Metallo-β-lactamase enzymes confer resistance to a wide variety of commonly used antibiotics; this resistance poses a significant threat to human health where it arises in a pathogenic bacterium. Here, the structure of the novel metallo-β-lactamase NDM-1, which is capable of providing resistance against a broad spectrum of antibacterial agents, has been solved.
Keywords: metallo-β-lactamases, New Delhi MBL 1, antibiotic resistance
Abstract
Antibiotic resistance in bacterial pathogens poses a serious threat to human health and the metallo-β-lactamase (MBL) enzymes are responsible for much of this resistance. The recently identified New Delhi MBL 1 (NDM-1) is a novel member of this family that is capable of hydrolysing a wide variety of clinically important antibiotics. Here, the crystal structure of NDM-1 from Klebsiella pneumoniae is reported and its structure and active site are discussed in the context of other recently deposited coordinates of NDM-1.
1. Introduction
The rapid increase in resistance to β-lactam-derived antibiotics is a major clinical and epidemiological concern, as these antibiotics have long been the mainstay for the treatment of serious bacterial infections. The β-lactamases are a major cause of this resistance, as they are capable of hydrolysing the amide bond within the β-lactam ring to render the antibiotic inactive (Bebrone, 2007 ▶). The β-lactamases fall into four classes, of which classes A, C and D employ an active-site serine residue to effect catalysis, while class B, also called the metallo-β-lactamases (MBLs), require either one or two metal ions for their activity. The MBLs are of particular concern since not only are they located on highly transmissible plasmids (Yong et al., 2010 ▶), but they also have a broad spectrum of activity against almost all β-lactam antibiotics, including the carbapenems, which are often considered to be the ‘last line of defence’ against bacterial infection. Moreover, MBLs are insensitive to clinically available inhibitors (Drawz & Bonomo, 2010 ▶).
Sequence alignment allows the MBLs to be further subdivided (Garau et al., 2004 ▶). Subclass B1 MBLs include BclI from Bacillus cereus (Hussain et al., 1985 ▶), IMP-1 from Serratia marcescens (Osano et al., 1994 ▶) and Vim-2 from Pseudomonas aeruginosa (Poirel et al., 2000 ▶). These enzymes are active with one zinc ion in the active site, but display increased activity on the binding of a second metal ion (Davies & Abraham, 1974 ▶). Class B2 enzymes are only active with a single zinc ion in the active site and the binding of a second metal ion has been shown to inhibit turnover (Hernandes Valladares et al., 1997 ▶), while class B3 enzymes can only perform hydrolysis when two zinc ions are present (Bebrone, 2007 ▶).
The B1 subclass MBLs for which three-dimensional structures are known all share the same αβ/βα sandwich fold, with two central β-sheets surrounded by four α-helices (Garcia-Saez et al., 2008 ▶). The active site, with the two zinc ions, is located at the bottom of a solvent-accessible groove bounded by several loops. The first metal ion is coordinated with a tetragonal geometry by the imidazole groups of three histidine residues, with the fourth coordination site occupied by a water molecule. This water molecule also forms part of the coordination shell around the second metal ion, together with the side chains of a histidine, a cysteine and an aspartic acid residue and two additional water molecules. The water molecule shared between the two metal ions is likely to exist as a hydroxide ion, which can act as the nucleophile during enzymatic turnover (Concha et al., 1996 ▶).
Here, we present the structure of New Delhi MBL 1 (NDM-1) from Klebsiella pneumoniae, a novel class B1 MBL of great clinical significance owing to its capacity to hydrolyse a wide range of β-lactam antibiotics.
2. Materials and methods
2.1. Cloning, expression and purification
A synthetic gene encoding K. pneumoniae NDM-1 with codons optimized for expression in Escherichia coli (Geneart, Regensberg, Germany) was used as the template for construct design. A construct spanning amino acids 42–270 was amplified by PCR using the forward primer 5′-AAGTTCTGTTTCAGGGCCCGGGTGATCAGCGTTTTGGTGAT-3′ and the reverse primer 5′-ATGGTCTAGAAAGTCTTAACGCAGTTTATCTGCCATAC-3′ and the product was inserted into pOPINF via the In-Fusion cloning system (Clontech–Takara Bio Europe, Saint-Germain-en-Laye, France). The resulting construct contained an N-terminal hexahistidine tag that was cleavable from the NDM-1 construct using rhinovirus 3C protease.
