Abstract
Little is known about bacteria associated with Lepidoptera, the large group of mostly phytophagous insects comprising the moths and butterflies. We inventoried the larval midgut bacteria of a polyphagous foliivore, the gypsy moth (Lymantria dispar L.), whose gut is highly alkaline, by using traditional culturing and culture-independent methods. We also examined the effects of diet on microbial composition. Analysis of individual third-instar larvae revealed a high degree of similarity of microbial composition among insects fed on the same diet. DNA sequence analysis indicated that most of the PCR-amplified 16S rRNA genes belong to the γ-Proteobacteria and low G+C gram-positive divisions and that the cultured members represented more than half of the phylotypes identified. Less frequently detected taxa included members of the α-Proteobacterium, Actinobacterium, and Cytophaga/Flexibacter/Bacteroides divisions. The 16S rRNA gene sequences from 7 of the 15 cultured organisms and 8 of the 9 sequences identified by PCR amplification diverged from previously reported bacterial sequences. The microbial composition of midguts differed substantially among larvae feeding on a sterilized artificial diet, aspen, larch, white oak, or willow. 16S rRNA analysis of cultured isolates indicated that an Enterococcus species and culture-independent analysis indicated that an Entbacter sp. were both present in all larvae, regardless of the feeding substrate; the sequences of these two phylotypes varied less than 1% among individual insects. These results provide the first comprehensive description of the microbial diversity of a lepidopteran midgut and demonstrate that the plant species in the diet influences the composition of the gut bacterial community.
Microorganisms play important and often essential roles in the growth and development of many insect species. Endosymbionts contribute to insect reproduction, digestion, nutrition, and pheromone production (7, 13, 14, 20, 50, 53, 55). Symbiotic relationships between insects and their gut bacteria have been studied extensively in several systems, particularly in termites and aphids, which feed on wood and plant phloem content, respectively (8, 17, 31). However, relatively little is known about microbial associates in other insect groups, particularly those that feed on foliage.
Describing relationships between insects and their associated bacteria has proved challenging because many of the microbial associates are not readily culturable. The availability of culture-independent tools, such as PCR, provides an opportunity to detect and classify microorganisms that cannot be cultured by existing methods (74). PCR has revealed the phylogeny of endosymbionts and associates of a number of insect species (1, 4, 15, 19, 33, 55, 57, 62, 64).
Little is known about the bacteria associated with Lepidoptera, a primarily phytophagous group that is one of the largest insect orders, containing over 150,000 species (65). A few studies indicate that lepidopterans harbor midgut bacteria (14, 49). These studies suggest the possibility that microorganisms provide essential nutrients or assist in important biochemical functions. However, an understanding of the types of microbes present in the midgut and of their roles in insect development and function is lacking. Several attributes of Lepidoptera larvae make their bacteria of particular interest. In particular, the high alkalinity (typically pH 8 to 10) of most lepidopteran midguts and the diverse chemistry of the midgut generated by the unusually broad feeding range of some species make this a particularly challenging environment for microorganisms (5, 22, 29, 44). Furthermore, the Lepidoptera include some of the most damaging agricultural and forest pests worldwide, including species that overcome existing control measures, such as synthetic insecticides, microbial insecticides, genetic host plant resistance, and genetically modified plants (60, 70). Knowledge of the gut bacteria of the Lepidoptera and the roles they may play in larval biology could lead to new targets for pest management.
As an initial step toward understanding relationships between Lepidoptera and their midgut bacteria, we surveyed the midgut of the gypsy moth, Lymantria dispar (L.), which is an invasive species of Eurasian origin and the most damaging defoliator of deciduous trees in North America (51). The gypsy moth alters natural ecosystems, causes economic losses to the forest industry and homeowners, elicits allergic reactions in humans, and is highly polyphagous with a reported host range of 300 species (41). We examined the effects of diet and insect source on the microbial composition of third-instar midguts.
MATERIALS AND METHODS
Sources and treatments of gypsy moth larvae. (i) Larvae from diverse populations feeding on a common substrate.
Gypsy moth egg masses were obtained from the culture New Jersey Standard Strain (NJSS) at the U.S. Department of Agriculture (USDA)-Animal and Plant Inspection Service (APHIS) laboratory at the OTIS Air National Guard Base, Cape Cod, Mass. (MA colony) and the Beneficial Insects Introduction and Research Laboratory, U.S. Department of Agriculture-Agricultural Research Services, Newark, Del. (DE colony). In addition, egg masses were collected from field sites in Wisconsin (WI field) and Michigan (MI field). Wisconsin egg masses were collected on 24 March 1999 from five trees (four oak trees [Quercus sp. L.] and one maple tree [Acer sp. L.]) on a rural property in York Township, Dane County (R11E, T9N). The Michigan egg masses were collected 23 March 1999 from a newly established population in a mixed oak and aspen (Quercus sp. and Populus sp.) stand in Lincoln Township, Clare County (R5W, T18N). Field-collected egg masses were kept in plastic bags at 4°C from the time of collection until they were used in assays. Assays were timed to coincide with Wisconsin field larval emergence (around 5 May 1999).
Gypsy moth larvae were reared as described in Broderick et al. (10). Briefly, egg masses were surface sterilized with a solution of Tween 80 (polyoxyethylene sorbitan monooleate), bleach, and distilled water. Larvae were reared in 17-cm-diameter petri dishes on a sterile artificial diet (USDA Hamden Formula) under a 16 h:8 h (light:dark) photoperiod at 25°C. Colony (MA and DE) larvae were reared in a quarantine facility at the University of Wisconsin—Madison Department of Entomology. To reduce the potential for introduction of pathogens into the quarantine facility, field egg masses and larvae (WI and MI) were reared in a separate campus quarantine facility at the University of Wisconsin—Madison Biotron under the same conditions as above. Larvae were provided with artificial diet after emergence and provided a new diet every 48 h.
(ii) Larvae from a common source feeding on multiple tree species.
Gypsy moth egg masses from culture NJSS at the USDA-APHIS laboratory (MA colony) were reared according to methods described previously (10, 11, 18).
