Structures of the nucleocapsid protein of human respiratory syncytial virus have been obtained from two crystal forms. The packing in one crystal form suggests that ionic interactions contribute to the stability of the native helical form of HRSVN.
Keywords: HRSVN, human respiratory syncytial virus, nucleocapsid proteins
Abstract
Respiratory syncytial virus (RSV) is a frequent cause of respiratory illness in infants, but there is currently no vaccine nor effective drug treatment against this virus. The RSV RNA genome is encapsidated and protected by a nucleocapsid protein; this RNA–nucleocapsid complex serves as a template for viral replication. Interest in the nucleocapsid protein has increased owing to its recent identification as the target site for novel anti-RSV compounds. The crystal structure of human respiratory syncytial virus nucleocapsid (HRSVN) was determined to 3.6 Å resolution from two crystal forms belonging to space groups P212121 and P1, with one and four decameric rings per asymmetric unit, respectively. In contrast to a previous structure of HRSVN, the addition of phosphoprotein was not required to obtain diffraction-quality crystals. The HRSVN structures reported here, although similar to the recently published structure, present different molecular packing which may have some biological implications. The positions of the monomers are slightly shifted in the decamer, confirming the adaptability of the ring structure. The details of the inter-ring contacts in one crystal form revealed here suggest a basis for helical packing and that the stabilization of native HRSVN is via mainly ionic interactions.
1. Introduction
Human respiratory syncytial virus (HRSV) is the leading cause of respiratory-tract diseases in infants and young children (Collins & Crowe, 2007 ▶). Since HRSV is highly contagious, most children less than two years old have been infected at least once and this is one of the most frequent reasons for hospitalization within this age group (Boyce et al., 2000 ▶; Glezen et al., 1986 ▶). Additionally, HRSV is thought to be associated with recurrent wheezing and asthma in later life (Openshaw et al., 2004 ▶; Stein et al., 1999 ▶). HRSV is also an important pathogen for elderly adults (Falsey, 1998 ▶) and immunocompromised transplant patients; indeed, these populations are very susceptible to bronchiolitis and pneumonia.
Unfortunately, to date there are no available vaccines or approved small-molecule drugs for therapeutic treatment, although humanized monoclonal antibodies are available (Welliver, 2010 ▶). Ribavirin has been prescribed in the past against RSV, but its level of efficacy has been controversial (Guerguerian et al., 1999 ▶; Long et al., 1997 ▶). Thus, there is a real need to develop antiviral drugs as well as vaccines against this pathogen.
HRSV is an enveloped virus with a negative-sense single-stranded RNA genome which has been classified within the Pneumovirus genus of the Paramyxoviridae family. The RNA genome contains ten genes and codes for 11 proteins, amongst them the nucleoprotein (N), phosphoprotein (P), matrix protein (M) and large protein (L).
The HRSV genome is contained by the human respiratory syncytial virus N protein (HRSVN) within a helical nucleocapsid (Bhella et al., 2002 ▶). HRSVN has also been shown to form decameric and undecameric rings (Tawar et al., 2009 ▶). The nucleocapsid acts to protect the viral genetic information and also serves as a template for viral replication.
Studies of antiviral compounds such as RSV604 (Chapman et al., 2007 ▶) revealed that HRSVN could be very promising as an anti-HRSV drug target. Indeed, targeting the genome protection or replication template might be possible novel strategies. Therefore, structural data are likely to be important in helping to improve the existing antiviral lead compounds and in designing new ones.
The structure of HRSVN has recently been reported at 3.3 Å resolution (Tawar et al., 2009 ▶) from monoclinic crystals belonging to space group C2 with two decameric rings in the asymmetric unit. We have previously described two further crystal forms of HRSVN (El Omari et al., 2008 ▶) and in this work we detail their structure determination. We observed distinct crystal packing for the orthorhombic form (space group P212121) and a triclinic form (space group P1), which contain one and four decameric rings per asymmetric unit, respectively. The packing of stacked rings in the orthorhombic form gives some indications of the molecular interactions in the native HRSVN helical structure.
