Abstract
A simple and reliable method for precipitating protein from bacterial culture supernatants based on a pyrogallol red-molybdate-methanol (PRMM) protocol has been developed and applied for the analysis of proteins secreted by a bacterial type III secretion system. PRMM-based precipitation has been shown to be more efficient and robust than are conventional protocols.
Recently, bacterial mechanisms for protein secretion have become an important focus of research due to the observation that key virulence factors of bacterial pathogens are secreted into the environment or even translocated into targeted eukaryotic cells. Gram-negative pathogens affecting animals, humans, and plants manipulate targeted host cells by secretion of effector proteins by type III secretion systems (5, 7). Our particular interest is the analysis of protein secretion by Salmonella enterica serovar Typhimurium via the type III secretion system (T3SS) encoded on the Salmonella pathogenicity island 2 (SPI2) (13). Protein secretion via the different T3SSs has been reported to depend on specific environmental factors, and techniques have been established to induce protein secretion into culture supernatants for a variety of T3SSs in vitro (3, 10). However, the standard procedures for protein precipitation of voluminous liquid samples such as culture supernatants harbor several drawbacks, including the handling of large amounts of culture supernatant, toxic chemicals, requirement of high centrifugation forces, low efficiency of precipitation at low protein concentrations, or interfering agents (1, 16). Here, we report on the development of a simple and reliable method for precipitating proteins from bacterial culture supernatant based on a pyrogallol red-molybdate-methanol (PRMM) protocol.
Using standard trichloroacetic acid (TCA) and acetone-based protocols for protein precipitation (6, 14), we were unable to reproducibly detect the S. enterica serovar Typhimurium SseB protein, a protein known to be secreted by the S. enterica serovar Typhimurium SPI2-encoded T3SS (2, 11), in culture supernatant (CS) and surface-detached (SD) preparations with satisfactory reliability. In vitro, this particular T3SS is inducible by applying low-pH and magnesium starvation culture conditions (2, 3), but the total amount of Salmonella-secreted proteins under these particular conditions is relatively small. Furthermore, individual protein concentrations within this fraction can vary substantially (11).
To optimize the protein yield in precipitates, a recently described method for protein precipitation from chromatographic samples (1, 8) was adapted to our experimental setup. For induction of protein secretion by the SPI2-encoded S. enterica serovar Typhimurium T3SS, the strains to be analyzed were grown as described previously (11) to an optical density at 600 nm of approximately 1.2. To obtain the CS fraction, the cultures were centrifuged for 15 min at 5,000 × g, and the supernatant of each sample was collected and filtered though a 0.22-μm-pore-size filter to remove residual bacterial cells. To obtain the SD fraction, proteins secreted but remaining bound to the cell surface, the sedimented cells were resuspended in 30 ml of phosphate-buffered saline, and after the addition of 5 ml of N-hexadecane that was prewarmed to 37°C, the samples were vortexed vigorously for 30 s. The suspensions were centrifuged for 15 min at 6,000 × g at 4°C and then placed on ice until the N-hexadecane solidified. The liquid phase of each sample was collected and filtered though a 0.22-μm-pore-size filter. For the precipitation of secreted proteins, an equal volume of a PRMM solution (0.05 mM pyrogallol red, 0.16 mM sodium molybdate, 1.0 mM sodium oxalate, 50.0 mM succinic acid, 20% [vol/vol] methanol in H2O, adjusted to pH 2.0 with HCl) was added to the CS and SD fractions. Subsequently, the pH of the solution was adjusted to 2.8 ± 0.1, and the proteins were allowed to precipitate for 1 to 2 h at room temperature, followed by an overnight incubation at 4°C. To achieve optimal precipitation, the methanol concentration in the PRMM solution was adjusted to 20% (vol/vol) (10% [vol/vol] methanol in the sample). The addition of methanol might reduce the dielectric constant of the aqueous solution to augment the efficiency of the precipitation process. Furthermore, the final pH of the sample should lie within a finite range of 2.8 ± 0.1, and a prolonged time for precipitation was also found to favor the efficacy of the protocol. The precipitate was sedimented by centrifugation at 10,000 × g for 1 h, and the supernatant was carefully removed. The precipitate was repeatedly rinsed with 1 ml of acetone, and the acetone was removed by evaporation at room temperature. To solubilize the precipitate, 100 μl of 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (25% glycerol, 8% SDS, 4% β-mercaptoethanol, 0.02% bromophenol blue, 100 mM Tris-HCl [pH 6.8]) and 100 μl of a saturated Tris-H2O solution were added to the samples, followed by an incubation at 95°C for 15 min.