Two-year-old white oak (Q. alba L.), larch [Larix laricina (Du Roi) K. Koch], and quaking aspen (P. tremuloides Michaux) trees were obtained from the Wisconsin Department of Natural Resources Nursery, Hayward. Two-year-old scrub willow (Salix fragilis L.) trees were obtained from the Iowa State Nursery, Ames. The trees were chilled at 4°C for 20 days after receipt to ensure good bud development. Following the chilling period, the trees were planted in 12-liter pots in Sunshine Mix LC1 (Sungro Horticulture, Bellevue, Wash.) and flood irrigated. The trees were grown in a greenhouse at 25°C under a 16 h:8 h (light:dark) photoperiod. Trees were watered every 5 days until bud development; following bud development the trees were watered every 2 to 3 days.
Larvae were provided with a sterilized artificial diet for 24 h after emergence and then were divided into 40 groups of 200 larvae each. These 40 groups were distributed among treatments such that 10 groups of larvae fed on leaves of one of the four tree species in petri dishes lined with a piece of moistened sterilized filter paper. For each tree species 10 trees were sampled, with one group of 200 larvae feeding on leaves collected from each tree (a total of 2,000 larvae for each tree species). Whole leaves were provided, with the petioles inserted in a microcentrifuge tube containing distilled water to prevent desiccation. Foliage was replaced every 48 h until the larvae were used for experiments.
Culture-independent methods. (i) Midgut separation and DNA extraction.
Third-instar larvae were surface sterilized for 5 s in 95% ethanol prior to dissection. Dissecting scissors were used to cut laterally behind the head capsule, and the gut was removed from the cuticle with larval forceps. The crop and midgut were collected and placed in a 1.5-ml microcentrifuge tube for processing. Samples were placed in a −80°C freezer.
Gypsy moth midguts were analyzed individually in all experiments. Total microbial DNA was extracted from individual gypsy moth crops and midguts by using a protocol modified from the method described by Ausubel et al. (3). Microcentrifuge tubes, each containing a single gut, were thawed, and 600 μl of Tris-EDTA (TE) (10 mM Tris-HCl [pH 8.0], 1 mM EDTA) was added to each tube. The contents of the tube were then sonicated (50 to 60 Hz, 117 V, 1.0 A; Branson Ultrasonics, Danbury, Conn.) for 30 s to separate bacterial cells from the gut wall, and 537 μl of TE was removed and placed in a new 1.5-ml microcentrifuge tube. The sample was sonicated under the same conditions for 45 s to break open bacterial cells and was mixed thoroughly with 60 μl of 10% sodium dodecyl sulfate and 3 μl of 50 mg of proteinase K/ml and was incubated for 1.5 h at 37°C. Each tube was mixed with 100 μl of 5 M NaCl prior to the addition of 80 μl of 10% cetyltrimethylammonium bromide-5 M NaCl. The sample was mixed thoroughly and incubated at 65°C for 30 min. DNA was extracted with equal volumes of chloroform-isoamyl alcohol (CIA) (24:1 [vol/vol]) and phenol CIA (25:24:1 [vol/vol/vol]). DNA was precipitated with isopropanol and recovered by centrifugation. Pellets were resuspended in 100 μl of TE buffer. DNA concentration was determined by absorbance ratio at 260/280 nm, and the DNA suspension was stored at −20°C until it was used for PCR and further analysis.
(ii) PCR amplification.
Bacterial 16S rRNA was amplified by PCR from total DNA by using primers 27F and 1492R (37). For terminal-restriction fragment length polymorphism (T-RFLP) analysis, the 5′ primer (27F) was labeled with the dye 6-carboxyfluorescein (6-FAM) (16, 42). All oligonucleotide primers were synthesized by the Biotechnology Center at the University of Wisconsin—Madison. PCR contents for a 50-μl volume were 25 ng of template DNA, 0.5 μl of 20 μM forward (27F) and reverse (1492R) primers, 0.8 μl of 10 mM deoxynucleoside triphosphates (Gibco BRL, Grand Island, N.Y.), 5 μl of 10× Taq polymerase buffer, and 0.5 μl of Taq DNA polymerase (2.5 U; Promega, Madison, Wis.). The thermal cycling conditions were an initial 3-min denaturation step at 94°C, 30 cycles at 94°C for 30 s, 55°C for 90 s, and 72°C for 2.5 min, with a final 5-min extension at 72°C. A hot-start protocol (34) was used with Taq polymerase added to each reaction after 1 min of the 3-min initial 94°C denaturation step. All reactions were carried out in 0.2-ml reaction tubes in a Robocycler 96 (Stratagene, La Jolla, Calif.). Each PCR was examined by electrophoresis in a 0.8% agarose gel, and gels were visualized by staining with ethidium bromide.
(iii) T-RFLP analysis.
Samples of PCR amplification products from 10 individual larvae exposed to each diet type and from each population source were digested (37°C for 3 h) with the restriction endonucleases AluI and HhaI (Promega). Digestion reactions (10 μl total volume) contained 5 μl of PCR product, 0.3 μl of enzyme, and 0.3 μl of the appropriate incubation buffer according to the synthesis protocol. Samples (5 μl) of the digestion reactions were mixed with 19 μl of deionized formamide and 1 μl of an internal standard (CST ROX 200-2000; BioVentures, Inc., Murfreesboro, Tenn.), denatured for 5 min at 95°C, and kept on ice until they were loaded into an autosampler. The samples were analyzed by capillary electrophoresis in an ABI DNA sequencer (model 310; Perkin-Elmer, Foster City, Calif.) (39). Data from individual samples were analyzed with GeneScan 3.1 (Perkin-Elmer) and were compared to other samples by using the Ribosomal Database Project II (RDP II) T-RFLP Online Analysis software (46). To assess the similarity of microbial composition among midguts, T-RFLP profiles from replicate samples were aligned and analyzed by pairwise comparison (40). Similarity values, Sab, were determined by using the equation 2 Nab / (Na + Nb), where Nab is the number of peaks in common between samples and Naand Nbare the number of total peaks in each sample.
(iv) Cloning 16S rRNA genes.
Total DNA from pools of 10 larvae from all population and diet treatment samples was PCR-amplified separately by using the bacterial primers 27F and 1492R. Clone libraries were constructed by ligating PCR product into pGEM-T Vector system (Promega) according to the manufacturer's instructions and by transforming competent Escherichia coli cells (DH5α) by electroporation (61). Plasmid DNA was isolated from transformants by using QIAprep Spin Plasmid Miniprep kits (QIAGEN, Valencia, Calif.). All clones in libraries of approximately 250 clones from each diet type were sequenced.