2. Materials and methods
Cloning, expression, purification and crystallization of HRSVN were carried out as described previously (El Omari et al., 2008 ▶). Briefly, the HRSVN gene was cloned in the pET3d expression vector (Novagen). The latter plasmid codes for N-terminally His6-tagged HRSVN. HRSVN was expressed in Rosetta(DE3)pLysS cells after induction with 1 mM IPTG at 303 K overnight. Protein purification was carried out on nickel-affinity (HiTrap chelating column, GE Healthcare) and gel-filtration columns (S200, GE Healthcare). 10 mg pure HRSVN was routinely obtained from 1 l bacterial culture.
Initial crystallization trials were performed by the sitting-drop vapour-diffusion method at 292 K with HRSVN concentrated to 18 mg ml−1. Crystals belonging to space group P212121 grew in Hampton Research Natrix condition No. 36 [10%(w/v) PEG 400, 0.1 M KCl, 0.05 M HEPES pH 7.0, 0.01 M CaCl2] and crystals belonging to space group P1 grew in Natrix condition No. 43 [10%(v/v) MPD, 0.05 M ammonium acetate, 0.05 M Tris pH 7.5, 0.01 M MgCl2]. Crystal growth and morphology were very variable. HRSVN crystals were prepared for cryoprotection by transferring them to reservoir solution containing 20%(v/v) glycerol.
HRSVN crystals generally diffracted poorly, but after extensive crystal testing 3.6 Å resolution data sets were collected on BM14 and ID29 at the ESRF (Grenoble, France) as described previously (El Omari et al., 2008 ▶). The data were processed with HKL-2000 (Otwinowski & Minor, 1996 ▶; Table 1 ▶). Unfortunately, for the orthorhombic form a completeness of only 81% was achieved owing to anisotropic diffraction and spot overlaps. Matthews coefficients predicted ten or 11 monomers in the asymmetric unit, which would correspond to one decameric ring or one undecameric ring, with solvent contents of 57 and 52%, respectively. Tenfold symmetry rather than 11-fold symmetry was confirmed by calculating the self-rotation function in CNS (Brünger et al., 1998 ▶) and by inspecting the self-Patterson peaks in GROPAT (R. Esnouf, unpublished program). The second crystal form was originally satisfactorily indexed and integrated in the monoclinic space group P21. However, the work reported here shows this to be a pseudo-space group and it in fact belongs to the lower symmetry triclinic space group P1 with four HRSVN rings per asymmetric unit.
Table 1. Data-collection and refinement statistics.
Values in parentheses are for the last shell.
| Data collection | ||
| Beamline | ESRF BM14 | ESRF ID29 |
| Wavelength (Å) | 0.9785 [peak] | 0.9797 |
| Resolution range (Å) | 50.0–3.6 (3.73–3.60) | 55.0–3.6 (3.73–3.60) |
| Space group | P212121 | P1 |
| Unit-cell parameters (Å, °) | a = 133.6, b = 149.9, c = 255.1 | a = 164.3, b = 175.7, c = 241.9, α = 90.1, β = 89.9, γ = 89.9 |
| Monomers in the asymmetric unit | 10 | 40 |
| No. of observed reflections | 592190 | 620513 |
| No. of unique reflections | 53027 (4253) | 330486 (33168) |
| Multiplicity | 11.2 (10.9) | 1.9 (1.8) |
| Rmerge (%) | 14.5 (85.6) | 11.5 (66.9) |
| Completeness (%) | 81.7 (66.2) | 97.3 (97.7) |
| 〈I/σ(I)〉 | 14.1 (2.5) | 6.7 (1.2) |
| Refinement | ||
| Resolution (Å) | 30–3.6 | 50–3.6 |
| Rwork/Rfree (%) | 19.6/22.4 | 22.9/26.5 |
| R.m.s.d. bonds (Å) | 0.007 | 0.008 |
| R.m.s.d. angles (°) | 1.10 | 1.17 |
| Mean B factor (Å2) | 54.7 | 150.5 |
Although crystals of selenomethionine-labelled HRSVN were successfully obtained, MAD data collection did not yield sufficient anomalous signal to allow structure determination (El Omari et al., 2008 ▶). Thus, the structure of the P212121 crystal form was determined by molecular-replacement methods with the program MOLREP (Vagin & Teplyakov, 2011 ▶) using the atomic coordinates of HRSVN (PDB entry 2wj8; Tawar et al., 2009 ▶) as a search template. The search model consisted of the decameric ring stripped of its RNA. REFMAC5 (Murshudov et al., 2011 ▶) was used to carry out initial rigid-body optimization and restrained refinement. During the refinement process, NCS restraints were applied to each monomer present in the asymmetric unit. Model rebuilding was performed using Coot (Emsley & Cowtan, 2004 ▶). The structure of the P1 crystal form was solved and refined in the same manner using the coordinates of the HRSVN orthorhombic form as the search model.
