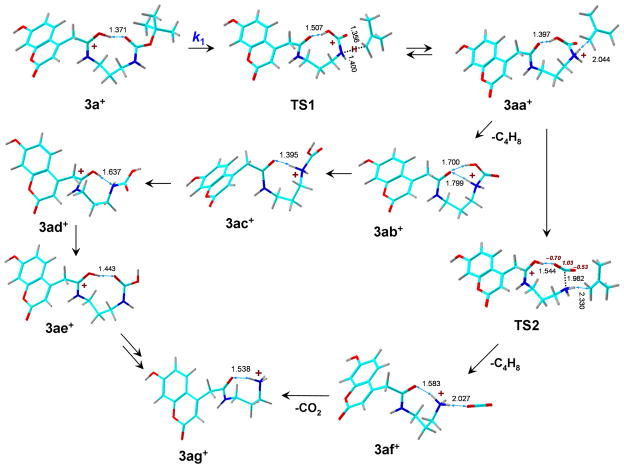
The elimination of isobutylene was studied for the coumarin-derived ion 3a+. The elimination must involve cleavage of the t-butyl C—O bond with a transfer of one of the t-butyl hydrogen atoms onto the product ion. The hydrogen acceptor groups and the timing of the bond cleavage and proton migration were a priori unknown and were investigated by mapping the potential energy surface along two pathways. The pertinent relative energies are listed in Table 2 and arrayed in the potential energy diagram (Figure 7a). Pathway A (Scheme 2) commenced with t-butyl hydrogen atom transfer onto the carbamate nitrogen through TS1 in which the t-butyl C—O bond was practically completely broken at d(C—O) = 3.121 Å. Concomitantly, a proton from the coumarin amide group migrated to the carbamate moiety to reform the COOH group. The migration required a TS energy of 145 kJ mol−1 (Table 2). The intermediate in this pathway was an ion-molecule complex between an N-protonated carbamic acid and neutral isobutylene (3aa+) which was 112 kJ mol−1 above 3a+. Direct elimination of i-C4H8 from 3aa+ leads to a high-energy tautomer of the m/z 321 ion which is the N-protonated carbamic acid 3ab+. The loss of i-C4H8 from 3aa+ was presumed to occur at the thermochemical threshold which was 142 kJ mol−1 relative to 3a+ (Table 2). If formed, 3ab+ can be expected to isomerize by exothermic proton migrations and chain rotations to more stable ions 3ac+, 3ad+, and 3ae+ (Table 2).
Table 2.
Ion Dissociation and Transition State Energies.
| Reaction | Relative Energya,b |
|||
|---|---|---|---|---|
| B3LYP | B3LYP | MP2 | B3-MP2c | |
| 6-31+G(d,p) | 6-311++G(2d,p) | 6-311++G(2d,p) | ||
| 3a+ → 3aa+ | 118 | 106 | 118 | 112 |
| 3a+ → TS1 | 141 | 133 | 157 | 145 |
| 3a+ → 3ab+ + i-C4H8 | 144 | 130 | 154 | 142 |
| 3ab+ → 3ac+ | −27 | −23 | −23 | −23 |
| 3ab+ → 3ad+ | −36 | −33 | −28 | −31 |
| 3ab+ → 3ae+ | −85 | −82 | −67 | −74 |
| 3a+ → 3ae+ + i-C4H8 | 59 | 48 | 87 | 68 |
| 3a+ → TS2 | 118 | 103 | 122 | 112 |
| 3a+ → 3af+ + i-C4H8 | 21 | 2 | 26 | 14 |
| 3a+ → 3ag+ + i-C4H8 + CO2 | 40 | 19 | 49 | 34 |
| 3a+ → TS3 | 100 | 97 | 124 | 110 |
| 3a+ → 3ba+ | 83 | 74 | 93 | 84 |
| 3a+ → 3bb+ + i-C4H8 | 117 | 107 | 143 | 125 |
| 3a+ → TS4 | 128 | 118 | 146 | 132 |
| 3a+ → 3bc+ | 27 | 18 | 47 | 33 |
| 3bc+ → 3ae+ + i-C4H8 | 31 | 30 | 41 | 35 |
| 3ae+ → TS5 | 53 | 54 | 39 | 46 |
| 3ae+ → 3bc+ | 58 | 59 | 44 | 51 |
| 3ae+ → TS6 | 103 | 99 | 88 | 93 |
| 3ae+ → 3ag+ + CO2 | −18 | −29 | −39 | −34 |
| 3af+ → 3ag+ + CO2 | 19 | 17 | 23 | 20 |
In units of kJ mol−1.
Including B3LYP/6-31+G(d,p) zero-point energies and referring to 0 K.
From averaged B3LYP and MP2 single point energies.
Figure 7.