Expression trials were performed in E. coli Rosetta pLysS cells using Overnight Express instant TB medium (Merck, Nottingham, England) supplemented with 50 µg ml−1 ampicillin and 32 µg ml−1 chloramphenicol. Cells were grown in 2 l culture flasks at 310 K for 4 h; the temperature was then lowered to 298 K and the cells were incubated for a further 20 h. The cells were harvested by centrifugation for 15 min at 6000g, resuspended in 50 mM Tris pH 7.5, 500 mM NaCl, 0.2% Tween 20, 10 µg ml−1 DNAse and an EDTA-free protease-inhibitor cocktail tablet (Roche) and lysed using a Basic Z Cell Disruptor (Constant Systems Ltd, Daventry, England) at a pressure of 207 MPa. Cellular debris was removed by centrifugation at 30 000g for 30 min. The supernatant was loaded onto a 1 ml HisTrap FF column (GE Healthcare, Amersham, England) followed by extensive washing with 50 mM Tris pH 7.5, 500 mM NaCl, 20 mM imidazole prior to elution of NDM-1 in 50 mM Tris pH 7.5, 500 mM NaCl, 500 mM imidazole. The protein was injected onto a 16/60 HiLoad Superdex 200 column (GE Healthcare, Amersham, England) and eluted with 20 mM Tris pH 7.5, 200 mM NaCl. Protein-containing fractions were analysed by SDS–PAGE (NuPage, Invitrogen). The N-terminal tag was removed by overnight incubation at 277 K with His-tagged 3C protease, which was then removed together with any uncleaved protein by a further round of nickel-affinity chromatography. The protein purity and quality were analysed by SDS–PAGE and electrospray mass spectrometry, with a typical preparation yielding 5 mg pure NDM-1 per litre of culture.
2.2. Crystallization and data collection
Crystals of NDM-1 were grown using the sitting-drop vapour-diffusion method by mixing 100 nl NDM-1 solution at a concentration of 35 mg ml−1 with 100 nl 5 mM CoCl2, 5 mM NiCl2, 5 mM MgCl2, 5 mM CdCl2, 100 mM HEPES pH 7.5, 12%(w/v) PEG 3350 using a Cartesian MicroSys crystallization robot (Digilab Ltd, Huntingdon, England) followed by incubation at 294 K for three weeks. Crystals were cryoprotected by the addition of 25% glycerol to the reservoir solution followed by flash-cooling in liquid nitrogen.
Diffraction data were collected to a maximum resolution of 1.9 Å at station I04-1 (Diamond Light Source, England) at a wavelength of 0.9163 Å using a Pilatus 2M hybrid pixel-array detector. Additional data sets were collected at station I02 at wavelengths corresponding to the K edges of cobalt (1.6052 Å), nickel (1.4881 Å) and zinc (1.2828 Å) to aid the identification of the metal ions present in the structure. All data were collected at a temperature of 100 K and all data reduction was performed using xia2 (Winter, 2010 ▶).
2.3. Structure solution and refinement
Initial phase estimates were calculated by molecular replacement using the program Phaser (McCoy et al., 2007 ▶) with the coordinates of the homologous protein Vim-4 (PDB entry 2whg; Lassaux et al., 2011 ▶), with metal ions and solvent molecules removed, as a search model. The resulting maps allowed automatic chain tracing with ARP/wARP (Langer et al., 2008 ▶), followed iteratively by cycles of refinement in REFMAC (Murshudov et al., 2011 ▶) and manual rebuilding in Coot (Emsley et al., 2010 ▶). Large residual peaks were visible in F o − F c density maps at the positions expected to be occupied by zinc ions in the active site and also at five of the crystal contacts. These peaks were all interpreted as metal ions and those at the active site were initially modelled as zinc, while the identities of the other peaks remained ambiguous since the crystals were grown from a solution containing four additional divalent metal ions. The identity of the ions in the other peaks was ascertained by comparison of peak heights in anomalous difference Fourier maps calculated using data collected at wavelengths corresponding to the K edges of cobalt, nickel or zinc. Metals were manually added to the model using Coot and were included in subsequent rounds of refinement.
