Abstract
Nicotianamine synthase (NAS), the key enzyme in the biosynthetic pathway for the mugineic acid family of phytosiderophores, catalyzes the trimerization of S-adenosylmethionine to form one molecule of nicotianamine. We purified NAS protein and isolated the genes nas1, nas2, nas3, nas4, nas5-1, nas5-2, and nas6, which encode NAS and NAS-like proteins from Fe-deficient barley (Hordeum vulgare L. cv Ehimehadaka no. 1) roots. Escherichia coli expressing nas1 showed NAS activity, confirming that this gene encodes a functional NAS. Expression of nas genes as determined by northern-blot analysis was induced by Fe deficiency and was root specific. The NAS genes form a multigene family in the barley and rice genomes.
Graminaceous plants that adopt the Strategy II mechanism of Fe acquisition (Römheld, 1987) secrete Fe chelators, called phytosiderophores, from their roots to solubilize sparingly soluble Fe in the rhizosphere. The amount of phytosiderophore secreted increases dramatically under Fe-deficiency stress. To our knowledge, the MA is the only class of phytosiderophore so far identified in plants (Takagi, 1976). Tolerance to Fe deficiency in graminaceous plants is thought to depend on the quantity of MAs secreted by plants under Fe-deficiency stress (Takagi et al., 1984; Römheld and Marschner, 1986; Marschner et al., 1987; Mori et al., 1987, 1988; Kawai et al., 1988; Mihashi and Mori, 1989; Singh et al., 1993). Of the graminaceous plants, rice secretes the least MAs and is the species most susceptible to Fe deficiency in calcareous soils. Transgenic rice overexpressing the genes of MA biosynthesis under Fe deficiency should secrete greater amounts of MAs and tolerate Fe deficiency.
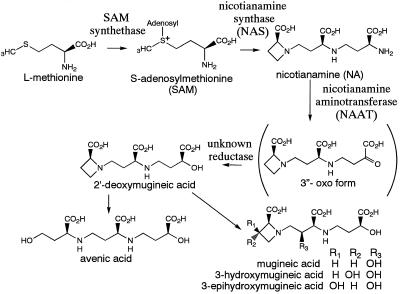
The biosynthetic pathway for MAs has been determined (Fig. 1). SAM is synthesized from Met by SAM synthetase. Subsequently, three molecules of SAM are combined to form one molecule of NA by NAS. NA is then converted to [3“-keto acid] by NAAT, and deoxymugineic acid is synthesized by the subsequent action of a reductase. A further series of hydroxylation steps produces the other MA members from deoxymugineic acid (Fig. 1; Mori and Nishizawa, 1987; Shojima et al., 1989, 1990; Ma and Nomoto, 1993). Three cDNAs encoding SAM synthetase from barley (Hordeum vulgare L.) roots have been cloned (Takizawa et al., 1996; accession nos. D63835, D85273, and D85238), but these genes are not induced by Fe deficiency. Recently, NAAT was purified and two NAAT cDNAs (naat-A and naat-B) were cloned from Fe-deficient barley roots (Takahashi et al., 1997). naat-A expression was shown to be specifically induced in Fe-deficient roots. A clone encoding the putative mugineic acid synthase Ids3, which converts deoxymugineic acid to mugineic acid and is strongly induced by Fe deficiency, was cloned from the barley genome using differential hybridization (Nakanishi et al., 1993; accession no. D37796).
Figure 1.
Biosynthetic pathway of the MAs of phytosiderophores.
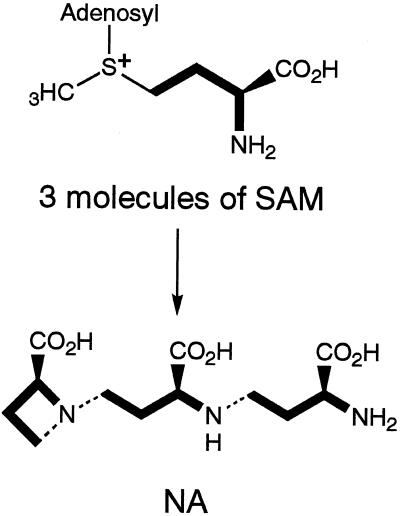
The synthesis of NA from SAM is similar to polyamine synthesis from decarboxy-SAM. In contrast to spermidine synthase (Pajula et al., 1979), however, NAS catalyzes the polymerization of three SAM molecules with the release of adenine and the azetidine ring formation at the same time (Fig. 2). Therefore, NAS is a novel type of enzyme. Previously, we reported the partial purification of NAS from the roots of Fe-deficient barley and showed that NAS activity was induced under Fe-deficiency stress (Higuchi et al., 1994, 1995; Kanazawa et al., 1995). Since NAS is vulnerable to degradation by proteases using the purification method previously described, it has been difficult to purify sufficient quantities to determine its partial amino acid sequence. In this paper we describe an improved purification procedure for NAS that allowed us to obtain sufficient quantities for amino acid sequencing. Subsequently, we cloned and characterized several genes encoding NAS or NAS-like proteins.
Figure 2.
NAS catalyzes trimerization of SAM and ring formation to synthesize NA. Bold lines indicate the unit incorporated in NA. Hatched lines indicate newly formed N—C bonds.
MATERIALS AND METHODS
Preparation of Plant Material
Barley (Hordeum vulgare L. cv Ehimehadaka no. 1) was grown in hydroponic culture as previously described (Higuchi et al., 1994). Two weeks after Fe-deficiency treatment, when severe Fe chlorosis appeared on the fourth or fifth leaf, the roots were harvested, crushed in liquid N2, and stored at −80°C until use.
Assay of NAS Activity
Enzyme solutions were equilibrated with reaction buffer (50 mm Tris, 1 mm EDTA, 3 mm DTT, 10 μm p-APMSF, and 10 μm E-64, pH 8.7) and concentrated by ultrafiltration using Ultrafree C3LGC NMWL10000 (Millipore). The details of the method used for detecting NAS activity were described by Higuchi et al. (1996a).