The final R work and R free were 19.6% and 22.4%, respectively, for the P212121 crystal form and 22.9% and 26.5%, respectively, for the P1 crystal form. The geometry of the models was checked with MolProbity (Chen et al., 2010 ▶). Refinement statistics are summarized in Table 1 ▶. The final coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 2yhm for the orthorhombic form and 2yhp and 2yhq for the triclinic form. Figures were prepared with PyMOL (DeLano, 2002 ▶).
3. Results and discussion
In this work, we report the structure determination of two further crystal forms of HRSVN. These forms diffracted to a resolution (3.6 Å) which, in combination with the significant level of noncrystallographic symmetry, was sufficient to allow refinement. However, the presence of a partner protein, HRSVP, used in the first structure determination of HRSVN was not required in the current study. The phosphoprotein was reported to be necessary to reproducibly obtain diffraction-quality crystals in spite of a lack of ordered density for this protein in the refined structure (Tawar et al., 2009 ▶). Although we experienced high variability in crystal quality, it was clear that for the orthorhombic and triclinic crystal forms the presence of the phosphoprotein in the purification or crystallization steps was not essential to yield diffraction-quality crystals.
We solved the structure of HRSVN in two crystal forms (P212121 and P1) by molecular replacement to 3.6 Å resolution using the previously reported structure (Tawar et al., 2009 ▶). The primitive triclinic crystals were originally indexed as P21 (El Omari et al., 2008 ▶), but structure refinement with reasonable statistics could not be carried out in the latter space group. This lattice is unlikely to be twinned, so the data were re-indexed in the triclinic space group P1, resulting in rapid lowering of R work and R free. The asymmetric unit of the P212121 form contains one annular ring composed of ten HRSVN monomers, in agreement with the self-rotation function, whereas the P1 crystal form asymmetric unit contains four rings with a total of 40 monomers. Structural analyses were carried out on the primitive orthorhombic form because the quality of the maps was superior despite the nominal resolutions of both data sets being identical.
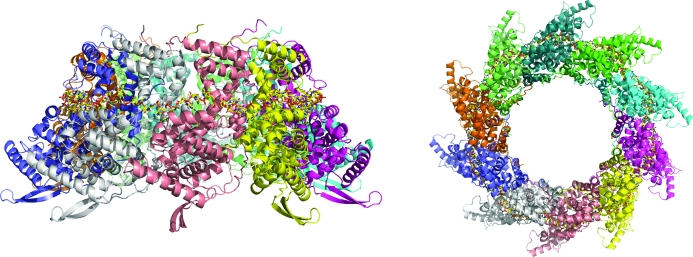
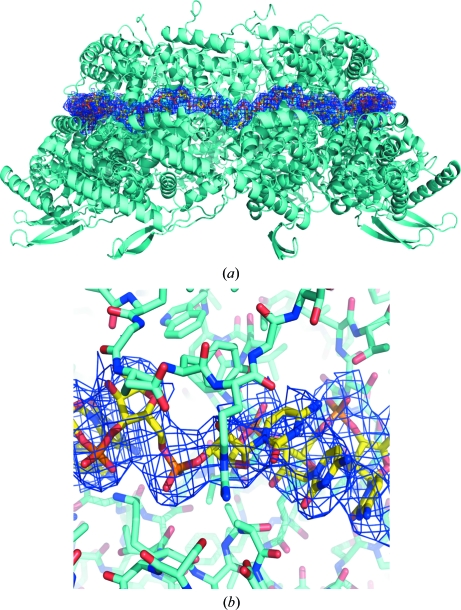
The overall HRSVN structure (Fig. 1 ▶) is very similar to that previously reported (PDB entry 2wj8). Clear electron density could be seen for HRSVN from the first residue to residues 360–369 depending on the particular monomer. The RNA, which forms a continuous belt of electron density around the ring, is also very well defined. It has previously been suggested that this continuous electron density for RNA is a consequence of the averaging out of the polymer ends (Tawar et al., 2009 ▶). In the HRSVN structure the RNA is located on the outside of the ring, with seven ribonucleotides contacting each protein monomer, whereas in the cases of the vesicular stomatitis virus (VSV; Green et al., 2006 ▶) and rabies nucleocapsid (Albertini et al., 2006 ▶) proteins the RNA is located inside the ring and nine ribonucleotides are bound per monomer. Poly-rC was modelled into the RNA electron density because of the random positions of the RNA bases bound to the nucleoprotein (Tawar et al., 2009 ▶; Fig. 2 ▶). As described previously, the RNA is arranged in alternating rows of four and three stacked bases that are exposed and buried, respectively, within a protein groove (Tawar et al., 2009 ▶). HRSVN monomers are composed of two domains, N-terminal and C-terminal (NTD and CTD); the RNA binds in a groove at the interface of these two domains. The NTD and CTD have extensions named the ‘N-arm’ and ‘C-arm’, respectively (Tawar et al., 2009 ▶). The ‘arms’ make contact with the adjacent monomer and seem to be important for the integrity of the ring. The area between adjacent HRSVN monomers in the ring varies from approximately 2400 to 2600 Å2 and is composed of salt bridges, hydrogen bonds and hydrophobic interactions.