To compare the optimized PRMM protocol with conventional TCA- and acetone-based precipitation protocols, the CS and SD fractions were prepared from CREA0300 (S. enterica serovar Typhimurium, ATCC 14028), CREA0253 (CREA0300 sseC; T. Sieck, unpublished results), and CREA0300(pLRC5T). The plasmid vector pLRC5T encodes a C-terminally truncated SseD protein that is translationally fused to the nanopeptide reporter epitope hemagglutinin (HA) of the human influenza virus, yielding a protein of approximately 21 kDa (R. B. Caldwell, unpublished data). The recombinant sseD gene is transcriptionally controlled by the promoter of the sseA-G operon, PsseA (R. B. Caldwell, unpublished) (4).
CS and SD fractions were prepared from CREA0300, CREA0253, and CREA0300(pLRC5T) as described above, and the samples were divided into three sets to directly compare the PRMM protocol (set A) to TCA (final concentration, 10%; set B) and acetone (3 volumes acetone/volume of CS fraction; 4 volumes acetone/volume of SD fraction; set C)- based precipitation. The obtained protein precipitates were resuspended in 100 μl of 2× SDS sample buffer and 100 μl of saturated Tris-H2O. Equal amounts of the samples corresponding to the amount of protein secreted by approximately the amount of cells corresponding to an optical density at 600 nm of 1.0 were separated by SDS-PAGE (12.5% polyacrylamide) and subjected to Western blot analysis with specific rabbit antisera raised against the SseC protein (kind gift of M. Hensel) and with a monoclonal antibody specific for the HA tag (Roche diagnostics, Penzberg, Germany) in order to visualize the SseD-HA fusion protein encoded on pLRC5T. Western blots were processed by using enhanced chemiluminescence (Amersham Biosciences).
The efficacy of the novel protocol was compared to that of the standard protocols for protein precipitation employing TCA and acetone (Fig. 1). By the PRMM protocol, SseC migrating to 52.8 kDa (Fig. 1A) could be precipitated in the CS fraction and in the SD fraction of CREA0300 and CREA0300(pLRC5T). As expected, no SseC protein could be detected in CREA0253, whereas SseB was found in the SD fractions of all strains (data not shown). The same pattern of SseC and SseB secretion could be observed, albeit with much less efficiency, by acetone precipitation. Interestingly, the TCA protocol failed to precipitate the SseC protein in the CS fraction and showed only slight amounts of SseC in the SD fraction in CREA0300. In addition, the recombinant SseD-HA fusion protein encoded by pLRC5T was efficiently precipitated by the PRMM protocol, whereas only a faint band appeared in the acetone-prepared sample—and then only with prolonged photographic exposure time. The recombinant SseD (Fig. 1A) and SseB (data not shown) proteins were not precipitated by TCA. To confirm the absence of disrupted bacterial cells in the processed samples, the same membranes were reprobed for Hsp60 by using a monoclonal antibody raised against Yersinia enterocolitica Hsp60 (kind gift of I. B. Autenrieth) (12) that also binds to Salmonella Hsp60 (Fig. 1B). The CS fraction showed no binding activity, and the SD fraction showed only faint Hsp60-specific bands, a finding that indicated the presence of no-to-minimal levels of Hsp60 protein in the processed samples. Thus, lysis of bacterial cells was not the reason for the presence of recombinant SseD-HA or native SseC proteins in CS or SD fractions. The superior efficacy of the PRMM protocol was further confirmed by parallel silver staining of the SDS-PAGE-separated CS and SD fractions that we analyzed (Fig. 1C).
FIG. 1.
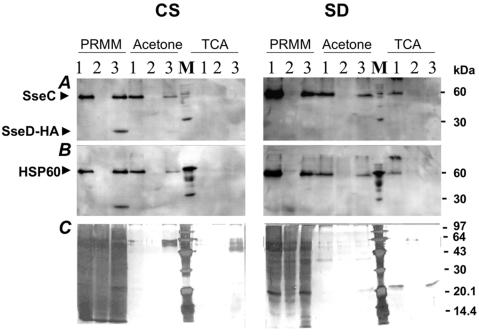
Efficient precipitation of proteins from CS and SD fractions by the PRMM protocol. Equal amounts of samples from CS or SD fractions prepared from cultures of strains CREA0300 (1), CREA0253 (2), or CREA300(pLRC5T) (3), respectively, were separated by SDS-12.5% PAGE and subjected to sequential Western blot analysis with an αSseC antiserum and a monoclonal antibody against the HA tag (A) and in a second step as a control with a monoclonal antibody against Hsp60 protein (B). (C) Silver staining of a duplicate gel of the procedure depicted in panel A. M, low-molecular-weight marker (Pharmacia) with added control protein Hsp60-HA migrating to 60 kDa. The data shown in this figure are representative of data from a series of three independent experiments.
Pyrogallol red-molybdate has also been studied for use as a method for total protein determination in urine samples based on differential absorbance at 600 nm of the pyrogallol red-molybdate-protein bound complex (9, 15). Preliminary results indicate that the method presented here may also provide a quick and simple tool that can be used to quantify the total amount of secreted protein in bacterial culture supernatants. Using the parameters specified in these articles, we found a qualitative difference in the absorbance at 600 nm in the PRMM-precipitated CS and SD fractions of a wild-type strain of Salmonella and that of an SPI2 secretion mutant (R. B. Caldwell, unpublished), suggesting that the difference in the total amount of total protein present in such samples might be quantified.