Purification of NAS Proteins
The roots were crushed into a fine powder in liquid N2 and then homogenized in a household juicer with 200 mL of extraction buffer (0.2 m Tris, 10 mm EDTA, 5% [v/v] glycerol, 10 mm DTT, 0.1 mm E-64, 0.1 mm p-APMSF, and 5% [w/v] insoluble PVP, pH 8.0) per 100 g fresh weight. The homogenate was centrifuged for 30 min at 22,500g. Ammonium sulfate was added to the supernatant to yield a final concentration of 400 mm. After 1 h on ice, the precipitate was removed by centrifugation for 30 min at 22,500g.
The supernatant was loaded onto a TSK gel Butyl Toyopearl 650M column (10-mL bed volume for every 100 g fresh weight roots; Fractogel TSK Butyl-650M, Merck, Darmstadt, Germany) that had been equilibrated with a loading buffer (20 mm Tris, 1 mm EDTA, 3 mm DTT, 400 mm (NH4)2SO4, and 0.1 mm p-APMSF, pH 8.0). NAS was eluted with elution buffer (10 mm Tris, 1 mm EDTA, 3 mm DTT, 0.1 mm p-APMSF, 5% glycerol, and 0.05% Chaps, pH 8.0).
KCl was added to the active fraction to give a final concentration of 0.4 m, and 1 m potassium phosphate buffer (pH 8.0) was added to a final concentration of 1 mm. The NAS fraction was loaded onto a hydroxyapatite column (100 to approximately 350 mesh; Nacalai Tesque, Kyoto, Japan; 10-mL bed volume for each 100 mg of protein in the NAS fraction) equilibrated with loading buffer (1 mm potassium phosphate buffer, 10 mm KCl, 3 mm DTT, and 0.1 mm p-APMSF, pH 8.0). NAS activity was eluted in the void volume.
The resulting NAS fraction was then loaded onto another TSK gel Butyl Toyopearl 650M column (1-mL bed volume for each 10 mg of protein in the NAS fraction) and eluted in the manner described above.
The active fraction was loaded onto a DEAE-Sepharose fast-flow column (5-mL bed volume for each 25 mg of protein in the NAS fraction, Pharmacia) equilibrated with the loading buffer (20 mm Tris, 1 mm EDTA, 3 mm DTT, and 0.1 mm p-APMSF, pH 8.0). The column was eluted using a stepwise gradient with increasing KCl concentration in the elution buffer (20 mm Tris, 1 mm EDTA, 3 mm DTT, 0.1 mm p-APMSF, and 0.05% Chaps, pH 8.0, with 50, 100, 150, or 200 mm KCl). The fraction with NAS activity was eluted with the buffer that contained 150 mm KCl.
The active fraction was loaded onto an Ether Toyopearl 650M column (0.5-mL bed volume; Fractogel TSK Butyl-650M, Merck) equilibrated with loading buffer (20 mm Tris, 1 mm EDTA, 3 mm DTT, 0.1 mm p-APMSF, and 1.2 m [NH4]2SO4, pH 8.0). NAS did not bind to the column and was eluted in the void volume. This unbound fraction was subsequently loaded onto a Butyl Toyopearl 650M column (bed volume, 0.3 mL) equilibrated with loading buffer (20 mm Tris, 1 mm EDTA, 3 mm DTT, 0.1 mm p-APMSF, and 1.2 m [NH4]2SO4, pH 8.0). The column was washed with buffer (20 mm Tris, 1 mm EDTA, 3 mm DTT, 0.4 m [NH4]2SO4, and 0.1 mm p-APMSF, pH 8.0) and the NAS activity was then eluted with the elution buffer (10 mm Tris, 1 mm EDTA, 3 mm DTT, 0.1 mm p-APMSF, 5% glycerol, and 0.05% Chaps, pH 8.0).
The proteins in the active fraction were separated by SDS-PAGE at 4°C using 11% acrylamide slab gels. After SDS-PAGE the gel was stained with 0.3 m CuCl2 (Dzandu et al., 1988), and then the stained bands were cut out. The gel fragments were destained with 0.25 m EDTA/0.25 m Tris (pH 9.0) and then homogenized with 1% SDS, 25 mm Tris, and 192 mm Gly. Each homogenate was placed in an elution cup and polypeptides were electroeluted with SDS-free buffer containing 25 mm Tris and 192 mm Gly. The NAS fractions from all the steps described above were stored at 4°C with 10 μm E-64.
Gel Filtration
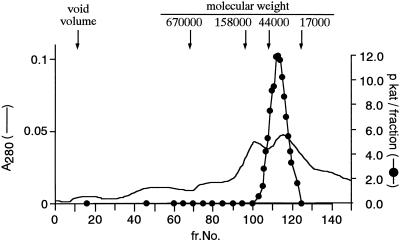
The active NAS fraction after hydroxyapatite chromatography was applied to a Sephacryl S300HR column (1.5 × 71 cm, 125 mL; Pharmacia) equilibrated with buffer (50 mm Tris, 1 mm EDTA, 100 mm KCl, 0.05% Chaps, 0.1 mm p-APMSF, and 3 mm DTT, pH 8.0). The column was calibrated with standard proteins: thyroglobulin (Mr 670,000), γ-globulin (Mr 158,000), ovalbumin (Mr 44,000), and myoglobin (Mr 17,000). Linear flow was 10 cm h−1.
Determination of Partial Amino Acid Sequence
Proteins were digested with CNBr using the method of Gross (1967) with the following modifications. NAS peptide isolated on SDS-PAGE gels was eluted from the gel by homogenizing in a 10-fold volume of 70% (v/v) formic acid containing 1% (w/v) CNBr in a 1.5-mL tube and by subsequent overnight incubation at 4°C. The supernatant was collected, dried under a partial vacuum, resuspended in the SDS-PAGE sample buffer, and incubated overnight at room temperature. After the proteins were digested, the small peptides were separated by electrophoresis using Tricine SDS-PAGE (Schägger and Jagow, 1987) in 16.5% (w/v) acrylamide gels. The peptides were transferred onto a PVDF membrane by electroblotting (Towbin et al., 1979) and stained with amido black. Each band on the PVDF membrane was cut out and the amino acid sequence was determined by automated Edman degradation in a gas-phase sequencer (model 492A protein sequencer, model 785A programmable absorbance detector, and model 140C microgradient system, Applied Biosystems).