(v) 16S rRNA sequencing and data analysis.
Sequencing reactions were performed by using the BigDye reaction mix (Perkin-Elmer Corp.) according to the manufacturer's instructions. Purified plasmid DNA was initially sequenced by using the plasmid primers T7 and SP6, which flank the insert DNA in pGEM-T, and additionally sequenced with 704F and 787R to obtain full 1,500-bp sequences (37). DNA from cultured organisms was purified by using the PCR product Pre-sequencing Kit (USB, Cleveland, Ohio) and was sequenced by using either 27F, 1492R, 704F, or 787R (37). Two representatives of each colony morphology and a total of 400 clones (approximately 45 to 115 per diet treatment) were analyzed. All 16S rRNA sequences (1,450 to 1,490 bp) were compiled by using the SeqMan program from the DNAStar software package (DNASTAR, Inc., Madison, Wis.) and were compared to available databases by use of BLAST to determine approximate phylogenetic affiliations (2). Individual sequences of isolates and clones that were ≥98% identical to each other were considered the same phylotype and were combined for analysis. All sequences were tested for possible chimeric structures by using RDP Check_Chimera (39).
(vi) Culturing and enumeration of midgut bacteria.
Twenty-third-instar larvae from each of the five diet treatments were dissected for culturing studies following a 24-h starvation period to allow gut clearing. Midguts were each placed in 1 ml of phosphate-buffered solution (PBS) in a 1.5-ml microcentrifuge tube. Ten larvae feeding on foliage from two trees of each species were used. Individual guts in PBS were sonicated for 45 s and then were dilution plated on 1/10 strength tryptic soy agar (TSA) at neutral pH and were incubated at 28°C. Colonies were classified based on morphological parameters of shape, color, margins, elevation, and texture. Colonies of each designated morphology were counted after 24, 48, and 72 h. Isolates of two representatives of each colony morphology were purified for individual colonies, and cellular morphology of each isolate was examined with 1,000× magnification. DNA was extracted with the EasyDNA kit (Invitrogen, Carlsbad, Ohio) from 14-h-old cultures of each isolate in 1/2-strength tryptic soy broth. DNA was quantified, amplified, and sequenced as described above.
Ten individual third-instar midguts from larvae feeding on artificial diet were dissected, sonicated, and stained with 4,6-diamino-2-phenylindole (DAPI) to determine the total number of bacterial cells per gut (72). For each individual midgut, twenty randomly selected fields were counted by epifluorescence microscopy (Olympus America Inc., Melville, N.Y.).
Nucleotide sequence accession numbers. The compiled 16S rRNA sequences have been deposited in the GenBank database under accession numbers AY39005 to AY39035.
RESULTS
Bacterial diversity in third-instar gypsy moth midguts.
Evaluation of the midgut bacteria of gypsy moth larvae using a combination of 16S rRNA analysis of the cultured bacteria and culture-independent PCR amplification of 16S rRNA sequences led to the identification of 23 phylotypes (Table 1). Approximately 65% of these phylotypes are newly reported sequences (<98% identity to any of the bacterial 16S rRNA sequences within GenBank). The majority (70%) of these bacterial sequences fell within the low G+C gram-positive and γ-Proteobacteria divisions. Actinobacterium, Cytophaga/Flexibacter/Bacteroides, and α-Proteobacteria accounted for 17, 9, and 4% of the sequences, respectively.
TABLE 1.
Bacterial phylotypes identified by culturing and culture-independent analysis of third-instar gypsy moth midguts based on 16S rRNA gene sequence analysis
| Sequence identy and identification method | Bacterial division | Genus (≥95% identity) | Database matches (≥98% identity) | Detection in gypsy moth fed ona:
|
||||
|---|---|---|---|---|---|---|---|---|
| Artificial diet | Larch | Willow | Aspen | White oak | ||||
| Identified by culturing and culture-independent analysis | ||||||||
| NAB1 | γ-Proteobacteria | Pseudomonas | P. putida | 1.8 × 105 | 1.1 × 107 | 3.6 × 105 | 3.5 × 105 | |
| NAB2 | γ-Proteobacteria | Pseudomonas | 2.2 × 106 | |||||
| NAB3 | γ-Proteobacteria | Enterobacter | Uncultured soil bacterium F42326.1 | 4.8 × 106 | 7.0 × 107 | 8.9 × 107 | ||
| NAB4 | γ-Proteobacteria | Pantoea | P. agglomerans | 2.5 × 105 | 3.7 × 104 | 1.5 × 107 | ||
| NAB5 | γ-Proteobacteria | Serratia | S. marcescens | 4.6 × 104 | 7.5 × 108 | 4.0 × 104 | ||
| NAB6 | low G+C gram positive | Bacillus | 4.0 × 108 | |||||
| NAB7 | low G+C gram positive | Staphylococcus | S. lentus | 8.1 × 104 | 9.1 × 106 | 4.5 × 106 | ||
| NAB8 | low G+C gram positive | Staphylococcus | S. cohnii | 2.9 × 106 | 4.7 × 106 | 1.3 × 107 | ||
| NAB9 | low G+C gram positive | Staphylococcus | S. xylosus | 1.3 × 106 | 1.07 × 105 | |||
| NAB10 | low G+C gram positive | Paenibacillus | 2.6 × 106 | |||||
| NAB11 | low G+C gram positive | Enterococcus | E. faecalis | 1.5 × 108 | 1.9 × 106 | 3.6 × 108 | 1.4 × 104 | 2.1 × 107 |
| NAB12 | low G+C gram positive | Enterococcus | 4.8 × 107 | 4.1 × 106 | 2.4 × 105 | |||
| NAB13 | Actinobacterium group | Rhodococcus | 1.2 × 107 | |||||
| NAB14 | Actinobacterium group | Microbacterium | 5.2 × 108 | 2.2 × 106 | ||||
| NAB15 | CFBb group | 7.4 × 104 | ||||||
| Identified by culture-independent analysis only | ||||||||
| NAB16 | α-Proteobacteria | Agrobacterium | + | + | + | |||
| NAB17 | γ-Proteobacteria | Enterobacter | + | + | + | + | + | |
| NAB3 | γ-Proteobacteria | Enterobacter | + | |||||
| NAB18 | γ-Proteobacteria | + | + | |||||
| NAB19 | γ-Proteobacteria | + | + | |||||
| NAB20 | low G+C gram positive | + | + | + | ||||
| NAB21 | Actinobacterium group | Micrococcus | + | |||||
| NAB22 | Actinobacterium group | + | ||||||
| NAB23 | CFB group | + | ||||||
For cultured cells the numbers represent the average population of phylotype based on colony morphology in 20 individual larvae per treatment (CFU/milliliter of gut material). For noncultured samples, a plus indicates presence in guts of insects in diet treatment.
