The crystal structure of recombinant UbiX has been determined to 1.5 Å resolution.
Keywords: Pseudomonas aeruginosa, PA4019, aromatic acid decarboxylases
Abstract
The ubiX gene (PA4019) of Pseudomonas aeruginosa has been annotated as encoding a putative 3-octaprenyl-4-hydroxybenzoate decarboxylase from the ubiquinone-biosynthesis pathway. Based on a transposon mutagenesis screen, this gene was also implicated as being essential for the survival of this organism. The crystal structure of recombinant UbiX determined to 1.5 Å resolution showed that the protein belongs to the superfamily of homo-oligomeric flavine-containing cysteine decarboxylases. The enzyme assembles into a dodecamer with 23 point symmetry. The subunit displays a typical Rossmann fold and contains one FMN molecule bound at the interface between two subunits.
1. Introduction
Pseudomonas aeruginosa is an opportunistic pathogen that causes persistent infections in humans. The limited reservoir of antibacterial drugs against Gram-negative bacteria, inherent difficulties in the clinical treatment of P. aeruginosa infections and the onset of antibiotic resistance emphasize the need to identify novel drug targets against these pathogens. Randomized transposon mutagenesis screens (Jacobs et al., 2003 ▶) indicated several genes from the ubiquinone-biosynthetic pathway in P. aeruginosa as being essential, suggesting the corresponding gene products as potential targets for antibacterial drugs. One of these genes is denoted ubiX and is proposed to encode an enzyme that is involved in the decarboxylation of the metabolite 3-octaprenyl-4-hydroxybenzoate (Howlett & Bar-Tana, 1980 ▶; Gulmezian et al., 2007 ▶). UbiX is related in sequence to the phenylacrylic acid decarboxylases that form the PaD family of FMN-dependent enzymes. Decarboxylation of phenylacrylic acids by these enzymes during fermentation is an important process in the flavouring of wine and beer (Mukai et al., 2010 ▶). The PaD family in turn belongs to the HFCD (homo-oligomeric flavine-containing cysteine decarboxylase) superfamily (Steinbacher et al., 2003 ▶). The bona fide HFCD decarboxylases catalyze the oxidative decarboxylation of C-terminal cysteine residues in, for example, coenzyme A (Aberhart et al., 1985 ▶) and lantibiotic (Kupke et al., 2000 ▶) biosynthesis. The structures of members of the HFCD branch of this superfamily are known for the enzymes AtHAL3 from coenzyme A biosynthesis in Arabidopsis thaliana (Steinbacher et al., 2003 ▶; Kupke et al., 2001 ▶), EpiD from epidermin biosynthesis in Staphylococcus epidermidis (Blaesse et al., 2000 ▶) and MrsD from mersacidin biosynthesis in Bacillus sp. HIL-Y85/54278 (Blaesse et al., 2003 ▶). However, these proteins do not show any detectable overall sequence similarity to the branch of the superfamily containing the phenylacrylic acid decarboxylases and UbiX-like enzymes.
Based on genetic studies, the gene ubiX from Escherichia coli and its paralogue pad1p from yeast have been invoked in the decarboxylation of the metabolic intermediate 3-octaprenyl-4-hydroxybenzoate in ubiquinone biosynthesis (Gulmezian et al., 2007 ▶; Howlett & Bar-Tana, 1980 ▶). Also based on genetic evidence, another gene, ubiD, has been annotated as encoding a 3-octaprenyl-4-hydroxybenzoate decarboxylase (Gulmezian et al., 2007 ▶). This enzyme is not related in sequence to UbiX and it has been proposed that the two proteins represent redundant decarboxylases in this pathway or that they act together in the decarboxylation step (Gulmezian et al., 2007 ▶). It is noteworthy that there are no in vitro biochemical data demonstrating that decarboxylation of benzoic acid derivates is catalyzed by either of these two enzymes. In addition to ubiX, several strains of pathogenic E. coli contain another gene that encodes a related enzyme denoted Pad1. Despite the lack of overall sequence identity, the structure of the enzyme subunit of E. coli Pad1 is similar to those of other members of the HFCD superfamily, but the packing of the subunits in the homododecamers is different (Rangarajan et al., 2004 ▶). However, no decarboxylase activity was confirmed for E. coli Pad1 using a variety of phenylacrylic acids as potential substrates.