Cloning of nas Genes
A pYH23 cDNA library prepared from the poly(A+) RNA of Fe-deficient barley roots was screened with a PCR product corresponding to the sequence of the NAS homolog of rice, expressed sequence tag cDNA clone RICR2562A (D24790), and RICR0168A (D23792). The primers used were:5′-ATGGAGGCTCAGAACCAAGAGGTCGC-3′(N-terminal forward primer) and 5′-GGATGAGCTCCTCCCGCATCGCCT-3′ (N-terminal reverse primer) from D24790, and 5′-CAACCTGAGCAAGCTGGAGTACGACC-3′ (internal forward primer) and 5′-TTCTTCTCGGCCGCCATGCCCACGA-3′ (internal reverse primer) from D23792 (Fig. 5). The probe obtained by PCR amplification was labeled with [α-32P]dATP using the random primer-labeling kit version 2 (TaKaRa, Shiga, Japan). The labeled DNA was purified in a ProbeQuant G-50 microcolumn (Pharmacia). The isolated cDNA clones were sequenced by the Thermo Sequenase Cycle Sequencing Kit (Shimadzu, Tokyo, Japan), following the protocol of the manufacturer, using a DNA sequencer (model DSQ-1000L, Shimadzu).
Figure 5.
Elution pattern of NAS activity from the gel-filtration column. The black circles indicate NAS activity. The solid line indicates protein concentration monitored at 280 nm. The active NAS fraction after hydroxyapatite chromatography was applied to a Sephacryl S300HR column (1.5 × 71 cm, 125 mL). The column was calibrated with standard proteins: thyroglobulin (Mr 670,000), γ-globulin (Mr 158,000), ovalbumin (Mr 44,000), and myoglobin (Mr 17,000). The linear flow was 10 cm h−1.
Expression in Escherichia coli
An EcoRI site was introduced near the first ATG of the nas1 cDNA, and a PstI site was introduced near the first stop codon of the nas1 cDNA by PCR mutagenesis. The primers used were 5′-GAGAGAGAGAATTCGCCATGGATGCCCAGAACAAGGAG-3′ and 5′-GAGAGAGAGGATCCCTGCAGCTTCAATCAAAAGGCCAGCTC-3′ (EcoRI and PstI sites are underlined). An EcoRI-PstI fragment containing the nas1-coding sequence was excised from the PCR product and cloned into pMAL-c2 (New England Biolabs) to give pMAL-NAS1, which was introduced into E. coli XL1-Blue, and the recombinant bacteria were cultured in Luria-Bertani medium containing 100 μg mL−1 ampicilllin and 20 μg mL−1 tetracyclin at 37°C until the A600 of the culture reached 0.5. At this time isopropyl β-d-thiogalactopyranoside was added to a final concentration of 0.3 mm. After 4 h a crude extract from the cells was prepared as described by the manufacturer of the pMAL kit.
Northern-Blot Analysis
The hybridization probe used for northern blots was a HindIII-NotI restriction fragment containing the full-length nas1 cDNA labeled with [α-32P]dATP. The labeled DNA was purified in a ProbeQuant G-50 microcolumn (Pharmacia). Total RNA was isolated from the roots or leaves of barley according to the procedure of Naito et al. (1988). RNA (5 μg per lane) was separated on 1.4% (w/v) agarose gels containing 5% (v/v) formaldehyde and blotted onto membranes (Hybond-N+, Amersham). The membrane was hybridized with the labeled probe in 0.5 m Church phosphate buffer (Church and Gilbert, 1984), 1 mm EDTA, and 7% (w/v) SDS with 100 μg mL−1 salmon sperm DNA at 65°C overnight. After hybridization the blot was washed twice with 40 mm Church phosphate buffer and 1% (w/v) SDS at 65°C for 10 min and at high stringency with 2× SSPE and 0.1% (w/v) SDS at 65°C for 10 min, and radioactivity was then detected using a BAS-2000 image analyzer (Fuji, Tokyo, Japan).
Southern-Blot Analysis
Genomic DNA prepared from leaves of barley and rice was digested with BamHI, EcoRI, or HindIII, separated on a 0.8% (w/v) agarose gel (10 μg per lane), and alkali transferred onto a Hybond-N+ membrane. The membrane was hybridized with the same probes under the same conditions as described above.
RESULTS
Improvement of the Extraction and Column Chromatography Procedure Used to Purify NAS from Fe-Deficient Barley Roots
We previously reported the partial purification of NAS (Higuchi et al., 1994), but we were unable to obtain sufficient amounts of protein to determine its partial amino acid sequence. Subsequently, we discovered that E-64, a thiol protease inhibitor, was very effective in protecting NAS from degradation (Higuchi et al., 1996a). In this study frozen roots were crushed to a fine powder in liquid N2 and then rapidly homogenized with buffer containing 0.1 mm E-64 to avoid degradation of NAS. This improvement increased the recovery of NAS about 20-fold.
In the previous work NAS activity was detected in the 30- to 35-kD protein fraction recovered from the SDS-polyacrylamide gel after the removal of SDS (Higuchi et al., 1994), but the rate of recovery was very low. Therefore, we further improved the column chromatography procedures. NAS is relatively hydrophobic and a buffer containing Chaps effectively increased the rate of recovery and the resolution of the column chromatography. Several ion-exchange chromatography media were tested, and DEAE-Sepharose fast-flow and DEAE Sephacel were found to be the most effective. Both Butyl Toyopearl and another hydrophobic chromatography medium, Ether Toyopearl, effectively removed impurities from the 30- to 35-kD fraction.