(a) B3-MP2/6-311++G(2d,p) potential energy surface for dissociations of 3a+ and (b,c) RRKM kinetics of elimination of i-C4H8. (b) Rate constants for elimination through TS1 (k1) and TS3 (k3); (c) Calculated fractions of non-dissociating 3a+ at the indicated reaction times: Black circles: 1 ms; black squares: 100 μs; blue circles: 42 μs.
Scheme 2.
Pathway A for dissociations of ion 3a+. The atoms are color-coded as follows: Green = C; red = O; blue = N, gray = H.
An alternative branch of pathway A from 3aa+ was considered that involved carbamate proton migration to the amide carbonyl in the ion-molecule complex, accompanied by cleavage of the carbamate N—CO2 bond through TS2 which was practically isoenergetic with 3aa+ (Scheme 2). The resulting ion-molecule complex (3af+) contained a weakly bound CO2 molecule which required only 20 kJ mol−1 to be eliminated to give the m/z 277 product ion (3ag+). The fact that intermediates 3aa+ and TS2 have nearly identical energies which are lower than that of TS1 (Table 2) suggests that the isobutylene elimination step, 3aa+ → TS2 → 3af+, should be followed by fast elimination of CO2 from complex 3af+ which does not involve a high TS energy. These features would have a large effect on the dissociation kinetics, as discussed later.
Pathway B involves an initial transfer of a t-butyl H atom to the carbamate oxygen atom (Scheme 3). The pertinent TS (TS3) showed a nearly complete cleavage of the t-butyl C—O bond and led to an ion-molecule complex (3ba+) which was 84 kJ mol→1 above 3a+ (Table 2). Elimination of i-C4H8 from complex 3ba+ can occur endothermically to form ion 3bb+ at 125 kJ mol→1 above 3a+. An alternative branch involves rotation of the carbamate COOH group through TS4 (ETS4 = 132 kJ mol→1 relative to 3a+) to give intermediate 3bc+ which then loses i-C4H8 to form the m/z 321 ion as structure 3ae+ which is 57 kJ mol→1 lower energy than 3bb+. The elimination of CO2 from both 3ae+ and 3bb+ must involve proton migration. One possible branch of pathway B proceeds through TS5 to transfer the amide proton onto the carbamate nitrogen and weaken the N—CO2 bond in intermediate 3ac+ which is 51 kJ mol→1 above 3ae+. Ion 3ac+ reacts by transferring the carbamate COOH proton onto the amide carbonyl with concomitant cleavage of the N—CO2 bond in TS6 which is 95 kJ mol−1 above 3ae+ and represents the highest energy point in the entire pathway B.
Scheme 3.
Pathway B for dissociations of ion 3a+. The atoms are color-coded as follows: Green = C; red = O; blue = N, gray = H.
Pathways A and B differ in the initial step of t-butyl hydrogen migration which forms isomeric intermediates 3aa+ and 3ba+ through TS1 and TS3, respectively. These are the high points on the potential energy surface for the elimination of i-C4H8. We used RRKM calculations to investigate the unimolecular kinetics for loss of i-C4H8 through TS1 and TS3, as shown in Figure 7b and 7c. The pathway B dissociation was ≥3 orders of magnitude faster than the pathway A dissociation in the entire energy interval and both showed substantial kinetic shifts (Figure 7b). The ion dissociation time scale was limited by the ion drift time through the collision cell which was estimated to be 75 μs for m/z 377 ions at ELAB = 5 eV. A broader range of up to 1 ms was considered in the calculations to account for loss of kinetic energy. Under the conditions of the 1 ms dissociation time, the precursor ion showed no substantial (>1%) depletion up to ca. 2.07 eV internal energy even when the dissociation proceeded through the kinetically more favorable pathway B (Figure 7c). The RRKM dissociation onset taken at 1% dissociation (2.07 eV) is still higher than the upper energy estimate from the CID experiments (1.86 eV, vide supra). This indicates that the CID measurements involved multiple collisions that increased the ion internal energy to 2.07 eV needed for 1% dissociation to be observed on a 1 ms time scale. The consequent elimination of CO2 from the m/z 321 intermediate should occur spontaneously if proceeding through pathway A. This is incompatible with the observation of two separate dissociation onsets for the elimination of i-C4H8 and CO2. The calculated TS for the loss of CO2 (95 kJ mol−1 in TS6 from from 3ae+, Table 2) indicates that additional internal energy is required in the precursor ion to drive the two-step dissociation.