3. Results and discussion
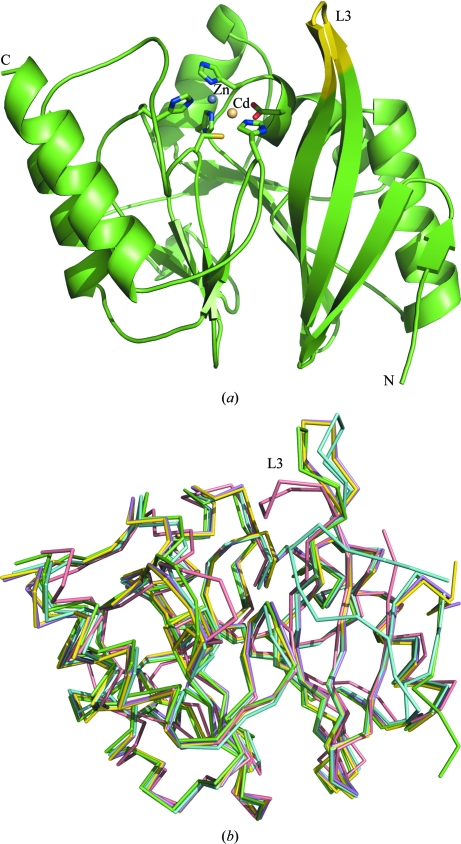
Crystals of NDM-1 belong to space group I422 and contain one molecule per asymmetric unit. The final model contained amino acids 42–270 of NDM-1, two additional residues (Gly and Pro) which remained at the N-terminus of the construct after cleavage with 3C protease, seven metal ions and 115 water molecules, and was refined to R = 18.0% and R free = 20.9%. Data-collection and refinement statistics are shown in Table 1 ▶. The amino-acid sequence of NDM-1 is markedly different from those of many other subclass B1 MBLs (Yong et al., 2010 ▶), with the closest homologue, Vim-4, sharing only 38% sequence identity. However, the overall fold of the enzyme is the classic αβ/βα sandwich that has been observed for homologous MBLs, in which a central core of two twisted β-sheets pack against one another via their inner faces, with their outer faces each flanked by two α-helices (Fig. 1 ▶ a).
Table 1. X-ray data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell.
| Data set | Native | Zinc edge | Cobalt edge | Nickel edge |
|---|---|---|---|---|
| X-ray source | I04-1 | I02 | I02 | I02 |
| Space group | I422 | I422 | I422 | I422 |
| Unit-cell parameters (Å) | a = b = 120.2, c = 88.1 | a = b = 120.3, c = 88.3 | a = b = 120.3, c = 88.2 | a = b = 120.5, c = 88.3 |
| Wavelength (Å) | 0.9173 | 1.2828 | 1.6052 | 1.4852 |
| Temperature (K) | 100 | 100 | 100 | 100 |
| Resolution (Å) | 28.8–1.91 (1.96–1.91) | 28.84–2.68 (2.75–2.68) | 28.8–2.47 (2.54–2.47) | 28.6–2.91 (2.98–2.91) |
| Unique reflections | 25258 (3611) | 22682 (3295) | 22231 (3242) | 22404 (3265) |
| Completeness (%) | 99.8 (97.6) | 99.8 (98.0) | 99.7 (96.7) | 99.3 (92.6) |
| Multiplicity | 13.3 (12.2) | 10.1 (9.3) | 8.7 (7.4) | 9.8 (9.1) |
| 〈I/σ(I)〉 | 26.3 (3.3) | 26.6 (3.1) | 27.5 (2.9) | 27.5 (2.9) |
| Rmerge† (%) | 6.7 (76) | 7.5 (69.1) | 5.9 (66.4) | 7.1 (51.6) |
| Refinement | ||||
| Rwork/Rfree‡ (%) | 18.0/20.9 | |||
| Model | ||||
| Protein atoms | 1722 | |||
| Water molecules | 117 | |||
| Metal ions | 7 | |||
| R.m.s.d. from ideal | ||||
| Bond lengths (Å) | 0.018 | |||
| Bond angles (°) | 1.576 | |||
| Ramachandran favoured | 97.4 | |||
| Ramachandran outliers | 0.4 | |||
R
merge = 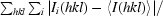
 , where I
i(hkl) is the intensity of reflection hkl and
, where I
i(hkl) is the intensity of reflection hkl and 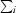 is the sum over all i measurements of reflection hkl.
is the sum over all i measurements of reflection hkl.
R
work = 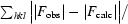
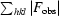 , where F
obs and F
calc are the observed and calculated structure factors, respectively. R
free is calculated in the same manner but using a random subset (5%) of reflections that are excluded from refinement.