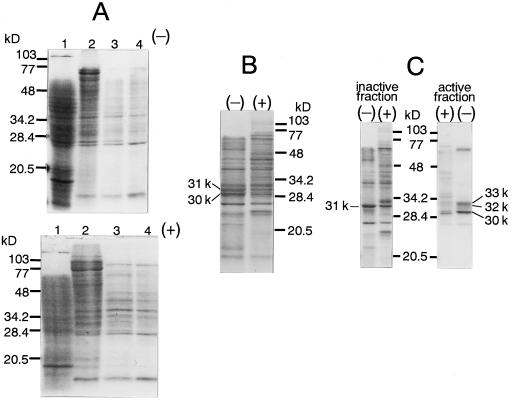
Comparison of the Peptides on SDS-PAGE in the Purified NAS Fraction from Fe-Deficient Barley Roots and Control Barley Roots
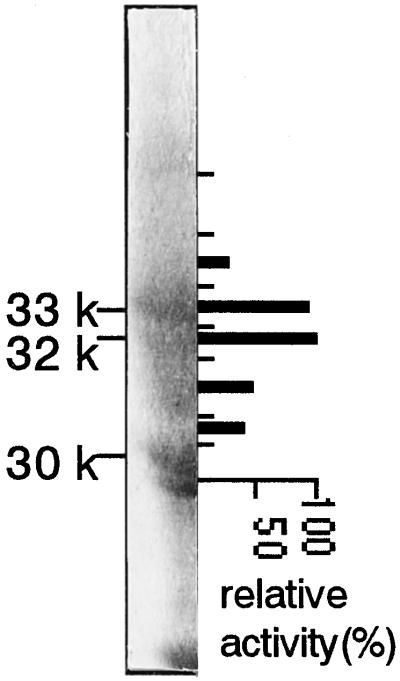
In general, NAS activity was detected as a broad peak on the SDS-polyacrylamide gel from 30 to 35 kD. To identify the NAS protein, we compared the peptides in the NAS fraction obtained from Fe-deficient barley roots with those from Fe-sufficient (control) barley roots. NAS was purified from 200 g of Fe-deficient and control roots. The NAS activity of the control roots was about one-quarter of the Fe-deficient roots at each purification step. The active NAS fraction from each purification step was analyzed by SDS-PAGE (Fig. 3). An almost identical pattern was observed in both Fe-deficient and control roots before the DEAE-Sepharose step (Fig. 3A). After the DEAE-Sepharose step it became clear that the 30- and 31-kD peptides were induced by Fe deficiency (Fig. 3B). After the Ether Toyopearl step, the 31-kD peptide was eliminated from the active NAS fraction. Two more Fe-deficiency-inducible peptides that were 32 and 33 kD in size were also detected in the active NAS fraction (Fig. 3C). Both the 32- and 33-kD peptides had NAS activity, but the 30-kD peptide was inactive (Fig. 4).
Figure 3.
Comparison of NAS purification from Fe-deficient (−) and Fe-sufficient (control, +) barley roots. SDS-PAGE was carried out using 12.5% acrylamide slab gels (Laemmli, 1970). Gels were stained with Coomassie brilliant blue. A, top, From Fe-deficient barley roots; bottom, from control barley roots. Lanes 1, Crude extract, 200 μg of protein. Lanes 2, After Butyl Toyopearl 650M, 100 μg of protein. Lanes 3, After hydroxyapatite, 20 μg of protein. Lanes 4, After Butyl Toyopearl 650M, 15 μg of protein. B, After DEAE-Sepharose fast flow, 25 μg of protein. C, After Ether Toyopearl 650M, one-quarter of each fraction.
Figure 4.
Preparative SDS-PAGE was carried out using 11% acrylamide slab gels. A portion of the gel in this figure was stained with Coomassie brilliant blue and the rest of the gel was stained with Cu. The gel containing proteins between 30 and 35 kD in size was cut into seven fragments (indicated by the short lines). The recovery was low but NAS activity was reproducibly detected by elimination of SDS and Cu during electroelution of peptide from the gels. The thick bars indicate relative NAS activity of peptides from each gel fragment. The activity of the most active fraction was set at 100%.
Estimation of the Mr of NAS Using Gel Filtration
According to previous data, the Mr of NAS is between 40,000 and 50,000 (Higuchi et al., 1994), but this did not correspond with the value estimated by SDS-PAGE. In this study the buffer containing Chaps effectively increased the recovery of active enzyme and the resolution of the peak pattern of the column. Consequently, the Mr of NAS was more accurately determined to be 35,000 (Fig. 5) and corresponds well to the value estimated by SDS-PAGE.
Determination of the Partial Amino Acid Sequence of NAS
The active NAS fraction was purified from 1 kg of Fe-deficient barley roots. Because 32- and 33-kD peptides from the preparative SDS-PAGE gel could not be completely separated from each other, they were digested together with CNBr, whereas the 30-kD peptide was digested separately. The partial amino acid sequences of the fragments obtained from the 32- plus 33-kD peptides and the 30-kD peptide were homologous to each other (Fig. 6). The Mr of the 33- plus 32-kD-1 fragment (Fig. 6) was almost the same as the original; thus, we speculated that this sequence corresponded to the N-terminal region of NAS. A search of the database revealed that these putative NAS amino acid sequences were similar to the translation products of rice expressed sequence tag cDNA clones of unknown function (accession nos. D23792 and D24790), with 80.0% identity in a 33-amino acid overlap in the former and 68.4% identity in a 19-amino acid overlap in the latter (Fig. 6).
Figure 6.
Partial amino acid sequence of combined 32- plus 33-kD peptides and that of the 30-kD peptide. The amino acid sequences of two rice homologs are also shown. The arrows indicate PCR primers (for the DNA sequences of primers, see Methods). IF, Internal forward primer; IR, internal reverse primer; NF, N-terminal forward primer; NR, N-terminal reverse primer.