CFB, Cytophaga/Flavobacteria/Bacteroides.
Variation in bacterial composition among individual larvae.
Comparisons of the T-RFLP profiles of the bacteria in replicate larvae from either the same population source or the same diet treatment showed a high similarity index of between 0.96 and 1.00 (Tables 2 and 3). These kinds of similarities were also reflected in the types of colony morphologies obtained in the culture-dependent analyses. The majority of isolates examined were found in >90% of the larvae examined and all were isolated from >50% of the larvae in a given treatment.
TABLE 2.
Pairwise comparisons for similarity of T-RFLPs from the midgut of gypsy moth from lab and field populations feeding on an artificial diet
| Population | Coefficient within population | Coefficient between populations
|
||
|---|---|---|---|---|
| MA colony | DE colony | WI field | ||
| MA colony | 0.996 | |||
| DE colony | 0.997 | 0.952 | ||
| WI field | 0.996 | 0.994 | 0.958 | |
| MI field | 0.998 | 0.955 | 0.992 | 0.953 |
TABLE 3.
Pairwise comparisons for similarity of T-RFLPs from the midgut of gypsy moth fed various diets
| Diet | Coefficient within group | Coefficient between diet groups
|
|||
|---|---|---|---|---|---|
| Artificial diet | Larch | Aspen | Willow | ||
| Artificial diet | 0.993 | ||||
| Larch | 0.962 | 0.382 | |||
| Aspen | 0.975 | 0.139 | 0.029 | ||
| Willow | 0.968 | 0.077 | 0.053 | 0.083 | |
| White Oak | 0.989 | 0.045 | 0.028 | 0.038 | 0.091 |
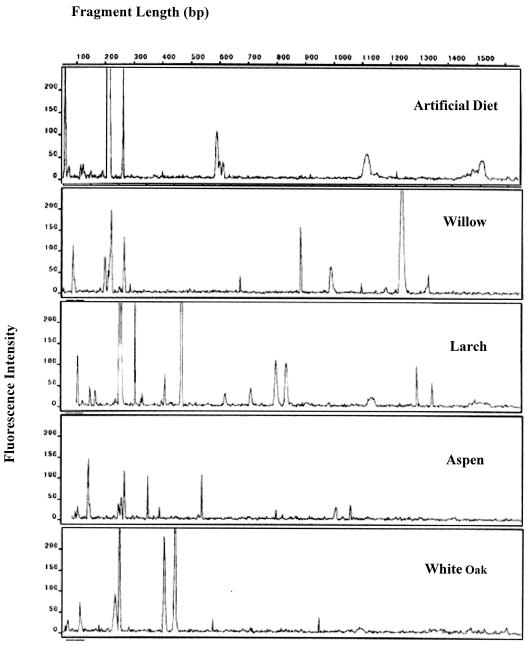
To identify factors associated with variation in microbial composition, we compared PCR-amplified 16S rRNA genes isolated from the midguts of gypsy moth from lab and field populations by using T-RFLP analysis. The T-RFLP profiles of the MA colony and WI field samples and those of the DE colony and MI field all fed artificial diet were very similar (Table 2). Moreover, these pairs differed only slightly (less than 6%) from each other (Table 2), indicating that egg source did not have a significant effect on the bacteria of the gypsy moth midgut. Given these similarities, only the MA colony population was used to examine the variation in bacterial diversity due to diet. Diet greatly affected the T-RFLP profiles (Fig. 1), as indicated by the lower similarity values of T-RFLP profiles from different diets (Table 3).
FIG. 1.
T-RFLP profiles of third-instar midguts of gypsy moth feeding on diverse diets. Each profile is representative of 10 individual larvae.
Comparison of bacterial diversity revealed by culturing and culture-independent analysis.
Culturing yielded 15 distinguishable types of bacteria across all diet treatments from gypsy moth midguts based on cellular and colony morphological traits; 93% of these isolates could be assigned to a genus, while only 47% could be assigned to a species by using 16S rRNA analysis (Table 1). DAPI staining and microscopic enumeration indicated that there were 4 × 108 viable bacterial cells/gut. The phylotypes indicated by culture-independent methods exhibited greater divergence and diversity than phylotypes recovered by culturing. In addition to the 15 phylotypes recovered by culturing, 9 phylotypes were indicated based on 16S rRNA analysis in gypsy moth midguts across all diet treatments; only four of these phylotypes could be assigned definitively to a genus.
Effect of diet on bacterial composition.
Two phylotypes, Enterococcus faecalis and an uncultivated Enterobacter sp. (NAB17) were found in all larvae, regardless of treatment. In comparing the relative diversity of gut bacteria associated with each diet treatment, we found that 8 bacterial phylotypes were detected in only one of the diet treatments (Table 1). Larvae feeding on larch contained the greatest diversity of bacterial phylotypes (15, of which 11 were cultivated). Larvae feeding on aspen contained the highest proportion of uncultivated bacterial phylotypes of all the diet treatments. Among the 13 phylotypes identified from this group, over 50% of the phylotypes were obtained by using culture-independent methods only. On average, 51% of phylotypes in each diet treatment were previously reported sequences.
Comparison of T-RFLP profiles and 16S rRNA sequences.
To assess the ability of T-RFLP analysis to describe the bacterial composition of the midgut, 16S rRNA sequences derived from midguts were analyzed in silico by using the RDP II T-RFLP Analysis Program (43) and were compared to actual T-RFLPs from the same samples. In general, 16S rRNA sequences showed the predicted peak pattern, and all sequences could be accounted for in the T-RFLP. However, this analysis also demonstrated that multiple fragments were found at the same peak position regardless of the restriction enzyme, especially among similar 16S sequences. These results are consistent with previous indications of the resolution limits of T-RFLP analysis in assessing the full extent of diversity (23). However, these results also indicate that T-RFLP analysis is useful as a first step to assess sample diversity and identify treatments or conditions for further investigation.