As part of a larger effort to identify novel targets in P. aeruginosa (http://www.aeropath.eu/), we have cloned the gene annotated as coding for UbiX in this organism (PA4019), produced the enzyme in soluble form and determined the crystal structure of the FMN-containing holoenzyme to high resolution.
2. Materials and methods
2.1. Cloning, expression and purification of PA4019
The PA4019 gene was amplified from P. aeruginosa O1 genomic DNA by PCR and cloned into the pET-based vector pNIC-Bsa4 (GenBank No. EF198106) using ligation-independent cloning (LIC; Gräslund et al., 2008 ▶). Briefly, the PCR product was incubated with T4 polymerase (NEB) in the presence of dCTP to create single-stranded overhangs and the linearized vector was treated with T4 polymerase in the presence of dGTP, creating complementary single-stranded overhangs. The two fragments were annealed and transformed into E. coli DH5α competent cells. The correct DNA sequence of the expression construct was verified by sequencing.
The plasmid pNIC-Bsa4-PA4019 was transformed into E. coli BL21 (DE3) cells for protein production. An overnight culture was diluted 100-fold into LB broth and grown at 310 K until the OD600 reached 0.6. At this point IPTG was added to a final concentration of 0.2 mM, the temperature was lowered to 291 K and the culture was continued for approximately 12 h.
The E. coli cells were harvested by centrifugation at 3000g for 10 min. The cells were resuspended in lysis buffer (50 mM Tris–HCl, 500 mM NaCl, 10 mM imidazole pH 8.0), lysozyme was added to 0.5 mg ml−1 and the cells were lysed by sonication. The cell debris was removed by centrifugation at 40 000g for 30 min at 277 K and the supernatant was passed through 1 ml Ni–NTA agarose (Qiagen) packed in a gravity-flow column. The affinity resin was washed with 40 ml lysis buffer followed by 25 ml lysis buffer containing 50 mM imidazole. PA4019 was eluted with 10 ml elution buffer (50 mM Tris–HCl, 500 mM NaCl, 250 mM imidazole pH 8.0). The His tag was cleaved overnight with 1 mg TEV protease in dialysis cassettes, at the same time reducing the high imidazole concentration. The tagged and processed proteins were separated on Ni–NTA resin. The flowthrough was collected and the protein was further purified on a HiPrep Sephacryl S-200 (GE Healthcare) size-exclusion chromatography column using a buffer consisting of 20 mM Tris pH 8, 200 mM NaCl and run with the same buffer at 0.5 ml min−1. The purified protein was concentrated to 7 ml min−1 before storage. In preparations used for crystallization experiments, FMN was added to a final concentration of 0.5 mM before the gel-chromatography step to ensure saturation of the cofactor-binding sites.
2.2. Crystallization and data collection
Crystals of PA4019 were obtained by mixing 1 µl protein solution (7 mg ml−1 protein in 10 mM Tris–HCl, 150 mM NaCl, 1 mM DTT pH 8.0) with 1 µl of a reservoir solution consisting of 1.2 M ammonium sulfate, 200 mM NaCl and 100 mM cacodylate pH 6.3. The drops were equilibrated against 1 ml of the same buffer using the hanging-drop vapour-diffusion method. Protein crystals appeared overnight at 293 K.
X-ray diffraction data were collected to 1.5 Å resolution on beamline ID14-2 at the ESRF (Grenoble, France) at 100 K. Before data collection, crystals of the holoenzyme were quickly soaked in cryoprotection solution consisting of 0.8 M ammonium sulfate, 200 mM NaCl, 100 mM cacodylate pH 6.3 and 20% glycerol. Soaking of the crystals for longer time periods in the cryosolution led to the diffusion of bound FMN, as indicated by a loss of the yellow colour. The data were integrated using iMOSFLM and scaled with SCALA within the CCP4 program package (Winn et al., 2011 ▶). The crystals belonged to space group F23, with unit-cell parameter a = 141.8 Å. Evaluation of the Matthews coefficient indicated the presence of one subunit of PA4019 in the asymmetric unit. A summary of the statistics of data collection is presented in Table 1 ▶.