, where F
obs and F
calc are the observed and calculated structure factors, respectively. R
free is calculated in the same manner but using a random subset (5%) of reflections that are excluded from refinement.
Figure 1.
(a) The structure of NDM-1 shows that the protein contains the αβ/βα sandwich common to all MBLs despite the low sequence identity between NDM-1 and other metallo-β-lactamases. The metal-chelating residues within the active site are displayed as sticks. The zinc ion is coloured blue and the cadmium ion is coloured fawn. Flexible loop L3, which is proposed to play a role in substrate binding, is highlighted in yellow. (b) Superposition of our coordinates (green) with other models of NDM-1 (PDB entries 3q6x, cyan; 3rkj, violet; 3rkk, yellow; 3s0z, pink) shows the protein structure to be rather rigid with one key region of flexibility, loop L3. All figures were produced using PyMOL (Schrödinger LLC).
Several sets of coordinates for NDM-1 have recently been deposited in the PDB (PDB entries 3rkj, 3rkk, 3s0z and 3q6x), the first three corresponding to protein alone (Guo et al., 2011 ▶; Y. Kim, C. Tesar, R. Jedrzejczak, T. A. Binkowski, G. Babnigg, J. Sacchettini & A. Joachimiak, unpublished work) and the last to NDM-1 in complex with hydrolysed ampicillin (Zhang & Hao, 2011 ▶). When superposed, the independently solved structures of NDM-1 showed a high degree of structural similarity (Fig. 1 ▶ b and Table 2 ▶). For example, the Cα atoms across the entire backbone of 3q6x superpose well on our coordinates, with an r.m.s.d. of 0.63 Å. However, structures 3rkj, 3rkk and 3s0z show a greater variation in r.m.s.d. across the protein backbone. 3rkj and 3rkk do not contain metal ions in their active sites and there are local differences in the loops responsible for coordinating the metal ions. In structure 3s0z a loop spanning residues 163–176 is involved in crystal contacts and adopts a very different conformation from those observed in the other four structures.
Table 2. R.m.s.d. in Cα positions between the structure described in this work and other deposited NDM-1 structures.
R.m.s.d. calculations were performed with LSQKAB using the following data ranges: all backbone, residues 47–270; omitting loop L3, residues 47–62 and 74–270; loop L3 alone, residues 63–73. The r.m.s.d. calculated for loop L3 represents the r.m.s.d. for residues 63–73 inclusive after the entire backbone has been superposed and was calculated from the individual r.m.s.d. values for these residues after such a superposition.
A detailed inspection of the r.m.s.d. variation for our coordinates and 3q6x reveals that loop L3 (residues 63–73), which is adjacent to the active site, is the region of greatest variation (Fig. 1 ▶ b), with much greater deviation than that observed for the rest of the protein (Table 2 ▶). L3 also displays the greatest deviations in structures 3rkj and 3s0z, adopting four clearly different conformations when the structures are superposed. This loop has been shown to be involved in substrate binding and its increased hydrophobicity compared with other proteins of this subclass has been proposed as the basis for the broad substrate specificity of NDM-1 (Zhang & Hao, 2011 ▶). It is likely that the flexibility of this loop is also important in the binding of structurally diverse substrates, since its reorientation could increase the range of molecules accepted into the active site and furthermore allow the enzyme to form the optimally stable complexes with these substrates.