Conclusions
Protonation and dissociations of three t-butyl carbamates that are used in newborn screening of lysosomal storage disorders were investigated by experiment and theory. t-Butyl carbamates containing aminophenol amide and tertiary carbamate groups (5) were calculated to have greater gas-phase basicities than compounds containing a coumarin amide and secondary carbamate groups (3 and 4). This was consistent with the higher ionization efficiencies in electrospray observed for the first group of compounds. The favorable protonation sites in type 3 and 4 compounds were at the coumarin amide oxygens in both the gas phase and methanol solution. In contrast, the favorable protonation sites in type 5 compounds differed in the gas phase and methanol solution. Collision-induced dissociation of type 3 ions showed a sequential dissociation by loss of i-C4H8 and CO2. Computational investigation of reaction pathways identified several transition states and intermediates and allowed us to propose a plausible mechanism for this practically important ion dissociation.
Acknowledgments
Support of this research by the NIH-National Institute of Diabetes, Digestive and Kidney Diseases (Grant R01 DK067859) and the National Science Foundation (Grant CHE-1055132) is gratefully acknowledged. The Department of Chemistry Computational Center has been supported jointly by the NSF and University of Washington.
References
- 1.Pearson AJ, Roush WR. Handbook of Reagents for Organic Synthesis - Activating Agents and Protecting Groups. John Wiley & Sons; 1999. p. 83. online version http://www.knovel.com/web/portal/browse/display?_EXT_KNOVEL_DISPLAY_bookid=2129&VerticalID=0. [Google Scholar]
- 2.Ashworth IW, Cox BG, Meyrick B. Kinetics and mechanism of N-Boc cleavage: Evidence of a second-order dependence upon acid concentration. J Org Chem. 2010;75:8117–8125. doi: 10.1021/jo101767h. [DOI] [PubMed] [Google Scholar]
- 3.Garner GV, Gordon DB, Tetler LW, Sedgwick RD. Fast atom bombardment mass spectrometry of butyloxycarbonyl protected (BOC) amino acids. Org Mass Spectrom. 1988;18:486–488. [Google Scholar]
- 4.Gelb MH, Tureček F, Scott CR, Chamoles NA. Direct multiplex assay of enzymes in dried blood spots by tandem mass spectrometry for the newborn screening of lysosomal storage disorders. J Inherit Metabol Dis. 2006;29:397–404. doi: 10.1007/s10545-006-0265-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tureček F, Scott CR, Gelb MH. Methods in Molecular Biology. Totowa, NJ, United States: 2007. Tandem mass spectrometry in the detection of inborn errors of metabolism for newborn screening; p. 359. [DOI] [PubMed] [Google Scholar]; Quantitative Proteomics by Mass Spectrometry. pp. 143–157. [Google Scholar]
- 6.Li Y, Scott CR, Chamoles NA, Ghavami A, Pinto BM, Tureček F, Gelb MH. Direct multiplex assay of lysosomal enzymes in dried blood spots for newborn screening. Clin Chem. 2004;50:1785–1796. doi: 10.1373/clinchem.2004.035907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duffey TA, Bellamy G, Diaker J, Elliott S, Glass M, Scott CR, Tureček F, Gelb MH. Triplex tandem mass spectrometry for newborn screening of Fabry, Pompe and mucopolysaccharidosis I (Hurler Syndrome): Results of a pilot study. Clin Chem. 2010;56:1854–1861. doi: 10.1373/clinchem.2010.152009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spáčil Z, Elliott S, Reeber SL, Gelb MH, Scott CR, Tureček F. Comparative fast liquid chromatography: Application to newborn screening of Pompe, Fabry, and Hurler diseases. Anal Chem. 2011;84:4822–4828. doi: 10.1021/ac200417u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blanchard S, Sadílek M, Scott CR, Tureček F, Gelb MH. Tandem mass spectrometry for the direct assay of lysosomal enzymes in dried blood spots: Application to screening newborns for mucopolysaccharidosis I (Hurler-Scheie syndrome) Clin Chem. 2008;54:2067–2070. doi: 10.1373/clinchem.2008.115410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang D, Wood T, Sadilek M, Scott CR, Turecek F, Gelb MH. Tandem mass spectrometry for the direct assay of enzymes in dried blood spots: Application to newborn screening for mucopolysaccharidosis II (Hunter disease) Clin Chem. 2007;53:137–140. doi: 10.1373/clinchem.2006.077263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolfe B, Blanchard S, Sadílek M, Scott CR, Tureček F, Gelb MH. Tandem mass spectrometry for the direct assay of lysosomal enzymes in dried blood spots: Application to screening newborns for mucopolysaccharidosis