Table 1. Data-collection and refinement statistics for PA4019.
Values in parentheses are for the highest resolution shell.
| Data collection | |
| Wavelength () | 0.933 |
| Space group | F23 |
| Unit-cell parameter () | a = 141.8 |
| Resolution () | 81.851.5 (1.581.50) |
| No. of observed reflections | 166122 (24020) |
| No. of unique reflections | 35844 (5474) |
| No. of reflections in the R free data set | 1885 |
| Completeness (%) | 99.9 (100) |
| R merge † (%) | 6.9 (49.3) |
| Mean I/(I) | 14.4 (2.8) |
| Multiplicity | 4.4 (4.4) |
| B factor from Wilson plot (2) | 21.6 |
| Refinement | |
| R ‡/R free § (%) | 17.1/19.4 |
| No. of non-H atoms | |
| Protein | 1770 |
| FMN | 31 |
| Water | 139 |
| Sulfate | 10 |
| B factors (2) | |
| Protein, average | 14.6 |
| FMN | 14.7 |
| Water | 22.5 |
| Sulfate | 20.7 |
| R.m.s.d. bond lengths () | 0.019 |
| R.m.s.d. bond angles () | 1.92 |
| Ramachandran plot (%) | |
| Favoured | 100 |
| Allowed | 0 |
| Disallowed | 0 |
R
merge = 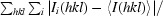
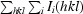 , where Ii(hkl) is the intensity measured for a given reflection and I(hkl) is the average intensity for multiple measurements of this reflection.
, where Ii(hkl) is the intensity measured for a given reflection and I(hkl) is the average intensity for multiple measurements of this reflection.
R = 
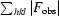 , where F
obs and F
calc are the observed and calculated structure-factor amplitudes, respectively, for 95% of the reflection data used in the refinement.
, where F
obs and F
calc are the observed and calculated structure-factor amplitudes, respectively, for 95% of the reflection data used in the refinement.
R
free = 
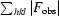 for 5% of the reflection data that were excluded during refinement.
for 5% of the reflection data that were excluded during refinement.
2.3. Structure determination
The structure of PA4019 was solved by molecular replacement using the program Phaser (McCoy et al., 2007 ▶) with the coordinates of E. coli Pad1 (PDB entry 1sbz; Rangarajan et al., 2004 ▶) as the search model. Refinement was carried out by cycles of manual rebuilding using Coot (Emsley et al., 2010 ▶) and refinement with REFMAC5 (Murshudov et al., 2011 ▶). The refined model of the holoenzyme consisted of a polypeptide chain comprising residues 1–205. Electron density for the remaining four C-terminal residues could not be observed, most likely owing to disorder of this part of the polypeptide chain. The model further contained one FMN molecule, two sulfate ions and 139 water molecules. The final model was evaluated with the program MolProbity (Chen et al., 2010 ▶) and model and refinement statistics are summarized in Table 1 ▶. The crystallographic data for PA4019 were deposited in the PDB under accession code 3zqu.
3. Results and discussion
PA4019 was purified to homogeneity as assessed by SDS–PAGE in a two-step procedure consisting of affinity chromatography followed by gel filtration. The protein eluted at a volume corresponding to a molecular mass of approximately 270 kDa, indicating an oligomeric structure, most likely made up of dodecamers considering the molecular weight of the PA4019 monomer (25 kDa). After cleavage of the His-tagged enzyme by TEV protease, the construct gives an enzyme preparation consisting of wild-type enzyme comprising residues 1–209 with one additional serine residue at the N-terminus. The final preparation was yellow in colour and the absorption spectrum showed maxima at 380 and 460 nm indicative of bound flavin.