Cloning and Nucleotide Sequences of cDNA Clones Encoding NAS
PCR amplification of total cDNA prepared from Fe-deficient barley roots using degenerate primers designed from the partial amino acid sequence of barley NAS was not successful. We then used partial sequences from the rice expressed sequence tag clones (Fig. 6, arrows) as the primers. The resulting 205-bp amplification product using N-terminal forward and N-terminal reverse primers and a 274-bp amplification product using internal forward and internal reverse primers were used as probes for hybridization. A cDNA library prepared using poly(A+) RNA from Fe-deficient barley roots was screened and 19 positive clones using the 205-bp probe and 88 positive clones using the 274-bp probe were obtained.
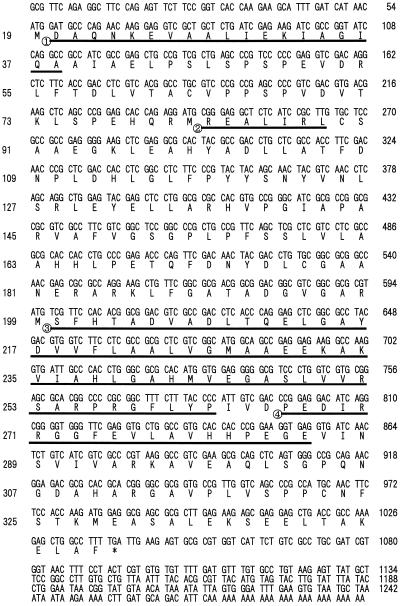
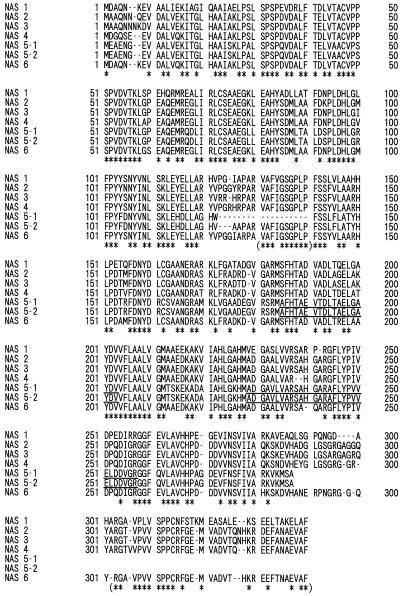
Nucleotide sequence analysis of one of these positive clones, designated nas1, revealed an open reading frame of 985 bp encoding a 328-amino acid polypeptide with a predicted Mr of 35,144. This corresponded well with the size of the NAS polypeptide estimated by SDS-PAGE. The partial amino acid sequences of the 32- plus 33-kD peptide matched the portions of the deduced amino acid sequence of nas1 (Fig. 7, underlined). The predicted pI of 5.2 matched the value estimated by native IEF electrophoresis well (data not shown). Another six NAS-like clones, nas2, nas3, nas4, nas5-1, nas5-2, and nas6, were also obtained (Table I; Fig. 8). The nucleotide sequences for the NAS genes from barley have been deposited in the database and given the following accession numbers: AB010086 (nas1), AB011265 (nas2), AB011264 (nas3), AB011266 (nas4), AB011267 (nas5-1), AB011268 (nas5-2), and AB011269 (nas6). The partial amino acid sequences of the 30-kD peptide matched portions of the deduced amino acid sequence of nas5-1 and nas5-2 (Fig. 8, underlined). The 5′ and 3′ noncoding region of these seven clones differed from each other, except nas5-1 and nas5-2. Sequences of nas5-1 and nas 5-2 were identical, with the exception of the 122 to 140 amino acid residues of nas5-2. nas5-1 may be a deletion clone of nas5-2, although we did not determine whether this deletion clone is an artifact.
Figure 7.
Nucleotide and deduced amino acid sequence of the barley nas1 cDNA clone. The underlined sequences indicate four partial amino acid sequences of fragments from combined 32- plus 33-kD peptides. The nucleotide sequence is numbered from the 5′ end of the cDNA clone and indicated to the right of each row. Amino acid numbers are indicated on the left of each row.
Table I.
Properties of deduced peptides from nas clones
| Clone | Amino Acid Residues | Mr | pI | Identity to nas1 | Identity to nas2 | Identity to nas4 |
|---|---|---|---|---|---|---|
| no. | % | |||||
| nas1 | 328 | 35,144 | 5.20 | — | ||
| nas2 | 336 | 35,839 | 5.07 | 72 | — | |
| nas3 | 336 | 36,013 | 5.47 | 72 | 95 | |
| nas4 | 330 | 35,396 | 4.91 | 73 | 89 | — |
| nas5-1 | 268 | 28,802 | 5.32 | 57 | 58 | 56 |
| nas5-2 | 283 | 30,148 | 5.22 | 61 | 61 | 59 |
| nas6 | 329 | 35,350 | 5.07 | 74 | 89 | 88 |
pI values were predicted by the DNASIS program (Hitachi, Tokyo, Japan).
Figure 8.
Comparison of the deduced amino acid sequences of barley proteins NAS 1 to NAS 6. Asterisks indicate identical amino acid residues in all sequences. Asterisks in parentheses indicate identical amino acid residues in all sequences except NAS 5-1 and 5-2. The underlined sequences indicate the partial amino acid sequences of fragments from 30-kD peptide.
Expression of NAS1 in E. coli
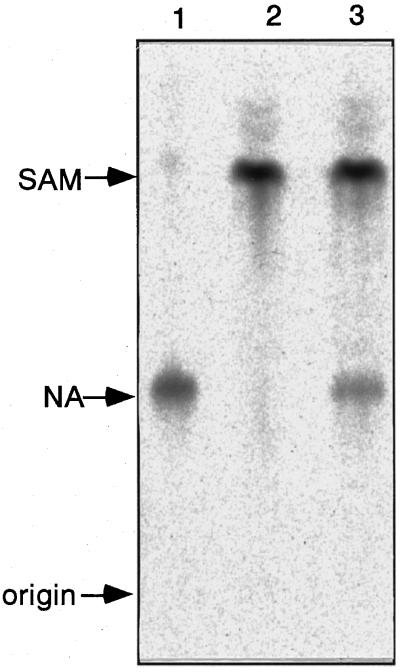
To confirm the enzymatic function of the gene product, nas1 was expressed as a maltose-binding protein fusion in E. coli. The bacterial strains containing the NAS1 expression vector or the empty vector were induced with isopropyl β-d-thiogalactopyranoside, and the crude extracts were analyzed for NAS activity by TLC (Fig. 9). The crude extract from the strain transformed with pMAL-NAS1 had NAS activity, whereas the crude extract from the strain transformed with the vector had no NAS activity.