In conclusion, the method presented here represents a simple and reliable protocol for the qualitative precipitation of proteins from culture supernatants. As corroborated by the selective detection of several proteins secreted by the SPI2-encoded T3SS in CS and SD fractions, the PRMM protocol allows the reproducible precipitation of proteins from samples with low protein content more effectively than with either TCA- or acetone-based protocols. Therefore, we think that this protocol will facilitate the analysis of protein samples with various protein concentrations in a number of systems.
Acknowledgments
We thank M. Hensel and I. B. Autenrieth for the kind gift of antisera, and we also thank J.-M. Buerstede and H. Apfel for continuous support.
REFERENCES
- 1.Aguilar, R. M., J. J. Bustamante, P. G. Hernandez, A. O. Martinez, and L. S. Haro. 1999. Precipitation of dilute chromatographic samples (ng/ml) containing interfering substances for SDS-PAGE. Anal. Biochem. 267:344-350. [DOI] [PubMed] [Google Scholar]
- 2.Beuzon, C. R., G. Banks, J. Deiwick, M. Hensel, and D. W. Holden. 1999. pH-dependent secretion of SseB, a product of the SPI-2 type III secretion system of Salmonella typhimurium. Mol. Microbiol. 33:806-816. [DOI] [PubMed] [Google Scholar]
- 3.Deiwick, J., T. Nikolaus, S. Erdogan, and M. Hensel. 1999. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol. Microbiol. 31:1759-1773. [DOI] [PubMed] [Google Scholar]
- 4.Hensel, M., J. E. Shea, S. R. Waterman, R. Mundy, T. Nikolaus, G. Banks, A. Vazquez-Torres, C. Gleeson, F. C. Fang, and D. W. Holden. 1998. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol. Microbiol. 30:163-174. [DOI] [PubMed] [Google Scholar]
- 5.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hwang, B. J., and G. Chu. 1996. Trichloroacetic acid precipitation by ultracentrifugation to concentrate dilute protein in viscous solution. BioTechniques 20:982-984. [DOI] [PubMed] [Google Scholar]
- 7.Lahaye, T., and U. Bonas. 2001. Molecular secrets of bacterial type III effector proteins. Trends Plant Sci. 6:479-485. [DOI] [PubMed] [Google Scholar]
- 8.Marshall, T., N. J. Abbott, P. Fox, and K. M. Williams. 1995. Protein concentration by precipitation with pyrogallol red prior to electrophoresis. Electrophoresis 16:28-31. [DOI] [PubMed] [Google Scholar]
- 9.Marshall, T., and K. M. Williams. 2000. Total protein determination in urine: elimination of a differential response between the Coomassie blue and pyrogallol red protein dye-binding assays. Clin. Chem. 46:392-398. [PubMed] [Google Scholar]
- 10.Michiels, T., P. Wattiau, R. Brasseur, J. M. Ruysschaert, and G. Cornelis. 1990. Secretion of Yop proteins by yersiniae. Infect. Immun. 58:2840-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nikolaus, T., J. Deiwick, C. Rappl, J. A. Freeman, W. Schroder, S. I. Miller, and M. Hensel. 2001. SseBCD proteins are secreted by the type III secretion system of Salmonella pathogenicity island 2 and function as a translocon. J. Bacteriol. 183:6036-6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noll, A., and I. B. Autenrieth. 1996. Yersinia-Hsp60-reactive T cells are efficiently stimulated by peptides of 12 and 13 amino acid residues in a MHC class II (I-Ab)-restricted manner. Clin. Exp. Immunol. 105:231-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shea, J. E., M. Hensel, C. Gleeson, and D. Holden. 1996. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 93:2593-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sivaraman, T., T. K. Kumar, G. Jayaraman, and C. Yu. 1997. The mechanism of 2,2,2-trichloroacetic acid-induced protein precipitation. J. Protein Chem. 16:291-297. [DOI] [PubMed] [Google Scholar]
- 15.Watanabe, N., S. Kamei, A. Ohkubo, M. Yamanaka, S. Ohsawa, K. Makino, and K. Tokuda. 1986. Urinary protein as measured with a pyrogallol red-molybdate complex, manually and in a Hitachi 726 automated analyzer. Clin. Chem. 32:1551-1554. [PubMed] [Google Scholar]
- 16.Yeang, H. Y., F. Yusof, and L. Abdullah. 1995. Precipitation of Hevea brasiliensis latex proteins with trichloroacetic acid and phosphotungstic acid in preparation for the Lowry protein assay. Anal. Biochem. 226:35-43. [DOI] [PubMed] [Google Scholar]