II (Hunter syndrome) Anal Chem. 2011;83:1152–1156. doi: 10.1021/ac102777s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khaliq T, Sadilek M, Scott CR, Tureček F, Gelb MH. Tandem mass spectrometry for the direct assay of lysosomal enzymes in dried blood spots: Application to screening newborns for mucopolysaccharidosis IVA (Morquio A) Clin Chem. 2011;57:128–131. doi: 10.1373/clinchem.2010.149880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duffey TA, Sadilek M, Scott CR, Tureček F, Gelb MH. Tandem mass spectrometry for the direct assay of lysosomal enzymes in dried blood spots: Application to screening newborns for mucopolysaccharidosis VI (Maroteaux-Lamy syndrome) Anal Chem. 2010;82:9587–9591. doi: 10.1021/ac102090v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Remko M. Ab initio MO study of protonation of carbamic acid, methyl carbamate and methyl N-methylcarbamate. Collect Czech Chem Commun. 1988;53:1141–1148. [Google Scholar]
- 15.Gregersen JA, Hao C, Turecek F. Electron super-rich radicals. III. On the peculiar behavior of the aminodihydroxymethyl radical in the gas phase. J Phys Chem A. 2009;113:5855–5864. doi: 10.1021/jp9019987. [DOI] [PubMed] [Google Scholar]
- 16.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenberg JL, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Montgomery JA, Jr, Peralta JE, Ogliaro F, Bearpark M, Heyd JJ, Brothers E, Kudin KN, Staroverov VN, Kobayashi R, Normand J, Raghavachari K, Rendell A, Burant JC, Iyengar SS, Tomasi J, Cossi M, Rega N, Millam JM, Klene M, Knox JE, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Martin RL, Morokuma K, Zakrzewski VG, Voth GA, Salvador P, Dannenberg JJ, Dapprich S, Daniels AD, Farkas O, Foresman JB, Ortiz JV, Cioslowski J, Fox DJ. Gaussian 09, Revision A.02; Gaussian, Inc; Wallingford CT: 2009. [Google Scholar]
- 17.(a) Becke AD. New mixing of Hartree-Fock and local density-functional theories. J Chem Phys. 1993;98:1372–1377. [Google Scholar]; (b) Becke AD. Density functional thermochemistry. III. The role of exact exchange. J Chem Phys. 1993;98:5648–5652. [Google Scholar]; (c) Stephens PJ, Devlin FJ, Chabalowski CF, Frisch MJ. Ab Initio calculation of vibrational absorption and circular dichroism spectra using density functional force fields. J Phys Chem. 1994;98:11623–11627. [Google Scholar]
- 18.Rauhut G, Pulay P. Transferable scaling factors for density functional derived vibrational force fields. J Phys Chem. 1995;99:3093–3100. [Google Scholar]
- 19.Møller C, Plesset MS. A note on an approximation treatment for many-electron systems. Phys Rev. 1934;46:618–622. [Google Scholar]
- 20.Tureček F. Proton affinity of dimethyl sulfoxide and relative stabilities of C2H6OS molecules and C2H7OS+ ions. A comparative G2(MP2) ab initio and density functional theory study. J Phys Chem A. 1998;102:4703–4713. [Google Scholar]
- 21.Čížek J, Paldus J, Šroubková L. Cluster expansion analysis for delocalized systems. Int J Quantum Chem. 1969;3:149–167. [Google Scholar]
- 22.Purvis GD, Bartlett RJ. A full coupled-cluster singles and doubles model. The inclusion of disconnected triples. J Chem Phys. 1982;76:1910–1918. [Google Scholar]
- 23.Curtiss LA, Raghavachari K, Pople JA. Gaussian-2 theory using reduced Møller-Plesset orders. J Chem Phys. 1993;98:1293–1298. [Google Scholar]
- 24.Tomasi J, Mennucci B, Cammi R. Quantum mechanical continuum solvation models. Chem Rev. 2005;105:2999–3093. doi: 10.1021/cr9904009. [DOI] [PubMed] [Google Scholar]
- 25.Tureček F, Cramer CJ. Thermochemistry of simple enols and enol cation-radicals revisited. A G2(MP2) ab initio study. J Am Chem Soc. 1995;117:12243–12253. [Google Scholar]
- 26.Gilbert RG, Smith SC. Theory of Unimolecular and Recombination Reactions. Blackwell Scientific Publications; Oxford: 1990. pp. 52–132. [Google Scholar]
- 27.Zhu L, Hase WL. Quantum Chemistry Program Exchange. Indiana University; Bloomington: 1994. Program No. QCPE 644. [Google Scholar]
- 28.Frank AJ, Sadílek M, Ferrier JG, Tureček F. Sulfur oxyacids and radicals in the gas phase. A variable-time neutralization-photoexcitation-reionization mass spectrometric and ab initio/RRKM study. J Am Chem Soc. 1997;119:12343–12353. [Google Scholar]
- 29.Chupka WA. Effect of unimolecular decay kinetics on the interpretation of appearance potentials. J Chem Phys. 1959;30:191–211. [Google Scholar]
- 30.Ervin KM. Experimental techniques in gas-phase ion thermochemistry. Chem Rev. 2001;101:391. doi: 10.1021/cr990081l. [DOI] [PubMed] [Google Scholar]