Figure 1.
Ribbon diagrams showing orthogonal views of the overall structure of the HRSVN decameric ring (in the orthorhombic crystal form) with bound RNA.
Figure 2.
(a) Ribbon diagram of decameric HRSVN (orthorhombic crystal form) coloured in cyan. Electron density for the RNA is contoured in dark blue at 1σ, with poly-C shown in standard atom colours. (b) Close-up view of the RNA–HRSVN interaction.
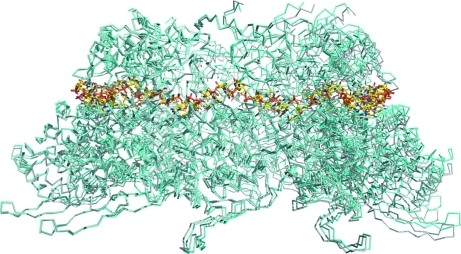
Superimposition of the HRSVN rings revealed that each monomer does not overlap exactly (Fig. 3 ▶). A maximum shift of 4 Å could be seen between the monomers compared with those used for superimposition. This movement explains why rigid-body refinement was crucial to achieve good statistics after initial molecular-replacement solution using a decameric ring as a search template. This result also indicates that the decameric ring is not totally rigid and is able to undergo small rigid-body movements whilst maintaining its integrity. This adaptability is not surprising as HRSVN can form helical structures; in addition, the N–N interaction has been reported to be stabilized by the RNA belt and by extension of the N-arm of one subunit into the adjacent subunit (Tawar et al., 2009 ▶).
Figure 3.
Superimposition of HRSVN from the orthorhombic crystal form (shown in cyan) with the previously solved HRSVN structure (grey; PDB entry 2wj8). The RNA is shown in standard atom colours.
The crystal packings within the two forms reported here are rather different from that of the previously published structure. In the latter crystal form, which belongs to space group C2, two rings are present in the asymmetric unit with their respective tenfold axes almost perpendicular to one another (Tawar et al., 2009 ▶). The P1 crystal form is composed of decameric rings positioned along the b axis all pointing in the same direction. The crystal lattice consists of parallel layers of HRSVN stacked in a brick-wall pattern. Within a layer, the HRSVN rings make contact through their N-terminal domains and the layers are stacked through contacts between the HRSVN N-terminal domains from one layer and the C-terminal domains of a lower layer.