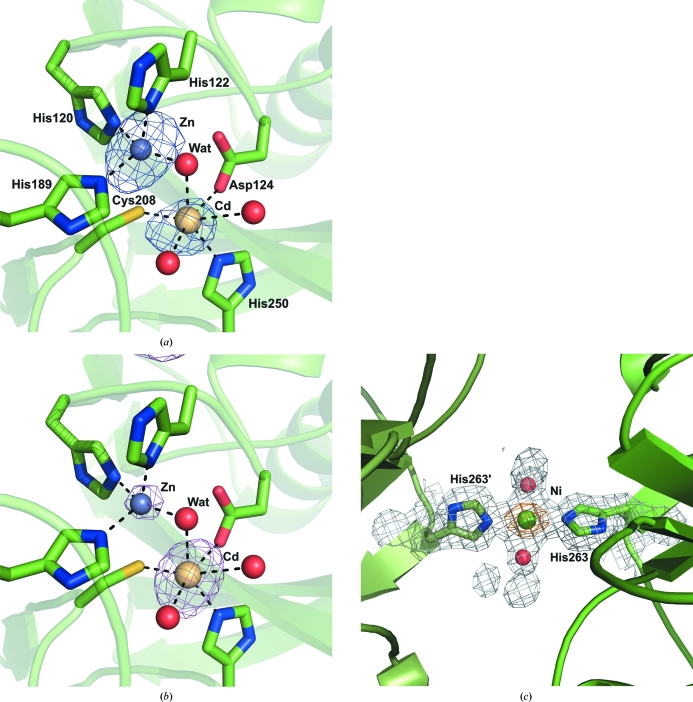
The active site contains two metal ions, with the first (site 1) tetrahedrally coordinated by histidine residues 120, 122 and 189 at coordination distances of 2.16, 1.99 and 2.19 Å, respectively, and a water molecule (termed the catalytic water and labelled Wat in Figs. 2 ▶ and 3 ▶). The metal ion in site 2 is octahedrally coordinated by three protein-derived ligands, Asp124 (2.42 Å), Cys208 (2.52 Å) and His250 (2.35 Å), and three water molecules, one of which is the catalytic water (Fig. 2 ▶ a). The structures of other MBLs of the same class typically show two zinc ions in the active site. The coordination geometry and distances for the site 1 metal strongly suggest that zinc is present in this position (Dokmanić et al., 2008 ▶); however, the distances and geometry observed for the ion in site 2 are more characteristic of cadmium. The metal-ion composition of the active site was further analysed by comparison of peak heights in anomalous difference density maps calculated using data collected at the K edges of zinc, cobalt and nickel (Table 1 ▶, Figs. 2 ▶ a and 2 ▶ b). The zinc-edge anomalous difference map contained a strong 0.16 e (15.5σ) peak at site 1, which was much smaller in maps calculated at the cobalt and nickel edges, with values of 0.06 e (6.5σ) and 0.04 e (4.1σ), respectively. The metal ion in site 2 displayed different anomalous scattering properties, with peak heights of 0.1 e (9.5σ), 0.14 e (14.8σ) and 0.12 e (10.9σ) for data collected at the zinc, cobalt and nickel edges, respectively. This scattering behaviour, and the observed coordination geometries, suggests the active site contains a zinc ion in site 1. The identity of the metal ion in site 2 is less obvious as the scattering profile and octahedral coordination suggests a cobalt or cadmium ion in site 2. However, the metal–ligand distances observed for site 2 are much more similar to those observed for cadmium (Dokmanić et al., 2008 ▶). Additionally, a residual peak of 0.38 e (8.1σ) remained in the F o − F c map when the metal ion in site 2 was modelled as cobalt which was absent when the metal was modelled as cadmium, suggesting that cadmium is the metal ion present in site 2. It has been reported that zinc in site 2 can readily be replaced by other metal ions such as cobalt (Badarau et al., 2007 ▶) and cadmium (Paul-Soto et al., 1999 ▶), so it is likely that cadmium from the crystallization medium has replaced the zinc ion usually present in site 2. The anomalous difference maps also allowed the unambiguous identification of one cobalt ion and four nickel ions in crystal contacts (Fig. 2 ▶ c).
Figure 2.
(a) The active site of NDM-1 contains a zinc ion (blue) in site 1 and a cadmium ion (fawn) in site 2, both of which are coordinated by the water molecule (labelled Wat) responsible for hydrolysis of the antibiotic. An anomalous difference Fourier map calculated using data collected at the Zn K edge shows a much larger peak for the metal ion in site 1 relative to that in site 2. The anomalous maps shown are all contoured at a level of 0.05 e (5σ). (b) An anomalous difference Fourier map calculated using data collected at the Co K edge shows a stronger peak for the metal ion at site 2. The relative peak heights are consistent with the assignment of the metal ions in the active site and at crystal contacts. (c) An example of a nickel ion (green) located at a crystal contact between His263 and the equivalent amino acid, labelled His263′, in an adjacent protein molecule. The anomalous difference map calculated using data collected at the Ni K edge is shown in orange.