DISCUSSION
These results demonstrate that the microbial diversity of the gypsy moth midgut is relatively simple, with 15 phylotypes at its most complex and 7 phylotypes at its simplest. The relative simplicity of this community is especially apparent compared to other gut environments, such as the human intestine and termite hindguts, which contain at least 500 and 50 phylotypes, respectively (54, 76). We expect that the census presented here is fairly complete given that we can account for more than 40% of the viable cells by culturing techniques, and all of the peaks in the T-RFLP analysis are accounted for in the PCR-generated 16S rRNA sequences. However, there may well be minor species whose presence is obscured because they were not cultured by our methods and were not detected by either T-RFLP analysis or 16S rRNA sequence analysis. Diet had a significant impact on midgut microbial diversity, as shown by culturing and sequence analysis of 16S rRNA genes amplified directly from larval midguts. Molecular, culture-independent methods revealed that many of the amplified 16S rRNA genes from gypsy moth midguts consisted of sequences not previously described. Perhaps the most surprising result of this study was the culturing of numerous bacterial species whose 16S rRNA sequences were as yet undescribed in public databases and, in some cases, diverged deeply from known species or genera.
Molecular characterization of cultured isolates and culture-independent methods identified E. faecalis in all midguts. While the 16S rRNA sequences from these isolates match most closely to E. faecalis, preliminary analysis revealed that these isolates differ physiologically (data not shown) from the clinical isolates of E. faecalis reported previously (21, 52). However, other E. faecalis isolates from environmental samples consistently diverge from the clinical type strains in certain physiological characteristics, including colony color, carbohydrate metabolism profiles, and growth temperature profiles (26, 47). Further analysis is required to determine the implications of these differences for species definition within the genus Enterococcus (63).
The novelty of phylotypes found in the gypsy moth midguts was anticipated, based on the insect's physiology. While the caterpillar lives in a common terrestrial macroenvironment as an external phytophage, the microenvironment in which these midgut bacteria reside is chemically extreme. Most insect midguts are generally neutral to acidic (pH 4 to 7), while lepidopteran midguts are typically pH 8 to 10. However, values as high as 12.4 have been reported for gypsy moth (5, 22, 29). In addition, the diverse plants on which this insect feeds contain an array of allelochemicals, such as phenolics, tannins, and terpenoids, many of which are toxic to microorganisms (28). Therefore, this extreme environment may have selected for unusual phenotypes, resulting in unusual phylotypes.
The microbial midgut inhabitants seem likely to benefit the insect host, based on observations of other insect systems (7, 9, 14, 31). Examples of such benefits may include modifying midgut pH, detoxifying plant allelochemicals, and maintaining the midgut microbial community structure. Midgut pH is important from the perspective of both the microbes and the insect. A predominant member of the midgut microbial community, E. faecalis, is commonly found at higher pH (8 to 9) and acidifies its environment through its metabolism (45). This could confer some advantage to the insect host, as some microbial toxins of lepidoptera, such as Bacillus thuringiensis toxin, are activated by alkaline conditions (75). Thus, E. faecalis might help protect this insect from B. thuringiensis toxin by decreasing the midgut pH. Previous work has shown that larvae that are more susceptible to B. thuringiensis toxin have smaller populations of E. faecalis in their midguts (11). Bacteria may also contribute to their own survival by managing midgut pH to enhance their own growth or exclude competing microorganisms. While numerous studies have investigated microbial diversity in neutral and acidic environments (6, 24, 27, 32, 56, 66), few have examined bacteria in highly alkaline environments, particularly in nonaquatic ecosystems (25, 30). Further analysis of these bacteria may identify adaptations that permit them to function in this extreme environment, and whether they have a role in maintaining it.
The broad range of phytochemicals consumed by this polyphagous herbivore may present a challenge for both the gypsy moth and its associated bacteria. Toxic compounds may select for bacteria that can metabolize them, and these bacteria may degrade ingested compounds that are otherwise toxic to the insect (41). For example, the 16S rRNA gene sequence of one cultured isolate from the gypsy moth (NAB13) was most similar to a Rhodococcus species, and recent work identified an enzyme produced by R. erythropolis DCL14 (73) that degrades monoterpenes, a widespread class of phytochemicals that can be toxic to many insect larvae and adults (38). The gypsy moth shows relatively higher tolerance to monoterpenes than many other insects (58). If the gypsy moth isolate of Rhodococcus species also produces this enzyme, it might contribute to the detoxification of monoterpenes. Identifying traits that facilitate bacterial survival in the gypsy moth midgut may lead to novel chemistry and degradation processes that allow bacteria to live in other extreme environments.
Establishment and maintenance of microbial assemblages within the larval midgut may contribute to insect health, perhaps by suppression of pathogens. For example, one cultured isolate from the gypsy moth midgut (NAB5) is highly similar (>99%) to a sequence found in the gut of the sheep mite, Psoroptes ovis (48), and both match Serratia marcesens to the species level (>98%). Similar sequences have been found in most insects and other arthropods (12, 35, 67). Serratia species are generally considered pathogens and can cause disease in insect production facilities (35, 68). The frequency at which this bacterium occurs across diverse arthropods suggests an important relationship with members of this phylum. Serratia might be an opportunistic pathogen living in the insect midgut at subclinical levels, causing disease only when the community structure is altered. The bacteria in the community may also contribute to each others' survival by providing nutrients or processing wastes.
Despite the relatively simple composition of the gypsy moth midgut community, there are challenges to elucidating the roles of various bacteria in gypsy moth biology and in maintenance of species composition within the midgut. In particular, there are several possibilities concerning the establishment of the bacterial midgut composition. For example, it is not known whether midgut bacteria are established through ingestion of plant material and whether different host plant chemistries select for specific bacterial genotypes. The plants selected for this study represent a diversity of secondary plant metabolites that the gypsy moth commonly encounters in the field and may contribute to the observed differences in the midgut bacterial composition. In addition, individual gypsy moth larvae routinely switch host plants in the field with only minor direct effects on the larvae (69, 70). However, host plant switching can alter the performance of gypsy moth parasitoids and could likewise affect the composition of the bacterial community (36). The relative stability of the gut bacterial community is also unknown. Further analysis of this system, including identification of the source of microbial inoculum in insects fed on sterilized artificial diet, use of methods, such as metagenomics (59), that link function to phylogeny, and improved culturing techniques will advance our understanding of the role of microbes in the gypsy moth midgut.