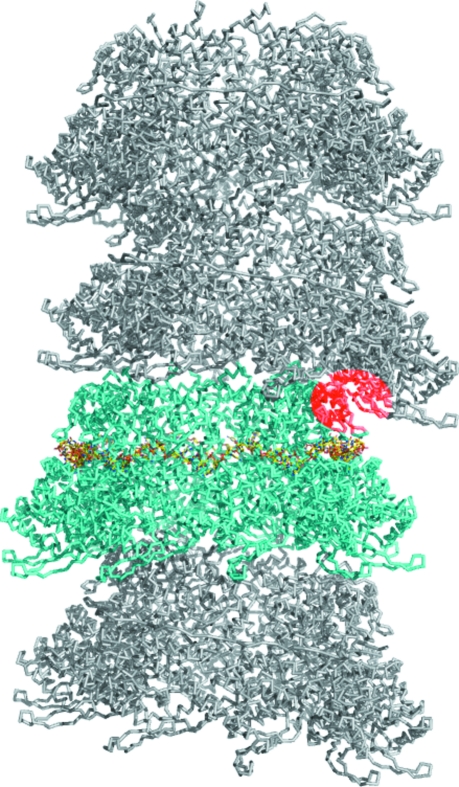
In contrast, the P212121 structure reported in this current work has only one ring in the asymmetric unit. The lattice is composed of rings stacked on top of each other, intriguingly resembling the HRSVN helical structure (Fig. 4 ▶). The tenfold axes of the HRSVN rings are tilted by approximately 13° between consecutive rings and are parallel to each other in every other ring; hence, the N–N area surface contacts between rings are very different, although the C4 helix is always at the interface. Each pseudo-helical structure is surrounded by six others, with only two being orientated in the same direction. The packing is tighter between antiparallel pseudo-helices than between parallel pseudo-helices; indeed, in the latter crystal contacts are only made through the N-terminal domain. A model for RSVN helix assembly has been proposed (Tawar et al., 2009 ▶).
Figure 4.
Ribbon diagram of HRSVN from the orthorhombic crystal form (cyan, with bound RNA shown in standard atom colours) together with three symmetry-related molecules (coloured grey). The interface between chains D and I′ is highlighted in red.
It has been postulated that the HRSVN nucleocapsid is very flexible, which would allow helical forms varying from a pitch of 69 Å to more than 100 Å whilst maintaining N–N interactions and RNA connectivity (Tawar et al., 2009 ▶). This flexibility might also be functionally important in allowing the polymerase complex to read the RNA bases without disassembling the helix.
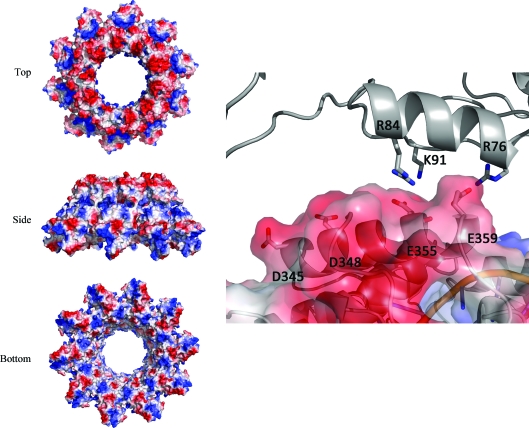
In the structure presented here (P212121 form), the monomers with the closest surface contact between rings [chain D and chain I from the symmetry-related molecule (I′)] could be representative of the interactions present in certain HRSVN helical structures, although the helix pitch (55 Å) is smaller than the minimum pitch (69 Å) to avoid clashes with the subsequent helical turn (Tawar et al., 2009 ▶). Residues involved in the interaction of chains D and I′ were determined using PISA (Krissinel & Henrick, 2007 ▶) as Glu355 and Glu359 of chain D and Arg76 and Arg84 of chain I′ (Figs. 4 ▶ and 5 ▶). The latter chains share a rather small interface area of 190 Å2. Surface-charge distribution analysis highlights a strong negatively charged surface at the top of the ring, whereas the base of the ring contains areas of both positive and negative charge. Overall, very few hydrophobic residues are present at the ring interface (Fig. 5 ▶); therefore, it would be very likely that the stability of the HRSVN helical form is largely guided by electrostatic interactions. The main residues responsible for the positively charged patches at the ring interface are Arg76, Arg84 and Lys91, whereas the negative surface charge results from the acidic side chains of Asp345, Asp348, Glu355 and Glu359 (Fig. 5 ▶). The residues located on the negative and positively charged surfaces but not selected by PISA (Asp345, Asp348 and Lys91) might nevertheless play a role in the helical stability if conformational changes take place, thereby allowing helical deformation whilst maintaining the interactions. The interactions described above almost certainly only reflect one of the possible conformations of the HRSVN helix and their details might change as a function of helix pitch and/or when the polymerase complex is perturbing the helix during threading through the RNA. HRSVN helical plasticity is likely to be essential for protecting the RNA and forming complexes with the polymerase, this being achieved by the malleability of consecutive N–N interactions (Tawar et al., 2009 ▶) and the plasticity of stacked HRSVN. Inhibiting the formation of HRSVN rings in the first case or the assembly into helices in the second case might represent a potential target for structure-based drug design.