3.1. Structure of the subunit
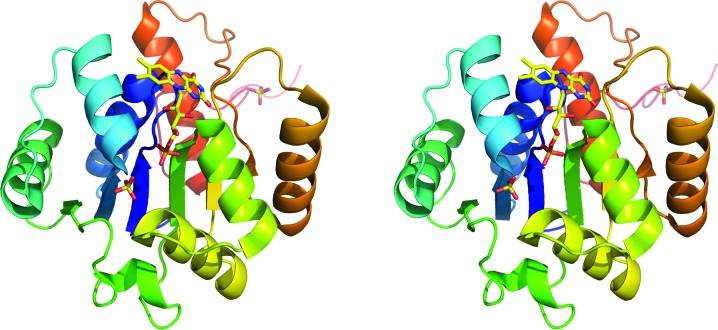
The asymmetric unit of the crystals contained one subunit of PA4019. The subunit is built up of one domain, which displays a typical Rossmann fold (Fig. 1 ▶). The central β-sheet consists of six parallel β-strands with strand order 321456 and the sheet is flanked on each side by three α-helices. An additional α-helix following strand β2 is located at the C-terminal end of the sheet and its N-terminus forms part of the binding site for the phosphate group of the cofactor. The C-terminal residues of the subunit (Pro198–Leu205) extend from the globular protein and interact with an adjacent subunit in the oligomer. In addition to the bound FMN molecule (see below), two sulfate ions are associated with each subunit. One of the sulfate ions was located in the vicinity of the phosphate group of the FMN. It is held in place by a number of hydrogen bonds to the side chains of Asn117 and Gln81, the main-chain N atom of Lys40 and a water molecule. Another sulfate ion was observed in an internal cavity close to the isoalloxazine ring of the FMN. This sulfate ion forms a tight network of hydrogen bonds with the side chains of residues Ser90, Lys129, Arg139, Glu140, Tyr169 and Arg185 and the main-chain N atom of Gly91. Loss of FMN does not result in any large-scale conformational changes and the structures of the apoenzyme subunit and the apo dodecamer are very similar to that of the holoenzyme (data not shown).
Figure 1.
Stereo representation of the overall structure of a subunit of PA4019. The colour changes from blue at the N-terminus to red at the C-terminus. FMN and sulfate ions are shown in stick representation. This figure was produced using PyMOL (DeLano, 2002 ▶).
3.2. Quaternary structure
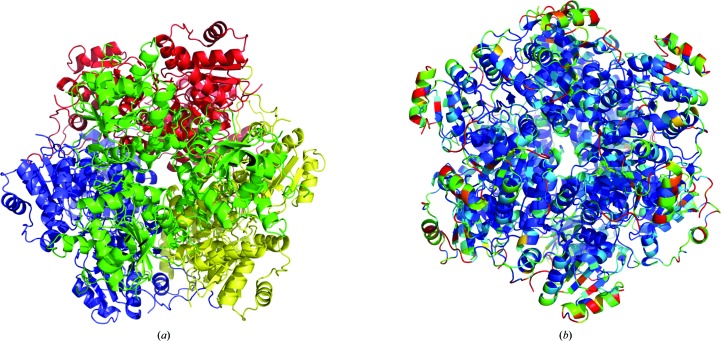
The single polypeptide chain in the asymmetric unit forms tight interactions with adjacent chains in the crystal lattice and a dodecameric molecule is generated by the crystallographic symmetry. This quaternary structure is consistent with the results from gel chromatography, which also indicated a multi-subunit arrangement of PA4019 in solution. Three subunits are related by a threefold axis and form a trimeric building block (Fig. 2 ▶ a). Amino-acid residues from helix 118–131 of subunit A form hydrogen bonds to loop 140–143 of subunit B, while the carbonyl O atom of Thr113 of subunit A interacts via hydrogen bonds with Ser109 and Asn150 from subunit B. These interactions are repeated following the threefold symmetry. The dodecamer is formed through a tetrahedral arrangement of four of these trimers (Fig. 2 ▶ a). Strand β6 and its flanking loop regions form hydrogen bonds to loop 139–145 and residues Ser15 and Thr48 of the second trimer. Analysis of amino-acid sequence conservation among aromatic acid decarboxylases using the ConSurf server (Ashkenazy et al., 2010 ▶) showed that all these interactions are conserved and that the least conserved residues are solvent-exposed (Fig. 2 ▶ b).
Figure 2.
Schematic view of the dodecamer of PA4019. (a) Three subunits assemble into trimeric building blocks (shown in different colours) that pack into the dodecamer. (b) Amino-acid sequence conservation in aromatic acid decarboxylases is shown using the structure of PA4019. The colour code varies from blue (conserved) to red (variable).