Figure 9.
TLC analysis of NAS activity assay mixture from E. coli expressing NAS1. Lane 1, Standard NA; lane 2, pMAL only; lane 3, pMAL-NAS1.
Northern-Blot Analysis
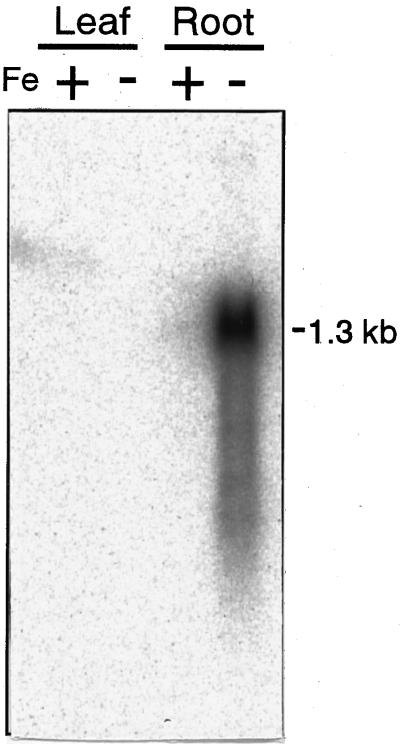
Northern hybridization analysis revealed that nas1-mRNA was not detected in either the leaves or roots of the control plants. In contrast, nas1 was highly expressed in the roots but not in the leaves of Fe-deficient plants (Fig. 10). This result corresponds well with the previously described expression pattern of NAS activity (Higuchi et al., 1994).
Figure 10.
Northern-blot analysis of nas1. RNA was extracted from Fe-deficient (−) and control (+) roots and leaves. Each lane was loaded with 5 μg of RNA. Total RNA was extracted after 1 week of Fe-deficiency treatment.
Southern-Blot Analysis
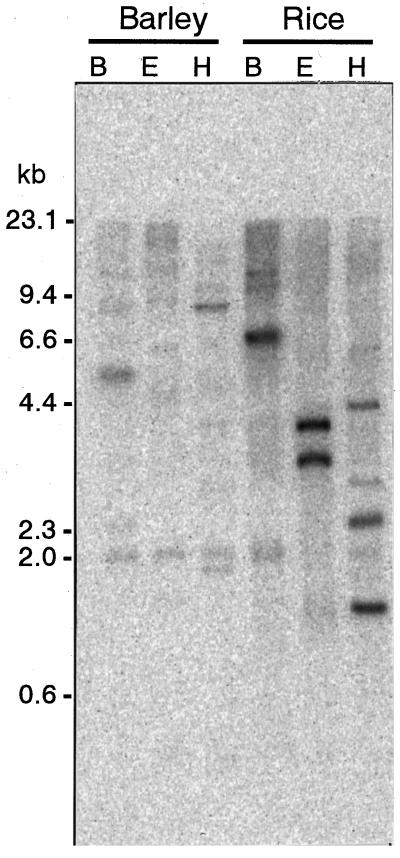
The number of nas and nas-like genes in the barley and rice genomes was assessed by Southern-blot analysis (Fig. 11). With the barley genome DNA, cutting with BamHI and EcoRI each produced 7 bands, which were detected on Southern blots, whereas cutting with HindIII produced 10 bands. None of the seven clones obtained in this work have BamHI or EcoRI sites, and only nas2 and nas4 have HindIII sites. This result corresponds well with the existence of at least seven nas cDNA clones.
Figure 11.
Southern-blot analysis of nas-like genes. Genomic DNA from barley and rice were digested with BamHI (lanes B), EcoRI (lanes E), and HindIII (lanes H) and probed with nas1 under high-stringency conditions.
DISCUSSION
In this paper we describe the partial purification of several NAS proteins induced by Fe deficiency (Fig. 4) and their corresponding genes (Fig. 8). The enzymatic function of the nas1 product was confirmed (Fig. 9). In addition, a 30-kD peptide and its gene, nas5-1 or nas5-2, which are homologous to NAS but lack NAS activity, were also found (Fig. 4). Several NAS proteins and their corresponding genes were found, but it remains unclear whether each NAS protein functions differently. It is possible that a heteropolymer composed of these homologous 30-, 32-, and 33-kD peptides forms the native enzyme. However, the Mr of NAS estimated by gel filtration was 35,000 (Fig. 5), and we did not observe increased NAS activity by combining the 30-, 32-, and 33-kD peptides in vitro (data not shown). Therefore, we tentatively conclude that native NAS exists as a monomer.
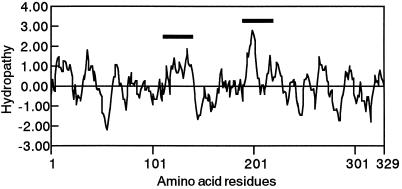
Another possibility is that native NAS exists as a complex of 30-, 32-, and 33-kD peptides, because NAS proteins are relatively hydrophobic, indicating that they may be membrane bound and localized in some organelle (Fig. 12). This may allow the efficient catalysis of trimerization of SAM. We have already shown that radioactivity from [1-14C]Met, the precursor of MAs, is localized in “particular vesicles” in the Fe-deficient barley root cell (Nishizawa and Mori, 1987; Nishizawa et al., 1990). This suggests that biosynthesis of MAs occurs inside particular vesicles.
Figure 12.