Acknowledgments
We thank Frank Martin (USDA-APHIS), Roger Fuester (Beneficial Insects Introduction and Research Laboratory, USDA), Dave Schumacher (WI-DATCP), and Roger Mech (MI-DNR) for their assistance in obtaining gypsy moth egg masses. We also thank Trenten Marty (WI-DNR) for providing the trees. We are grateful to Madeline Fischer and Angela Kent (University of Wisconsin—Madison) for their assistance with T-RFLP analysis.
This work was funded by the Wisconsin Department of Natural Resources, McIntire-Stennis Projects 4054 and 4529, the Howard Hughes Medical Institute, and Hatch Projects 4038 and 5234 from the College of Agricultural and Life Sciences at the University of Wisconsin—Madison.
REFERENCES
- 1.Aksoy, S. 1995. Molecular characteristics of the endosymbionts of the tsetse flies: 16S rRNA locus and over-expression of chaperonins. Insect Molec. Biol. 4:23-29. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Meyers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Molec. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., B. Roger, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1994. Current protocols in molecular biology, p. 2.4.1-2.4.5. John Wiley & Sons, Inc., New York, N.Y.
- 4.Bandi, C., D. Damiani, L. Magrassi, A. Grigolo, R. Fani, and L. Sacchi. 1994. Flavobacteria as intracellular symbionts in cockroaches. Proc. R. Soc. Lond. 257:43-48. [DOI] [PubMed] [Google Scholar]
- 5.Berenbaum, M. 1980. Adaptive significance of the midgut pH in larval Lepidoptera. Am. Nat. 115:138-146. [Google Scholar]
- 6.Bintrim, S. B., T. J. Donohue, J. Handelsman, G. P. Roberts, and R. M. Goodman. 1997. Molecular phylogeny of Archaea from soil. Proc. Natl. Acad. Sci. USA 94:277-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brand, J. M., J. W. Bracke, A. J. Markovetz, D. L. Wood, and L. E. Browne. 1975. Production of verbenol pheromone by a bacterium isolated from bark beetles. Nature 254:136-137. [DOI] [PubMed] [Google Scholar]
- 8.Breznak, J. A. 1982. Intestinal microbiota of termites and other xylophagous insects. Annu. Rev. Microbiol. 36:323-343. [DOI] [PubMed] [Google Scholar]
- 9.Breznak, J. A., and J. M. Switzer. 1986. Acetate synthesis from H2 plus CO2 by termite gut microbes. Appl. Environ. Microbiol. 52:623-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Broderick, N. A., R. M. Goodman, K. F. Raffa, and J. Handelsman. 2000. Synergy between zwittermicin A and Bacillus thuringiensis subsp. kurstaki against gypsy moth (Lepidoptera: Lymantriidae). Environ. Entomol. 29:101-107. [Google Scholar]
- 11.Broderick, N. A., R. M. Goodman, J. Handelsman, and K. F. Raffa. 2003. Effect of host diet and insect source on synergy of gypsy moth (Lepidoptera: Lymantriidae) mortality to Bacillus thuringiensis subsp. kurstaki by zwittermicin A. Environ. Entomol. 32:387-391. [Google Scholar]
- 12.Bucher, G. E. 1963. Nonsporulating bacterial pathogens, p. 117-147. In E. A. Steinhaus (ed.), Insect pathology. Academic Press, Inc., New York, N.Y.
- 13.Buchner, P. 1965. Endosymbionts of animals with plant microorganisms. John Wiley & Sons, Inc., New York, N.Y.
- 14.Campbell, B. C. 1989. On the role of microbial symbiotes in herbivorous insects. In E. Bernays (ed.), Insect-plant interactions. CRC Press, Boca Raton, Fla.
- 15.Cazemier, A. E., J. H. P. Hackstein, H. J. M. Op den Camp, J. Rosenberg, and C. van der Drift. 1997. Bacteria in the intestinal tract of different species of arthropods. Microb. Ecol. 33:189-197. [DOI] [PubMed] [Google Scholar]
- 16.Chehab, F. F., and Y. W. Kan. 1989. Detection of specific DNA sequences by fluorescence amplification: a color complementation assay. Proc. Natl. Acad. Sci. USA 86:9178-9180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen, D.-Q., and A. H. Purcell. 1997. Occurrence and transmission of facultative endosymbionts in aphids. Curr. Microbiol. 34:220-225. [DOI] [PubMed] [Google Scholar]
- 18.Chenot, A. B., and K. F. Raffa. 1998. Effects of parasitoid strain and host instar on the interaction of Bacillus thuringiensis subsp. kurstaki with the gypsy moth larval parasitoid Cotesia melanoscela. Environ. Entomol. 27:137-147. [Google Scholar]
- 19.Clark, M. A., L. Baumann, M. A. Munson, P. Baumann, B. C. Campbell, J. E. Duffins, L. S. Osborne, and N. A. Moran. 1992. The eubacterial endosymbionts of whiteflies (Homoptera: Aleyrodidae) constitute a major lineage distinct from the endosymbionts of aphids and mealybugs. Curr. Microbiol. 25:119-123. [Google Scholar]
- 20.Dasch, G. A., E. Weiss, and K. P. Chang. 1984. Endosymbionts of insects, p. 811-833. In N. R. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology. Williams & Wilkins, Baltimore, Md.
- 21.Devriese, L. A., M. D. Collins, and R. Wirth. The genus Enterococcus, p. 1465-1487. In A. Barlows, H. G. Truper, M. Dworkin, W. Harder, and K. H. Schliefer (ed.), The prokaryotes. Springer-Verlag, New York, N.Y.