3.3. FMN binding
PA4019 contains one FMN molecule per subunit, which is bound at the interface between two subunits. The isoalloxazine ring is inserted into a pocket of the enzyme and is not accessible from the bulk solution. The si-face of the ring is packed tightly with loops at the C-terminal ends of two β-strands from the central β-sheet, in particular residues Gly13–Ser15. The O2 O atom of the isoalloxazine ring interacts with the side chain of Thr105, the O4 O atom forms hydrogen bonds to the main-chain N atom of Ser15 and the side chain of Arg139, and the N3 atom of the ring interacts with the main-chain O atom of Arg139. The ribityl moiety is bound at the interface between two adjacent subunits in the dodecamer and interacts with residues from both subunits. The O2′ hydroxyl forms a hydrogen bond to the carbonyl O atom of Gly13, the O3 hydroxyl is linked through a water-mediated system of hydrogen bonds to the side chain of Thr105, and the O4′ hydroxyl group interacts with the carbonyl group of Cys116 from a neighbouring subunit. In addition, it also forms a water-mediated intra-subunit hydrogen bond to the main-chain O atom of Asn117. The phosphate moiety is anchored to the N-terminus of an α-helix by a hydrogen bond to the side chain of Ser39 and forms additional interactions with the main-chain N atoms of Gly13 and Ser15 and the side chain of Thr105. The flavin ring is bent, with a ‘butterfly angle’ between the planes separated by the N5–N10 axis of 163°, indicating that the flavin is reduced, most likely as a result of radiation exposure during data collection (Røhr et al., 2010 ▶).
On the re-face of the flavin ring a small hydrophilic cavity is formed from residues of three adjacent subunits and this cavity is closed off from the bulk solution, in particular by the side chains of Met85 and Trp200. Access of external ligands to this cavity would require conformational changes of these side chains and possibly also a larger part of the C-terminal residues. The presence of a bound sulfate ion in this cavity suggests that such movements of enzyme residues are possible.
3.4. Comparison to related proteins
A DALI (Holm & Rosenström, 2010 ▶) search of the PDB using the atomic coordinates for PA4019 identified several proteins which are structurally similar to PA4019, all of them members of the HFCD superfamily. These include enzymes from the bona fide branch of the HFCD superfamily: phosphopantothenoylcysteine decarboxylase from A. thaliana (PDB entries 1mvn and 1e20; Steinbacher et al., 2003 ▶; Kupke et al., 2001 ▶) and human (PDB entry 1qzu; Manoj & Ealick, 2003 ▶), EpiD from S. epidermidis (PDB entry 1g63; Blaesse et al., 2000 ▶) and MrsD from Bacillus sp. HIL-Y85/54728 (PDB entry 1p3y; Blaesse et al., 2003 ▶). The r.m.s.d. values between aligned individual subunits of P. aeruginosa PA4019 and these enzymes are typically in the range 1.4–3 Å, although they do not show any significant overall sequence homology.
In addition, two structures from the PaD branch of the superfamily were identified, E. coli Pad1 (PDB entry 1sbz; Rangarajan et al., 2004 ▶), which was used as a search model, and an FMN-free putative aromatic acid decarboxylase from Aquifex aeolicus (PDB entry 2ejb; B. Bagautdinov & N. Kunishima, unpublished work). Not unexpectedly, these two structures gave the highest Z scores (28.1 and 27.7, respectively) and this is reflected in the r.m.s.d. of aligned Cα atoms, which was in the range 1.1–1.5 Å. The two enzymes also showed significant overall sequence identity to PA4019 (40 and 46%, respectively).
3.5. Enzymatic activity
Since the putative natural substrate of UbiX, 3-octaprenyl-4-hydroxybenzoate, was not available, we tested simple analogues such as 4-hydroxybenzoic acid, vanillic acid and 3-carboxymethyl aminomethyl-4-hydroxybenzoic acid as potential substrates. In none of these cases could decarboxylase activity be detected. This is similar to the findings with the UbiX analogue Pad1 from E. coli, which was also unable to decarboxylate a series of phenylacrylic acids (Rangarajan et al., 2004 ▶). However, these observations are not yet conclusive as the enzyme might only be active with the physiological substrate 3-octaprenyl-4-hydroxybenzoate or might require additional cofactors and/or metabolites for enzymatic activity. The reductive decarboxylation of aromatic carboxylic acids such as p-coumaric or ferulic acid to their corresponding 4-ethyl derivatives by phenylacrylic decarboxylases would be consistent with the presence of a flavine as a cofactor. However, it is not obvious why the decarboxylation of benzoate derivates would require the presence of FMN, unless FMN acts as an acid/base catalyst and not as a redox cofactor. Since enzymatic activity for PA4019 from P. aeruginosa or its homologue Pad1 from E. coli has not yet been demonstrated, the function of these enzymes remains enigmatic.