Hydropathy profile of NAS1. Hydrophobicity was analyzed by the DNASIS program, as described by Kyte and Doolittle (1982), with a window size of 10 amino acid residues. Hydrophilic domains are below the zero line. Bold lines indicate hydrophobic regions.
The catalytic mechanism of NA synthesis from SAM by NAS may be similar to that of methyl transferase using SAM as a methyl donor or those of spermidine synthase and spermine synthase using decarboxy-SAM as a substrate (Pajula et al., 1979). The common catalytic domain of these enzymes has been discussed in relation to amino acids occupying similar positions in the secondary and tertiary structures (Schluckebier et al., 1995; Hashimoto et al., 1998). If the 30-kD peptide lacking NAS activity has SAM-binding activity, comparison of NAS protein and the inactive NAS homolog may reveal the catalytic or the binding domain structure of NAS. Because NA is contained in all plants surveyed so far, NAS may be widespread among plant species. The translation products of the putative rice NAS homologs, D23792 and D24790, were very similar to NAS1 protein, with 80.2% identity in a 111-amino acid overlap in the former and 78.9% identity in a 71-amino acid overlap in the latter. Putative Arabidopsis NAS homologs, AC003114 (function unknown) and AB005245 (function unknown), were also found with 45% identity in the former and 46% identity in the latter to NAS1. Therefore, comparison of NAS protein homology among many species may also reveal information about its catalytic domain.
Since full-length nas1 used as a probe could also detect other nas genes, a 1.3-kb band smear was seen on northern blots (Fig. 10), and many bands were detected on Southern blots (Fig. 11). Therefore, we conclude that nas belongs to a multigene family in the barley genome and probably also in the rice genome. The number of copies should be confirmed by Southern-blot analysis using conserved sequences of nas as the probes.
On northern blots a smeared band produced the strongest signal, but weak and short bands were also detected. These may represent degradation products of nas1 mRNA (Fig. 10). Since NAS proteins are vulnerable to degradation, it is quite possible that the expression of the nas gene in barley roots is strictly controlled to avoid overproduction of MAs. For instance, resupplying Fe to Fe-deficient barley decreased NAS activity within 24 h (Higuchi et al., 1996a).
We could tentatively categorize the seven nas clones into several types based on homology of amino acid sequences: type 1, nas1; type 2, nas2, 3, 4, and 6; and type 3, nas5 (Table I). Nineteen independent nas1 clones were obtained from a cDNA library prepared using poly(A+) RNA from Fe-deficient barley roots, but only one nas4 clone was obtained. The numbers of clones of nas2, nas3, nas5-1, nas5-2, and nas6 were two, four, five, three, and three, respectively. These results suggest that the expression of nas genes under Fe deficiency differs from one nas gene family to the next. Northern-blot analysis using a specific probe for each nas gene will reveal the precise expression pattern of each gene and suggest the function of each nas gene under different stress conditions.
Until now, NA, the reaction product of NAS, has been thought to function only as an intermediate substrate for MAs synthesis in graminaceous plants (Fig. 1). Since the nas gene belongs to a multigene family, NA has a function in the survival of graminaceous plants other than as a precursor of MAs. In Strategy I plants, which lack the ability to produce MAs, it has been proposed that NA plays a key role as an endogenous chelator of divalent metal cations, such as Fe2+, Cu2+, Zn2+, and Mn2+, in the xylem and phloem and that it contributes to the homeostasis of those metals in plants (Stephan et al., 1994; Higuchi et al., 1996b). NA may also play a role in the metabolism of Fe or other metals in Strategy II plants. The fact that NAS activity is not enhanced by Fe deficiency in Strategy I plants (Higuchi et al., 1995, 1996b) suggests that expression of the nas gene is not regulated by Fe in these plants. Transgenic plants with suppressed NAS expression will help clarify the role of NA in both Strategy I and Strategy II plants.
ACKNOWLEDGMENTS
We thank Dr. Naimatullah Bughio, Mr. Nobuyuki Sato, and Dr. Michiko Takahashi for the preparation of Fe-deficient plant material. We are thankful to Dr. Emmanuel Delhaize, Commonwealth Scientific and Industrial Research Organization, Australia, for editing.
Abbreviations:
- Chaps
3-[(3-cholamidopropyl)dimethyl-ammonio]propanesulfonic acid
- CNBr
cyanogen bromide
- E-64
trans-epoxysuccinyl-leucylamido-(4-guanidino)butane
- MAs
mugineic acid family
- NA
nicotianamine
- NAAT
nicotianamine aminotransferase
- NAS
nicotianamine synthase
- p-APMSF
(p-amidinophenyl)methanesulfonyl fluoride
- SAM
S-adenosylmethionine
LITERATURE CITED
- Church M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzandu JK, Johnson JF, Wise GE. Sodium dodecyl sulfate-gel electrophoresis: staining of polypeptide using heavy metal salts. Anal Biochem. 1988;174:157–167. doi: 10.1016/0003-2697(88)90531-3. [DOI] [PubMed] [Google Scholar]
- Gross E. The cyanogen bromide reaction. Methods Enzymol. 1967;11:238–255. [Google Scholar]
- Hashimoto T, Tamaki K, Suzuki K, Yamada Y. Molecular cloning of plant spermidine synthases. Plant Cell Physiol. 1998;39:73–79. doi: 10.1093/oxfordjournals.pcp.a029291. [DOI] [PubMed] [Google Scholar]
- Higuchi K, Kanazawa K, Nishizawa NK, Chino M, Mori S. Purification and characterization of nicotianamine synthase from Fe-deficient barley roots. Plant Soil. 1994;165:173–179. [Google Scholar]