- 22.Dow, J. A. T. 1992. pH gradients in lepidopteran midgut. J. Exp. Biol. 172:255-375. [DOI] [PubMed] [Google Scholar]
- 23.Dunbar, J., L. O. Ticknor, and C. R. Kuske. 2001. Phylogenetic specificity and reproducibility and new method for analysis of terminal restriction fragment profiles of 16S rRNA genes from bacterial communities. Appl. Environ. Microbiol. 67:190-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Felske, A, A. Wolterink, R. van Lis, W. M. de Vos, and A. D. L. Akkermans. 1999. Searching for predominant soil bacteria: 16S rRNA cloning versus strain cultivation. FEMS Microbiol. Ecol. 30:137-145. [DOI] [PubMed] [Google Scholar]
- 25.Fry, N. K., J. K. Fredrickson, S. Fishbain, S. Wagner, and D. A. Stahl. 1997. Population structure of microbial communities associated with two deep, anaerobic, alkaline aquifers. Appl. Environ. Microbiol. 63:1498-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geldrich, E. E., B. A. Kenner, and P. W. Kabler. 1964. Occurrence of coliforms, fecal coliforms, and streptococci in vegetation and insects. Appl. Microbiol. 12:63-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goebel, B. M., and E. Stackebrandt. 1994. Cultural and phylogenetic analysis of mixed microbial populations found in natural and commercial bioleaching environments. Appl. Environ. Microbiol. 60:1614-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Govenor, H. L., J. C. Schultz, and H. M. Appel. 1997. Impact of dietary allelochemicals on gypsy moth (Lymantria dispar) caterpillars: importance of midgut alkalinity. J. Insect Physiol. 43:1169-1175. [DOI] [PubMed] [Google Scholar]
- 29.Gringorten, J. L., D. N. Crawford, and W. R. Harvey. 1993. High pH in the ectoperitrophic space of the larval lepidopteran midgut. J. Exp. Biol. 183:353-359. [DOI] [PubMed] [Google Scholar]
- 30.Gsell, T. C., W. E. Holben, and R. M. Ventullo. 1997. Characterization of the sediment bacterial community in groundwater discharge zones of an alkaline fen: a seasonal study. Appl. Environ. Microbiol. 63:3111-3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Houk, E. J. 1987. Symbionts, p. 123-129. In A. K. Minks and P. Harrewijn (ed.), Aphids; their biology, natural enemies, and control. Elsevier Biomedical Press, Amsterdam, The Netherlands.
- 32.Hugenholtz, P., C. Pitulle, K. L. Hershberger, and N. R. Pace. 1998. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180:366-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hypsa, V., and S. Aksoy. 1997. Phylogenetic characterization of two transovarially transmitted endosymbionts of the bedbug Cimex lectularius (Heteroptera: Cimicidae). Insect Molec. Biol. 6:301-304. [DOI] [PubMed] [Google Scholar]
- 34.Kebelmann-Betzing, C., K. Seeger, S. Dragon, G. Schmitt, A. Moricke, T. A. Schild, G. Henze, and B. Beyermann. 1998. Advantages of a new Taq DNA polymerase in multiplex and time-release PCR. BioTechniques 24:154-158. [DOI] [PubMed] [Google Scholar]
- 35.Krieg, A. 1987. Diseases caused by bacteria and other prokaryotes, p. 323-355. In J. R. Fuxa and Y. Tanada (ed.), Epizootiology of insect diseases. John Wiley & Sons, Inc., New York, N.Y.
- 36.Kruse, J. J., and K. F. Raffa. 1999. Effect of food plant switching by an herbivore on its parasitoid: Cotesia melanoscela development in Lymantria dispar exposed to reciprocal dietary crosses. Ecolog. Entomol. 24:37-45. [Google Scholar]
- 37.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Inc., New York, N.Y.
- 38.Langheim, J. H. 1994. Higher-plant terpenoids—a phytocentric overview of their ecological roles. J. Chem. Ecol. 20:1223-1280. [DOI] [PubMed] [Google Scholar]
- 39.Larsen, N., G. J. Olsen, B. L. Maidak, M. J. McCaughey, R. Overbeek, T. J. Macke, T. L. Marsh, and C. R. Woese. 1993. The ribosomal database project. Nucleic Acids Res. 21:3021-3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lesser, T. D., R. H. Lindecrona, T. K. Jensen, B. B. Jensen, and K. Moller. 2000. Changes in bacterial community structure in the colon of pigs fed different experimental diets and after infection with Bracyspira hyodysenteriae. Appl. Environ. Entomol. 66:3290-3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liebhold, A. M., K. W. Gottschalk, R. M. Muzika, M. E. Montgomery, R. Young, K. O'Day, and B. Kelley. 1995. Suitability of North American tree species to the gypsy moth: a summary of field and laboratory tests. U.S. Department of Agriculture Forest Service NE Forest Experimental Station General Technical Bulletin NE-211. U.S. Department of Agriculture, Washington, D.C.
- 42.Liu, W.-T., T. L. Marsh, H. Chang, and L. J. Forney. 1997. Characterization of microbial diversity by determining terminal restriction length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, P. R. Saxman, J. M. Stredwick, G. M. Garrity, B. Li, G. J. Olsen, S. Pramanik, T. M. Schmidt, and J. M. Tiedje. 2000. The RDP (Ribosome Database Project) continues. Nucleic Acids Res. 28:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Makkar, H. P. S., and K. Becker. 1996. Effect of pH, temperature, and time on inactivation of tannins and possible implications of detannification studies. J. Agric. Food Chem. 44:1291-1295. [Google Scholar]
- 45.Manero, A., and A. R. Blanch. 1999. Identification of Enterococcus spp. with a biochemical key. Appl. Environ. Micobiol. 65:4425-4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marsh, T. L., P. Saxman, J. Cole, and J. Tiedje. 2000. Terminal restriction fragment length polymorphism analysis program, a web-based research tool for microbial community analysis. Appl. Environ. Microbiol. 66:3616-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martin, J. D., and J. O. Mundt. 1972. Enterococci in insects. Appl. Microbiol. 24:575-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mathieson, B. R. F., and M. J. Lehane. 1996. Isolation of the Gram negative bacterium Serratia marcesens from the sheep scab mite Psoroptes ovis. Vet. Rec. 138:210-211. [DOI] [PubMed] [Google Scholar]
- 49.McKillip, J. L., C. L. Small, J. L. Brown, J. F. Brunner, and K. D. Spence. 1997. Sporogenous midgut bacteria of the leafroller, Pandemis pyrusana (Lepidoptera: Tortricidae). Environ. Entomol. 26:1475-1481. [Google Scholar]
- 50.Mittler, T. E. 1988. Applications of artificial feeding techniques for aphids, p. 145-170. In A. K. Minks and P. Harrewijn (ed.), Aphids: their biology, natural enemies, and control. Elsevier Biomedical Press, Amsterdam, The Netherlands.