Supplementary Material
PDB reference: PA4019, 3zqu
Acknowledgments
We acknowledge access to synchrotron radiation at the ESRF, Grenoble, France and thank the staff for support at the beamline. This work was supported by the European Commission through the 7th Framework program, grant agreement HEALTH-F3-2008-223461.
References
- Aberhart, D. J., Ghoshal, P. K., Cotting, J. A. & Russell, D. J. (1985). Biochemistry, 24, 7178–7182. [DOI] [PubMed]
- Ashkenazy, H., Erez, E., Martz, E., Pupko, T. & Ben-Tal, N. (2010). Nucleic Acids Res. 38, W529–W533. [DOI] [PMC free article] [PubMed]
- Blaesse, M., Kupke, T., Huber, R. & Steinbacher, S. (2000). EMBO J. 19, 6299–6310. [DOI] [PMC free article] [PubMed]
- Blaesse, M., Kupke, T., Huber, R. & Steinbacher, S. (2003). Acta Cryst. D59, 1414–1421. [DOI] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- DeLano, W. L. (2002). PyMOL http://www.pymol.org.
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Gräslund, S., Sagemark, J., Berglund, H., Dahlgren, L. G., Flores, A., Hammarström, M., Johansson, I., Kotenyova, T., Nilsson, M., Nordlund, P. & Weigelt, J. (2008). Protein Expr. Purif. 58, 210–221. [DOI] [PubMed]
- Gulmezian, M., Hyman, K. R., Marbois, B. N., Clarke, C. F. & Javor, G. T. (2007). Arch. Biochem. Biophys. 467, 144–153. [DOI] [PMC free article] [PubMed]
- Holm, L. & Rosenström, P. (2010). Nucleic Acids Res. 38, W545–W549. [DOI] [PMC free article] [PubMed]
- Howlett, B. J. & Bar-Tana, J. (1980). J. Bacteriol. 143, 644–651. [DOI] [PMC free article] [PubMed]
- Jacobs, M. A., Alwood, A., Thaipisuttikul, I., Spencer, D., Haugen, E., Ernst, S., Will, O., Kaul, R., Raymond, C., Levy, R., Chun-Rong, L., Guenthner, D., Bovee, D., Olson, M. V. & Manoil, C. (2003). Proc. Natl Acad. Sci. USA, 100, 14339–14344. [DOI] [PMC free article] [PubMed]
- Kupke, T., Hernandez-Acosta, P., Steinbacher, S. & Culianez-Macia, F. A. (2001). J. Biol. Chem. 276, 19190–19196. [DOI] [PubMed]
- Kupke, T., Uebele, M., Schmid, D., Jung, G., Blaesse, M. & Steinbacher, S. (2000). J. Biol. Chem. 275, 31838–31846. [DOI] [PubMed]
- Manoj, N. & Ealick, S. E. (2003). Acta Cryst. D59, 1762–1766. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Mukai, N., Masaki, K., Fujii, T., Kawamukai, M. & Iefuji, H. (2010). J. Biosci. Bioeng. 109, 564–569. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Rangarajan, E. S., Li, Y., Iannuzzi, P., Tocilj, A., Hung, L.-W., Matte, A. & Cygler, M. (2004). Protein Sci. 13, 3006–3016. [DOI] [PMC free article] [PubMed]
- Røhr, Å., Hersleth, H.-P. & Andersson, K. K. (2010). Ang. Chem. Int. Ed. 49, 2324–2327. [DOI] [PubMed]
- Steinbacher, S., Hernández-Acosta, P., Bieseler, B., Blaesse, M., Huber, R., Culiáñez-Macià, F. A. & Kupke, T. (2003). J. Mol. Biol. 327, 193–202. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: PA4019, 3zqu