- Higuchi K, Kanazawa K, Nishizawa NK, Mori S. The role of nicotianamine synthase in response to Fe nutrition status in Gramineae. Plant Soil. 1996a;178:171–177. [Google Scholar]
- Higuchi K, Nishizawa NK, Römheld V, Marschner H, Mori S. J Plant Nutr. 1996b;19:1235–1239. [Google Scholar]
- Higuchi K, Nishizawa NK, Yamaguchi H, Römheld V, Marschner H, Mori S. Response of nicotianamine synthase activity to Fe-deficiency in tobacco plants as compared with barley. J Exp Bot. 1995;289:1061–1063. [Google Scholar]
- Kanazawa K, Higuchi K, Fushiya S, Nozoe S, Nishizawa NK, Chino M, Mori S (1995) Induction of two enzyme activities involved in the biosynthesis of mugineic acid in Fe deficient barley roots. In Abadia J, eds, Iron Nutrition in Soils and Plants. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 37–41
- Kawai S, Itoh K, Takagi S, Iwashita T, Nomoto K. Studies on phytosiderophores: biosynthesis of mugineic acid and 2′-deoxymugineic acid in Hordeumvulgare L. var. Minorimugi. Tetrahedron Lett. 1988;29:1053–1056. [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Ma JF, Nomoto K. Two related biosynthetic pathway of mugineic acids in Granineous plants. Plant Physiol. 1993;102:373–378. doi: 10.1104/pp.102.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner H, Römheld V, Kissel M. Localization of phytosiderophore release and of iron uptake along with intact barley roots. Physiol Plant. 1987;71:157–172. [Google Scholar]
- Mihashi S, Mori S. Characterization of mugineic acid-Fe transporter in Fe-deficient barley roots using the multi-compartment transport box method. Biol Metals. 1989;2:146–154. [Google Scholar]
- Mori S, Hachisuka M, Kawai S, Takagi S, Nishizawa NK. Peptides related to phytosiderophore secretion by Fe-deficient barley roots. J Plant Nutr. 1988;11:653–662. [Google Scholar]
- Mori S, Nishizawa N. Methionine as a dominant precursor of phytosiderophores in graminaceae plants. Plant Cell Physiol. 1987;28:1081–1092. [Google Scholar]
- Mori S, Nishizawa N, Kawai S, Sato S, Takagi S. Dynamic state of mugineic acid and analogous phytosiderophores in Fe-deficient barley. J Plant Nutr. 1987;10:1003–1011. [Google Scholar]
- Naito S, Dube PH, Beachy RN. Differential expression of conglycinin alpha' and beta subunit gene in transgenic plants. Plant Mol Biol. 1988;11:109–124. doi: 10.1007/BF00015664. [DOI] [PubMed] [Google Scholar]
- Nakanishi H, Okumura N, Umehara Y, Nishizawa NK, Chino M, Mori S. Expression of a gene specific for iron deficiency (Ids3) in the roots of Hordeum vulgare. Plant Cell Physiol. 1993;34:401–410. [PubMed] [Google Scholar]
- Nishizawa N, Mori S. The particular vesicle appearing in barley root cells and its relation to mugineic acid secretion. J Plant Nutr. 1987;10:1012–1020. [Google Scholar]
- Nishizawa NK, Shojima S, Mori S (1990) The ultrastructural studies on mugineic acid secretion by barley roots under the Fe-deficient stress (abstract no. IV-416). Transactions of 14th International Congress of Soil Science Society, Kyoto, Japan.
- Pajula RL, Raina A, Eloranta T. Polyamine synthesis in mammalian tissues. Eur J Biochem. 1979;101:619–626. doi: 10.1111/j.1432-1033.1979.tb19756.x. [DOI] [PubMed] [Google Scholar]
- Römheld V. Different strategies for iron acquisition in higher plants. Physiol Plant. 1987;70:231–234. [Google Scholar]
- Römheld V, Marschner H. Evidence for a specific uptake system for iron phytosiderophores in roots of grasses. Plant Physiol. 1986;70:175–180. doi: 10.1104/pp.80.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate- polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 kDa to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Schluckebier G, O'Gara M, Saenger W, Cheng X. Universal catalytic domain structure of AdoMet-dependent methyl transferases. J Mol Biol. 1995;247:16–20. doi: 10.1006/jmbi.1994.0117. [DOI] [PubMed] [Google Scholar]
- Shojima S, Nishizawa NK, Fushiya S, Nozoe S, Irifune T, Mori S. Biosynthesis of phytosiderophores. Plant Physiol. 1990;93:1497–1503. doi: 10.1104/pp.93.4.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shojima S, Nishizawa NK, Mori S. Establishment of a cell-free system for the biosynthesis of nicotianamine. Plant Cell Physiol. 1989;30:673–677. [Google Scholar]
- Singh K, Chino M, Nishizawa NK, Ohata T, Mori S (1993) Genotypic variation among Indian graminaceous species with respect to phytosiderophore secretion. In Randall PJ, eds, Genetic Aspects of Plant Mineral Nutrition. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 335–339
- Stephan UW, Schmidke I, Pich A. Phloem translocation of Fe, Cu, Mn and Zn in Ricinus seedlings in relation to the concentrations of nicotianamine, an endogenous chelator for divalent metal ions, in different seedling parts. Plant Soil. 1994;165:181–188. [Google Scholar]
- Takagi S. Naturally occurring iron-chelating compounds in oat- and rice-root washing. I. Activity measurement and preliminary characterization. Soil Sci Plant Nutr. 1976;22:4232–4233. [Google Scholar]
- Takagi S, Nomoto K, Takemoto S. Physiological aspect of mugineic acid, a possible phytosiderophore of graminaceous plants. J Plant Nutr. 1984;7:469–477. [Google Scholar]
- Takahashi M, Yamaguchi H, Nakanishi H, Kanazawa K, Shioiri T, Nishizawa NK, Mori S (1997) Purification, characterization and sequencing of nicotianamine aminotransferase (NAAT-III) expressed in Fe-deficient barley roots. In Ando T, eds, Plant Nutrition—For Sustainable Food Production and Environment. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 279–280
- Takizawa R, Nishizawa N, Nakanishi H, Mori S. Effect of iron deficiency on S-adenosylmethionine synthetase in barley roots. J Plant Nutr. 1996;19:1189–1200. [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]