- 51.Montgomery, M. E., and W. E. Wallner. 1989. The gypsy moth: a westward migrant, p. 353-373. In A. A. Berryman (ed.), Dynamics of forest insect populations: patterns, causes, implications. Plenum Press, New York, N.Y.
- 52.Mundt, J. O. 1986. Enterococci, p. 1063-1065. In P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology. Williams & Wilkins, Baltimore, Md.
- 53.Nolte, D. J. 1977. The action of locustol. J. Insect Physiol. 23:899-903. [Google Scholar]
- 54.Okhuma, M., and T. Kudo. 1996. Phylogenetic diversity of the intestinal bacterial community in the termite Reticulitermes speratus. Appl. Environ. Microbiol. 62:461-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O'Neill, S. L., R. Giordano, A. M. E. Colbert, T. L. Karr, and H. M. Robertson. 1992. 16S rRNA phylogenetic analysis of the bacterial endosymbionts associated with cytoplasmic instability in insects. Proc. Natl. Acad. Sci. USA 89:2699-2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ovreas, L., S. Jensen, F. L. Daae, and V. Torsvik. 1998. Microbial community changes in a perturbed agricultural soil investigated by molecular and physiological approaches. Appl. Environ. Microbiol. 64:2739-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paster, B. J., F. E. Dewhirst, S. M. Cooke, V. Fussing, L. K. Poulsen, and J. A. Breznak. 1996. Phylogeny of not-yet-cultured spirochetes from termite guts. Appl. Environ. Microbiol. 62:347-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Powell, J. S., and K. F. Raffa. 1999. Effects of selected Larix laricina terpenoids on Lymantria dispar (Lepidoptera: Lymantriidae) development and behavior. Environ. Entomol. 28:148-154. [Google Scholar]
- 59.Rondon, M. R., P. R. August, A. D. Bettermann, S. F. Brady, T. H. Grossman, M. R. Liles, K. A. Loiacono, B. A. Lynch, I. A. MacNeil, C. Minor, C. L. Tiong, M. Gilman, M. S. Osburne, J. Clardy, J. Handelsman, and R. M. Goodman. 2000. Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl. Environ. Microbiol. 66:2541-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roush, R. T. 1987. Ecological genetics of insecticide and acaricide resistance. Annu. Rev. Entomol. 32:361-380. [DOI] [PubMed] [Google Scholar]
- 61.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 62.Santo Domingo, J. W., M. G. Kaufman, M. J. Klug, and J. M. Tiedje. 1998. Characterization of the cricket hindgut microbiota with fluorescently labeled rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64:752-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schleifer, K. H., and R. Kiepper-Balz. 1984. Transfer of Streptococcus faecalis and Streptococcus faecium to the genus Enterococcus nom. rev. as Enterococcus faecalis comb. nov. and Enterococcus faecium comb. nov. Int. J. Syst. Bact. 34:31-34. [Google Scholar]
- 64.Schroder, D., H. Deppisch, M. Obermayer, G. Krohne, E. Stackebrandt, B. Holldobler, W. Goebel, and R. Gross. 1996. Intracellular endosymbiotic bacteria of Camponotus species (carpenter ants): systematics, evolution, and ultrastructural characterization. Molec. Microbiol. 21:479-489. [DOI] [PubMed] [Google Scholar]
- 65.Scoble, M. J. 1992. The Lepidoptera: form, function, and diversity. Oxford University Press, New York, N.Y.
- 66.Shiomi, Y., M. Nishiyama, T. Onizuka, and T. Marumoto. 1999. Comparison of bacterial community structures in the rhizoplane of tomato plants grown in soils suppressive and conducive towards bacterial wilt. Appl. Environ. Microbiol. 65:3996-4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sri-Arunotai, S., P. P. Sikorowski, and W. W. Neel. 1975. Study of pathogens of pecan weevil larvae. Environ. Entomol. 4:790-792. [Google Scholar]
- 68.Steinhaus, E. A. 1959. Serratia marcescens Bizio as an insect pathogen. Hilgardia 28:351-380. [Google Scholar]
- 69.Stoyenoff, J. L., J. A. Witter, M. E. Montgomery, and C. A. Chilcote. 1994. Effects of host switching on gypsy moth (Lymantria dispar (L)) under field conditions. Oecologia 97:143-157. [DOI] [PubMed] [Google Scholar]
- 70.Stoyenoff, J. L., J. A. Witter, and M. E. Montgomery. 1994. Nutritional indices in the gypsy moth (Lymantria dispar (L)) under field conditions and host switching situations. Oecologia 97:158-170. [DOI] [PubMed] [Google Scholar]
- 71.Tabashnik, B. E. 1994. Evolution of insect resistance to Bacillus thuringiensis. Annu. Rev. Entomol. 39:47-79. [DOI] [PubMed] [Google Scholar]
- 72.Tholen, A., B. Schink, and A. Brune. 1997. The gut microflora of Reticulitermes flaipes, its relation to oxygen, and evidence for oxygen-dependent acetogenesis by the most abundant Enterococcus sp. FEMS Microbiol. Ecol. 24:137-149. [Google Scholar]
- 73.van der Vlugt-Bergmans, C. J. B., and M. J. van der Werf. 2001. Genetic and biochemical characterization of a novel monoterpene ɛ-lactone hydrolase from Rhodococcus erythropolis DCL14. Appl. Environ. Microbiol. 67:733-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ward, D. M., R. Weller, and M. M. Bateson. 1990. 16S rRNA reveal numerous uncultured microorganisms in a natural community. Nature 345:63-65. [DOI] [PubMed] [Google Scholar]
- 75.Wilson, G. R., and T. G. Benoit. 1993. Alkaline pH activated Bacillus thuringiensis spores. J. Invert. Pathol. 62:87-89. [Google Scholar]
- 76.Wilson, K. H., and R. B. Blitchington. 1996. Human colonic biota studied by ribosomal DNA sequence analysis. Appl. Environ. Microbiol. 62:2273-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]