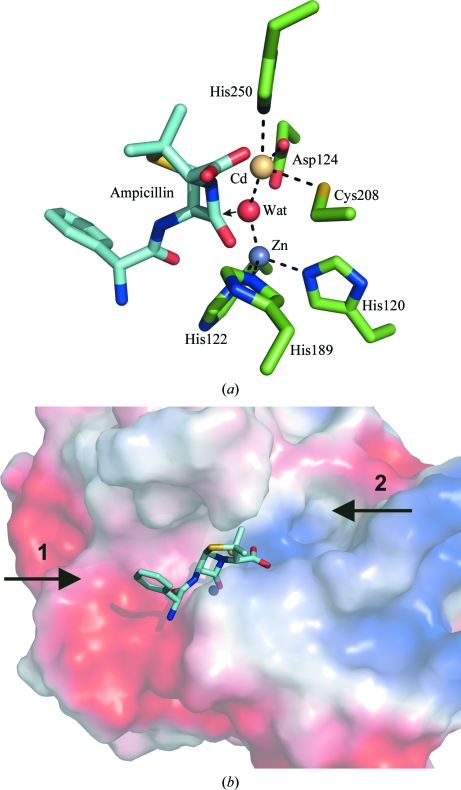
Figure 3.
(a) An ampicillin molecule modelled into the active site of NDM-1 shows that the shared catalytic water molecule is ideally positioned to attack the β-lactam carbonyl group, as indicated by the arrow. (b) A representation of the van der Waals surface of NDM-1 shows that the substrate-binding groove on both sides of the metal ions is large enough to accommodate sizeable ligands, suggesting that β-lactam antibiotics carrying very large substituent groups could still be bound and hydrolysed by NDM-1. The point of variation between the penicillin-derived β-lactams is indicated by arrow 1 and the main point of variation in the carbepenem antibiotics is shown by arrow 2, both of which point into the large substrate-binding groove.
Although our structure contains a nonphysiological metal ion, it is likely to represent the structure of the catalytically active protein since other MBLs have been shown to retain β-lactam hydrolysis activity when substituted with Co or Cd (Badarau et al., 2007 ▶). The active site is also similar to that observed in structure 3s0z (r.m.s.d. of 1.1 Å calculated for the Cα positions of the active-site residues), which contains two zinc ions separated by a similar distance (3.77 Å) as the zinc and cadmium ions in our structure (3.64 Å). The metal-ion separation is greater (4.58 Å) in the structure of product-bound NDM-1 (PDB entry 3q6x), with a slight outward flexing of histidines 120 and 122. A similar range of metal-separation distances has been observed for the other class B1 MBLs BclI and CcrA (Badarau et al., 2007 ▶) and was attributed to an inherent flexibility in the active site that is required to accept a wide variety of substrates. However, it could also represent a flexing of the active site required for catalytic turnover, since EXAFs measurements performed on β-lactamase L3 (a subclass B3 MBL from Stenotrophomonas maltophilia) have also shown an increase in zinc-ion separation from 3.42 to 3.72 Å during substrate binding and turnover (Breece et al., 2009 ▶).
Ampicillin (idealized coordinates obtained from the HIC-UP database; Kleywegt, 2007 ▶) was modelled into the active site (Fig. 3 ▶) by manual superposition of the lactam ring with the equivalent ring in the hydrolysis product in structure 3q6x. This places the C atom of the β-lactam ring adjacent to the catalytic water molecule, which is ideally positioned for nucleophilic attack on the ampicillin carbonyl group. The O atom of this group is also able to coordinate the zinc ion in site 1, increasing the polarization of the carbonyl bond to further facilitate hydrolysis (Fig. 3 ▶ a). Furthermore, the groove in which the active site is located is large enough, on both sides of the bound metal ions, to accommodate the binding of very large ligands. Since the major differences in the penicillin and carbepenem antibiotics are based on variation of the substituent groups at the position of the phenyl ring (position 1 in Fig. 3 ▶ b) or methyl groups (position 2 in Fig. 3 ▶ b) and there is sufficient space to accommodate these differences in the substrate-binding pocket, it is probable that this also contributes to the broad substrate range of this enzyme.
The structure of NDM-1 provides clues to the basis of the broad substrate specificity of this enzyme and modelling of the substrate strongly suggests the molecular basis for antibiotic hydrolysis, further increasing our understanding of this medically important class of enzymes. Owing to its clinical importance, NDM-1 represents an essential target for the design of novel antibiotics as a consequence of its ability to hydrolyse and deactivate many commonly prescribed antibacterial drugs. This presents a significant challenge for drug design, since its broad substrate range requires new drugs to be based around a novel skeleton that is immune to hydrolysis.