Figure 5.
Surface-charge representations of HRSVN (orthorhombic crystal form) are shown in three orientations on the left. Details of the interaction of chain D (surface representation) with chain I′ (grey) are shown on the right.
The comparison of the two HRSVN structures presented here with the recently published structure highlights the flexibility of the decameric ring formed by the viral nucleocapsid, an analysis that could provide insights into the stability of the HRSVN helical structure.
Supplementary Material
PDB reference: HRSVN, 2yhm
PDB reference: 2yhp
PDB reference: 2yhq
Acknowledgments
We thank Dr Felix Rey and his colleagues at the Pasteur Institute, Paris, France for releasing the coordinates of HRSVN prior to publication. We would also like to thank the staff of beamlines BM14 and ID29, ESRF, Grenoble, France for help with data collection.
References
- Albertini, A. A., Wernimont, A. K., Muziol, T., Ravelli, R. B., Clapier, C. R., Schoehn, G., Weissenhorn, W. & Ruigrok, R. W. (2006). Science, 313, 360–363. [DOI] [PubMed]
- Bhella, D., Ralph, A., Murphy, L. B. & Yeo, R. P. (2002). J. Gen. Virol. 83, 1831–1839. [DOI] [PubMed]
- Boyce, T. G., Mellen, B. G., Mitchel, E. F., Wright, P. F. & Griffin, M. R. (2000). J. Pediatr. 137, 865–870. [DOI] [PubMed]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- Chapman, J. et al. (2007). Antimicrob. Agents Chemother. 51, 3346–3353. [DOI] [PMC free article] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Collins, P. L. & Crowe, J. E. (2007). Fields Virology, edited by D. M. Knipe & P. M. Howley, pp. 1601–1646. Philadelphia: Lippincott, Williams & Wilkins.
- DeLano, W. L. (2002). PyMOL http://www.pymol.org.
- El Omari, K., Scott, K., Dhaliwal, B., Ren, J., Abrescia, N. G. A., Budworth, J., Lockyer, M., Powell, K. L., Hawkins, A. R. & Stammers, D. K. (2008). Acta Cryst. F64, 1019–1023. [DOI] [PMC free article] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Falsey, A. R. (1998). Vaccine, 16, 1775–1778. [DOI] [PubMed]
- Glezen, W. P., Taber, L. H., Frank, A. L. & Kasel, J. A. (1986). Am. J. Dis. Child. 140, 543–546. [DOI] [PubMed]
- Green, T. J., Zhang, X., Wertz, G. W. & Luo, M. (2006). Science, 313, 357–360. [DOI] [PubMed]
- Guerguerian, A. M., Gauthier, M., Lebel, M. H., Farrell, C. A. & Lacroix, J. (1999). Am. J. Respir. Crit. Care Med. 160, 829–834. [DOI] [PubMed]
- Krissinel, E. & Henrick, K. (2007). J. Mol. Biol. 372, 774–797. [DOI] [PubMed]
- Long, C. E., Voter, K. Z., Barker, W. H. & Hall, C. B. (1997). Pediatr. Infect. Dis. J. 16, 1023–1028. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Openshaw, P. J., Yamaguchi, Y. & Tregoning, J. S. (2004). J. Allergy Clin. Immunol. 114, 1275–1277. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1996). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Stein, R. T., Sherrill, D., Morgan, W. J., Holberg, C. J., Halonen, M., Taussig, L. M., Wright, A. L. & Martinez, F. D. (1999). Lancet, 354, 541–545. [DOI] [PubMed]
- Tawar, R. G., Duquerroy, S., Vonrhein, C., Varela, P. F., Damier-Piolle, L., Castagné, N., MacLellan, K., Bedouelle, H., Bricogne, G., Bhella, D., Eléouët, J. F. & Rey, F. A. (2009). Science, 326, 1279–1283. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Welliver, R. C. (2010). Curr. Opin. Pharmacol. 10, 289–293. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: HRSVN, 2yhm
PDB reference: 2yhp
PDB reference: 2yhq