Supplementary Material
PDB reference: New Delhi metallo-β-lactamase 1, 3zr9
Acknowledgments
We would like to thank the beamline staff at stations I02 and I04-1 at Diamond Light Source, L. Bird for help with protein expression and B. Dhaliwal for advice on crystallization. This research was supported by the Medical Research Council.
References
- Badarau, A., Damblon, C. & Page, M. I. (2007). Biochem. J. 401, 197–203. [DOI] [PMC free article] [PubMed]
- Bebrone, C. (2007). Biochem. Pharmacol. 74, 1686–1701. [DOI] [PubMed]
- Breece, R. M., Hu, Z., Bennett, B., Crowder, M. W. & Tierney, D. L. (2009). J. Am. Chem. Soc. 131, 11642–11643. [DOI] [PMC free article] [PubMed]
- Concha, N. O., Rasmussen, B. A., Bush, K. & Herzberg, O. (1996). Structure, 4, 823–836. [DOI] [PubMed]
- Davies, R. B. & Abraham, E. P. (1974). Biochem. J. 143, 129–135. [DOI] [PMC free article] [PubMed]
- Dokmanić, I., Šikić, M. & Tomić, S. (2008). Acta Cryst. D64, 257–263. [DOI] [PubMed]
- Drawz, S. M. & Bonomo, R. A. (2010). Clin. Microbiol. Rev. 23, 160–201. [DOI] [PMC free article] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Garau, G., García-Sáez, I., Brebone, C., Anne, C., Mercuri, P., Galleni, M., Frère, J.-M. & Dideberg, O. (2004). Antimicrob. Agents Chemother. 48, 2347–2349. [DOI] [PMC free article] [PubMed]
- Garcia-Saez, I., Docquier, J. D., Rossolini, G. M. & Dideberg, O. (2008). J. Mol. Biol. 375, 604–611. [DOI] [PubMed]
- Guo, Y., Wang, J., Niu, G., Shui, W., Sun, Y., Zhou, H., Zhang, Y., Yang, C., Lou, Z. & Rao, Z. (2011). Protein Cell, 2, 384–394. [DOI] [PMC free article] [PubMed]
- Hernandez Valladares, M., Felici, A., Weber, G., Adolph, H. W., Zeppezauer, M., Rossolini, G. M., Amicosante, G., Frère, J.-M. & Galleni, M. (1997). Biochemistry, 36, 11534–11541. [DOI] [PubMed]
- Hussain, M., Carlino, A., Madonna, M. J. & Lampen, J. O. (1985). J. Bacteriol. 164, 223–229. [DOI] [PMC free article] [PubMed]
- Kleywegt, G. J. (2007). Acta Cryst. D63, 94–100. [DOI] [PMC free article] [PubMed]
- Langer, G., Cohen, S. X., Lamzin, V. S. & Perrakis, A. (2008). Nature Protoc. 3, 1171–1179. [DOI] [PMC free article] [PubMed]
- Lassaux, P., Traoré, D. A., Loisel, E., Favier, A., Docquier, J.-D., Sohier, J. S., Laurent, C., Bebrone, C., Frère, J.-M., Ferrer, J.-L. & Galleni, M. (2011). Antimicrob. Agents Chemother. 55, 1248–1255. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Osano, E., Arakawa, Y., Wacharotayankun, R., Ohta, M., Horii, T., Ito, H., Yoshimura, F. & Kato, N. (1994). Antimicrob. Agents Chemother. 38, 71–78. [DOI] [PMC free article] [PubMed]
- Paul-Soto, R., Zeppezauer, M., Adolph, H. W., Galleni, M., Frère, J.-M., Carfi, A., Dideberg, O., Wouters, J., Hemmingsen, L. & Bauer, R. (1999). Biochemistry, 38, 16500–16506. [DOI] [PubMed]
- Poirel, L., Naas, T., Nicolas, D., Collet, L., Bellais, S., Cavallo, J. D. & Nordmann, P. (2000). Antimicrob. Agents Chemother. 44, 891–897. [DOI] [PMC free article] [PubMed]
- Winter, G. (2010). J. Appl. Cryst. 43, 186–190.
- Yong, D., Toleman, M. A., Giske, C. G., Cho, H. S., Sundman, K., Lee, K. & Walsh, T. R. (2010). Antimicrob. Agents Chemother. 53, 5046–5054. [DOI] [PMC free article] [PubMed]
- Zhang, H. M. & Hao, Q. (2011). FASEB J. 25, 2574–2578. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: New Delhi metallo-β-lactamase 1, 3zr9